Abstract
The integrin α (ITGA) subfamily genes play a fundamental role in various cancers. However, the potential mechanism and application values of ITGA genes in colon adenocarcinoma (COAD) remain elusive. The present study investigated the significance of the expression of ITGA genes in COAD from the perspective of diagnosis and prognosis. A COAD RNA-sequencing dataset was obtained from The Cancer Genome Atlas. The present study investigated the biological function of the ITGA subfamily genes through bioinformatics analysis. Reverse transcription-quantitative polymerase chain reaction was applied to investigate the distribution of integrin α8 (ITGA8) expression in COAD tumors and adjacent normal tissues. Bioinformatics analysis indicated that ITGA genes were noticeably enriched in cell adhesion and the integrin-mediated signaling pathway, and co-expressed with each other. It was also revealed through observation that the majority of gene expression was significantly low in tumor tissues (P<0.05), and diagnostic receiver operating characteristic curves revealed that most of the genes could serve as significant diagnostic markers in COAD (P<0.05), especially ITGA8 which had a high diagnostic value with an area under curve (AUC) of 0.989 [95% confidence interval (CI) 0.980–0.997] in COAD (P<0.0001). In addition, ITGA8 expression was verified in clinical samples and it was revealed that it was higher in adjacent normal tissues (P=0.041) compared to COAD tissues, and the AUC was 0.704 (95% CI, 0.577–0.831; P<0.0085). Multivariate survival analysis indicated that integrin α (ITGA5) may be an independent prognostic indicator for COAD overall survival. Gene set enrichment analysis indicated that ITGA5 may participate in multiple biological processes and pathways. The present study revealed that ITGA genes were associated with the diagnosis and prognosis of COAD. The mRNA expression of ITGA8 may be a potential diagnosis biomarker and ITGA5 may serve as an independent prognosis indicator for COAD.
Keywords: integrin α, mRNA, colon adenocarcinoma, prognosis, diagnosis
Introduction
Colon cancer (CRC) is the fourth leading cause of cancer-related deaths throughout the world after lung, breast, and prostate cancer. Based on the GLOBOCAN data, more than 1.8 million newly diagnosed carcinoma cases and 881,000 deaths related to this disease occurred in 2018 worldwide (1). Colon adenocarcinoma (COAD) is one of the most common pathological types of colon cancer (2). In recent years, colon adenocarcinoma has a significant upward trend in morbidity and mortality (3), especially in Western developed countries and Asian developing countries (4). Although, there are many treatments, including surgery and chemotherapy, the five-year survival rate of COAD is still not promising (5). Late diagnosis, unreliable biomarkers and therapeutic targets have become major obstacles in the treatment of colon adenocarcinoma (6). Therefore, early diagnosis and treatment are essential for the improvement of the prognosis and quality of life of the patients. Finding new targets in COAD may provide new alternatives and insights for comprehensive management strategies for COAD patients.
Integrins belong to heterodimeric surface receptors, which are composed of non-covalently associated α and β subunits, and as far as we know, the integrin family consists of 18α and 8β members (7–11). ITGA, a subfamily of integrins, has an α subunit composed of a seven-bladed β-propeller, a thigh, and two calf domains (12). There is an I domain (also called A domain), composed of ~200 amino acids inserted between blades 2 and 3 in the β-propeller, and contained in nine of the 18 integrin α chains (13). There are also domains that bind Ca2+ on the lower side of the blades facing away from the ligand-binding surface which are contained in the last three or four blades of the β propeller. Ligand binding is influenced by Ca2+ binding to these sites allosterically (14,15). Previous research has revealed that the integrin family mediates signal transduction by binding to the extracellular matrix via adhesion receptors on its surface (16). Each integrin has multiple activation states (12), and exerts effects through cascaded amplification of various paths (17). Extensive studies have revealed that integrins could function as signaling molecules through the cell membrane in either direction: ‘inside-out signaling’ caused by extracellular stimulation that causes intracellular linin and kindlin to bind to the cytoskeleton, leaving the extracellular domain in a high affinity state (8,18–20); and ‘outside-in signaling’, a complicated process in which the heterodimeric adhesion receptors of the integrins mediate cell adhesion to the extracellular matrix (ECM), then activate integrins to engage and interact with the cytoskeleton in order to activate a variety of intracellular signaling pathways (12), which enhance binding of activated integrin ligands and allow for the perception of the intracellular environment (9,20,21). These integrins could control cell attachment, movement, growth and differentiation, as well as survival (12,22).
Integrins modulate muititudinal human pathologies including thrombotic diseases, infectious diseases, inflammation, fibrosis, and cancer (17). In cancer, members of the integrin family of pattern recognition receptors participate in many cellular processes in the body, including adhesion, metastatic spread of tumor cells, and identification (22). In addition to altering the interaction of cells with the surrounding environment, the proliferation, survival and differentiation of cancer cells can be promoted by integrins through growth factors such as EGFR, VEGFR interaction, or tyrosine kinase receptors (23). Integrins, as cell adhesion receptors, are also observed and have been reported in various types of cancer, such as multiple myeloma (24), NSCLC (25), glioma (26), ovarian cancer (27) and oral squamous cell carcinoma (28). However, the potential mechanism and application value of ITGA genes remain elusive. Therefore, the aim of the present study was to explore the potential application of ITGA genes of COAD in the perspective of diagnosis and prognosis by using an RNA-sequencing (RNA-Seq) dataset from The Cancer Genome Atlas (TCGA; http://tcga-data.nci.nih.gov/).
Materials and methods
Patient information
TCGA databases were accessed on October 30, 2018, and a total of 456 COAD patient clinical parameters which consisted of 480 tumor and 41 adjacent normal tissue samples were collected. Clinical parameter information including sex, age, survival time (days), survival status and tumor-node-metastasis (TNM) stage were obtained.
Bioinformatics analysis of ITGA genes
To study the biological enrichment function of the ITGA subfamily, the online tool Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/; version 6.8; accessed January 5, 2019) (29,30), containing gene ontology (GO) enrichment functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (31,32) was used. An enrichment P-value <0.05 was considered as significant from a statistical perspective. Gene ontology includes 3 independent modules, biological processes (BP), molecular functions (MF), and cellular components (CC) (31). GO terms of ITGA genes were also obtained using the Biological Networks Gene Ontology tool (BiNGO) in Cytoscape_version 3.6.1 (33). Gene-gene interactions of ITGA genes were then investigated using Gene Multiple Association Network Integration Algorithm (GeneMANIA, http://www.genemania.org/, accessed December 25, 2018) (34). Protein-protein interactions (PPI) of ITGA genes were performed using the Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/, accessed November 19, 2018) (35,36).
mRNA expression levels of ITGA genes and diagnostic receiver operating characteristic (ROC) curves
The mRNA expression levels of ITGA genes were presented by box plots and scatter plots. A box plot of ITGA genes was downloaded from the Metabolic gEne RApid Visualizer (MEARV) (http://merav.wi.mit.edu/, accessed January 21, 2019) (37), while a scatter plot was generated from the TCGA dataset to integrate cancer and adjacent normal tissues of mRNA expression levels at 75% cut-off values. Diagnostic ROC curves investigated the statistically significant expression of tumor tissues and adjacent normal tissues in TCGA cohort.
Verification of the first affiliated hospital of Guangxi medical university cohort
COAD patient tissue samples
COAD patient tissues, 30 in all, including tumor tissues and paired adjacent normal tissues, were collected (from April to June 2018) at the Department of Colorectal and Anal Surgery of The First Affiliated Hospital of Guangxi Medical University (Nanning, China). All patients signed an informed consent form, and the experimental protocol was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University. [No. 2019(KY-E-001)]. Immediately after surgery, the tissue was placed in RNA protection solution and transferred to a −80°C refrigerator for preservation. The postoperative pathological diagnosis was COAD.
Detection of ITGA8 expression by reverse transcription- quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to assess ITGA8 expression in COAD tissue samples, including tumor and adjacent normal tissues. TRIzol® reagent (15596026; Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from tissues. Total RNA concentration was detected by NanoDrop One (Thermo Fisher Scientific, Inc.). And the RNA was reverse-transcribed (20-µl reaction system) applying a reverse transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) to create cDNA. Then in accordance with the FastStart Universal SYBR Green Master (ROX) kit (Roche Diagnostics) and the Applied Biosystems Quantsudio™ Real-Time PCR System (Q6) operation guide (Applied Biosystems; Thermo Fisher Scientific, Inc.), the reaction procedure was set up. The reaction conditions used were as follows: Pre-denaturation at 95°C for 10 min; then denaturation at 95°C for 15 sec, 60°C extension for 60 sec, 40 cycles; finally denaturation at 95°C for 15 sec, 60°C for 1 min, 95°C for 30 sec, and 60°C for 15 sec. GAPDH was used as an internal reference gene, and the primer sequences of ITGA8 and GAPDH were synthesized by Sangon Biotech Co., Ltd. The primer sequences were as follows: GAPDH forward, 5′-TGGTCCCTGCTCCTCTAAC-3′ and reverse, 5′-GGCTCAATGGCGTACTCTC-3′; and ITGA8 forward, 5′-GCTGCTGGGGAGTTTACTGG-3′ and reverse, 5′-GATGCCATCTGTTCTCCCGTG-3′. The gene expression level in the present study was calculated using the 2−∆∆Cq method (38).
Survival analysis
In TCGA database, 438 COAD patients were categorized into two groups namely a high and low-expression group, which were based on the 75% cut-off value of gene expression. Kaplan-Meier survival analysis was performed for sex, age, and stage, respectively. Then overall survival (OS) was determined to evaluate the prognostic value of COAD patients. Furthermore, sex, age, and TNM stage were adjusted using Cox proportional hazards regression model in TCGA database.
Gene set enrichment analysis (GSEA)
To further explore the potential value of biological processes and pathways, multivariate prognostic significance of the ITGA5 gene was grouped into low and high expression categories based on the 75% cut-off value of the expression levels. GSEA (http://software.broadinstitute.org/gsea/index.jsp, downloaded January 20, 2018) (39,40) was conducted to investigate underlying mechanisms by using the Molecular Signatures Database (MSigDB) c2 (c2.cp.kegg.v6.2.symbols.gmt) and c5 (c5.all.v6.2.symbols.gmt) (41). The enrichment gene sets in GSEA were identified as statistically significant when a nominal P-value <0.05 and a false discovery rate (FDR) <0.25 were attained.
Statistical analysis
Kaplan-Meier survival analysis and the log-rank test were conducted to assess different subgroups categorized by clinical and gene variables. Adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) were obtained using univariate and multivariate Cox proportional hazards models. TNM stage was selected to set up a Cox proportional hazard regression model. The paired t-test was applied for comparison of data between COAD tumors and adjacent normal tissues. A P<0.05 indicated that the differences exhibited statistical significance. The FDR in GSEA was adjusted for multiple testing according to the Benjamini-Hochberg procedure (42,43). All of the aforementioned statistical analyses were performed with SPSS Statistics software version 20.0 (IBM Corp.). Vertical scatter plots, ROC and survival curves were plotted using GraphPad Prism v.7.0 (GraphPad Software, Inc.).
Results
Baseline patient characteristics in TCGA
The expression of the ITGA subfamily of related genes was included from the TCGA RNAseq database. Firstly, information concerning tumor and adjacent normal tissues was isolated. Then clinical information was integrated with gene expression. In addition, cases that had no clinical prognostic information and people who had a survival time of 0 were excluded. Finally, information on the 438 COAD patient tumor samples and 41 adjacent normal tissue samples was obtained. Detailed baseline characteristics of the 438 COAD patients from the TCGA database are summarized in Table I. It was revealed that sex and age were not correlated with OS (all P>0.05). However, TNM stage was notably associated with OS (log-rank test P<0.001, adjusted P<0.001).
Table I.
Baseline patient characteristics in a TCGA cohort.
| OS | |||||
|---|---|---|---|---|---|
| Variables | Patients (n=438) | No. of events | MST (months) | HR (95% CI) | Log-rank P-value |
| Age (years) | |||||
| <65 | 168 | 30 | NA | 1 | 0.17 |
| ≥65 | 268 | 67 | 82.5 | 1.353 (0.879–2.081) | |
| Missinga | 2 | ||||
| Sex | |||||
| Male | 234 | 54 | 82.5 | 1 | 0.545 |
| Female | 204 | 44 | NA | 0.884 (0.593–1.318) | |
| Stage | |||||
| 1 and 2 | 240 | 34 | 101.4 | 1 | <0.001 |
| 3 and 4 | 187 | 59 | 62.7 | 2.684 (1.758–4.099) | |
| Missingb | 11 | ||||
Missing, information of age was unknown in 2 patients
Missing, information of TNM stage was not reported in 10 patients. TCGA, The Cancer Genome Atlas; OS, overall survival; MST, median survival time; 95% CI, 95% confidence interval; HR, hazards ratio; NA, not available.
Analysis of ITGA subfamily mRNA expression levels in TCGA databases
The 75% cut-off value of gene expression levels was used to categorize COAD patients into low-level groups and high-level groups. Then TNM stage was used for adjustment of these genes. Multivariate analysis indicated that ITGA5 exhibited statistical significance [P=0.016; HR (95% CI)=1.681 (1.100–2.570)] (Table II).
Table II.
Prognostic values of ITGA subfamily gene expression in COAD of a TCGA cohort.
| OS | |||||
|---|---|---|---|---|---|
| Gene | Patients (n=438) | No. of events | MST (days) | HR (95% CI) | Adjusted P-valuea |
| ITGA1 | |||||
| Low | 329 | 77 | 3,042 | 1 | 0.303 |
| High | 109 | 21 | 2,134 | 0.775 (0.477–1.259) | |
| ITGA2 | |||||
| Low | 329 | 78 | 2,532 | 1 | 0.176 |
| High | 109 | 20 | NA | 0.711 (0.434–1.165) | |
| ITGA2B | |||||
| Low | 329 | 74 | 2,475 | 1 | 0.792 |
| High | 109 | 24 | NA | 1.064 (0.670–1.691) | |
| ITGA3 | |||||
| Low | 329 | 70 | 2,532 | 1 | 0.898 |
| High | 109 | 28 | 2,047 | 0.971 (0.620–1.521) | |
| ITGA4 | |||||
| Low | 329 | 73 | 2,821 | 1 | 0.434 |
| High | 109 | 25 | 1,661 | 1.203 (0.757–1.912) | |
| ITGA5 | |||||
| Low | 329 | 73 | 2,821 | 1 | 0.016 |
| High | 109 | 25 | 2,047 | 1.681 (1.100–2.570) | |
| ITGA6 | |||||
| Low | 329 | 77 | 2,532 | 1 | 0.284 |
| High | 109 | 21 | NA | 0.767 (0.471–1.246) | |
| ITGA7 | |||||
| Low | 329 | 73 | 2,532 | 1 | 0.763 |
| High | 109 | 25 | 3,042 | 0.932 (0.59–1.472) | |
| ITGA8 | |||||
| Low | 329 | 80 | 2,475 | 1 | 0.206 |
| High | 109 | 18 | NA | 0.718 (0.430–1.199) | |
| ITGA9 | |||||
| Low | 329 | 62 | 2,821 | 1 | 0.165 |
| High | 109 | 36 | 2,047 | 1.340 (0.887–2.024) | |
| ITGA10 | |||||
| Low | 329 | 69 | 2,821 | 1 | 0.069 |
| High | 109 | 29 | 2,047 | 1.506 (0.969–2.343) | |
| ITGA11 | |||||
| Low | 329 | 70 | 2,821 | 1 | 0.641 |
| High | 109 | 28 | 1,910 | 1.111 (0.713–1.731) | |
| ITGAD | |||||
| Low | 329 | 73 | 2,475 | 1 | 0.801 |
| High | 109 | 25 | 3,042 | 1.060 (0.672–1.672) | |
| ITGAE | |||||
| Low | 329 | 80 | 2,475 | 1 | 0.438 |
| High | 109 | 18 | NA | 0.815(0.486–1.366) | |
| ITGAL | |||||
| Low | 329 | 72 | 2,532 | 1 | 0.173 |
| High | 109 | 26 | 2,134 | 1.370 (0.871–2.156) | |
| ITGAM | |||||
| Low | 329 | 72 | 2,821 | 1 | 0.382 |
| High | 109 | 26 | 2,134 | 1.222 (0.779–1.917) | |
| ITGAV | |||||
| Low | 329 | 81 | 2,532 | 1 | 0.327 |
| High | 109 | 17 | 2,047 | 0.768(0.452–1.303) | |
| ITGAX | |||||
| Low | 329 | 76 | 2,532 | 1 | 0.665 |
| High | 109 | 22 | 3,042 | 1.111(0.689–1.793) | |
Adjusted P-value, adjustment for TNM stage. COAD, colon adenocarcinoma; ITGA, integrin α; OS, overall survival; MST, median survival time; 95% CI, 95% confidence interval; HR, hazards ratio; NA, not available.
Bioinformatics analysis of the ITGA genes
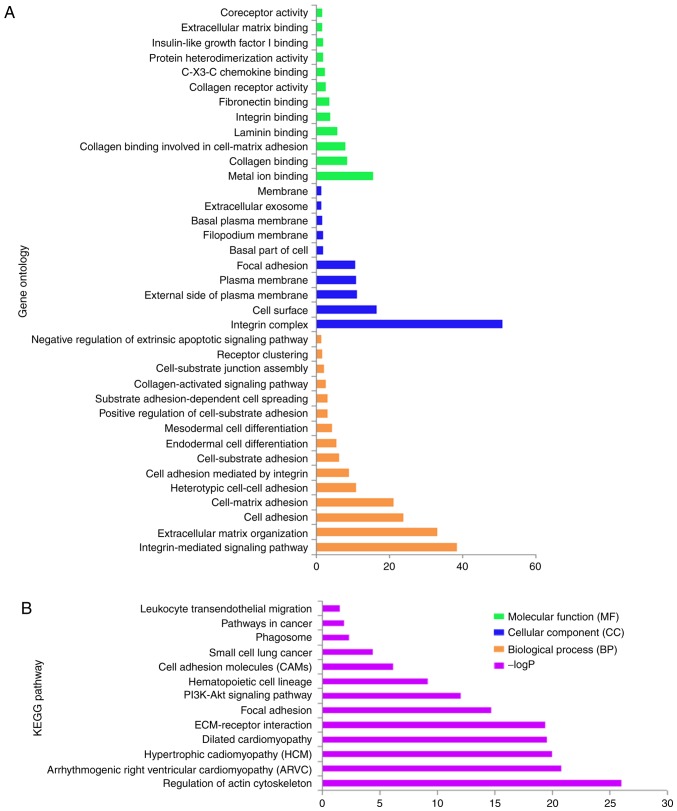
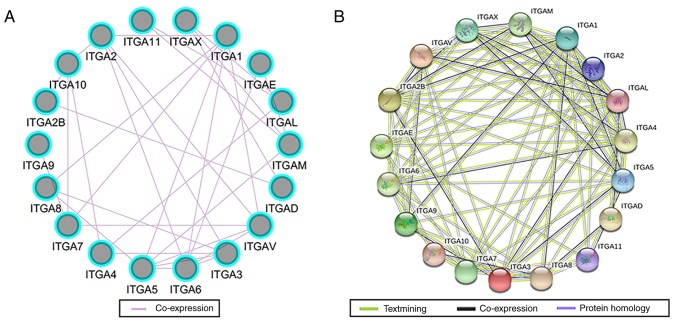
GO term enrichment analysis of ITGA genes revealed that biological processes mainly involved cell adhesion and the integrin-mediated signaling pathway (Fig. 1A and S1). KEGG pathway analysis mainly involved focal adhesion, the PI3K/AKt signaling pathway and regulation of actin cytoskeleton (Fig. 1B and S2). The interaction networks of gene-gene and protein-protein indicated that ITGA genes had co-expression with each other and with complex gene-gene and protein-protein interaction networks (Fig. 2A and B).
Figure 1.
GO term and KEGG analysis of the ITGA subfamily genes. (A) GO term enrichment. (B) KEGG enrichment. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ITGA, integrin α.
Figure 2.
Gene-gene and protein-protein interaction networks of ITGA genes. (A) GeneMANIA interaction networks. (B) Protein-protein interaction networks. ITGA, integrin α.
Analysis of ITGA subfamily gene expression levels in tumor and adjacent normal tissues based on TCGA
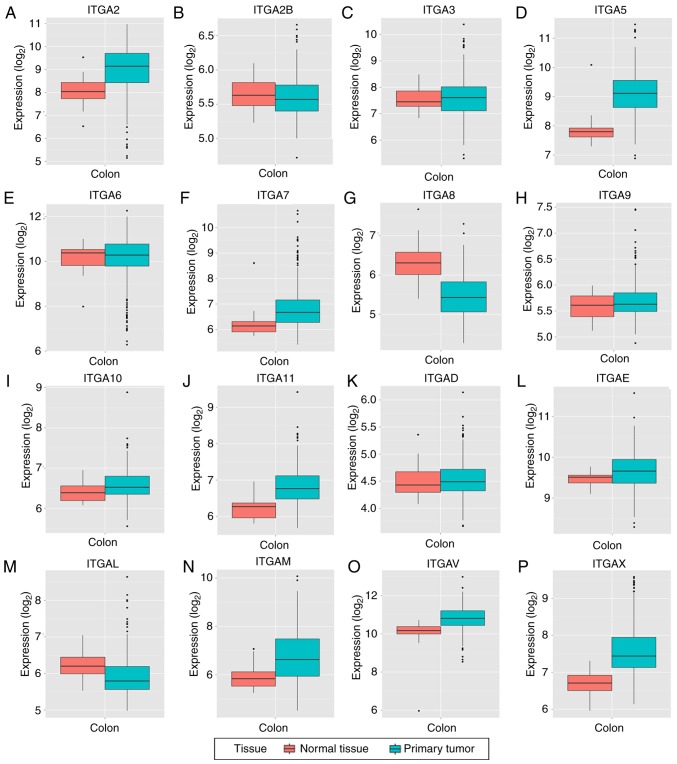
Box plots of the expression levels of 16 genes are presented in Fig. 3 (ITGA1 and ITGA4 are not presented). ITGA2B, ITGA6, ITGA8 and ITGAL were high in expression in adjacent normal tissues compared to tumor tissues, while the other 12 genes were high in tumor tissues compared to normal tissues.
Figure 3.
(A-P) The mRNA expression levels of ITGA genes in normal colon tissue and primary colon tumors. ITGA, integrin α.
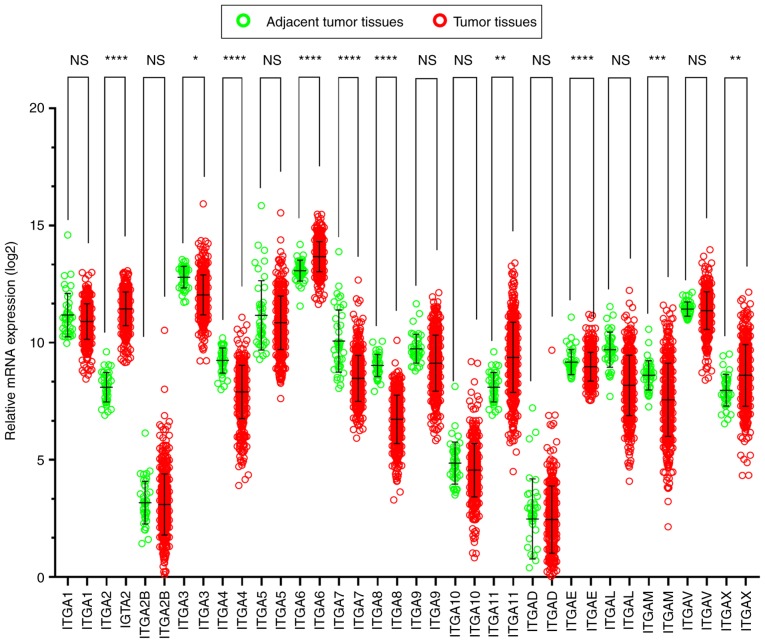
The scatter diagrams were used to present the expression between the tumor and adjacent tissues (Fig. 4) and the results revealed that ITGA2B, ITGA5, ITGA10, ITGAD, ITGAE and ITGAV exhibited no statistical significant differences, however the other genes significantly differed). It was also observed that the majority of genes were expressed at a significantly low level in tumor tissues, while the expression of adjacent normal tissues was high.
Figure 4.
Gene expression distribution of ITGA genes in TCGA. NS, not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. ITGA, integrin α; TCGA, The Cancer Genome Atlas.
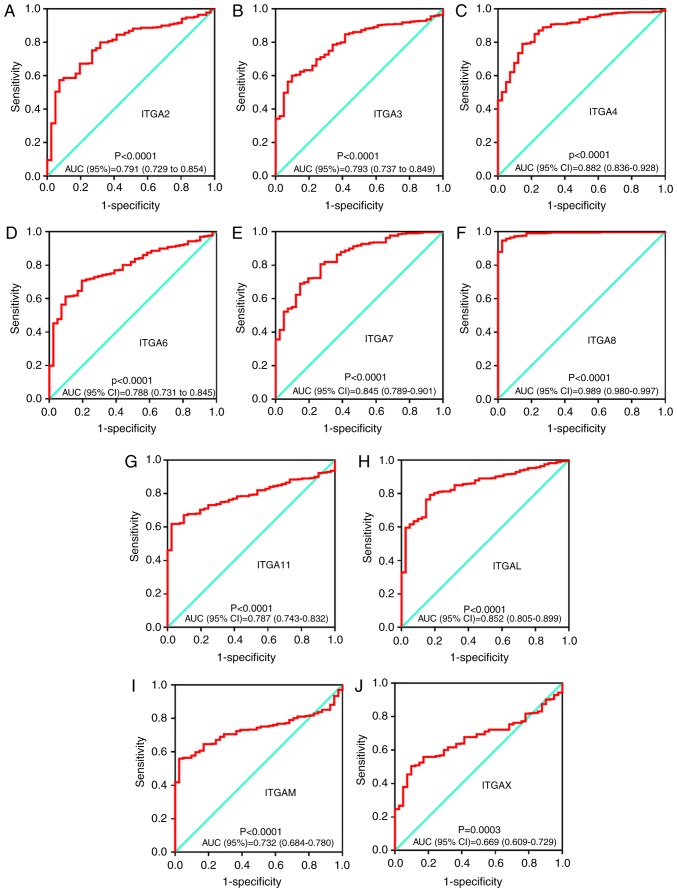
The possible potential application of ITGA genes in COAD tumor and adjacent tissues was further explored. The diagnostic ROC analysis of ITGA genes in the TCGA COAD cohort showed that ITGA2, ITGA3, ITGA4, ITGA6, ITGA7, ITGA8, ITGA11, ITGAL, ITGAM and ITGAX can serve as potential diagnostic biomarker for COAD (all P<0.05). Notably, ITGA8 [AUC (95% CI)=0.989 (0.980–0.997)] exhibited a high diagnostic value distinguishing tumor tissues and adjacent normal tissues of COAD (P<0.0001). All ROC curves are presented in Fig. 5.
Figure 5.
Diagnostic ROC curves of ITGA genes distinguishing COAD tumor tissues and adjacent normal tissues in a TCGA cohort. ROC curves of (A) ITGA2; (B) ITGA3; (C) ITGA4; (D) ITGA6; (E) ITGA7; (F) ITGA8; (G) ITGA11; (H) ITGAL; (I) ITGAM; and (J) ITGAX. ROC, receiver operating characteristic; ITGA, integrin α; COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; AUC, area under the curve.
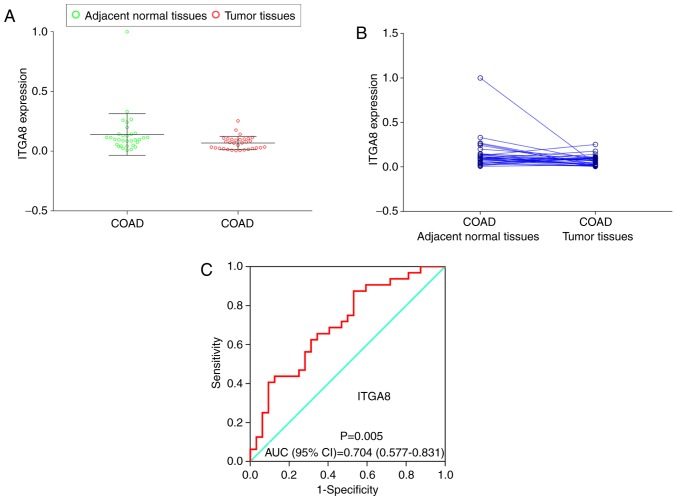
Validation of ITGA8 expression in clinical samples
To investigate and further validate the possible function of ITGA8 expression in the clinical sample cohort, the paired t-test was performed between COAD tumors and adjacent normal tissues (P=0.041), and a scatter diagram was selected to compare the expression levels of the clinical sample cohort and TCGA cohort (Fig. 6A and B). The results indicated that both cohorts exhibited a significantly high expression level in adjacent normal tissues. Then, the diagnostic ROC curve was used to study the underlying role of ITGA8 in clinical samples. The result revealed that ITGA8 had a significant value [P=0.005, AUC (95% CI)=0.704 (0.577–0.831)]; (Fig. 6C).
Figure 6.
RT-qPCR is used to assess the relative expression of ITGA8 in COAD adjacent normal tissues and tumor tissues. (A and B) Gene relative expression distribution of ITGA8. (C) Diagnostic ROC of ITGA8 relative expression distribution. ITGA, integrin α; COAD, colon adenocarcinoma; ROC, receiver operating characteristic; AUC, area under the curve.
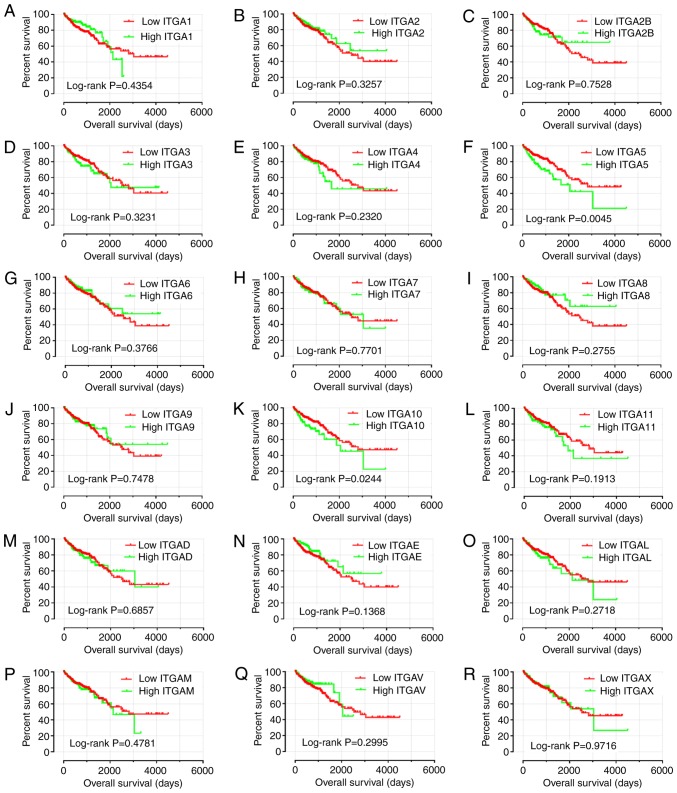
Prognostic survival analysis
To further explore the survival values, survival analysis curves were drawn according to gene expression (Fig. 7). Only ITGA5 and ITGA10 exhibited statistical significance (P<0.05). Consequently, it was observed that a high level of ITGA5 and ITGA10 expression were linked with poor prognosis for OS (log-rank test, P=0.0045 and P=0.0244).
Figure 7.
Kaplan-Meier survival curves for ITGA genes in COAD of TCGA cohort. OS stratified by (A) ITGA1, (B) ITGA2, (C) ITGA2B, (D) ITGA3, (E) ITGA4, (F) ITGA5, (G) ITGA6, (H) ITGA7, (I) ITGA8, (J) ITGA9, (K) ITGA10, (L) ITGA11, (M) ITGAD, (N) ITGAE, (O) ITGAL, (P) ITGAM, (Q) ITGAV, and (R) ITGAX. ITGA, integrin α; COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; OS, overall survival.
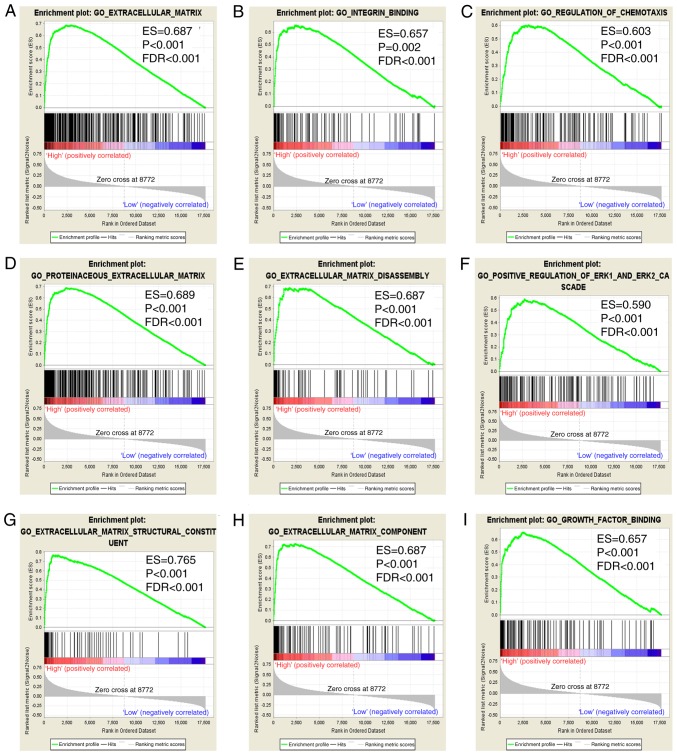
GSEA
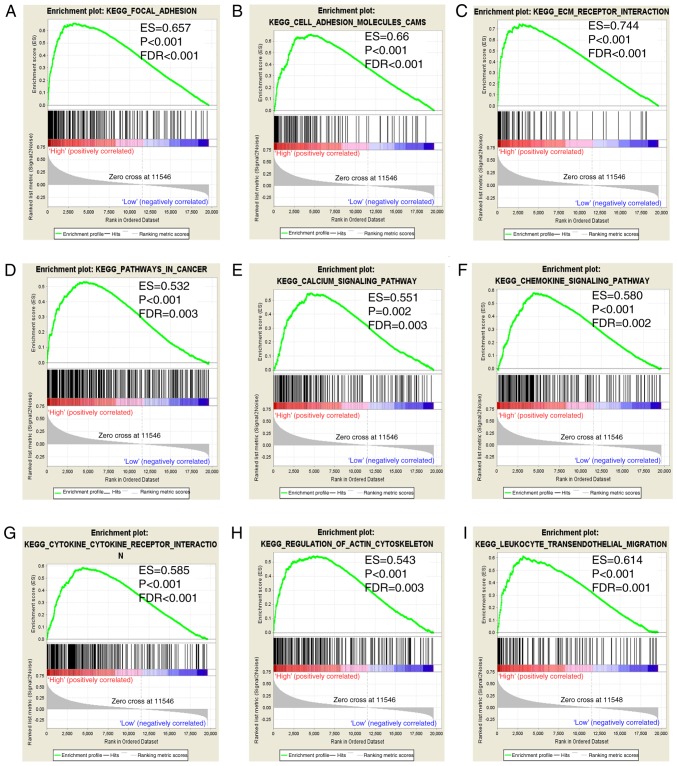
In the present study, prognostic value of ITGA5 was assessed to investigate its potential in GO terms and KEGG pathways in COAD prognosis. GSEA revealed that the c5 gene sets indicated that the high expression of ITGA5 may be mostly enriched in ECM (Fig. 8A-I). In addition, the c2 gene sets were significantly involved in focal adhesion, the chemokine signaling pathway, pathways in cancer and ECM receptor interaction (Fig. 9A-I).
Figure 8.
GSEA results of ITGA5 in TCGA COAD patients. (A-I) GSEA results of c5 reference gene sets for high-ITGA5 expression groups. GSEA, gene set enrichment analysis; ITGA5, integrin α5; TCGA, The Cancer Genome Atlas; COAD, colon adenocarcinoma.
Figure 9.
GSEA results of ITGA5 in TCGA COAD patients. (A-I) GSEA results of c2 reference gene sets for high-ITGA5 expression groups. GSEA, gene set enrichment analysis; ITGA5, integrin α5; TCGA, The Cancer Genome Atlas; COAD, colon adenocarcinoma.
Discussion
As is recognized, the occurrence and development of tumors are caused by multiple factors, and the homeostasis of the internal environment is crucial. Integrins are a family of cell adhesion proteins that can mediate cell-cell, cell-extracellular matrix (ECM), cell-pathogen interactions and signaling through adhesion receptors (7,12,44,45). The integrins are the main receptors for extracellular matrix proteins like collagen, fibronectin and laminin. In addition, integrins play a fundamental role in various biological processes via cellular adhesion mechanisms (10,46). The ITGA family is a subfamily of integrins, and certain previous studies had reported the relationship between the ITGA subfamily genes and colorectal cancer. Yang et al reported that ITGA2 was significantly overexpressed in both primary colon tumors and liver metastases with tissues from 43 patients as was determined by western blotting, immunohistochemistry and tissue microarray (47). The expression of ITGA3 was linked to other genes by cDNA Array and immunohistochemistry in colorectal cancer. It was revealed that ITGA3 was overexpressed in tumor tissues. In a study by Waisberg et al, the expression of ITGAV was assessed by PCR and immunohistochemistry in adult CRC patients (n=114), and the results indicated that the overexpression of ITGAV was associated with higher progression and spread of CRC (48). ITGA subfamily genes have also been reported in other types of cancer. ITGA1 was recently revealed to be associated with an invasive metastatic phenotype in hepatocellular and prostate cancers (49,50). Other studies revealed that ITGA2 was expressed in gastric cancer (51), pancreatic cancer (52) and pancreatic ductal adenocarcinoma (PDAC) (53). In addition, ITGA10 was expressed in B-cell lymphoma (54) and ITGA11 was expressed in breast cancer (55), lung squamous cancer (56) and neck squamous cell carcinoma (57).
However, there is little knowledge about the relationship between the ITGA subfamily genes and COAD. To the best of our knowledge, this was the first time that TCGA RNA sequencing dataset and PCR detection were used to investigate diagnostic and prognostic values of ITGA subfamily genes in COAD. The present results indicated that the mRNA expression levels of the ITGA subfamily genes were correlative with COAD in diagnosis and prognosis. Gene function enrichment analysis revealed that ITGA genes were significantly involved in biological processes, pathways of cell adhesion and the integrin-mediated signaling pathway. In addition, co-expression analysis revealed that ITGA genes were co-expressed with each other at both the gene and protein levels.
It was determined that ITGA2, ITGA6, ITGA11 and ITGAX were significantly expressed at a high level in cancer tissues, while ITGA1, ITGA3, ITGA4, ITGA7, ITGA8, ITGA9, ITGAL and ITGAM were significantly expressed at a high level in adjacent normal tissues in a TCGA cohort. The results of ROC curves revealed that ITGA8 had a high diagnostic value [AUC (95% CI)=0.989 (0.980–0.997)]. Kok-Sin et al reported that ITAGA8 was considered as a potential diagnostic marker, serving as a tumor suppressor gene as determined via DNA methylation and gene expression profiling assays, in colorectal cancer (58). In a study by Yang et al, the ITGA8 mRNA and protein levels were assessed in 483 LUAD tissues and 59 adjacent tissues, and the results indicated that the expression of ITGA8 was downregulated in LUAD (59). Then, to further validate the expression of the ITGA8 gene in cancer and adjacent tissues of COAD, RT-qPCR was performed, and the results revealed that ITGA8 was significantly expressed at a high level in adjacent normal tissues of COAD. Thus, it was hypothesized that ITGA8 may be a potential diagnostic marker in COAD.
Survival prognosis analysis results revealed that the high expression levels of ITGA5 and ITGA10 were associated with poor prognosis, while Kaplan-Meier curves from multivariate survival analysis revealed that the low expression of ITGA5 was linked to favorable prognosis of COAD OS in the TCGA cohort. Especially ITGA5 was an independent prognosis factor for OS of COAD patients. However, previous studies revealed that overexpression of ITGA5 indicated poor prognosis. A study by Shang et al revealed that low expression of ITGA5 indicated a good overall survival (OS) or relapse-free survival (RFS) of HBV-related HCC patients (60). Research by Haider et al revealed that high expression of ITGA5 was associated with a short survival time of pancreatic ductal adenocarcinoma (PDAC) patients (61). In addition, the results from a study by Yan et al indicated that the upregulated expression of ITGA5 reduced the overall survival of gastric cancer (GC) patients (62). Similar results were also reported in non-small cell lung cancer (NSCLC) (63) and glioblastoma cell invasion (64).
The results of GSEA in the present study indicated that ITGA5 (also known as FNRA, CD49e, VLA-5 and VLA5A) was markedly associated with the survival and progression of COAD, and the underlying mechanism of focal adhesion, ECM receptor interaction and extracellular matrix (ECM) were associated with its biological functions. Integrin α subunit and β subunit form heterodimeric integral membrane proteins that function in cell surface adhesion and signaling (16). Previous studies have reported that ITGA5 mediated cell adhesion and migration in human hepatocarcinoma cells by activating focal adhesion kinase (FAK) (65). A study by Yang and Wang revealed that ITGA5 participated in pathways involving focal adhesion and ECM-receptor interaction in osteosarcoma (66). In addition, ITGA5 may be involved in bladder cancer progression by extracellular matrix-receptor interaction and focal adhesion (67). In the present study, the results of GSEA indicated that ITGA5 may serve as an important adhesion molecule through its adhesion mechanism in COAD. To be specific, ITGA5 may act on COAD via the FAK signaling pathway and ECM receptor signaling pathway. However, these results require further research to be confirmed.
Although the present study was the first to reveal the role of the ITGA subfamily in the diagnosis and prognosis of COAD, it still has certain limitations. First, all the information was obtained from open databases, and the medical parameters were incomplete. Other potential influencing factors like tumor location, tumor size, lymphatic metastasis, and venous metastasis were not included. Second, disease-free survival should be listed as a factor to assess COAD prognosis. Third, the study required a larger multi-center and multi-regional as well as a multi-ethnic sample population. Fourth, the present study required further investigation at the protein level and COAD prognosis prediction, as well as further in vivo and in vitro experimental validation.
In conclusion, the present study revealed that the ITGA subfamily mRNA expression was associated with the diagnosis and prognosis of COAD. Combined with ROC curves and RT-qPCR verification, the ITGA8 expression level may be a potential diagnostic marker of COAD. In addition, survival analysis indicated that the expression of ITGA5 may serve as a prognostic biomarker of COAD. However, the present results still require further exploration and verification in the future.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the support of the National Key Clinical Specialty Programs (General Surgery and Oncology) and the Key Laboratory of Early Prevention and Treatment for Regional High-Incidence-Tumor (Guangxi Medical University), Ministry of Education, China. We would also like to thank The Cancer Genome Atlas (https://cancergenome.nih.gov/) for sharing the COAD dataset on open access.
Funding
The present study was sponsored in part by the 2018 Innovation Project of Guangxi Graduate Education (YCBZ2018036) and the Innovation Project of Guangxi Graduate Education (JGY2018037).
Availability of data and materials
The analyzed datasets generated during the study are available from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/).
Authors' contributions
YZG and GTR wrote the manuscript. YZG and FG made substantial contributions to the conception, design and intellectual content of the studies. YZG, GTR, XWL, XKW, CL and SW made key contributions to the analysis and interpretation of data. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients signed an informed consent form, and the experimental protocol was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University [no. 2019(KY-E-001)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Qian W, Wang S, Ji D, Wang Q, Li J, Peng W, Gu J, Hu T, Ji B, et al. Analysis of lncRNA-Associated ceRNA network reveals potential lncRNA biomarkers in human colon adenocarcinoma. Cell Physiol Biochem. 2018;49:1778–1791. doi: 10.1159/000495681. [DOI] [PubMed] [Google Scholar]
- 3.Thrumurthy SG, Thrumurthy SS, Gilbert CE, Ross P, Haji A. Colorectal adenocarcinoma: Risks, prevention and diagnosis. BMJ. 2016;354:i3590. doi: 10.1136/bmj.i3590. [DOI] [PubMed] [Google Scholar]
- 4.Jiang H, Du J, Gu J, Jin L, Pu Y, Fei B. A 65 gene signature for prognostic prediction in colon adenocarcinoma. Int J Mol Med. 2018;41:2021–2027. doi: 10.3892/ijmm.2018.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Li XJ, Li P, Guo XT. MicroRNA-145 regulates the proliferation, migration and invasion of human primary colon adenocarcinoma cells by targeting MAPK1. Int J Mol Med. 2018;42:3171–3180. doi: 10.3892/ijmm.2018.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukuda K, Tanino M, Soga H, Shimizu N, Shimizu K. A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun. 2000;278:653–658. doi: 10.1006/bbrc.2000.3839. [DOI] [PubMed] [Google Scholar]
- 7.Arnaout MA. Integrin structure: New twists and turns in dynamic cell adhesion. Immunol Rev. 2002;186:125–140. doi: 10.1034/j.1600-065X.2002.18612.x. [DOI] [PubMed] [Google Scholar]
- 8.Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin beta tails: A final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 9.Ginsberg MH, Partridge A, Shattil SJ. Integrin regulation. Curr Opin Cell Biol. 2005;17:509–516. doi: 10.1016/j.ceb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Takada Y, Ye X, Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:a004994. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 13.Larson RS, Corbi AL, Berman L, Springer T. Primary structure of the leukocyte function-associated molecule-1 alpha subunit: An integrin with an embedded domain defining a protein superfamily. J Cell Biol. 1989;108:703–712. doi: 10.1083/jcb.108.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oxvig C, Springer TA. Experimental support for a beta-propeller domain in integrin alpha-subunits and a calcium binding site on its lower surface. Proc Natl Acad Sci USA. 1998;95:4870–4875. doi: 10.1073/pnas.95.9.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desgrosellier JS, Cheresh DA. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman SL, Picard M. Integrins as therapeutic targets. Trends Pharmacol Sci. 2012;33:405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Shattil SJ, Kim C, Ginsberg MH. The final steps of integrin activation: The end game. Nat Rev Mol Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 20.Shen B, Delaney MK, Du X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr Opin Cell Biol. 2012;24:600–606. doi: 10.1016/j.ceb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz MA, Ginsberg MH. Networks and crosstalk: Integrin signalling spreads. Nat Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 23.Hehlgans S, Haase M, Cordes N. Signalling via integrins: Implications for cell survival and anticancer strategies. Biochim Biophys Acta. 2007;1775:163–180. doi: 10.1016/j.bbcan.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Ryu J, Koh Y, Park H, Kim DY, Kim DC, Byun JM, Lee HJ, Yoon SS. Highly expressed integrin-alpha8 induces epithelial to mesenchymal transition-like features in multiple myeloma with early relapse. Mol Cells. 2016;39:898–908. doi: 10.14348/molcells.2016.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo WH, Bian JJ, Tian GF, Lyu ZX, Gui YX, Ye L. Expression of fermintin family homologous protein 2 in non-small cell lung cancer and its clinical significance. Zhonghua Bing Li Xue Za Zhi. 2018;47:780–783. doi: 10.3760/cma.j.issn.0529-5807.2018.10.009. (In Chinese; Abstract available in Chinese from the publisher) [DOI] [PubMed] [Google Scholar]
- 26.Haas TL, Sciuto MR, Brunetto L, Valvo C, Signore M, Fiori ME, di Martino S, Giannetti S, Morgante L, Boe A, et al. Integrin alpha7 is a functional marker and potential therapeutic target in glioblastoma. Cell stem cell. 2017;21:35–50.e9. doi: 10.1016/j.stem.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Gong L, Zheng Y, Liu S, Peng Z. Fibronectin regulates the dynamic formation of ovarian cancer multicellular aggregates and the expression of integrin receptors. Asian Pac J Cancer Prev. 2018;19:2493–2498. doi: 10.22034/APJCP.2018.19.9.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang HW, Yen CY, Chen CH, Tsai JH, Tang JY, Chang YT, Kao YH, Wang YY, Yuan SF, Lee SY. Evaluation of the mRNA expression levels of integrins alpha3, alpha5, beta1 and beta6 as tumor biomarkers of oral squamous cell carcinoma. Oncol Lett. 2018;16:4773–4781. doi: 10.3892/ol.2018.9168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji J, Chen H, Liu XP, Wang YH, Luo CL, Zhang WW, Xie W, Wang FB. A miRNA combination as promising biomarker for hepatocellular carcinoma diagnosis: A study based on bioinformatics analysis. J Cancer. 2018;9:3435–3446. doi: 10.7150/jca.26101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gene Ontology Consortium, corp-author. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maere S, Heymans K, Kuiper M. BiNGO: A cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 34.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Yu T, Liao X, Yang C, Han C, Zhu G, Huang K, Yu L, Qin W, Su H, et al. The prognostic value of CYP2C subfamily genes in hepatocellular carcinoma. Cancer Med. 2018;7:966–980. doi: 10.1002/cam4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 41.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 44.Caswell PT, Norman JC. Integrin trafficking and the control of cell migration. Traffic. 2006;7:14–21. doi: 10.1111/j.1600-0854.2005.00362.x. [DOI] [PubMed] [Google Scholar]
- 45.Xiong J, Balcioglu HE, Danen EH. Integrin signaling in control of tumor growth and progression. Int J Biochem Cell Biol. 2013;45:1012–1015. doi: 10.1016/j.biocel.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Felding-Habermann B, Mueller BM, Romerdahl CA, Cheresh DA. Involvement of integrin alpha V gene expression in human melanoma tumorigenicity. J Clin Invest. 1992;89:2018–2022. doi: 10.1172/JCI115811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Q, Bavi P, Wang JY, Roehrl MH. Immuno-proteomic discovery of tumor tissue autoantigens identifies olfactomedin 4, CD11b, and integrin alpha-2 as markers of colorectal cancer with liver metastases. J Proteomics. 2017;168:53–65. doi: 10.1016/j.jprot.2017.06.021. [DOI] [PubMed] [Google Scholar]
- 48.Waisberg J, De Souza Viana L, Affonso Junior RJ, Silva SR, Denadai MV, Margeotto FB, De Souza CS, Matos D. Overexpression of the ITGAV gene is associated with progression and spread of colorectal cancer. Anticancer Res. 2014;34:5599–5607. [PubMed] [Google Scholar]
- 49.Liu X, Tian H, Li H, Ge C, Zhao F, Yao M, Li J. Derivate Isocorydine (d-ICD) suppresses migration and invasion of hepatocellular carcinoma cell by downregulating ITGA1 expression. Int J Mol Sci. 2017;18(pii):E514. doi: 10.3390/ijms18030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg EE, Prudnikova TY, Zabarovsky ER, Kashuba VI, Grigorieva EV. D-glucuronyl C5-epimerase cell type specifically affects angiogenesis pathway in different prostate cancer cells. Tumour Biol. 2014;35:3237–3245. doi: 10.1007/s13277-013-1423-6. [DOI] [PubMed] [Google Scholar]
- 51.Chuang YC, Wu HY, Lin YL, Tzou SC, Chuang CH, Jian TY, Chen PR, Chang YC, Lin CH, Huang TH, et al. Blockade of ITGA2 induces apoptosis and inhibits cell migration in gastric cancer. Biol Proced Online. 2018;20:10. doi: 10.1186/s12575-018-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong J, Lu X, Xu J, Xiong W, Zhang H, Yu X. Coexpression of UCA1 and ITGA2 in pancreatic cancer cells target the expression of miR-107 through focal adhesion pathway. J Cell Physiol. 2019;234:12884–12896. doi: 10.1002/jcp.27953. [DOI] [PubMed] [Google Scholar]
- 53.Lu Y, Li C, Chen H, Zhong W. Identification of hub genes and analysis of prognostic values in pancreatic ductal adenocarcinoma by integrated bioinformatics methods. Mol Biol Rep. 2018;45:1799–1807. doi: 10.1007/s11033-018-4325-2. [DOI] [PubMed] [Google Scholar]
- 54.Lemma SA, Kuusisto M, Haapasaari KM, Sormunen R, Lehtinen T, Klaavuniemi T, Eray M, Jantunen E, Soini Y, Vasala K, et al. Integrin alpha 10, CD44, PTEN, cadherin-11 and lactoferrin expressions are potential biomarkers for selecting patients in need of central nervous system prophylaxis in diffuse large B-cell lymphoma. Carcinogenesis. 2017;38:812–820. doi: 10.1093/carcin/bgx061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Y, Liu G, Yuan Y, Zhao J, Yang Y, Li Y. Analysis of differential gene expression profile identifies novel biomarkers for breast cancer. Oncotarget. 2017;8:114613–114625. doi: 10.18632/oncotarget.23061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang R, Zhang TT, Zhai GQ, Guo XY, Qin Y, Gan TQ, Zhang Y, Chen G, Mo WJ, Feng ZB. Evaluation of the HOXA11 level in patients with lung squamous cancer and insights into potential molecular pathways via bioinformatics analysis. World J Surg Oncol. 2018;16:109. doi: 10.1186/s12957-018-1375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parajuli H, The MT, Abrahamsen S, Christoffersen I, Neppelberg E, Lybak S, Osman T, Johannessen AC, Gullberg D, Skarstein K, Costea DE. Integrin alpha11 is overexpressed by tumour stroma of head and neck squamous cell carcinoma and correlates positively with alpha smooth muscle actin expression. J Oral Pathol Med. 2017;46:267–275. doi: 10.1111/jop.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kok-Sin T, Mokhtar NM, Ali Hassan NZ, Sagap I, Mohamed Rose I, Harun R, Jamal R. Identification of diagnostic markers in colorectal cancer via integrative epigenomics and genomics data. Oncol Rep. 2015;34:22–32. doi: 10.3892/or.2015.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang X, Deng Y, He RQ, Li XJ, Ma J, Chen G, Hu XH. Upregulation of HOXA11 during the progression of lung adenocarcinoma detected via multiple approaches. Int J Mol Med. 2018;42:2650–2664. doi: 10.3892/ijmm.2018.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shang L, Ye X, Zhu G, Su H, Su Z, Chen B, Xiao K, Li L, Peng M, Peng T. Prognostic value of integrin variants and expression in post-operative patients with HBV-related hepatocellular carcinoma. Oncotarget. 2017;8:76816–76831. doi: 10.18632/oncotarget.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haider S, Wang J, Nagano A, Desai A, Arumugam P, Dumartin L, Fitzgibbon J, Hagemann T, Marshall JF, Kocher HM, et al. A multi-gene signature predicts outcome in patients with pancreatic ductal adenocarcinoma. Genome Med. 2014;6:105. doi: 10.1186/s13073-014-0105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan P, He Y, Xie K, Kong S, Zhao W. In silico analyses for potential key genes associated with gastric cancer. PeerJ. 2018;6:e6092. doi: 10.7717/peerj.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng W, Jiang C, Li R. Integrin and gene network analysis reveals that ITGA5 and ITGB1 are prognostic in non-small-cell lung cancer. Onco Targets Ther. 2016;9:2317–2327. doi: 10.2147/OTT.S91796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mallawaaratchy DM, Buckland ME, McDonald KL, Li CC, Ly L, Sykes EK, Christopherson RI, Kaufman KL. Membrane proteome analysis of glioblastoma cell invasion. J Neuropathol Exp Neurol. 2015;74:425–441. doi: 10.1097/NEN.0000000000000187. [DOI] [PubMed] [Google Scholar]
- 65.Maschler S, Wirl G, Spring H, Bredow DV, Sordat I, Beug H, Reichmann E. Tumor cell invasiveness correlates with changes in integrin expression and localization. Oncogene. 2005;24:2032–2041. doi: 10.1038/sj.onc.1208423. [DOI] [PubMed] [Google Scholar]
- 66.Yang J, Wang N. Analysis of the molecular mechanism of osteosarcoma using a bioinformatics approach. Oncol Lett. 2016;12:3075–3080. doi: 10.3892/ol.2016.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang ZQ, Zang WD, Chen R, Ye BW, Wang XW, Yi SH, Chen W, He F, Ye G. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet Mol Res. 2013;12:1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed datasets generated during the study are available from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/).