Abstract
Fermentation of Theobroma cacao L. beans is the most critical stage in the production of cocoa products such as chocolates and its derivatives. There is a limited understanding of the complex response of microbial diversity during cocoa bean fermentation. The aim of the present study was to investigate microbial communities in the cocoa bean fermentation heap using a culture-independent approach to elucidate microbial diversity, structure, functional annotation and mapping unto metabolic pathways. Genomic DNA was extracted and purified from a sample of cocoa beans fermentation heap and was followed by library preparations. Sequence data was generated on Illumina Hiseq 2000 paired-end technology (Macrogen Inc). Taxonomic analysis based on genes predicted from the metagenome identified a high percentage of Bacteria (90.0%), Yeast (9%), and bacteriophages (1%) from the cocoa microbiome. Lactobacillus (20%), Gluconacetobacter (9%), Acetobacter (7%) and Gluconobacter (6%) dominated this study. The mean species diversity, measured by Shannon alpha-diversity index, was estimated at 142.81. Assignment of metagenomic sequences to SEED database categories at 97% sequence similarity identified a genetic profile characteristic of heterotrophic lactic acid fermentation of carbohydrates and aromatic amino acids. Metabolism of aromatic compounds, amino acids and their derivatives and carbohydrates occupied 0.6%, 8% and 13% respectively. Overall, these results provide insights into the cocoa microbiome, identifying fermentation processes carried out broadly by complex microbial communities and metabolic pathways encoding aromatic compounds such as phenylacetaldehyde, butanediol, acetoin, and theobromine that are required for flavour and aroma production. The results obtained will help develop targeted inoculations to produce desired chocolate flavour or targeted metabolic pathways for the selection of microbes for good aroma and flavour compounds formation.
Keywords: Bioinformatics, Biotechnology, Microbiology, Molecular biology
1. Introduction
Fresh cocoa bean pulp is rich in fermentable sugars such as glucose, fructose and sucrose, and has a low pH of 3.0–3.5, mainly due to the presence of citric acid. The activities of a variety of microorganisms result in a change in the chemical composition of the pulp during fermentation (Camu et al., 2008). The pulp is microbiologically sterile, when healthy undamaged pods are opened aseptically (Jespersen et al., 2005). However, sterile pulp gets inoculated with a variety of microorganisms from the machete, workers' hands, carrying-baskets, fermentation boxes, fruit flies and other insects (Jespersen et al., 2005; Camu et al., 2008). Cocoa bean fermentation is generally characterized by the production of metabolites, such as organic acids (lactic and acetic acid) and flavouring compounds, which are important components in defining cocoa bean quality for chocolate production (Camu et al., 2008; Afoakwa et al., 2009). The presence of various metabolites reflect the presence of related genes in the microbial population and give a more direct collective phenotypic view of the cocoa bean fermentation microbiome because of the products of gene expression (Antony and Chandra, 1997).
Yeasts are the initial colonisers of cocoa pulp and their presence is favoured by the relatively high acidity of pulp (pH 3.6), due to citric acid, and together with low oxygen levels to utilise pulp carbohydrates under both aerobic and anaerobic conditions (Urso et al., 2008). The activities of yeasts present in the pulp results in a changed fermentation environment that favours the development of lactic-acid bacteria (LAB). Yeasts convert sucrose, glucose, and fructose present in the pulp to ethanol and carbon dioxide. They also metabolise citric acid causing the pH value to increase in the pulp leading to enhanced growth of bacteria. Several yeasts isolates produce organic acids including acetic, oxalic, phosphoric, succinic, and malic acids (Nielsen et al., 2005; Daniel et al., 2009). They further produce volatile compounds important in the development of full chocolate flavour (Leal et al., 2008; Smid and Kleerebezem, 2014). LAB during fermentation, utilize glucose via the Embden-Meyerhof and the hexose monophosphate pathways yielding more than 85% and 50% lactic acid respectively. LAB is also involved in the production of ethanol, acetic acid, glycerol, mannitol, and carbon dioxide (Vandamme and Soetaert, 2002; Madorobaa et al., 2011). During cocoa bean fermentation, acetic acid bacteria (AAB) oxidize ethanol produced by yeasts into acetic acid that infiltrates into the bean, lowering the inner pH. Moreover, the energy-yielding reactions of AAB increase the temperature of the fermenting bean mass. The low inner pH of bean, coupled with the heat, triggers the activation of endogenous enzymes, mainly proteolytic enzymes (Noor-Soffalina et al., 2009; Yao et al., 2014), aminopeptidase, invertase, polyphenol oxidase and glycosidases (Camu et al., 2007, 2008). This leads to a series of reactions responsible for the final quality of the fermented bean and chocolate (Schwan and Wheals, 2004; Ouattara et al., 2008; Lefeber et al., 2010).
Flavour precursors of cocoa are developed during the primary processing of fermentation and drying of the cocoa beans. This development of flavour precursors involves the action of various microorganisms in the cocoa pulp and the action of endogenous enzymes on carbohydrates, proteins and polyphenols in the cocoa beans (Camu et al., 2008; Garcia-Armisen et al., 2010). Fermented, dried cocoa bean are roasted and this results in more than 500 volatile compounds such as aldehydes formed by Strecker degradation of amino acids, methylpropanal, 2- and 3-methylbutanal, phenylacetaldehyde and pyrazines that are considered as important contributors to the cocoa aroma (Smit et al., 2005a; Frauendorer and Schieberlie, 2006; Afoakwa et al., 2008). The amyl alcohols are common compounds in food flavour, which are also found in the cocoa fermentation process. These alcohols are used to evaluate cocoa flavour and degree of fermentation (Oberparleiter and Ziegleder, 1997; Smit et al., 2005b). In addition, compounds such as 3-methyl-1-butanol and phenylacetaldehyde have been reported as derivatives from amino acids catabolism released during fermentation (Afoakwa et al., 2008).
During the last decade, the microbial diversity of spontaneous cocoa bean fermentation processes have been investigated through the application of culture-dependent and culture-independent techniques (Papalexandratou et al., 2011a, 2011c; Illeghems et al., 2012). A culture-independent technique such as PCR-DGGE had been employed with the aim of identifying both cultivable and uncultivable but potentially important contributors in a microbial ecosystem using whole microbial community (metagenomic) DNA. However, this method gives a biased outcome for several reasons, as they are confronted with PCR challenges such as preferential DNA amplification (Illeghems et al., 2012). Currently, microbial ecological studies have seen the introduction of metagenomics that deals with the direct genetic analysis of genomes contained within an environmental sample has revealed several microbes that were previously unculturable (Tyson et al., 2004; Venter et al., 2004; Eisen, 2007; Thomas et al., 2012). This method provides access to the functional gene composition of microbial communities and thus gives a much broader description than phylogenetic surveys, which are often based only on the diversity of one gene, for instance the 16S rRNA gene for bacterial identification (Wilmes and Bond, 2006; Gilbert et al., 2008). However, this great potential of metagenomics has not been exploited in cocoa fermentation.
This current study reports utilizing a culture-independent approach to elucidate the microbial species diversity and structure as well as functional protein compositions in the cocoa bean fermentation microbiome using shotgun Illumina technology to overcome the limitations of the culture-dependent techniques.
2. Materials and methods
Matured ripped cocoa pods from mixed hybrid cocoa Criollo and Forastero tree plantations obtained from Cocoa Research Institute of Ghana farms (Long. 0*21′185”(W) and Lat. 6*13′26” (N)) located at Akyem Tafo in Ghana were harvested manually. These were transported in baskets, and used for fermentation on the same day in Ghana. Plantation workers cut the pods open with machetes, and the beans and surrounding pulp were manually scooped out. Approximately 250 kg of wet beans with pulp was heaped on banana leaves on the ground, resulting in a heap of 62 cm high and 101 cm diameter and which were covered with extra banana leaves completely and left to ferment. Fermentation was carried out for 4 days and 400 g of fermented beans was sampled at 24 hr, 48 hr, 72 hr and 96 hr. These samples were labelled as A1, A2, A3, and A4 respectively. At 24 hr the initial colonisers, yeasts species control fermentation, while LAB and AAB control fermentation at 48 and 72 hr. The samples were placed in sterile sample bags and transported on ice to the laboratory for genomic DNA extraction.
2.1. Genomic DNA extraction and Illumina sequencing
Whole-community metagenomic DNA was isolated in quadruplicate (A1, A2, A3 and A4), each time from 20 g of the sample being homogenized in a Stomacher® 400 (Seward Laboratory System Inc, Florida-USA) for 5 min, with 70 ml saline (mixture). The fluid (mixture) was decanted and subsequently centrifuged at 170 x g at 4 °C for 5 min to remove large particles. The supernatant was filtered through cheesecloth with an approximate pore size of 20-μm. The genomic DNA was extracted according to a modified method of Camu et al. (2008). For a purer DNA sample to meet the sequencing standard, 400 mL CTAB lyse buffer (AppliChem, GmbH, Darmstadt, Germany) was used for re-extraction to remove excessive lipids. The genomic DNA was visualized on a 1% Agarose gel with 1kb ladder (GeneScript) at a voltage of 80V for 45 minutes. The DNA quantity and quality was determined with Qubit. Shotgun whole genome library was constructed using the TruSeq Sample Prep kits (Illumina, California-USA) according to the manufacturer's protocol. The four samples A1, A2, A3 and A4 were pooled together for construction of the library. A paired-end sequencing of the library was run on one lane of Illumina HiSeq 2000 flow cell for whole genome shotgun metagenomic sequencing at Macrogen Inc. (Korea). The obtained reads were subjected to quality control checks using FASTQC v0.11.2 tool and normalisation of sequence data coming from the high throughput sequencing pipelines. Bad sequences and sequence reads less than 100 bases with a median quality score below 20 were removed. Host plant sequences were also screened out. Metagenome analysis was carried out using three consecutive steps: de novo read assembly (Luo et al., 2012), species binning, open reading frames (ORFs) identification within the sequence data using FragGeneScan version 1.19 (Rho et al., 2010). The ORFs were subsequently screened for potential protein-encoding genes (PEGs) via a BLASTX search against the SEED non-redundant database. The assembled reads were uploaded to the online server Metagenome Rapid Annotation Using Subsystem Technology (MG-RAST) (Meyer et al., 2008) under the name DSA Metagenome and was assigned the Metagenome ID: 4572378.3. The MG-RAST v.3.5 online server quality control pipeline was utilized to further remove reads of short length and poor quality before annotation and the analysis of metagenomic data (Meyer et al., 2008). The organisms were classified based on M5NR protein database, and functions annotated and classified based on SEED subsystem incorporated in the online server MG-RAST. The maximum e-value of 10−5, minimum 70% identity of, and a minimum alignment length of 30 were applied as the parameter settings in the analysis. The species alpha diversity was estimated using the Shannon diversity index (MG-RAST). The most abundant species were recruited using the MG-RAST server.
2.2. KEGG pathway assignment
The Kyoto Encyclopaedia of Genes and Genomes (KEGG) mapping method was used in recruiting and assigning metabolic pathways (Kanehisa and Goto, 2000). The sequences that mapped unto KEGG metabolic pathways were assigned enzyme commission (EC) numbers as obtained in the pathway database.
3. Results
3.1. Paired –end sequencing
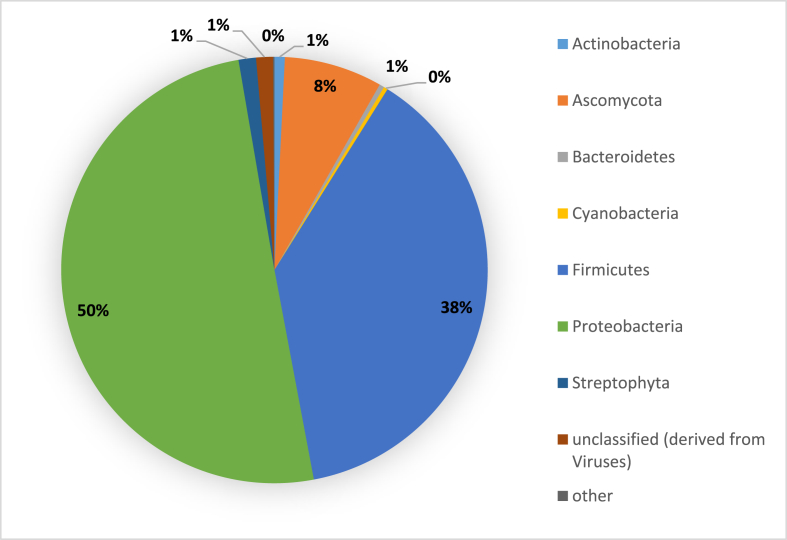
Paired-end sequencing of libraries generated 46 gigabytes (GB) (462,844,468 reads) of data. Quality filtering retained 373,289,474 reads (approximately 38 GB). The identified OTUs and ORFs were searched against databases such as RDP, M5NR/NCBI NR released 2/2/2011 (Sayers et al., 2009), using Basic Local Alignment Search Tool (BLAST) and BLAST-like alignment tool (BLAT) (Kent, 2002). The results indicated that the taxonomic assignment of the Bacteria domain was the most abundant (90%) within the cocoa beans fermentation metagenome sequences. The domains Eukaryota and Viruses accounted for 9% and 1% respectively of the total sequences reads. In the bacteria domain, the Proteobacteria phylum represented half of the sequences (50%) and was followed by the next largest phylum Firmicutes representing 38% of cocoa beans fermentation metagenome sequences. Present also in this metagenome sequences was the taxon Ascomycota that accounts for 8% and a small number of the sequences representing 1% were derived from Viruses (Fig. 1).
Fig. 1.
Phylum distribution within Theobroma cacao beans microbiome.
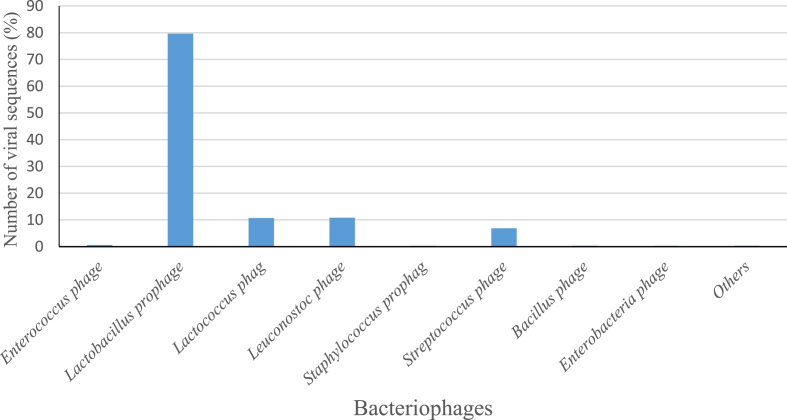
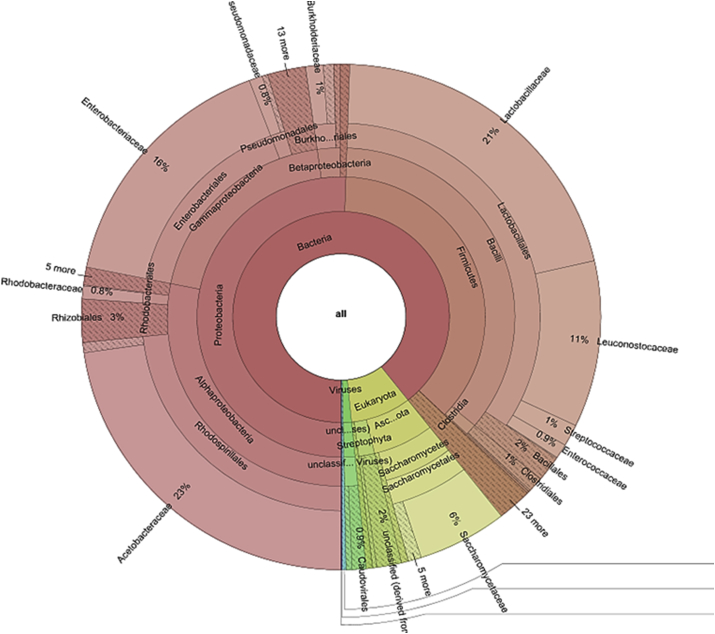
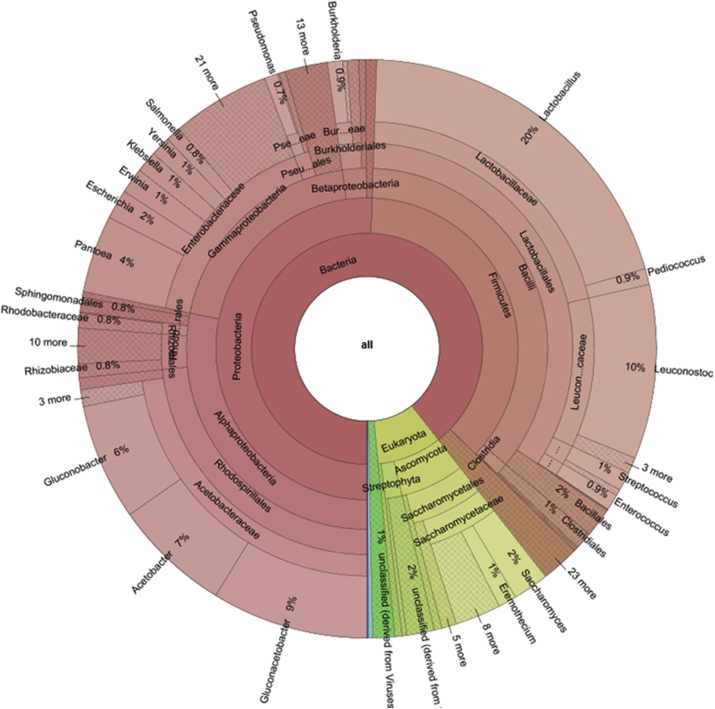
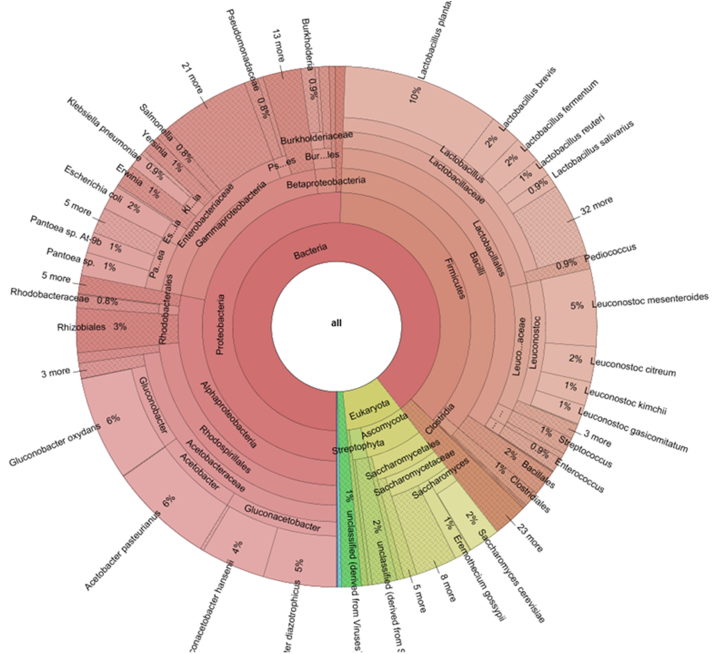
Most of the virus-derived sequences were identified with the virus family Siphoviridae. Within the total sequence reads of the family Siphoviridae, Lactobacillus bacteriophages were the highest occurring at 79.7%. Lactococcus bacteriophage and Leuconostoc bacteriophage were the other dominant bacteriophages at 10.7% and 10.8% respectively. Some phages were also derived from Bacillus, Pseudomonas, Staphylococcus, Enterococcus and Enterobacteria, but were the least represented (Fig. 2). Furthermore, the most abundant families were Acetobacteraceae (23%), Lactobacillaceae (21%), Enterobacteriaceae, Leuconostocaceae, and Saccharomycetaceae with the relative abundance of 16%, 11% and 6% respectively within the entire sequence data (Fig. 3). Subsequently, at the genus level, Lactobacillus was the most abundant at 20%. Additionally, three main genera of the Acetobacteraceae family were identified. These were Gluconacetobacter, Acetobacter and Gluconobacter with 9%, 7% and 6% abundance respectively. Rare and low abundance genera included Granulibacter, Acidiphilium and Roseomonas. Four genera of Leuconostocaceae family were represented, with the genus Leuconostoc (10%) being the most abundant within the sequence dataset. The others were Oenococcus, Weissella, and Fructobacillus. Worthy of note was the presence of Pantoea, Escherichia, and Saccharomyces with a relative abundance of 4%, 2%, and 2% respectively (Fig. 4).
Fig. 2.
Bacteriophage ORFs identified in the family Siphoviridae in the cocoa microbiome.
Fig. 3.
Taxonomic distribution of Family within cocoa fermentation metagenome.
Fig. 4.
Taxonomic distribution of genera within cocoa fermentation microbiome.
3.2. Species diversity indices
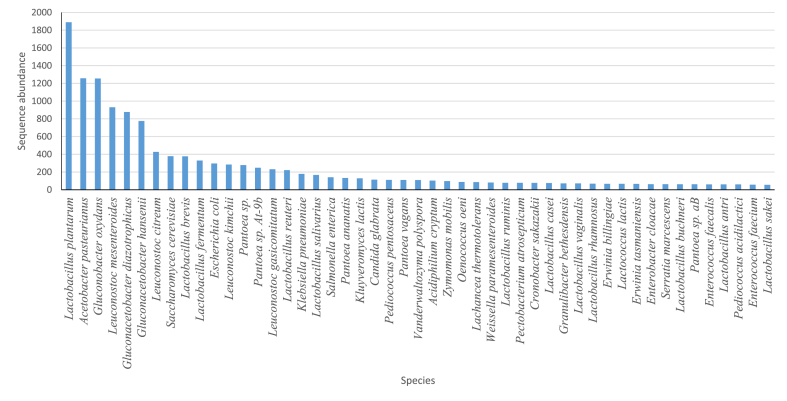
The fermentation of cocoa beans was observed to have a great diversity of microorganisms after species binning. The estimated Shannon alpha (α) diversity value from assembled contigs at the species-level annotations was 142.819. The most abundant species were Lactobacillus plantarum (10%), Acetobacter pasteurianus (6%), Gluconobacter oxydans (6%), Leuconostoc mesenteroides (5%), Gluconacetobacter diazotrophicus (5%), Gluconacetobacter hansenii (4%), Leuconostoc citreum (2%), Saccharomyces cerevisiae (2%), Lactobacillus brevis (2%), Lactobacillus fermentum (2%), and Escherichia coli (2%) (Figs. 5 and 6). Other species identified belong to the genera Streptococcus, Enterococcus, Pantoea, Erwinia, Yersinia, Klebsiella. The major yeasts species identified were Saccharomyces cerevisiae and Kluyveromyces lactis (Fig. 6).
Fig. 5.
Species diversity during cocoa fermentation based on MG-RAST annotation online web server.
Fig. 6.
Species diversity of the top fifty most abundant species in the cocoa fermentation microbiome.
3.3. Protein-coding genes in cocoa fermentation microbiome
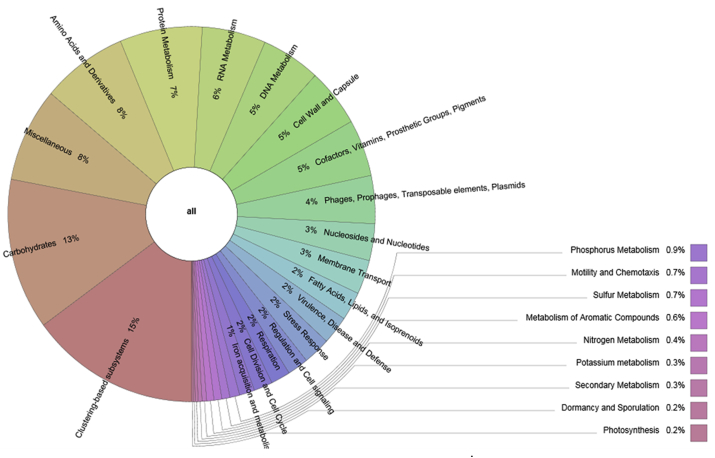
Several functional categories were identified based on the Subsystem annotation databases implemented in the MG-RAST server. The number of sequences representing the carbohydrate metabolism category was 13% of the total functions determined. Amino acids and derivatives, and proteins metabolisms sequences accounted for 8% and 7% of the functions identified respectively. Metabolism of aromatic compounds sequences was 0.6% of the entire metabolism process. Approximately, 5% of the entire cocoa beans fermentation metagenome sequences were assigned to the metabolism of cofactors, vitamins, prosthetic groups and pigments. Lipids, fatty acid and isoprenoid metabolism represented 2% abundance of the entire sequences. As much as 2% of the entire sequences was coding for stress response by these microbes (Fig. 7). DNA and RNA metabolism, cell wall and capsule, virulence, diseases and defense, phages and prophages are among several predicted protein function categories that were identified (Fig. 7).
Fig. 7.
Predicted protein coding genes in cocoa fermentation metagenome sequence data.
3.4. Common pathway for synthesis of aromatic compounds (DAHP synthase to chorismate)
Several aromatic genes sequences encoding for three types of DAHP synthase and functions of genes involved at each stage were identified (Table 1). These sequence reads are part of the common pathway for aromatic biosynthetic compounds that starts with 2-keto-3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) and proceeds through seven more steps to the major intermediate, chorismate. The first step of the pathway: DAHP synthase I alpha and DAHP synthase I beta are homologous proteins (AroAI alpha and AroAI beta), and DAHP synthase II (AroAII). The second step is 3-dehydroquinate synthase (AroB). The third step has two non-homologous types of 3-dehydroquinate dehydratase, (AroCI and AroCII). The aroma gene 3,7-dideoxy-D-threo-hepto-2,6-diulosonate synthase (AroB′) was also identified. Shikimate dehydrogenase involved in the fifth step, with its subgroups of four homologous groups (AroDI alpha, beta, gamma, delta) were also identified. The sixth step, shikimate kinase (AroEI and AroEII), 5-Enolpyruvylshikimate-3-phosphate synthase (EPSPS) (AroF), and chorismate synthase (AroG) were all identified in the cocoa fermentation microbiome (Table 1).
Table 1.
Identified fermentation genes in aromatic amino acids metabolic pathways annotated in SEED-subsystem.
| Genes | Functional Role |
|---|---|
| AroAIα | 2-keto-3-deoxy-D-arabino-heptulosonate-7-phosphate synthase I alpha (EC 2.5.1.54) |
| AroAIβ | 2-keto-3-deoxy-D-arabino-heptulosonate-7-phosphate synthase I beta (EC 2.5.1.54) |
| AroAII | 2-keto-3-deoxy-D-arabino-heptulosonate-7-phosphate synthase II (EC 2.5.1.54) |
| AroB | 3-dehydroquinate synthase (EC 4.2.3.4) |
| AroCI | 3-dehydroquinate dehydratase I (EC 4.2.1.10) |
| AroCII | 3-dehydroquinate dehydratase II (EC 4.2.1.10) |
| AroDIα | Shikimate 5-dehydrogenase I alpha (EC 1.1.1.25) |
| AroDIβ | Shikimate/quinate 5-dehydrogenase I beta (EC 1.1.1.282) |
| AroDIγ | Shikimate 5-dehydrogenase I gamma (EC 1.1.1.25) |
| AroDIδ | Quinate/shikimate 5-dehydrogenase I delta (EC 1.1.1.25) |
| AroEI | Shikimate kinase I (EC 2.7.1.71) |
| AroEII | Shikimate kinase II (EC 2.7.1.71) |
| AroEIII | Shikimate kinase III (EC 2.7.1.71) |
| AroF | 5-Enolpyruvylshikimate-3-phosphate synthase (EC 2.5.1.19) |
| AroG | Chorismate synthase (EC 4.2.3.5) |
| TyrR | Transcriptional repressor protein TyrR |
| TrpR | Transcriptional repressor protein Trp |
3.5. Aromatic amino acids and derivatives metabolism
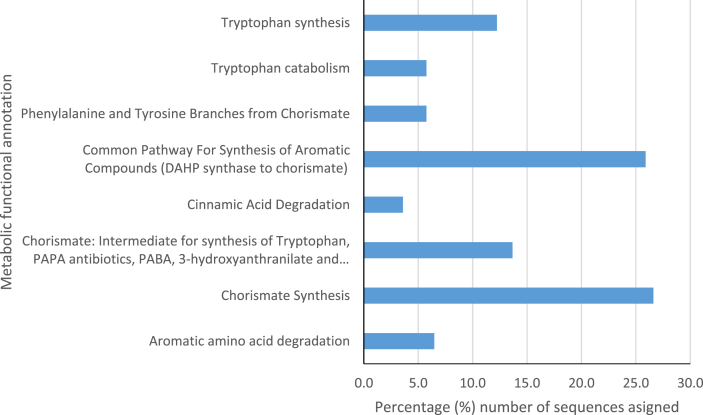
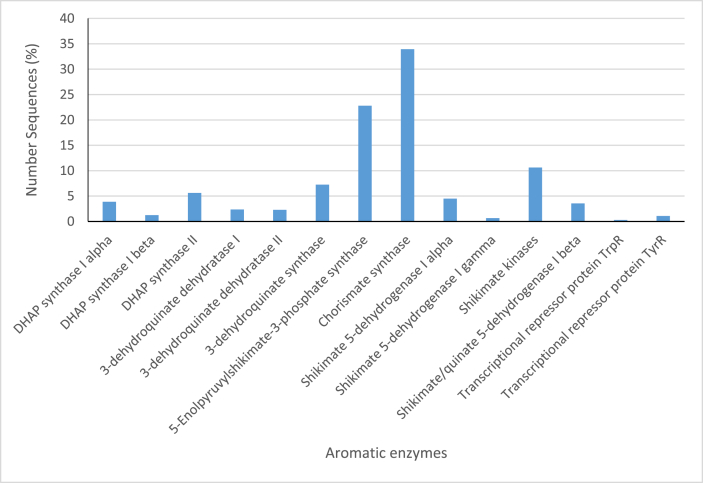
The aromatic amino acids and derivatives metabolism processes category had ORFs sequences encoding for enzymes in common pathways for the synthesis of aromatic compounds, that is, the 3-deoxy-D-arabinoheptulosonate-7-phosphate (DAHP) synthase to chorismate at 27% abundance. Sequence reads representing chorismate synthesis was 28%, Tryptophan catabolism and synthesis was 7% and 12% abundance respectively and aromatic amino acids degradation metabolic processes was 8% in abundance of the category aromatic amino acids and derivatives (Fig. 8). Some sequences encoding enzymatic reactions for the biosynthesis of aromatic compound phenylacetaldehyde (Fig. 9) and purine alkaloid theobromine (Fig. 10) through the metabolism of aromatic amino acids were also identified. Sequences reads encoding for the enzyme aromatic aminotransferase, which catalyses the transamination step in the conversion of the aromatic amino acids tyrosine, phenylalanine, and tryptophan to their corresponding alpha-keto acids, were detected and associated with L. plantarum, and L. brevis. The subcategory pathway for aromatic amino acid ORF sequences reads were annotated to enzymes such as 3-dehydroquinate synthase (7.2%), 5-enolpyruvylshikimate-3-phosphate synthase (22.8%), Shikimate kinases (10.6%), Chorismate synthase (33.9%). Furthermore, lactic and acetic acid bacteria significantly expressed shikimate 5-dehydrogenases (4.5%) and 2-keto-3-deoxy-D-arabino-heptulosonate-7-phosphate (DHAP) synthase II (5.6%) during the period of cocoa beans fermentation (Fig. 11).
Fig. 8.
Aromatic amino acids.
Fig. 9.
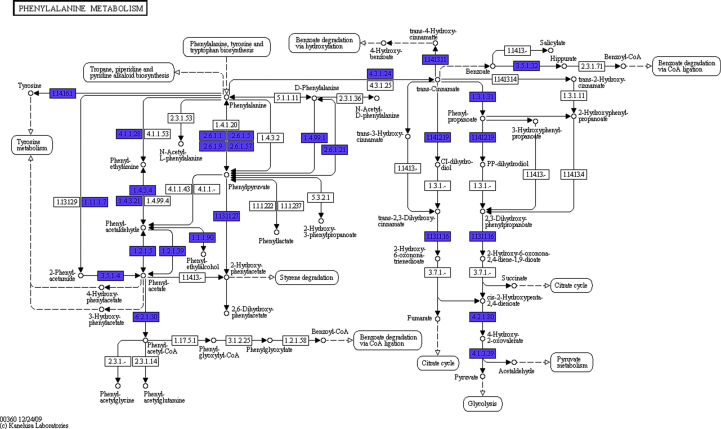
Metabolic biosynthesis of phenylacetaldehyde as obtained from Kegg database.
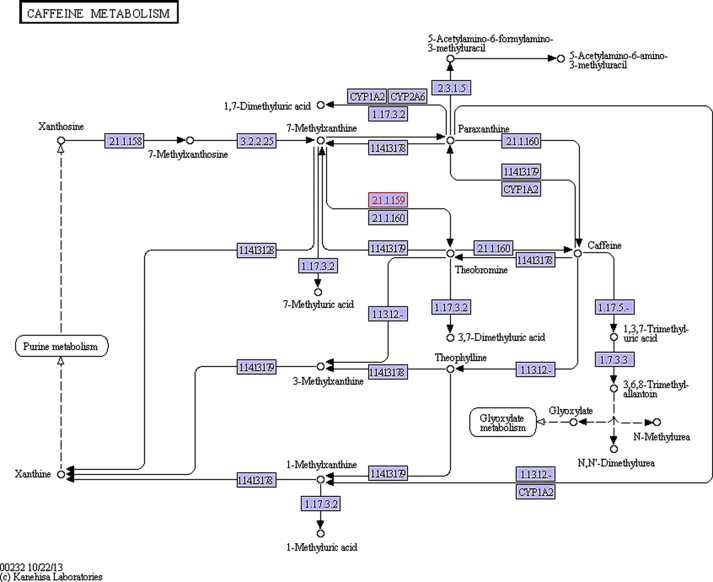
Fig. 10.
Metabolic biosynthesis of purine alkaloid theobromine as obtained from Kegg database. The blue coloured boxes (Figs. 9 and 10) with EC numbers indicate enzymes that were obtained from the cocoa beans fermentation sequence data, but the box EC number in red represent link to this KEGG map. The unidentified genes could be a result of the random fragmentation of the genomic DNA during library preparation. In this process there is the likelihood some of the genomic DNA segment could not be present in the library for sequencing.
Fig. 11.
Aromatic enzymes in common pathway for aromatic compounds.
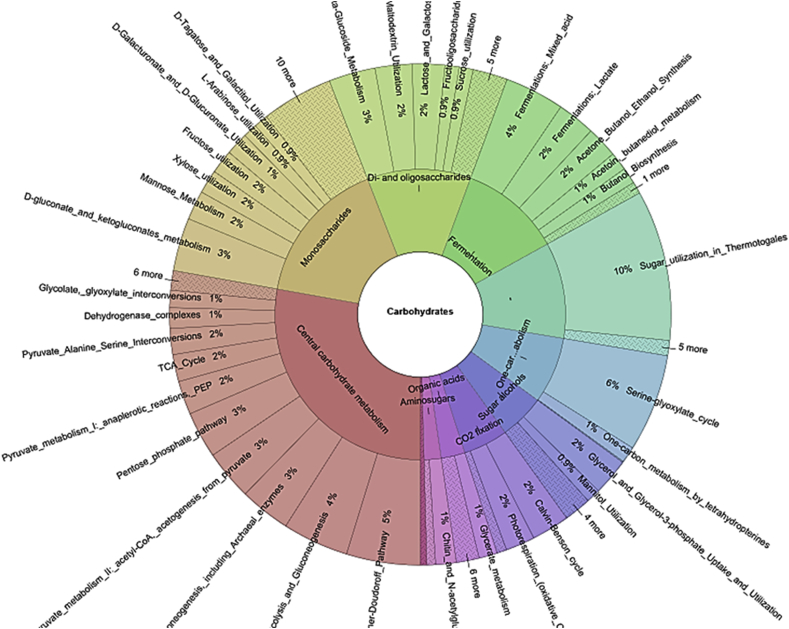
3.6. Carbohydrate metabolism
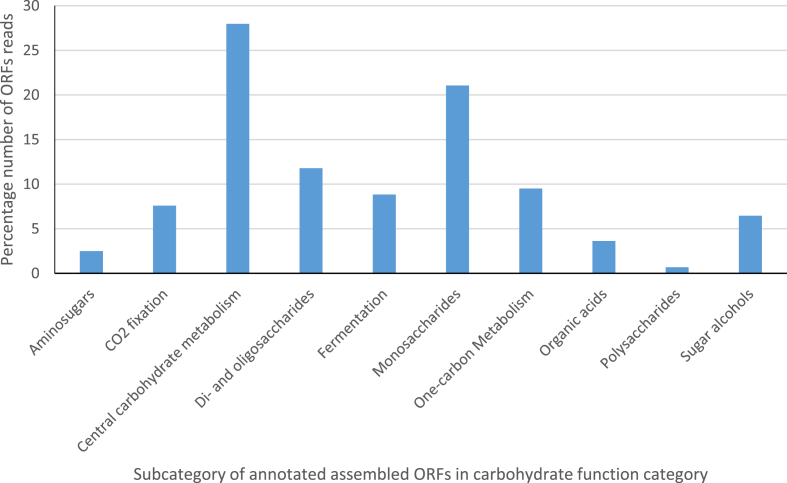
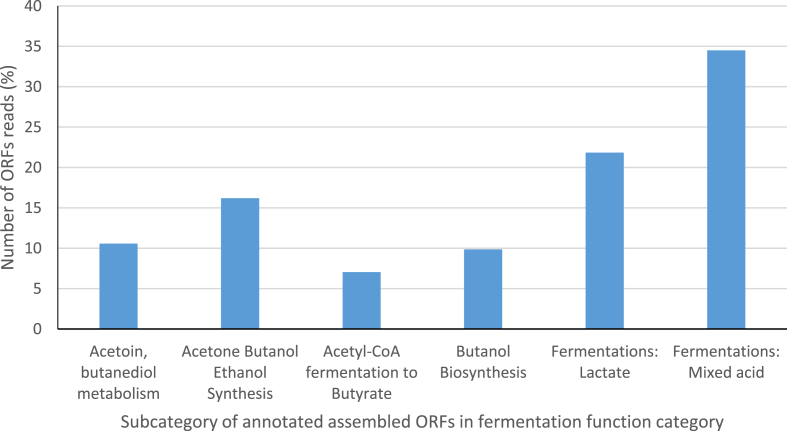
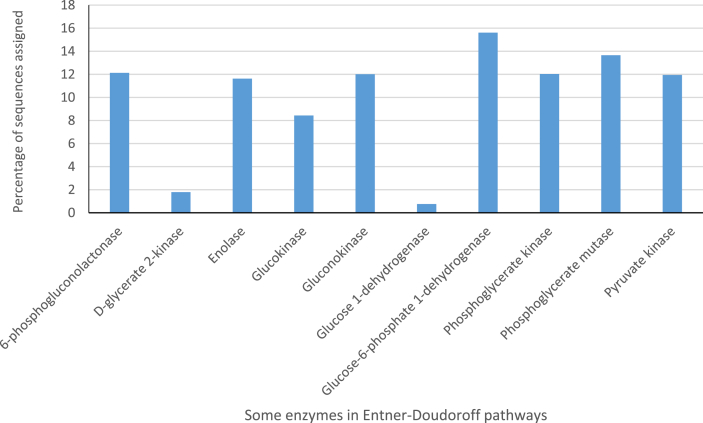
In relation to only the carbohydrate metabolic processes functional category, there were ORF sequences reads encoding for processes such as fermentation (8.8%), Di- and oligosaccharides (11.8%), a monosaccharide (21.1%) and other central carbohydrates metabolism (27.1%) (Fig. 12). Furthermore, in relation to the carbohydrate functional category, the subcategory monosaccharides metabolism included sequences for such processes as fructose (2%), xylose (2%), D-galacturonate and D-glucuronate (1%) utilization, Manose, (2%) and D-gluconate and ketogluconate metabolism (3%) abundance in sequence proportion (Fig. 13). The subcategory Di- and oligosaccharides metabolisms sequences had 2% of lactose and galactose uptake utilization processes with respect to the carbohydrate metabolism functional category (Fig. 13). The ORF reads that make-up only the fermentation metabolism functional category were mixed acid fermentation at 34.5%, fermentation lactate utilization process sequence reads was 21.8%, acetone butanol, ethanol synthesis was 16.2%, Acetoin butanediol metabolism was 10.6% and butanol biosynthesis was 9.6% (Fig. 14). A high number of ORF sequence reads encoding for enzymes involved in Entner-Doudoroff pathway, glycolysis and glucogenesis pathways, pyruvate metabolisms, pentose phosphate pathways among many others in the central carbohydrate metabolisms were identified (Fig. 13). Notable enzymes in the Entner-Doudoroff pathway were enolase, Gluconokinase, glucose-6-phosphate 1- dehydrogenase, pyruvate kinases, 6-Phosphogluconolactonase and Phosphoglycerate mutase (Fig. 15).
Fig. 12.
Carbohydrate category based on subsystem annotation in MG-RAST online server.
Fig. 13.
Carbohydrate category and subcategory functional annotation based on subsystem database implemented in MG-RAST online webserver.
Fig. 14.
Fermentation category based on subsystem annotation in MG-RAST online server.
Fig. 15.
Some enzymes of the Entner-Doudoroff pathway identified in a cocoa metagenomic data set related to fermentation based on SEED subsystems.
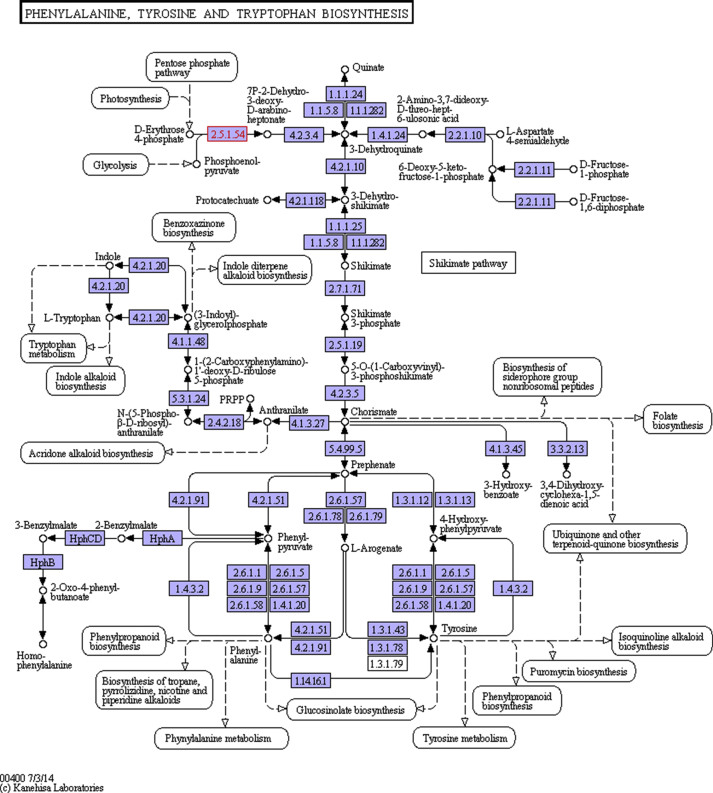
Most of the assembled ORFs annotated to protein gene tags of Lactobacillus plantarum, and Acetobacter pasteurianus in the subsystem database mapped to phenylalanine, tyrosine and tryptophan biosynthesis in the KEGG pathways database (Fig. 16). This indicated the roles played by these microbes for the formation of aromatic amino acid and derivatives biosynthesis processes.
Fig. 16.
Lactobacillus plantarum and Acetobacter pasteurianus sequences mapped to a metabolic pathway for phenylalanine, tyrosine and tryptophan biosynthesis based on KEGG database. The boxes in blue indicate enzymes that were mapped from the cocoa microbiome sequence data.
3.6.1. Lactobacillus plantarum
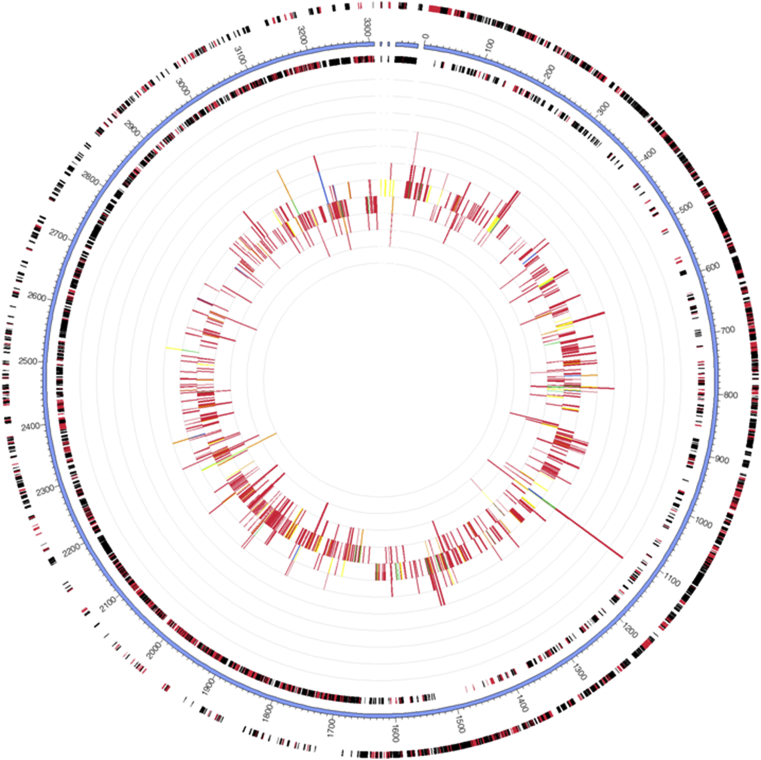
The recruited sequences for Lactobacillus plantarum when mapped to the reference genome of Lactobacillus plantarum ATCC 14917 downloaded from NCBI FTP site showed 6587 features were covered. The calculated GC content was 43.3%. The prominent features as annotated by RAST server were sequences encoding biotin, riboflavin, lipoic acid biosynthesis, phages and prophages elements. Sequences encoding for membrane transport were highly detected as well as fatty acid metabolisms. The recruited Lactobacillus plantarum showed a high number of sequence reads encoding for osmotic, oxidative, cold shock and detoxification stress responses (Fig. 17).
Fig. 17.
Recruitment plot of Lactobacillus plantarum from metagenome to the reference genome Lactobacillus plantarum ATCC 14917 in MG-RAST. The outmost circle represents forward strand genes (Red: protein, Black: RNA). The second circle is contigs for the reference genome. The third circle is the reverse strand genes (Red: protein, Black: RNA). The inner circle is abundant information of metagenome query microorganism based on color-coded by e-value exponent as: blue, −3 to −5; green, −5 to −10; yellow, −10 to −20; orange, −20 to −30; red, less than −30.
3.6.2. Acetobacter pasteurianus
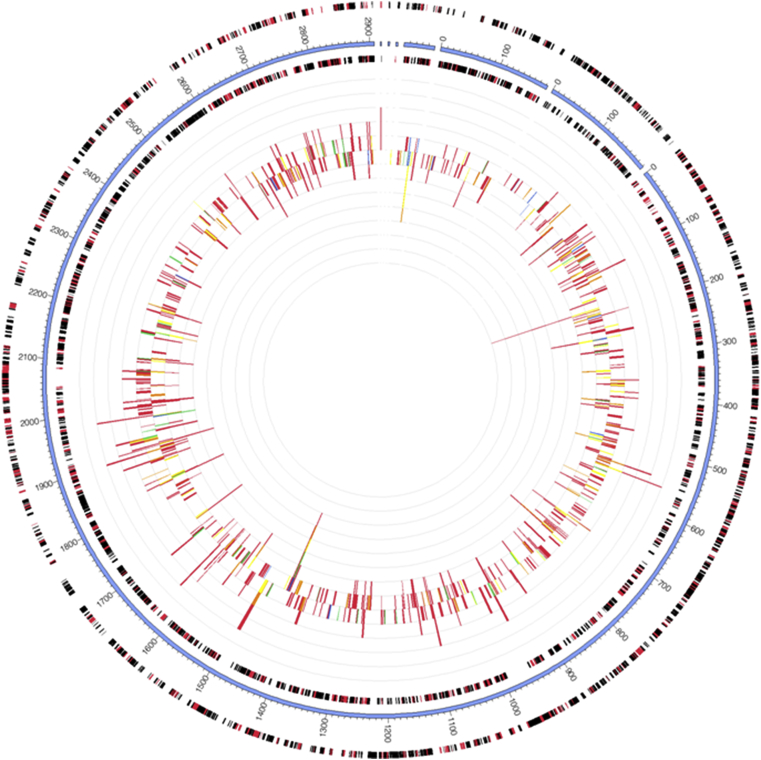
The recruited sequences for Acetobacter pasteurianus when mapped to the reference genome of Acetobacter pasteurianus IFO 3283-01 downloaded from NCBI FTP site on the RAST server for annotation showed 1127 contigs of protein-encoding genes (PEGs) (Fig. 18). The GC content calculated for this submitted sequences was 55.4%. The RAST server revealed several ORF sequence reads encoding for features such as biosynthesis of vitamins and pigment, cell wall and capsule components metabolisms, phages, prophages and transposable elements, respiration enzymes, osmotic and oxidative stress responses mapped unto Acetobacter pasteurianus during the fermentation period.
Fig. 18.
Recruitment plot of Acetobacter pasteurianus from metagenome to the reference genome Acetobacter pasteurianus IFO 3283-01 in MG-RAST. The outmost circle represents forward strand genes (Red: protein, Black: RNA). The second circle is contigs for the reference genome. The third circle is reverse strand genes (Red: protein, Black: RNA). The inner circle is abundant information of metagenome query microorganism based on color-coded by e-value exponent as: blue, −3 to −5; green, −5 to −10; yellow, −10 to −20; orange, −20 to −30; red, less than −30.
4. Discussion
High-throughput sequencing approach has the potential to identify rare microbial taxa responsible for shifts in the community structure (Hartmann et al., 2015). The dominant taxonomic phyla Proteobacteria and Firmicutes as observed were similar to findings in earlier studies carried out in Ghana (Camu et al., 2007) and Ecuador (Papalexandratou et al., 2011b). The co-dominance of Proteobacteria and Firmicute in cocoa is attributed to the rich source of carbohydrates, glucose, sucrose and glycerol in the pulp of cocoa beans (Camu et al., 2007). The substrate of fermentation environment and other environmental factors such as pH and temperature usually defines presence and succession of microbes (Nielsen et al., 2007). Aeration of the cocoa beans fermentation heap favours the growth of Acetobacter and Gluconobacter. These genera were significantly observed in this study are known to oxidize lactic acid to acetic acid and succinic acid. The presence of Bacillus pp in this study and filamentous fungi, such as Aspergillus, Penicillium are known to be potential microbes that cause off-tastes of the fermentation products if fermentation is allowed to continue beyond five days (Camu et al., 2007).
Additionally, the present study based on whole-genome shotgun sequencing revealed a rich microbial diversity with an alpha-diversity estimated at 142.81 with Lactobacillus plantarum and Acetobacter pasteurianus as dominant species as compared to a low alpha-diversity of 42.6 reported for kimchi (Park et al., 2009) which had Leuconostoc mesenteriodes as the most abundant species. Similarly, research on microbial diversity in Ghanaian cocoa fermentation had employed the method of culturing, biochemical test, PCR-DGGE and sequencing 16S rRNA amplicons but that resulted in only 16 species identified (Camu et al., 2007) as compared to the current research, which identified over fifty species. The low diversity associated with the 16S amplicons method can be attributed to the challenges of primer selectivity as compared to the metagenomic approach of directly sequencing genetic material from the environment. As noted in the present study, a higher number of species was achieved with a large dataset of sequence reads suggesting a contribution to the identification microbial species, especially low abundant and rare ones (Hasan et al., 2014; Sims et al., 2014). In an earlier study on cocoa fermentation, Acetobacter pasteurianus, Lactobacillus plantarum were identified as the most dominant (Camu et al., 2007). The current study concurred with that report with the identification of A. pasteurianus, L. plantarum, Gluconobacter oxydans, Gluconacetobacter diazotrophicus and Gluconacetobacter hansenii and, a further thirty-two more Lactobacillus species were identified as important microorganisms in cocoa bean fermentation. Previous studies (Nielsen et al., 2007; Papalexandratou et al., 2011a, b, c) failed to identify G. oxydans and G. diazotrophicus because these species cannot be grown easily under laboratory conditions, thereby supporting the need for use of metagenomics and shotgun sequencing (Schwan and Wheals, 2004; Camu et al., 2007; Hasan et al., 2014). The presence of Gluconobacter sp reflects their preference for ethanol and sugars as energy sources while the yeast population reported in this study metabolize sugars and citric acid for production of ethanol under anaerobic conditions below pH 4 in the carbohydrate-rich pulp (Koma et al., 2012). The presences of functional groups for vitamins, and pigment metabolism in this study can be correlated to microbial species such as yeast and LAB that have been widely employed in food fermentation processes for the biosynthesis of certain important products or metabolites. Yeasts and LAB are important probiotics because they are capable of synthesizing essential biomolecules such as vitamins or bioactive peptides to improve the nutritional and organoleptic properties of the final products. For example, S. cerevisiae is able to concentrate large quantities of thiamine, nicotinic acid and biotin and thus form enriched products (Patel et al., 2013). This study further reports the presence of bacteriophages in cocoa fermentation. Bacteriophages are of great economic importance in fermentation (Smid and Hugenholtz, 2010). They lyse bacterial cells leading to the release of bacterial content into the fermentation matrix thereby positively influencing the aroma profiles (Cardoso et al., 2012; Smid and Kleerebezem, 2014). The observation of ORFs encoding for peptidoglycan hydrolases (PGHs) and bacteriophages in the study was an indication of the lysing of lactic acid bacteria and other fermenting microorganisms cell wall and capsules that lead to the release of intracellular enzymes into the fermentation matrix. Many of these released enzymes further metabolize substrates in the food matrix with the accompanying release of aroma as noted in lactic acid fermentation (Smid and Hugenholtz, 2010; Smid and Kleerebezem, 2014). Although the lysing of bacteria by bacteriophage and expulsion of its contents may result in enhanced aroma profile, an excessive infestation of the fermenting mass with bacteriophage could lead to under-fermentation of the beans and thereby reducing the economic value of the beans. This phenomenon of under-fermentation caused by bacteriophages had also been observed in milk fermentation by LAB species (Marcó et al., 2012; Smid and Kleerebezem, 2014).
Predicting the function of the protein-coding genes enhances the understanding of microbial communities on how they are influenced by their environment (Weckx et al., 2011). DNA and RNA metabolism, membrane transport proteins, protein metabolism, regulation and cell signaling, respiration, nucleosides and nucleotides metabolisms among many others were functional proteins that were observed in this study. Nucleic acids are critical for the storage and use of genetic information, and its interpretation through the processes of transcription and protein biosynthesis. Coenzymes allow cells to use a small set of metabolic intermediates to carry chemical groups between different reactions. They act as a bridge between catabolism and anabolism. Membrane proteins are required to enable the specific passage or transport of selected substances across membranes (Klaenhammer et al., 2005; Weckx et al., 2011). They also act as receptors and signal sensors of environmental cues such as pH and temperature changes during cocoa bean fermentation. Similar observations were obtained for marine environment (Gilbert et al., 2008), kimchi (Park et al., 2011), snail gut (Cardoso et al., 2012) and human saliva (Hasan et al., 2014) microbiomes. Carbohydrate, amino acids and derivatives, aromatic compounds, are key functional categories that were identified in the current study. The observed results lend credence to the importance of LAB and AAB in the biosynthesis of carbohydrates, amino acids and their derivatives. This is evident in the presence of many ORFs reads annotated to LAB and AAB in accomplishing fermentation through the Glycolysis - Embden-Meyerhoff-Parnas pathway (EMP) and Pentose Phosphate Pathway (PPP) because the cocoa bean pulp is rich in glucose, sucrose, proteins, fatty acids that enhance metabolism (De Filippo et al., 2012). Furthermore, identification of ORFs sequence reads encoding for alcohol dehydrogenase, and series of enzymes sequence reads in the Entner-Doudoroff, glycolysis and pentose phosphate pathways by L. plantarum, A. pasteurianus and many other identified microbes were indicative of active utilization of glucose, sucrose, fructose, lactose, maltose and galactose by microorganism present during cocoa beans fermentation (Schwan and Wheals, 2004; Papalexandratou et al., 2011b). These bacteria synthesize pyruvate from glucose and generating cellular energy for cells metabolic activities. The pyruvate is converted to lactate by lactic acid bacteria and other mixed acid fermentation products of acetate, succinate, formate, and ethanol that are released into the fermenting mass to enhance flavour characteristics (Miguel et al., 2010; Nalbantoglu et al., 2014).
The observation of ORFs assigned to proteins, lipids, fats and fatty acid metabolism was highly indicative of the abundant aromatic compounds profile within cocoa beans fermentation, potentially impacting on the aroma profile of the fermented food product (Steen et al., 2005; Samson and Moineau, 2013; Smid and Kleerebezem, 2014). Additionally, the presences of ORFs encoding respiratory functional group in this study suggests the activities of aerobic acetic acid bacteria that is highly linked to the aerobic stage of the fermentation. Fermentation usually takes place in the absences of oxygen where it converts NADH and pyruvate produced in glycolysis into NAD+. However, in some instances during fermentation the availability of oxygen causes bacteria to use NADH and pyruvate to generate energy in respiration (Weckx et al., 2011; Illeghems et al., 2013). Periodic turning or mixing of beans for uniform fermentation results in the aeration of the heap and subsequently the onset of aerobic respiration due to the presence of acetic acid bacteria, Acetobacter and Gluconobacter sp.
The overlapping nature of microbial succession during cocoa fermentation confirms the evidence that most bacteria possess genes that enable them to survive at different times during the fermentation period. The presence of oxidative, osmotic and heat shock stress response sequences reads are suggestive of the adaptation mechanisms the microbes engage in to survive the varying changes in temperature, pH and metabolites during cocoa beans fermentation, which could play a role in detoxification of toxic compounds. The bacterial stress response enables bacteria to survive adverse and fluctuating conditions in their immediate surroundings. Various bacterial mechanisms recognize different environmental changes and mount an appropriate response. In addition, the observation of ORFs encoding for virulence, disease and defence and antibiotic resistance associated genes is expected in a heterogeneous and uncontrolled fermentation of cocoa beans fermentation. This is attributed to the microbial species succession whereby toxic compounds released by other bacteria in their defence towards survival and changing environmental conditions during fermentation which is the result of changing microbial community in the fermentation matrix. This inhibitory and defence mechanism is directed towards other species of the same genera or different genera by inhibiting cell wall synthesis and preventing RNase and DNase activities (Nes et al., 2007; Vaibhav et al., 2014). Further, this study also identified ORFs encoding for lactate fermentation, acetoin, and butanediol metabolism, which was consistent with previous studies (Park et al., 2011). This fermentation process produces aroma compounds. Functional enrichment of the microbiome showed that cocoa fermentation had high metabolic versatility with regard to amino acids, carbohydrate, amino acids metabolism and stress response. The functional distribution of protein-encoding ORFs in this study is similar to the findings reported in metagenomics study of kimchi (Park et al., 2011).
To the best of our knowledge, the present study is the first to concurrently, study the functional annotation of ORFs sequence reads in chorismate synthesis, shikimate pathways and biosynthesis of phenylacetaldehyde, acetoin and butanediol, and species diversity during cocoa fermentation. Furthermore, observation of sequence reads for chorismate synthesis involving identified series of aromatic enzymes such as chorismate mutase, chorismate synthase and DAHP synthase among others, in this study are involved in aromatic compound biosynthesis using shikimate pathway. These pathways lead to the production of organic acids and secondary metabolites and the release of aroma profiles (Koma et al., 2012). Chorismate biosynthetic pathway is central to most prokaryotes and several eukaryotes. Chorismate is an intermediate in the synthesis of L-phenylalanine, L-tyrosine and L-tryptophan. These amino acids serve as substrates and flavour precursors in other pathways for secondary metabolites such as alkaloids, flavonoids, and indole derivative compounds (Wegley et al., 2007). Microorganisms identified in this study were observed to use the Ehrlich pathways to produce aromatic compounds; diacetyl, acetaldehyde, 2,3-butanediol and acetoin. They transaminate the amine group to alpha-keto acids as was predicted in the present study, with the conversion of phenylpyruvate to phenylacetaldehyde and finally the aromatic compound. This property exhibited by these microorganisms is known to enhance the sensory characteristics of fermented food products (Smid and Kleerebezem, 2014).
Lactobacillus plantarum, Lactobacillus fermentum, Gluconobacter oxydans and Acetobacter pasteurianus among others in this study were observed to encode the common precursors of aromatic amino acids and secondary metabolites within the shikimate pathway with the transfer of phosphoryl catalysed by shikimate kinase enzyme within the shikimate pathway (Hartmann et al., 2006; Weckx et al., 2011). Similarly, the observation of ORFs encoding enzymes within the citrate metabolic pathway suggest the utilization and release of secondary metabolites and flavour compounds such as diacetyl, acetoin, butanediol, and acetaldehydes, which have a profound impact on the aroma of fermented food products. Acetaldehyde, an essential aroma compound synthesised from pyruvate is a precursor metabolite for other aroma compounds such as phenylacetaldehyde, acetate, ethanol and acetoin (Rademaker et al., 2007; Lachenmeier and Sohnius, 2008). These microorganisms usually secrete Acetoin, a colourless to pale yellow liquid with a pleasant, buttery odour, during fermentation. Its presence can be associated with the characteristic pleasant flavour observed during cocoa beans fermentation (Smid and Kleerebezem, 2014). The results in this study have shown that genes encoding for aromatic amino acids were found in the cocoa bean microbiome with the observation of ORFs encoding for enzymes within D-gluconate and ketogluconate, and L-ascorbate utilisation pathways. The flavour compounds originate from amino acid conversion to the corresponding alpha-keto acids through transamination with alpha-ketoglutarate as the most important amino acceptor (Schmeisser et al., 2007; Hehemann et al., 2010).
Among the significant findings of this study was the identification of ORFs encoding for an important purine alkaloid, theobromine synthesis that is related to caffeine. Cocoa plants accumulate theobromine instead of caffeine as the major purine alkaloid. The major sites of theobromine biosynthesis in cocoa plants are the young pericarp and cotyledons of fruits (Zheng et al., 2004). Theobromine is a known methylxanthine present in chocolate and other cocoa products which stimulates the heart, relaxes the smooth muscles and dilates blood vessels, and has specifically been used to treat high blood pressure and hardening of the arteries (Afoakwa et al., 2009).
In addition, metabolic pathways for various microbial biosynthesis of the substrate to secondary metabolites were elucidated based on protein-encoding ORFs in this study. Saccharomyces cerevisiae and L. plantarum metabolisms of sucrose and starch confirm the functional similarity in their metabolism. A similar result was observed in Lactobacillus plantarum, Acetobacter pasteurianus, Gluconobacter oxydans and Leuconostoc mesenteroides utilizing the phenylalanine, tyrosine and tryptophan biosynthesis pathways. A variety of microorganisms grow in the cocoa fermentation heap, but they do not all grow at the same time. Yeasts are the first to colonise the fermentation heap followed by LAB and finally, the presence of oxygen and the lower pH favours the growth of acetic acid bacteria such as Acetobacter and Gluconobacter. Their metabolic strategies describe their ecological role and usefulness in the industry (Hultman and Auvinen, 2010). Lactobacillus plantarum and Acetobacter pasteurianus were highly recruited when mapped to the reference genome. AAB are known to be actively involved in conversion of lactic acids produced by LAB to acetic acid (Camu et al., 2007). In conclusion, these results advocate the potential of a microorganism to synthesize flavour compounds and their derivatives via the aerobic and anaerobic pathways. Furthermore, the metabolic capabilities of these microorganisms can be harnessed and applied in the production of high quality starter cultures for controlled cocoa fermentation. High throughput sequencing enhances species detection and functional annotation of ORFs sequence reads.
Declarations
Author contribution statement
Daniel S. Agyirifo: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Mark Wamalwa: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Emmanuel Plas Otwe, Isaac Galyuon, Jemmy Takrama, Steven Runo, Joseph Ngeranwa: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Afoakwa E.O., Paterson A., Fowler M., Ryan A. Flavour formation and character in cocoa and chocolate: a critical review. Crit. Rev. Food Sci. Nutr. 2008;48:840–857. doi: 10.1080/10408390701719272. [DOI] [PubMed] [Google Scholar]
- Afoakwa E.O., Paterson A., Fowler M., Ryan A. Matrix effects on flavour volatiles release in dark chocolates varying in particle size distribution and fat content using GC-mass spectrometry and GC-olfactometry. Food Chem. 2009;113:208–215. [Google Scholar]
- Antony U., Chandra T.S. Microbial population and biochemical changes in fermenting finger millet (Eleusine coracana) World J. Microbiol. Biotechnol. 1997;13:533–537. [Google Scholar]
- Camu, De Winter T., Addo S.K., Takrama J.S., Bernaert H., De Vuyst L.D. Fermentation of cocoa beans: influence of microbial activities and polyphenol concentrations on flavour of chocolate. J. Sci. Food Agric. 2008;88:2288–2297. [Google Scholar]
- Camu, Winter T.D., Verbrugghe K., Cleenwerck I., Vandamme P., Takrama J.S., Vancanneyt M., Vuyst L.D. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 2007;73:1809–1824. doi: 10.1128/AEM.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso A.M., Cavalcante J.n.J.V., Cantão M.c.E., Thompson C.E., Flatschart R., Glogauer A., Scapin S.N., Sade Y.B., Beltrão P.M.S.I., Gerber A.L., Martins O.B., Garcia E.S., de Souza W., Vasconcelos A.T.R. Metagenomic analysis of the microbiota from the crop of an invasive snail reveals a rich reservoir of novel genes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel H.M., Vrancken G., Takrama J.F., Camu N., De Vos P., De Vuyst L. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009;9:774–783. doi: 10.1111/j.1567-1364.2009.00520.x. [DOI] [PubMed] [Google Scholar]
- De Filippo C., Ramazzotti M., Fontana P., Cavalieri D. Bioinformatic approaches for functional annotation and pathway inference in metagenomics data. Briefings Bioinf. 2012;13:696–710. doi: 10.1093/bib/bbs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauendorer F., Schieberlie P. Identification of the key aroma compounds in cocoa powder based on molecular sensory correlations. J. Agric. Food Chem. 2006;54:5521–5529. doi: 10.1021/jf060728k. [DOI] [PubMed] [Google Scholar]
- Garcia-Armisen T., Papalexandratou Z., Hendryckx H., Camu N., Vrancken G., Vuyst L.D., Cornelis P. Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16 S rRNA gene clone library. Appl. Microbiol. Biotechnol. 2010;87:2281–2292. doi: 10.1007/s00253-010-2698-9. [DOI] [PubMed] [Google Scholar]
- Gilbert J.A., Field D., Huang Y., Edwards R., Li W., Gilna P., Joint I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M., Frey B., Mayer J., Mader P., Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M.D., Bourenkov G.P., Oberschall A., Strizhov N., Bartunik H.D. Mechanism of phosphoryl transfer catalyzed by shikimate kinase from Mycobacterium tuberculosis. J. Mol. Biol. 2006;364:411–423. doi: 10.1016/j.jmb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hasan N.A., Young B.A., Minard-Smith A.T., Saeed K., Li H., Heizer E.M., McMillan N.J., Isom R., Abdullah A.S., Bornman D.M., Faith S.A., Choi S.Y., Dickens M.L., Cebula T.A., Colwell R.R. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann J.H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hultman J., Auvinen P. Metagenomics opens up new frontiers in microbiology. Duodecim. 2010;126:1278–1285. [PubMed] [Google Scholar]
- Illeghems K., De Vuyst L., Papalexandratou Z., Weckx S. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illeghems K., De Vuyst L., Weckx S. Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics. 2013;14:526–540. doi: 10.1186/1471-2164-14-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen L., Nielsen D.S., Hønholt S. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res. 2005;5:441–453. doi: 10.1016/j.femsyr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T.R., Barrangou R., Buck B.L., Azcarate-Peril M.A., Altermann E. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2005;29:393–409. doi: 10.1016/j.femsre.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Koma D., Yamanaka H., Moriyoshi K., Ohmoto T., Sakai K. Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl. Environ. Microbiol. 2012;78:6203–6216. doi: 10.1128/AEM.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenmeier D., Sohnius E.M. The role of acetaldehyde outside ethanol metabolism in the carcinogenicity of alcoholic beverages: evidence from a large chemical survey. Food Chem. Toxicol. 2008;46:2903–2911. doi: 10.1016/j.fct.2008.05.034. [DOI] [PubMed] [Google Scholar]
- Leal G.A., Gomes L.H., Efraim P., de Almeida Tavares F.C., Figueira A. Fermentation of cacao (Theobroma cacao L.) seeds with a hybrid Kluyveromyces marxianus strain improved product quality attributes. FEMS Yeast Res. 2008;8:788–798. doi: 10.1111/j.1567-1364.2008.00405.x. [DOI] [PubMed] [Google Scholar]
- Lefeber T., Janssens M., Camu N., De Vuyst L. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media to compose a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 2010;76:7708–7716. doi: 10.1128/AEM.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang H., Shi Y., Liu Y., Yu C., Wang B., Lu Y., Han C., Cheung D.W., Yiu S.-M., Peng S., Xiaoqian Z., Liu G., Liao X., Li Y., Yang H., Wang J., Lam T.-W., Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madorobaa E., Steenkamp E.T., Therona J., Scheirlinckc I., Cloetea T.E., Huys G. Diversity and dynamics of bacterial populations during spontaneous sorghum fermentations used to produce ting, a South African food. Syst. Appl. Microbiol. 2011;34:227–234. doi: 10.1016/j.syapm.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Marcó M.B., Moineau S., Quiberoni A. Bacteriophages and dairy fermentations. Bacteriophage. 2012;2:149–158. doi: 10.4161/bact.21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., Paarmann D., Souza M.D., Olson R., Glass E.M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R.A. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinf. 2008;9:1–8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel M.G.C.P., Cardoso P.G., Lago L.A., Schwan R.F. Diversity of bacteria present in milk kefir grains using culture-dependent and culture-independent methods. Food Res. Int. 2010;43:1523–1528. [Google Scholar]
- Nalbantoglu U., Cakar A., Dogan H., Abaci N., Ustek D., Sayood K., Can H. Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 2014;41:42–51. doi: 10.1016/j.fm.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Nes I.F., Yoon S., Diep D.B. Ribosomally synthesiszed antimicrobial peptides (bacteriocins) in lactic acid bacteria: a review. Food Sci. Biotechnol. 2007;16:675–690. [Google Scholar]
- Nielsen D.S., Honholt S., Tano-Debrah K., Jespersen L. Yeast populations associated with Ghanaian cocoa fermentations analysed using denaturing gradient gel electrophoresis (DGGE) Yeast. 2005;22:271–284. doi: 10.1002/yea.1207. [DOI] [PubMed] [Google Scholar]
- Nielsen D.S., Teniola O.D., Ban-Koffi L., Owusu M., Andersson T., Holzapfel W. The microbiology of Ghanaian cocoa fermentations analysed using culture dependent and culture independent methods. Int. J. Food Microbiol. 2007;114:168–186. doi: 10.1016/j.ijfoodmicro.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Noor-Soffalina S.S., Jinap S., Nazamid S., Nazimah S.A.H. Effect of polyphenol and pH on cocoa Maillard-related flavour precursors in a lipidic model system. Int. J. Food Sci. Technol. 2009;44:168–180. [Google Scholar]
- Oberparleiter S., Ziegleder G. Amyl acohols as compounds indicative of raw cocoa bean quality. Zeitschrift für Lebensmittel-Untersuchung-Forschung A. 1997;204:156–160. [Google Scholar]
- Ouattara H.G., Ban-Koffi L., Karou G.T., Sangare A., Niamke S.L., Diopoh J.K. Implication of Bacillus sp. in the production of pectinolytic enzymes during cocoa fermentation. World J. Microbiol. Biotechnol. 2008;24:1753–1760. [Google Scholar]
- Papalexandratou Z., Camu N., Falony G., De Vuyst L. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol. 2011;28:964–973. doi: 10.1016/j.fm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Papalexandratou Z., Falony G., Romanens E., Jimenez J.C., Amores F., Daniel H.-M., De Vuyst L. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl. Eniviron. Microbiol. 2011;77:7698–7714. doi: 10.1128/AEM.05523-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papalexandratou Z., Vrancken G., De Bruyne K., Vandamme P., De Vuyst L. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol. 2011;28:1326–1338. doi: 10.1016/j.fm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Park E.J., Chang H.W., Kim K.H., Nam Y.D., Roh S.W., Bae J.W. Application of quantitative real-time PCR for enumeration of total bacterial, archaeal, and yeast populations in kimchi. J. Microbiol. 2009;47:682–685. doi: 10.1007/s12275-009-0297-1. [DOI] [PubMed] [Google Scholar]
- Park E.J., Kim H., Guy C.J., Kim M., Roh S.W., Bae J. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 2011;77:1284–1291. doi: 10.1128/AEM.01859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Shah N., Prajapati J.B. Biosynthesis of vitamins and enzymes in fermented foods by lactic acid bacteria and related genera - a promising approach. Croat. J. Food Sci. Technol. 2013;5:85–91. [Google Scholar]
- Rademaker J.L., Herbet H., Starrenburg M.J., Naser S.M., Gevers D. Diversity analysis of dairy and nondairy Lactococcus lactisisolates, using a novel multilocus sequence analysis scheme and (GTG)5-PCR fingerprinting. Appl. Environ. Microbiol. 2007;73:128–137. doi: 10.1128/AEM.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho M., Tang H., Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson J.E., Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Ann. Rev. Food Sci. Technol. 2013;4:347–368. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- Sayers E.W., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Geer L.Y., Helmberg W., Kapustin Y., Lipman D.J., Madden T.L., Maglott D.R., Miller V., Mizrachi I., Ostell J., Pruitt K.D., Schuler G.D., Sequeira E., Sherry S.T., Shumway M., Sirotkin K., Souvorov A., Starchenko G., Tatusova T.A., Wagner L., Yaschenko E., Ye J. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser C., Steele H., Streit W.R. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 2007;75:955–962. doi: 10.1007/s00253-007-0945-5. [DOI] [PubMed] [Google Scholar]
- Schwan R.F., Wheals A.E. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 2004;44:205–221. doi: 10.1080/10408690490464104. [DOI] [PubMed] [Google Scholar]
- Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–133. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Smid E.J., Hugenholtz J. Functional genomics for food fermentation processes. Ann. Rev. Food Sci. Technol. 2010;1:497–519. doi: 10.1146/annurev.food.102308.124143. [DOI] [PubMed] [Google Scholar]
- Smid E.J., Kleerebezem M. Production of aroma compounds in lactic fermentations. Ann. Rev. Food Sci. Technol. 2014;5:313–326. doi: 10.1146/annurev-food-030713-092339. [DOI] [PubMed] [Google Scholar]
- Smit B.A., van Hylckama Vlieg J.E., Engels W.J., Meijer L., Wouters J.T., Smit G. Identification, cloning, and characterization of a Lactococcus lactis branched-chain α-keto acid decarboxylase involved in flavor formation. Appl. Environ. Microbiol. 2005;71:303–311. doi: 10.1128/AEM.71.1.303-311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit G., Smit B.A., Engels W.J. Flavor formation by lactic acid bacteria and biochemical flavor profiling of cheese products. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2005;29:591–610. doi: 10.1016/j.femsre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Steen A., Buist G., Horsburgh G.J., Venema G., Kuipers O.P., Foster S.J., Kok J. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 2005;272:2854–2868. doi: 10.1111/j.1742-4658.2005.04706.x. [DOI] [PubMed] [Google Scholar]
- Thomas T., Gilbert J., Meyer F. Metagenomics - a guide from sampling to data analysis. Microb. Inf. Exp. 2012;2:1–12. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.W., Chapman J., Hugenholtz P., Allen E.E., Ram R.J., Richardson P.M., Solovyev V.V., Rubin E.M., Rokhsar D.S., Banfield J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- Urso R., Rantsiou K., Dolci P., Rolle L., Comi G., Cocolin L. Yeast biodiversity and dynamics during sweet wine production as determined by molecular methods. FEMS Yeast Res. 2008;8:1053–1062. doi: 10.1111/j.1567-1364.2008.00364.x. [DOI] [PubMed] [Google Scholar]
- Vaibhav D.B., Anju P.K., Keyur D.B., Navin R.S., Chaitanya G.J. Analysis of virulence associated and antibiotic resistance genes of microbes in subclinical mastitis affected cattle milk by pyrosequencing approach. J. Vet Sci. Med. Diag. 2014;3:2–7. [Google Scholar]
- Vandamme E.J., Soetaert W. Bioflavours and fragrances via fermentation and biocatalysis. J. Chem. Technol. Biotechnol. 2002;77:1323–1332. [Google Scholar]
- Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J., Wu D., Paulsen I., Nelson K.E., Nelson W., Fouts D.E., Levy S., Knap A.H., Lomas M.W., Nealson K., White O., Peterson J., Hoffman J., Parsons R., Baden-Tillson H., Pfannkoch C., Rogers Y.H., Smith H.O. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Weckx S., Allemeersch J., Meulen R.V.d., Vrancken G., Huys G., Vandamme P., Hummelen P.V., De Vuyst L. Metatranscriptome analysis for insight into whole-ecosystem gene expression during spontaneous wheat and spelt sourdough fermentations. Appl. Eniviron. Microbiol. 2011;77:618–626. doi: 10.1128/AEM.02028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L., Breitbart M., Edwards R.A., Rohwer F. Metagenomic analysis of the microbial community associated with the coralPorites astreoides. Environ. Microbiol. 2007;9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- Wilmes P., Bond P.L. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Yao W., Ouattara H.G., Goualie B., Soumahoro S., Niamke S. Analysis of some functional properties of acetic acid bacteria involved in Ivorian cocoa fermentation. J. Appl. Biosci. 2014;75:6282–6290. [Google Scholar]
- Zheng X.Q., Koyama Y., Nagai C., Ashihara H. Biosynthesis, accumulation and degradation of theobromine in developing Theobroma cacao fruits. J. Plant Physiol. 2004;161:363–369. doi: 10.1078/0176-1617-01253. [DOI] [PubMed] [Google Scholar]