Abstract
Background
Bacterial infections following childbirth—so-called puerperal infections—cause morbidity in 5%–10% of all new mothers. At low frequency, the infection can spread to the blood, resulting in life-threatening sepsis known as puerperal sepsis. Pathogens causing puerperal sepsis include group A Streptococcus (GAS), and epidemiological analyses have identified isolates of a single serotype, M28, as being nonrandomly associated with cases of puerperal sepsis. The genomes of serotype M28 GAS isolates harbor a 36.3-kb mobile genetic element of apparent group B Streptococcus origin, termed region of difference 2 (RD2).
Methods
The phenotypic (determined via tissue culture and a vaginal colonization model) and regulatory (determined via RNA sequencing analysis) contributions of RD2 were assessed by comparing parental, RD2 deletion mutant, and complemented mutant serotype M28 GAS strains.
Results
RD2 affords serotype M28 isolates an enhanced ability to adhere to human vaginal epithelial cells and to colonize the female reproductive tract in a mouse model of infection. In addition, RD2 influences the abundance of messenger RNAs from >100 core chromosomal GAS genes.
Conclusions
The data are consistent with RD2 directly, via encoded virulence factors, and indirectly, via encoded regulatory proteins, modifying the virulence potential of GAS and contributing to the decades-old association of serotype M28 isolates with cases of puerperal sepsis.
Keywords: Streptococcus pyogenes, mobile genetic elements, phenotypic variation, puerperal sepsis, vaginal colonization, gene regulation
Serotype M28 GAS isolates are, for undefined reasons, overrepresented in cases of puerperal sepsis. Here, we show that a pathogenicity island within M28 GAS isolates enhances their adherence to human epithelial cells and colonization of the female reproductive tract.
Group A Streptococcus (GAS; Streptococcus pyogenes) is a human bacterial pathogen that causes diseases ranging from self-limiting pharyngitis, of which there are >600 million cases annually, to severely invasive necrotizing fasciitis, which has a mortality rate of 25%–45% [1]. Pregnant and postpartum women have a 20-fold increase in the incidence of invasive GAS infections, compared with nonpregnant women [2, 3], with a large portion of this difference being attributable to puerperal infections, which are infections that can initiate in the reproductive tract of women following childbirth [4]. Puerperal infections are the sixth-leading cause of death among new mothers globally [5]. While fatalities due to puerperal infections are rare in the United States, infection frequencies of up to 6% among new mothers have been reported [6, 7], and these can lead to major complications, such as infertility [8]. Puerperal infections can spread from the reproductive tract to the blood, resulting in life-threatening sepsis known as puerperal sepsis, which is responsible for approximately 75 000 maternal deaths globally per year [2, 9]. Several pathogens cause puerperal sepsis, with pathogens of the genus Streptococcus being prevalent [10]. While Streptococcus agalactiae (group B Streptococcus [GBS]) is the most common streptococcal species causing puerperal infections, GAS is the most common species causing severe puerperal infections [4].
GAS strains are divided into serotypes based on the sequence of the 5′ end of the emm gene [11]. That some GAS serotypes show nonrandom associations with particular disease manifestations has been known for >5 decades [12, 13]. Of importance to this work, serotype M28 strains are statistically significantly associated with cases of puerperal sepsis, as identified from multiple population-based case studies performed in the United States and Western Europe [14–17]. The molecular basis of this association is unknown [18]. However, comparative analysis of an M28 GAS genome sequence previously identified 2 foreign genetic elements, designated region of difference 1 (RD1) and 2 (RD2) [19]. RD1 is 11.1 kb long and contains several hypothetical genes, none of which are predicted to encode virulence factors [19]. RD2 is 36.3 kb long, encodes 7 putative virulence factors, and shares significant homology to 2 chromosomal regions of multiple GBS isolates (Figure 1) [19, 20]. GBS organisms are constituents of the normal human vaginal microflora in approximately 25% of women [21] and are also a major cause of neonatal invasive infections [22]. Consequently, it was hypothesized more than a decade ago, but never tested owing to technical issues associated with the creation of an RD2 deletion mutant strain, that the acquisition of RD2 from GBS enhances the ability of M28 GAS isolates to cause puerperal sepsis.
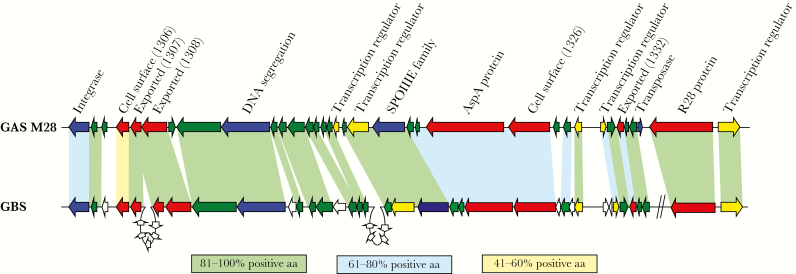
Figure 1.
Comparison of region of difference 2 (RD2) from serotype M28 group A Streptococcus (GAS) to genes present in group B Streptococcus (GBS). Genes are represented by block arrows and are color coded on the basis of their putative open reading frame function or the type of protein they encode (blue, mobility; yellow, gene regulators; red, extracellular proteins; and green, hypothetical). Genes unique to GBS are colored white. Shading between GAS and GBS genes is based on homology at the protein level. Percentages of shared amino acid were calculated by adding the percentages of identical and strongly similar amino acids together for each of the compared protein pairs. The 2 diagonal lines on the GBS part of the figure signify that the 2 genes are not contiguous with the other genes in GBS but are contiguous with the other genes in GAS.
Here, we describe an investigation of the phenotypic and regulatory consequences of RD2 acquisition by serotype M28 GAS. Our data are consistent with RD2 being a pathogenicity island that directly, via encoded virulence factors, and indirectly, via the transcriptional regulation of core chromosomally encoded genes, plays a key role in the association between serotype M28 GAS isolates and puerperal sepsis cases.
METHODS
Bacterial Strains and Growth Conditions
The GAS strains used in this study are summarized in Table 1. GAS were grown in Todd-Hewitt broth containing 0.2% yeast extract (hereafter, “THY broth”). When needed, spectinomycin or kanamycin were added to a final concentration of 150 µg/mL or 300 µg/mL, respectively.
Table 1.
Group A Streptococcus (GAS) Strains Used in This Study
| Strain | Description | Reference |
|---|---|---|
| MGAS2221 | A previously characterized serotype M1 GAS strain | [23] |
| PGAS951 | M11 strain from the CDC; isolated in 2016 from a blood specimen of a patient in CT; the patient did not have puerperal sepsis | This study |
| PGAS953 | M11 strain from the CDC; isolated in 2014 from a blood specimen of a patient in MN; the patient did not have puerperal sepsis | This study |
| PGAS955 | M11 strain from the CDC; isolated in 2017 from a blood specimen of a patient with puerperal sepsis in MN | This study |
| PGAS956 | M11 strain from the CDC; isolated in 2017 from a blood specimen of a patient with puerperal sepsis in NY | This study |
| PGAS958 | M28 strain from the CDC; isolated in 2015 from a blood specimen of a patient in CA; the patient did not have puerperal sepsis | This study |
| PGAS959 | M28 strain from the CDC; isolated in 2014 from a blood specimen of a patient in NY; the patient did not have puerperal sepsis | This study |
| PGAS961 | M28 strain from the CDC; isolated in 2013 from a blood specimen of a patient with puerperal sepsis in OR | This study |
| PGAS962 | M28 strain from the CDC; isolated in 2014 from a blood specimen of a patient with puerperal sepsis in NM | This study |
| MGAS6180 | A previously characterized serotype M28 GAS strain | [19] |
| M28.RD2SPEC | A derivative of MGAS6180 in which RD2 has been tagged with a spectinomycin-resistance cassette inserted between genes 1329 and 1330 | This study |
| M28ΔRD2-Kn | A derivative of MGAS6180 in which all copies of RD2 have been deleted, with a kanamycin-resistance cassette replacing them | This study |
| M28ΔRD2 | A derivative of M28ΔRD2-Kn in which the kanamycin resistance cassette was replaced with the natural RD2 integration site | This study |
| RD2Comp | A derivative of M28ΔRD2 in which RD2 from strain M28.RD2SPEC has been introduced | This study |
| M28ΔR28 | An r28 deletion mutant derivative of MGAS6180 | This study |
Abbreviations: CA, California; CDC, Centers for Disease Control and Prevention; CT, Connecticut; MN, Minnesota; NM, New Mexico; NY, New York; OR, Oregon.
Screening GAS Isolates for RD2
To test whether selected M11 and M28 GAS isolates harbored RD2, we performed polymerase chain reaction (PCR) analysis for detection of the RD2-encoded gene r28. PCR analyses included not only primers for the amplification of r28 but also for the amplification of the core chromosomally encoded gene ropB.
Tissue Culture Assays of Adherence
These assays were performed using a protocol that we described previously [24].
Construction of the RD2 Deletion Mutant–Derivative Strain, M28ΔRD2
To assess the contribution of RD2 to the virulence characteristics of serotype M28 GAS isolates, we created an RD2 deletion mutant strain (M28ΔRD2) via a 2-step procedure. The first step, which created strain M28ΔRD2-Kn, replaced most of RD2 with a kanamycin resistance cassette. Strain M28ΔRD2-Kn was created using a plasmid construct in which a kanamycin resistance cassette was flanked by 1-kb regions complementary to the 5′ and 3′ ends of RD2. See Supplementary Table 2 for the PCR primers used to create the plasmid construct. The second step removed the remaining fragments of RD2 and the kanamycin resistance cassette from strain M28ΔRD2-Kn. The resultant strain, M28ΔRD2, is a markerless and scarless RD2 deletion mutant strain that harbors an intact RD2 integration site (and, hence, can be complemented by reintroduction of RD2). Strain M28ΔRD2 was created from strain M28ΔRD2-Kn by recombinational replacement using the suicide vector pBBL740, and the protocol was similar to that which we have previously described [25]. Strain M28ΔRD2 was verified by tiling PCR, targeted sequencing, and Western blot analyses.
Complementation of Strain M28ΔRD2
To facilitate complementation of M28ΔRD2, we first created strain M28.RD2SPEC, which is a derivative of MGAS6180, in which a nonpolar spectinomycin resistance cassette was inserted between the RD2-encoded genes M28_1329 and M28_1330 [26]. Verification that insertion of the spectinomycin resistance cassette did not alter the abundance of M28_1329 or M28_1330 transcripts was gained via quantitative reverse transcription (RT)–PCR (Supplementary Figure 1). To complement strain, M28ΔRD2 we recovered total DNA from strain M28.RD2SPEC and used it to transform M28ΔRD2 via electroporation. Verification of the complemented strain, RD2Comp, was performed by PCR, targeted sequencing, and Western blot analyses. In addition, quantitative PCR analysis was performed to assay RD2 copy number (Supplementary Figure 2).
Construction of the r28 Deletion Mutant Strain M28ΔR28
To create an r28 deletion mutant derivative of MGAS6180, we replaced the r28 gene with a spectinomycin resistance cassette via a previously described protocol [26]. Verification of strain M28ΔR28 was performed by PCR, targeted sequencing, and Western blot analyses.
Tiling PCR Analysis
To assess for the presence or absence of RD2 in MGAS6180, M28ΔRD2, and M28Comp, we performed an overlapping PCR analysis of this region. Fifteen pairs of PCR primers were designed that were arrayed across RD2 (Supplementary Table 2), with average PCR size of approximately 3 kb, of which approximately 1 kb overlapped with adjacent PCR products. PCR reactions were performed for each of the tested GAS strains, using genomic DNA from each strain as a template.
Isolation of GAS Cell Wall Proteins
Cell wall protein fractions were isolated from mid-exponential phase (OD600 0.5) GAS cultures as previously described [24]. Proteins were separated on 7.5% Tris-HCl gels before they were transferred to membrane and analyzed by Western blot with a custom-made anti-R28 rabbit polyclonal antibody. After washing, Alexa Fluor 680 donkey anti-rabbit immunoglobulin G secondary antibody (Molecular Probes) was used (1:10 000 dilution), and the fluorescent signal was detected using a Li-Cor Odyssey Near-Infrared Imaging System. Note that the membrane was stained using the MemCode Reversible Proteins Stain Kit (Pierce) to serve as a loading control.
Quantitative RT-PCR Analyses
RNA samples from triplicate exponential phase cultures of each GAS strain under investigation were isolated, and converted into complementary DNA (cDNA), as previously described [23]. TaqMan primers and probes for the genes of interest, and the internal control gene proS, are shown in Supplementary Table 2.
Quantitative PCR Analyses
Total DNA samples from triplicate cultures of each GAS strain under investigation were isolated. The abundance of RD2 relative to that of MGAS6180 (which was previously shown to harbor approximately 8 copies of RD2 per genome [20]) was compared for each strain via use of primers and probes targeting the RD2-encoded gene r28 and the non–RD2-encoded gene proS. Data were normalized to proS DNA levels.
Mouse Model of Vaginal Colonization
We followed slightly modified versions of 2 previously described protocols [27, 28]. Briefly, 2 days before streptococcal infection, female CD-1 mice (12 weeks old; Charles River) were estrogenized by intraperitoneal injection of 0.5 mg of β-estradiol 17-valerate (Sigma-Aldrich) dissolved in 0.1 mL of sterile sesame oil (Sigma-Aldrich). On the day of inoculation (day 0), mice were inoculated in the vaginal lumen with 10 µL of GAS suspension that had been diluted to 1 × 106 colony-forming units (CFUs)/mL (hence, each mouse received 1 × 104 CFUs). Twenty mice were infected per GAS strain tested. Colonization levels were assayed on days 1, 3, 5, 7, and 10. To do so, the vaginal vaults of all mice were swabbed using FLOQSwabs (Copan Diagnostics). Each swab was subsequently transferred to a 1.5-mL microcentrifuge tube containing 500 µL of phosphate-buffered saline (PBS). The microcentrifuge tubes was thoroughly vortexed to release any GAS from the swab, and serial dilutions in PBS were plated onto Selective Strep Agar plates (Becton Dickinson) to enable determination of CFUs. This experiment was performed in accordance to the guidelines set forth in a protocol approved by the Institutional Animal Care and Use Committee of the University of Nevada–Reno.
Total RNA Isolation and RNA Sequencing (RNA-Seq) Analysis
Total RNA was isolated from duplicate THY broth cultures of MGAS6180 and M28RD2 in exponential phase, as previously described [23]. Ribosomal RNAs (rRNAs) were depleted using the Ribo-Zero Gram-Positive rRNA Removal Kit (Epicentre). The rRNA-depleted RNA was then used to generate cDNA libraries, using the ScriptSeq kit (Epicentre) [29]. To do this, RNA was fragmented, cDNA was synthesized using random hexamers containing a 5′ tagging sequence, RNA was hydrolyzed, and cDNA was tagged at the 3′ end. A limited number of PCR cycles (n = 14) were used to amplify the libraries via the 5′ and 3′ tags (the libraries were barcoded by using different primers), and the libraries were selected according to size (170–300 bp). The size-selected and barcoded libraries were run on an Illumina MiSeq flow cell system. Data were analyzed using CLC Genomics Workbench (Qiagen) and normalized to the overall sequencing depth, using total mapped read data. Statistical significance was tested using the Kal Z test with a false-discovery rate correction [30].
RNA-Seq data have been deposited in the Gene Expression Omnibus database at the National Center for Biotechnology Information (accession no. GSE116474; available at: http://www.ncbi.nlm.nih.gov/geo).
RESULTS
Serotype M28 GAS Isolates Show Greater Adherence to a Human Epithelial Cell Line Than Serotype M11 Isolates
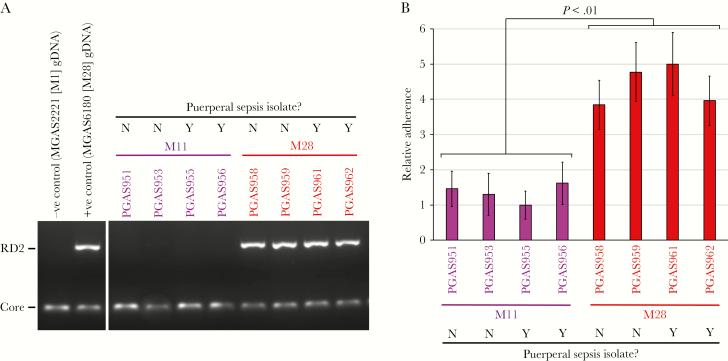
We hypothesize that the overrepresentation of M28 GAS isolates among cases of puerperal sepsis is in part a consequence of the RD2 element conferring an enhanced ability to adhere to vaginal epithelial cells. As a first step in testing this hypothesis, we compared adherence between 4 serotype M11 GAS isolates and 4 serotype M28 isolates. Note that, for each serotype, 2 isolates were recovered from patients with puerperal sepsis, while 2 were from patients with non–puerperal sepsis invasive infections. We chose to compare M28 isolates to M11 isolates because several studies have identified M11 isolates as also being recovered, albeit at lower frequency than M28, from cases of puerperal sepsis [15, 16]. In addition, serotype M11 isolates are not known to harbor the RD2 element, while all M28 isolates do [19], and this was consistent with a PCR-based screen of the isolates used in our study (Figure 2A). To test adherence, we used a tissue culture–based model, using the human epithelial cell line A431 [31]. As a group, the serotype M28 isolates adhered at a 3.4-fold higher level than the serotype M11 isolates (Figure 2B). No differences in adherence were observed between the puerperal sepsis and non–puerperal sepsis isolates, consistent with there being little to no within-serotype differences between isolates causing puerperal sepsis and those causing other diseases. While the data do not reveal whether the higher M28 GAS adherence was due to the presence of the RD2 element, they are consistent with this hypothesis.
Figure 2.
Serotype M28 group A Streptococcus (GAS) isolates have enhanced abilities to adhere to human epithelial cells, relative to M11 isolates. Four serotype M11 and 4 serotype M28 GAS isolates were compared. A, Polymerase chain reaction (PCR) analysis for detection of the region of difference 2 (RD2)–encoded gene r28 (RD2) revealed that the RD2 element is present in the tested M28 isolates but not in the tested M11 isolates. Amplification of a core chromosomally encoded gene, ropB (Core), served as a PCR control. B, A tissue culture–based adherence assay, using the human epithelial cell line A431. Data are presented relative to findings for the lowest adhering strain. The experiment was performed on 4 occasions, mean values (± standard errors of the mean) shown. Statistical significance was tested via a t test. Neg, negative; Pos, positive.
RD2 Can Be Deleted From M28 GAS
To facilitate investigation of whether RD2 modifies the virulence potential of GAS, we created strain M28ΔRD2, an RD2 deletion mutant derivative of the M28 isolate MGAS6180. Given the large size (36.3 kb), multicopy nature (n = approximately 8), and distribution of RD2 between both chromosomally integrated and extrachromosomal forms [20], the creation of M28ΔRD2 proved challenging, but we were ultimately successful. A complemented mutant derivative of M28ΔRD2, strain RD2Comp, was also constructed, by introducing a modified RD2 element that was tagged with a spectinomycin resistance cassette. One of several methods that were used to verify the absence of the RD2 element in strain M28ΔRD2 and the presence of the RD2 element in strain RD2Comp was tiling PCR. A series of 15 primer pairs were created that spanned RD2, with Figure 3A showing the approximate locations of each of the 15 resultant PCR products with regard to the RD2 element. PCR products of the expected size were gained using genomic DNA from the parental isolate and the complemented mutant derivative, strain while, as expected, no PCR products were gained for the RD2 deletion mutant strain (Figure 3B). The expected results were also obtained from a Western blot analysis of the RD2-encoded cell surface protein R28, with the parental and complemented strains but not the RD2 deletion mutant or an isogenic r28 deletion mutant strain (strain M28ΔR28) expressing this protein (Figure 3C).
Figure 3.
Verification of region of difference 2 (RD2) deletion and complementation in strains M28ΔRD2 and RD2Comp, respectively. Total DNA from the parental (M28), RD2 deletion mutant (M28ΔRD2), and complemented mutant (RD2Comp) strains was isolated and used in polymerase chain reaction (PCR) analysis with 15 pairs of primers (RD1 to RD30). A, Distribution of the primers and expected PCR products across RD2. Note that primers RD1 and RD30 are located outside of RD2. B, Generated PCR products for the 3 tested strains. Note that there is a 1-kb size difference between the RD1/2 PCR product of the parental and RD2Comp strains, and this is due to the presence of a 1-kb insertion sequence that flanks RD2 in MGAS6180 but not in the complemented strain. C, Western blot analysis of cell wall–associated R28 expression in the 4 indicated GAS strains. Note the characteristic ladder pattern of R28 [33]. The membrane was stained prior to Western blot analysis, to serve as a loading control.
RD2 Promotes GAS Adhesion to Human Epithelial Cells
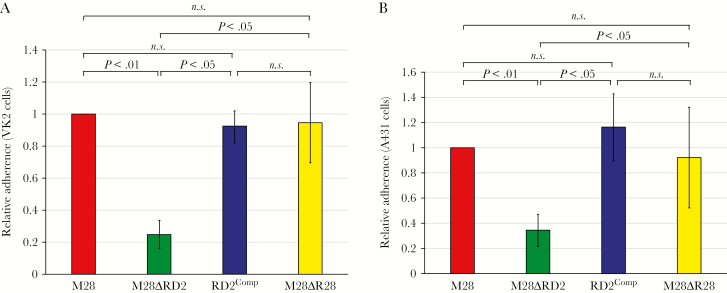
Using our newly constructed MGAS6180 derivatives, we tested the hypothesis that RD2 promotes the ability of GAS to adhere to epithelial cells. To reduce the possibility of our data being confounded by cell line–specific interactions, 2 cell lines were used: A431 and the human vaginal epithelial cell line VK2/E6E7 [32]. In each case, the parental and complemented isolates adhered at a higher level (by 3–4-fold) than the RD2 deletion mutant strain (Figure 4A and 4B). Thus, the RD2 element increased the ability of GAS to adhere to host cells. That the enhanced adherence phenotype was not simply attributable to the RD2-encoded R28 protein, which was previously shown to serve as an adhesin [33], was identified by including an r28 deletion mutant strain in our adherence assays. Strain M28ΔR28 showed levels of adherence similar to that of the parental strain (Figure 4A and 4B). Hence, although R28 may have a contributory role in the ability of RD2 to enhance GAS adherence, other RD2-encoded factors appear to play more significant roles.
Figure 4.
The region of difference 2 (RD2) element enhances the ability of serotype M28 group A Streptococcus (GAS) to adhere to human epithelial cells. The indicated strains were compared in tissue culture-based assays of adherence, using VK2/E6E7 (A) and A431 (B) human cell lines. Each experiment was performed on 4 occasions, with mean values (± standard errors of the mean) shown. For both data sets (A and B), P < .001 (via overall analysis of variance). Pair-wise strain comparisons were investigated using the Tukey multiple comparisons test, with P values shown. NS, not significant.
RD2 Enhances Colonization in a Murine Model of Vaginal Colonization
Given the adherence phenotype associated with RD2 and the association of serotype M28 isolates with puerperal sepsis, we hypothesized that the gain of RD2 by M28 isolates increases their ability to colonize the female reproductive tract. To test this hypothesis, we used a murine model of vaginal colonization [27, 28]. The parental, RD2 deletion mutant, and complemented strains were used to infect female mice that were pretreated with estradiol to synchronize their estrous cycle. Infections occurred by instilling 10 µL of PBS containing 1 × 104 GAS CFUs into the vaginal lumen. Colonization levels were calculated by swabbing the vaginal lumen of infected animals over time and determining the number of GAS CFUs on each swab. We identified 2–6-fold higher levels of CFUs after day 1 for the parental and complemented mutant strains, relative to the RD2 deletion mutant strain (Figure 5). Thus, the RD2 element confers onto serotype M28 GAS isolates an enhanced ability to colonize the female reproductive tract.
Figure 5.
Region of difference 2 (RD2) promotes the ability of serotype M28 group A Streptococcus (GAS) isolates to colonize the murine vagina. Mice treated with estradiol (0.5 mg) were vaginally challenged with 1 × 104 colony-forming units (CFUs) of the indicated GAS strains. Shown is the mean number (± the standard error of the mean) of CFUs recovered from vaginal swabs over time. A repeated-measure analysis indicated that a statistically significantly lower number of CFUs was recovered over time for the RD2 deletion mutant strain, relative to the parental and complemented mutant strains (P < .01).
RD2 Remodels the GAS Transcriptome
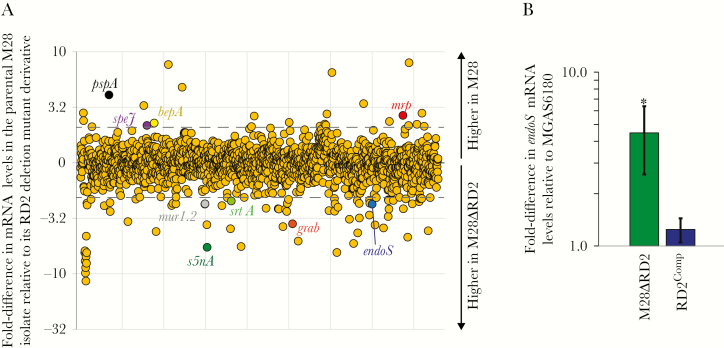
The RD2 element encodes 5 putative transcription regulators (Figure 1). While examples are rare, some studies have identified pathogenicity island–encoded regulators that have core chromosomal genes as regulatory targets [34–36]. To assess whether genes located outside of RD2 are differentially regulated between the presence and absence of this element, we performed an RNA-Seq–based transcriptome analysis. MGAS6180 and derivative M28ΔRD2 were compared from exponential phase THY broth cultures. Transcripts from 108 core chromosomal genes were identified as being differentially regulated, using a cutoff of ≥2-fold (Figure 6A and Supplementary Table 1). Transcripts that were altered in abundance in the presence of RD2 included several that encode virulence factors, such as the immunomodulatory endoglycosidase EndoS [37]. Verification of differential endoS mRNA levels between the presence and absence of RD2 was gained via quantitative RT-PCR and included use of the complemented strain RD2Comp (Figure 6B). Increasing and decreasing the abundance of different virulence factors, resulting in the generation of new virulence factor profiles, is key to the ability of GAS to cause distinct diseases [38–40]. Thus, the data are consistent with the phenotypic consequences of RD2 acquisition, which at a minimum is enhancement of vaginal colonization (Figure 5), in part being attributable to the RD2-mediated regulation of genes located outside of this element.
Figure 6.
Acquisition of the region of difference 2 (RD2) element alters the group A Streptococcus (GAS) transcriptome. A, Results of RNA sequencing analysis comparing the parental M28 strain MGAS6180 and its RD2-deletion mutant derivative (M28ΔRD2) during exponential phase growth in Todd-Hewitt broth containing 0.2% yeast extract. Each circle represents messenger RNA (mRNA) from a single gene. Genes are arranged in the same order as they appear in the chromosome. Select virulence factor–encoding mRNAs are colored and labeled. Dashed lines indicate ±2-fold. B, Findings of TaqMan-based quantitative reverse transcription polymerase chain reaction analysis. The abundance of endoS mRNA was determined from triplicate exponential phase GAS cultures of each indicated strain, run in duplicate. Shown is the mean (± standard deviation). P < .001 (via overall analysis of variance). Pair-wise strain comparisons with the parental isolate were investigated using the Tukey multiple comparisons test. *P < .05.
DISCUSSION
Phenotypic heterogeneity between isolates of a given bacterial species is a commonly observed phenomenon [41, 42]. In some instances, this heterogeneity is a consequence of the isolate-specific presence or absence of virulence factor–encoding mobile genetic elements. For example, cases of staphylococcal scaled skin syndrome are caused by only a subset of Staphylococcus aureus isolates—those that harbor a bacteriophage or plasmid that encode an exfoliative toxin [43]. Here, we investigated whether the presence of the RD2 genomic island in serotype M28 GAS isolates contributes to their nonrandom association with cases of puerperal sepsis. We identified that RD2 enhanced the ability of GAS to adhere to human epithelial cell lines and to colonize in a murine model of vaginal colonization (Figures 4 and 5) and, hence, that RD2 can appropriately be classified as a pathogenicity island. We also discovered that RD2 regulates the abundance of mRNAs from core chromosomally encoded genes (Figure 6), consistent with this element modifying the virulence potential of GAS through both direct and indirect mechanisms.
The RD2 pathogenicity island is distributed in the GAS population mostly along serotype-specific lines. All serotype M28 and most serotype M2 isolates tested to date contain RD2 [19]. Of note, serotype M2 GAS strains are associated with cases of vaginitis and urinary tract infections [14, 19], which we hypothesize may in part be due to RD2 modifying the disease potential of these isolates, similar to that described here for M28 GAS isolates and puerperal sepsis. RD2 has also been detected at low frequency in select isolates of other serotypes (ie, M4, M48, M53, M61, M77, and M87) [19]. A molecular explanation for the distribution of RD2 in the GAS population (ie, what are the barriers to the transfer and acquisition of this element?) is currently under investigation.
Our data are consistent with RD2 enhancing the adherence and colonization of M28 GAS isolates to the female reproductive tract (Figures 4 and 5), a precursor to the development of puerperal sepsis. Thus, we hypothesize that the presence of RD2 is a contributing factor in the enhanced frequency of M28 GAS recovery from cases of puerperal sepsis. Note that, although the development of puerperal sepsis may be enhanced by RD2, this element is not required for it, as evident from data showing that the non–serotype M28 puerperal sepsis isolates lack RD2 (Figure 2A) [44–46].
We hypothesize that the enhanced adherence phenotype conveyed by RD2 is, at least in part, a culmination of the activity of the 4 putative cell wall–anchored proteins encoded by this element (R28, AspA, M28_1306, and M28_1326; Figure 1). The R28 protein has previously been shown to function as an adhesin [33], although we saw no phenotypic consequences to deleting the R28-encoding gene in our tissue culture–based adherence assays (Figure 4A and 4B). Differences in the cell lines (ME180 vs VK2/E6E7 and A431) and protocols (eg, the length of time GAS organisms were incubated with the tissue culture cells before washing) may account for the disparate data with regard to the adhesin activity of R28. AspA also likely contributes to our observed adhesin phenotype, given previous data showing a role for this protein in binding to respiratory epithelial cells [47]. While the M28_1306 and M28_1326 proteins have not been studied in detail, they are both expressed during infection, as assessed by convalescent-phase serum but not acute-phase serum from infected patients harboring antibodies against these proteins [19]. Both proteins have LPXTG motifs toward their carboxy terminus, consistent with them being covalently anchored to the GAS cell wall. Note that, while the 4 proteins described above are the most obvious candidate proteins encoded within RD2 that promote adherence, we cannot rule out other RD2-encoded proteins directly or indirectly influencing this activity.
We identified that genes located outside of the RD2 element were differentially regulated in the presence of RD2 (Figure 6A). A molecular explanation for this regulatory phenotype remains to be investigated. However, our hypothesis is that RD2-encoded regulators target the transcription of core chromosomally encoded genes, and we aim to follow up on this hypothesis by creating and using M28 derivatives with mutations in one or more of the regulator-encoding genes. Consistent with this are data from a recent study that identified that enhancing transcription of the RD2-encoded regulator Spy1337 alters global transcript profiles and strain virulence [48].
In conclusion, we have identified that RD2 is a pathogenicity island conferring onto GAS an increased ability to colonize in a murine model of vaginal colonization. This enhanced colonization phenotype likely contributes to the association of RD2-containing serotypes, M2 and M28, with infections of the female urinary and/or reproductive tracts. Our findings expand insight into the importance of horizontal gene transfer as a mechanism driving strain emergence and phenotypic heterogeneity in a prevalent gram-positive pathogen.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr Izabela Sitkiewicz and Dr Nicole Green, for helpful discussions regarding this manuscript; and the Active Bacterial Core surveillance/Emerging Infections Programs Network, for providing GAS isolates that were vital for the completion of the work.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R03AI128290); the Nevada Women’s Health Initiative; the Nevada IDeA Networks of Biomedical Research Excellence (grant P20GM103440); Dr Mick Hitchcock (Hitchcock Scholar award to J. L. D.); and the University of Nevada–Reno Ronald E. McNair Postbaccalaureate Achievement Program (McNair Scholar award to J. M.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 8th ASM Conference on Streptococcal Genetics, 12–14 August 2016.
References
- 1. Sharkawy A, Low DE, Saginur R, et al. . Severe group a streptococcal soft-tissue infections in Ontario: 1992–1996. Clin Infect Dis 2002; 34:454–60. [DOI] [PubMed] [Google Scholar]
- 2. Hamilton SM, Stevens DL, Bryant AE. Pregnancy-related group a streptococcal infections: temporal relationships between bacterial acquisition, infection onset, clinical findings, and outcome. Clin Infect Dis 2013; 57:870–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Deutscher M, Lewis M, Zell ER, Taylor TH Jr, Van Beneden C, Schrag S; Active Bacterial Core Surveillance Team Incidence and severity of invasive Streptococcus pneumoniae, group A Streptococcus, and group B Streptococcus infections among pregnant and postpartum women. Clin Infect Dis 2011; 53:114–23. [DOI] [PubMed] [Google Scholar]
- 4. Mason KL, Aronoff DM. Postpartum group a Streptococcus sepsis and maternal immunology. Am J Reprod Immunol 2012; 67:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khan KS, Wojdyla D, Say L, Gülmezoglu AM, Van Look PF. WHO analysis of causes of maternal death: a systematic review. Lancet 2006; 367:1066–74. [DOI] [PubMed] [Google Scholar]
- 6. Yokoe DS, Christiansen CL, Johnson R, et al. . Epidemiology of and surveillance for postpartum infections. Emerg Infect Dis 2001; 7:837–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alexander AJ, Myers C, Beres SB, Olsen RJ, Musser JM, Mangino JE. Postpartum group A streptococcus case series: reach out to infection prevention! Open Forum Infect Dis 2018; 5:ofy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Callaghan WM, Creanga AA, Kuklina EV. Severe maternal morbidity among delivery and postpartum hospitalizations in the United States. Obstet Gynecol 2012; 120:1029–36. [DOI] [PubMed] [Google Scholar]
- 9. Maharaj D. Puerperal pyrexia: a review. Part I. Obstet Gynecol Surv 2007; 62:393–9. [DOI] [PubMed] [Google Scholar]
- 10. van Dillen J, Zwart J, Schutte J, van Roosmalen J. Maternal sepsis: epidemiology, etiology and outcome. Curr Opin Infect Dis 2010; 23:249–54. [DOI] [PubMed] [Google Scholar]
- 11. Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 2009; 9:611–6. [DOI] [PubMed] [Google Scholar]
- 12. Mitchell ES. Frequency of serotypes of Streptococcus pyogenes in different diseases. J Clin Pathol 1962; 15:231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilmers MJ, Cunliffe AC, Williams RE. Type-12 streptococci associated with acute haemorrhagic nephritis. Lancet 1954; 267:17–8. [DOI] [PubMed] [Google Scholar]
- 14. Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980–1990 and their association with disease. J Med Microbiol 1993; 39:165–78. [DOI] [PubMed] [Google Scholar]
- 15. Chuang I, Van Beneden C, Beall B, Schuchat A. Population-based surveillance for postpartum invasive group a streptococcus infections, 1995–2000. Clin Infect Dis 2002; 35:665–70. [DOI] [PubMed] [Google Scholar]
- 16. Byrne JL, Aagaard-Tillery KM, Johnson JL, Wright LJ, Silver RM. Group A streptococcal puerperal sepsis: initial characterization of virulence factors in association with clinical parameters. J Reprod Immunol 2009; 82:74–83. [DOI] [PubMed] [Google Scholar]
- 17. Darenberg J, Luca-Harari B, Jasir A, et al. . Molecular and clinical characteristics of invasive group A streptococcal infection in Sweden. Clin Infect Dis 2007; 45:450–8. [DOI] [PubMed] [Google Scholar]
- 18. Cao TN, Liu Z, Cao TH, et al. . Natural disruption of two regulatory networks in serotype M3 group A Streptococcus isolates contributes to the virulence factor profile of this hypervirulent serotype. Infect Immun 2014; 82:1744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Green NM, Zhang S, Porcella SF, et al. . Genome sequence of a serotype M28 strain of group a streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis 2005; 192:760–70. [DOI] [PubMed] [Google Scholar]
- 20. Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol 2011; 11:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patras KA, Wang NY, Fletcher EM, et al. . Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol 2013; 15:1154–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gendrin C, Merillat S, Vornhagen J, et al. . Diminished capsule exacerbates virulence, blood-brain barrier penetration, intracellular persistence, and antibiotic evasion of hyperhemolytic group B streptococci. J Infect Dis 2018; 217:1128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2006; 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Calfee G, Danger JL, Jain I, et al. . Identification and characterization of serotype-specific variation in group A streptococcus pilus expression. Infect Immun 2018; 86:e00792–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 2010; 78:1332–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lukomski S, Hoe NP, Abdi I, et al. . Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun 2000; 68:535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carey AJ, Weinberg JB, Dawid SR, et al. . Interleukin-17A contributes to the control of Streptococcus pyogenes colonization and inflammation of the female genital tract. Sci Rep 2016; 6:26836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watson ME Jr, Nielsen HV, Hultgren SJ, Caparon MG. Murine vaginal colonization model for investigating asymptomatic mucosal carriage of Streptococcus pyogenes. Infect Immun 2013; 81:1606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McClure R, Balasubramanian D, Sun Y, et al. . Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 2013; 41:e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jain I, Miller EW, Danger JL, Pflughoeft KJ, Sumby P. RocA is an accessory protein to the virulence-regulating CovRS two-component system in group A streptococcus. Infection and immunity 2017; 85:e00274–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wizemann TM, Moskovitz J, Pearce BJ, et al. . Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci U S A 1996; 93:7985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fichorova RN, Rheinwald JG, Anderson DJ. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol Reprod 1997; 57:847–55. [DOI] [PubMed] [Google Scholar]
- 33. Stålhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol Microbiol 1999; 33:208–19. [DOI] [PubMed] [Google Scholar]
- 34. Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci U S A 2005; 102:14249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 2007; 66:1174–91. [DOI] [PubMed] [Google Scholar]
- 36. Petrone BL, Stringer AM, Wade JT. Identification of HilD-regulated genes in Salmonella enterica serovar Typhimurium. J Bacteriol 2014; 196:1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collin M, Olsen A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J 2001; 20:3046–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cole JN, McArthur JD, McKay FC, et al. . Trigger for group A streptococcal M1T1 invasive disease. FASEB J 2006; 20:1745–7. [DOI] [PubMed] [Google Scholar]
- 39. Lynskey NN, Goulding D, Gierula M, et al. . RocA truncation underpins hyper-encapsulation, carriage longevity and transmissibility of serotype M18 group A streptococci. PLoS Pathog 2013; 9:e1003842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller EW, Danger JL, Ramalinga AB, Horstmann N, Shelburne SA, Sumby P. Regulatory rewiring confers serotype-specific hyper-virulence in the human pathogen group A Streptococcus. Mol Microbiol 2015; 98:473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sarkar P, Sumby P. Regulatory gene mutation: a driving force behind group a Streptococcus strain- and serotype-specific variation. Mol Microbiol 2017; 103:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krüger A, Lucchesi PM. Shiga toxins and stx phages: highly diverse entities. Microbiology 2015; 161:451–62. [DOI] [PubMed] [Google Scholar]
- 43. Kato F, Kadomoto N, Iwamoto Y, Bunai K, Komatsuzawa H, Sugai M. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect Immun 2011; 79:1660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Golińska E, van der Linden M, Więcek G, et al. . Virulence factors of Streptococcus pyogenes strains from women in peri-labor with invasive infections. Eur J Clin Microbiol Infect Dis 2016; 35:747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ben Zakour NL, Venturini C, Beatson SA, Walker MJ. Analysis of a Streptococcus pyogenes puerperal sepsis cluster by use of whole-genome sequencing. J Clin Microbiol 2012; 50:2224–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turner CE, Dryden M, Holden MT, et al. . Molecular analysis of an outbreak of lethal postpartum sepsis caused by Streptococcus pyogenes. J Clin Microbiol 2013; 51:2089–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Franklin L, Nobbs AH, Bricio-Moreno L, et al. . The AgI/II family adhesin AspA is required for respiratory infection by Streptococcus pyogenes. PLoS One 2013; 8:e62433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kachroo P, Eraso JM, Beres SB, et al. . Integrated analysis of population genomics, transcriptomics and virulence provides novel insights into Streptococcus pyogenes pathogenesis. Nat Genet 2019; 51:548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.