Abstract
The aim of the present study was to investigate the diagnostic and prognostic value of Wingless-type MMTV integration site (WNT) gene family expression in patients with hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). The clinical data of the patients and gene expression levels were downloaded from Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases. Receiver operating characteristic curve analysis was used to investigate the diagnostic value of WNT genes. Cox proportional hazard regression analysis and Kaplan-Meier survival analysis were performed to evaluate the association of WNT gene expression level with overall survival (OS) and recurrence-free survival (RFS). A nomogram was constructed for the prediction of prognosis. Hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated. Diagnostic receiver operating characteristic curve analysis suggested that WNT2 had a high diagnostic value, with an area under the curve (AUC) of >0.800 (P<0.0001, AUC=0.810, 95% CI: 0.767–0.852). Survival analysis indicated that the expression level of WNT1 was significantly associated with OS and RFS (adjusted P=0.033, adjusted HR=0.607, 95% CI: 0.384–0.960; and adjusted P=0.007, adjusted HR=0.592, 95% CI: 0.404–0.868, respectively). In the TCGA validation cohort, we also observed that WNT2 was significantly differentially expressed between HCC tissues and adjacent non-tumor tissues, and WNT1 was associated with both the OS and RFS of HCC. Therefore, through the GSE14520 HBV-related HCC cohort we concluded that WNT2 may serve as a diagnostic biomarker and WNT1 may serve as a prognostic biomarker. These results may also be extended to TCGA HCC verification cohort.
Keywords: hepatocellular carcinoma, WNT family genes, diagnosis, prognosis, biomarker
Introduction
Liver cancer is one of the most common lethal cancers worldwide. It has been reported that liver cancer ranks sixth among the most commonly diagnosed cancers worldwide, and was the fourth major cause of cancer-related deaths in 2018. Global cancer statistics indicate that ~841,000 new cases and 782,000 deaths occur annually (1). Hepatocellular carcinoma (HCC) is the major histological type of primary liver cancer, accounting for 75–85% of all cases. Infection with hepatitis virus [mainly hepatitis B virus (HBV) and hepatitis C virus], aflatoxin exposure, excessive alcohol consumption and tobacco smoking, are considered as the main risk factors for the development of HCC (2,3). In China, the predominant cause of HCC is chronic HBV infection, and it is estimated that ~70% of these patients have an established HBV infection history (4). Although the available therapies for HCC patients have greatly improved over the past decades, the clinical prognosis remains unfavorable, with a 5-year overall survival (OS) rate of ~30% following hepatic resection (2,5). Therefore, it is imperative to identify more sensitive diagnostic and prognostic biomarkers for HCC.
Wingless-type MMTV integration site (WNT) genes are a family of 19 genes that modulate both the canonical WNT signal transduction pathway (referred to as β-catenin-dependent) and non-canonical WNT signal transduction pathway (referred to as β-catenin-independent) (6). Previous studies indicated that WNT family genes are associated with various tumor biological processes, including cell proliferation (7,8), invasion (8–11), metastasis (12,13) and drug resistance (13–15). In addition, some researchers have reported that aberrant WNT expression levels are associated with diagnosis and prognosis prediction for certain tumors. Fu et al (16) proved that WNT2 can activate the WNT/β-catenin signal transduction pathway, which ultimately promotes esophageal cancer cell growth, and WNT2 enhances cell motility and invasiveness by inducing epithelial-to-mesenchymal transition. Furthermore, WNT2 expression level was found to be closely associated with the poor clinical performance status of patients with esophageal squamous cell carc WNT inoma.
However, the diagnostic and prognostic value of the WNT gene family expression in HBV-related HCC remains unclear. The primary goal of the present study was to investigate this association by collecting data from public databases and performing a series of bioinformatics analyses.
Materials and methods
Data sources
The clinical characteristics of patients with HBV-related HCC and the corresponding WNT gene family expression levels were downloaded from the GSE14520 dataset of Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520, accessed November 28, 2018) (17,18). The detailed process of GSE14520 genome-wide expression profile dataset processing has been described in our previous article (19). The Cancer Genome Atlas (TCGA) database HCC cohort was used as a validation cohort, and the data processing of RNA sequencing was described in our previous paper (19). The raw RNA sequencing dataset of TCGA was normalized by DESeq (20). The clinical data of these patients in the GSE14520 dataset included age, gender, serum alanine aminotransferase level, serum α-fetoprotein (AFP) level, cirrhosis, main tumor size, tumor number, Barcelona Clinic Liver Cancer (BCLC) stage, tumor-node-metastasis (TNM) stage, survival time, and survival status. The data for mRNA expression level of five WNT family genes (WNT3A, WNT8A, WNT9A, WNT9B and WNT10A) were unavailable in the Gene Expression Omnibus (GEO) database. Therefore, only 14 WNT genes were finally analyzed in the present study. As the dataset included in our research was obtained from a public database, the study did not require the approval of an ethics committee.
Bioinformatics analysis of WNT family genes
To explore the potential biological functions and possible pathways of WNT family genes, gene enrichment analyses, including Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, were conducted by applying the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics online tool, version 6.8 (https://david.ncifcrf.gov/, accessed December 2, 2018) (21). Statistically, an enrichment P-value of <0.05 was considered to indicate statistically significant differences. In order to further validate the results of enrichment analysis by DAVID, application package Biological Networks Gene Ontology tool (BiNGO) in the Cytoscape software (version 3.7.1) (22) was used to explore the GO terms of WNT family genes. Pearson's correlation coefficient was calculated to assess the relevance among WNT genes in the co-expression analysis. These data were visualized by the correlation plot package in the R platform (version 3.4.0). WNT gene-gene and protein-protein interactions were investigated by using the online resource GeneMANIA (http://genemania.org/, accessed December 6, 2018) (23) and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/cgi/input.pl, accessed December 6, 2018) (24,25), respectively.
Assessment of diagnostic value
The expression level of WNT family genes in tumor tissues and corresponding adjacent non-tumor tissues were compared via t-test statistical analysis. The public online resource Metabolic gEne RApid Visualizer (MERAV) (http://merav.wi.mit.edu/, accessed December 8, 2018) (26) was used to further validate that genes of the WNT family were differentially expressed between primary liver cancer tissues and normal liver tissues. Receiver operating characteristic (ROC) curve analysis was selected to determine the diagnostic value of these differentially expressed WNT family genes.
Survival analysis
The 212 patients with HBV-related HCC were categorized into high- and low-expression groups based on the median WNT gene expression level in tumor tissues. In order to investigate whether the expression level of WNT family genes was correlated with prognosis and outcome, Cox proportional hazard regression analysis and Kaplan-Meier survival analysis with log-rank tests were used to evaluate the association between WNT family gene expression, OS and recurrence-free survival (RFS).
Joint effects analysis of WNT family genes
Based on the results of multivariate Cox proportional hazard regression analysis and Kaplan-Meier survival analysis, only WNT1 and WNT6 were found to be significantly associated with RFS. Therefore, the combined effects of WNT1 and WNT6 were analyzed. The combinations were as follows: Low WNT1 expression and low WNT6 expression (group 1), low WNT1 expression and high WNT6 expression (group 2), high WNT1 expression and low WNT6 expression (group 3), and high WNT1 expression and high WNT6 expression (group 4).
Prognostic nomogram for survival prediction
All 212 patients with HBV-related HCC in the GSE14520 dataset of the GEO database were identified as the source population for nomogram construction. The variables that were related to prognosis outcome were selected to construct the nomogram, including sex, serum AFP level, cirrhosis, BCLC stage, tumor size and WNT gene expression. With each variable being assigned a score, the total point was calculated by summing up the scores of all the variables and located onto the scale. Therefore, the probabilities of the survival outcome could be predicted by drawing a vertical line to the total point.
Statistical analysis
All statistical analyses were performed with the SPSS software package, version 17.0 (SPSS Inc.). Comparison of WNT family gene expression levels between tumor tissues and corresponding adjacent non-tumor tissues was performed using t-tests. The Cox proportional hazards regression model was selected for univariate and multivariate analyses. By applying Kaplan-Meier survival analysis with log-rank test, the association of WNT family gene expression levels with OS and RFS time was observed. Vertical scatter plots, ROC curves and survival curves were plotted by GraphPad Prism software, version 7.0 (GraphPad Software, Inc.). P<0.05 was considered to indicate statistically significant differences.
Results
Characteristics of patients in the GEO database
In the GSE14520 dataset of the GEO database, there remained 212 patients with HBV-related HCC after excluding patients who had no reported HBV infection or any available survival information. Detailed characteristics of these patients are shown in Table SI. Serum AFP level, cirrhosis, tumor size, BCLC stage and TNM stage were found to be closely associated with OS (P<0.05), whereas sex, cirrhosis, BCLC stage and TNM stage were significantly associated with RFS (P<0.05). The remaining characteristics did not exhibit a significant association with OS or RFS (all P>0.05).
Bioinformatics analysis of WNT family genes
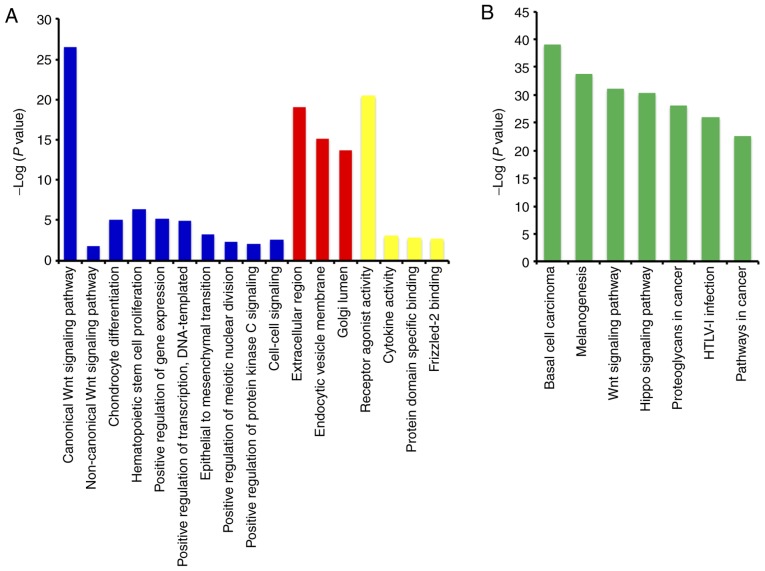
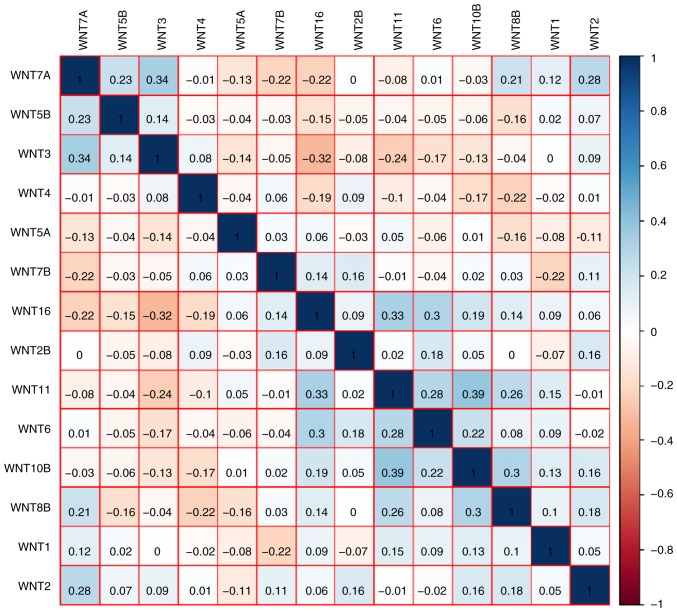
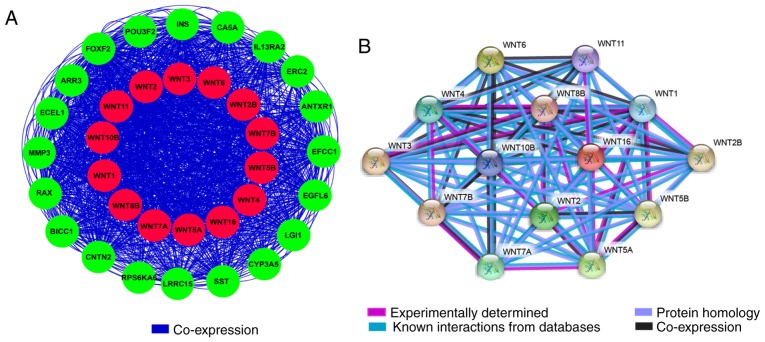
The GO function analysis indicated that WNT family genes were mainly enriched in the regulation of cell differentiation, cell proliferation, epithelial-to-mesenchymal transition, and modulation of the WNT signaling pathway (Figs. 1A, S1 and S2, and Table SII). The KEGG pathway analysis suggested that WNT family genes were associated with the WNT signaling pathway and other pathways (Fig. 1B and Table SIII). The Pearson's correlation coefficients of WNT family genes were calculated and used to assess whether these genes were correlated with each other. As shown in Fig. 2, the WNT family genes were correlated to some degree. The gene-gene and protein-protein interaction networks constructed by GeneMANIA and STRING, respectively, indicated that the WNT family genes were co-expressed and exhibited extensive homology at the protein level (Fig. 3A and B).
Figure 1.
Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of WNT family genes performed by DAVID online tool. (A) GO term enrichment of WNT family genes. (B) KEGG pathway enrichment of WNT family genes.
Figure 2.
Matrix graph of Pearson's correlations of WNT family gene expression levels in the Gene Expression Omnibus database.
Figure 3.
WNT family gene-gene and protein-protein interaction networks constructed by GeneMANIA and Search Tool for the Retrieval of Interacting Genes/Proteins databases. (A) Gene-gene interaction network of WNT family genes. (B) Protein-protein interaction network of WNT family genes.
Assessment of diagnostic value
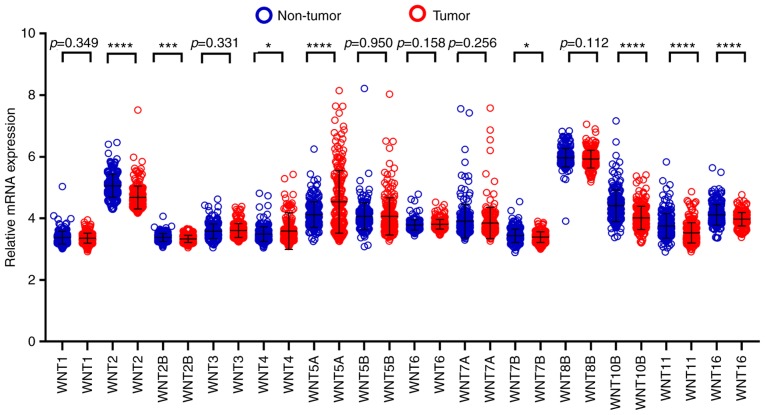
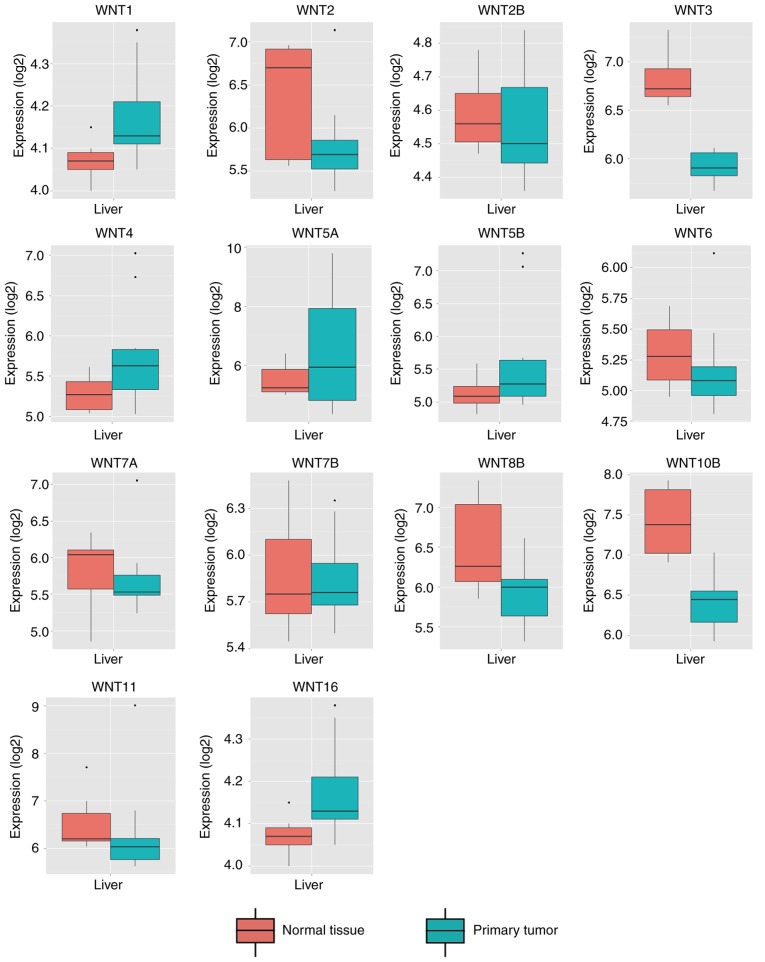
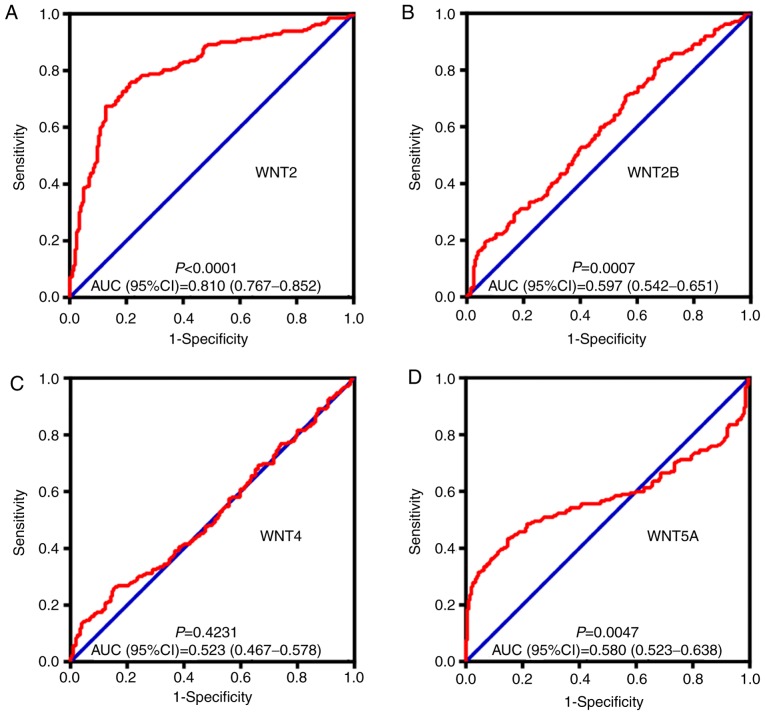
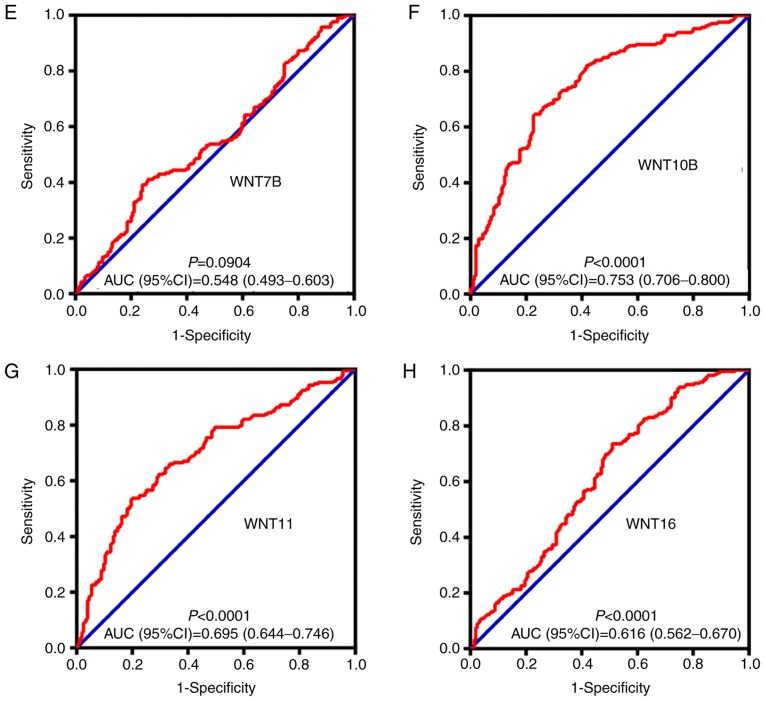
By comparing WNT family gene expression levels between tumor tissues and corresponding adjacent non-tumor tissues, a total of 8 WNT family genes (WNT2, WNT2B, WNT4, WNT5A, WNT7B, WNT10B, WNT11 and WNT16) were found to be differentially expressed in tumor and non-tumor tissues (Fig. 4) (P<0.05). The online resource MERAV was used to further validate genes of the WNT family that were differentially expressed in normal liver tissues and primary liver cancer tissues (Fig. 5). An ROC curve was constructed to further explore the diagnostic value of these 8 differentially expressed genes. As shown in Fig. 6, six WNT genes (WNT2, WNT2B, WNT5A, WNT10B, WNT11 and WNT16) had a potential prediction value, with all P-values <0.05 and area under the curve (AUC) >0.500; WNT2 in particular exhibited high accuracy in differentiating HCC tissues from non-tumor tissue (P<0.0001, AUC=0.810, 95% CI: 0.767–0.852).
Figure 4.
Relative expression of WNT family genes in 212 HCC tissues and 204 adjacent tissues in the Gene Expression Omnibus database. *P<0.05, ***P<0.001, ****P<0.0001.
Figure 5.
Expression levels of WNT family genes in normal liver and primary liver cancer tissues, as analyzed by MERAV.
Figure 6.
The receiver operating characteristics (ROC) curves of WNT family genes in distinguish HBV-related HCC tumor tissues and adjacent non-tumor tissues in GES14520. ROC curves of (A) WNT2; (B) WNT2B; (C) WNT4; (D) WNT5A; (E) WNT7B; (F) WNT10B; (G) WNT11; and (H) WNT16. AUC, area under the ROC curve; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
Survival analysis
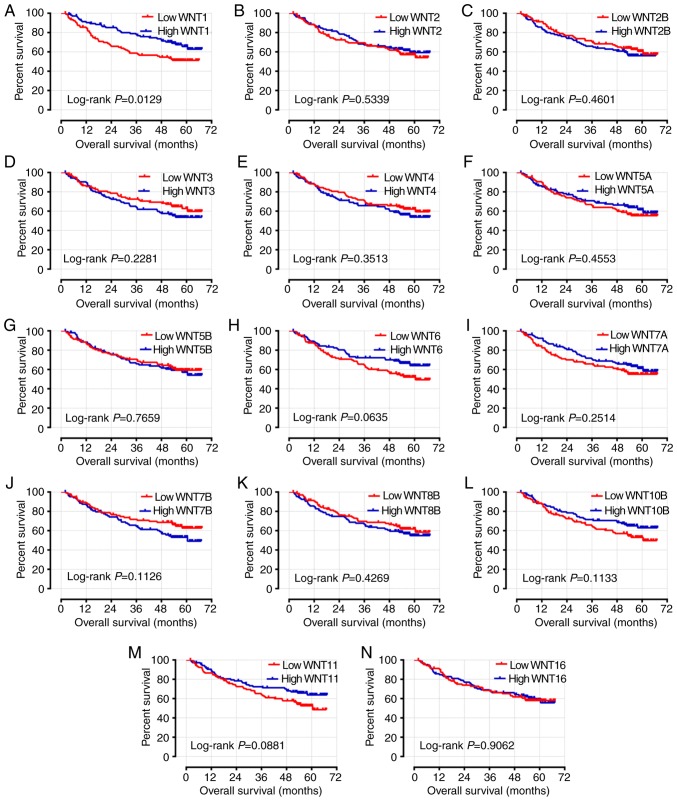
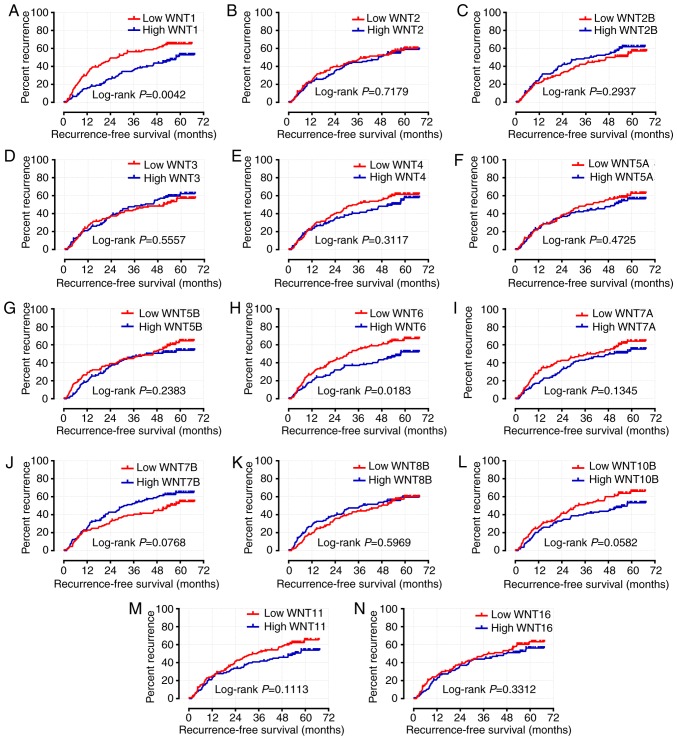
The characteristics associated with clinical prognostic outcome, including cirrhosis, tumor size and BCLC stage, were included in the multivariate Cox regression analysis. Following adjustment or these prognosis-related risk factors, the results indicated that the expression level of WNT1 was significantly associated with OS and RFS in the survival analysis (adjusted P=0.033, adjusted HR=0.607, 95% CI: 0.384–0.960 and adjusted P=0.007, adjusted HR=0.592, 95% CI: 0.404–0.868, respectively) (Table I; Figs. 7A and 8A). The expression level of WNT6 was closely associated with RFS (adjusted P=0.033, adjusted HR=0.665, 95% CI: 0.457–0.968), but WNT6 did not exhibit a significant association with OS (P>0.05) (Table I; Figs. 7H and 8H).
Table I.
Prognostic values of WNT gene expression in HBV-related HCC of the GSE14520 cohort.
| OS | RFS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene expression | Patient no. | No. of events | MST (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea | No. of events | MRT (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
| WNT1 | 0.007 | ||||||||||||
| Low | 106 | 49 | NA | 1 | 1 | 66 | 27 | 1 | 1 | ||||
| High | 106 | 33 | NA | 0.575 | 0.014 | 0.607 | 0.033 | 50 | 57 | 0.588 | 0.005 | 0.592 | |
| (0.370–0.895) | (0.384–0.960) | (0.406–0.850) | (0.404–0.868) | ||||||||||
| WNT2 | 0.444 | ||||||||||||
| Low | 106 | 43 | NA | 1 | 1 | 58 | 40 | 1 | 1 | ||||
| High | 106 | 39 | NA | 0.872 | 0.534 | 0.784 | 0.282 | 58 | 46 | 0.935 | 0.718 | 0.865 | |
| (0.565–1.345) | (0.503–1.222) | (0.650–1.346) | (0.597–1.254) | ||||||||||
| WNT2B | 0.091 | ||||||||||||
| Low | 106 | 38 | NA | 1 | 1 | 54 | 46 | 1 | 1 | ||||
| High | 106 | 44 | NA | 1.178 | 0.461 | 1.344 | 0.192 | 62 | 37 | 1.215 | 0.295 | 1.378 | |
| (0.763–1.818) | (0.862–2.093) | (0.844–1.751) | (0.950–1.997) | ||||||||||
| WNT3 | 0.622 | ||||||||||||
| Low | 106 | 37 | NA | 1 | 1 | 56 | 51 | 1 | 1 | ||||
| High | 106 | 45 | NA | 1.306 | 0.230 | 1.279 | 0.285 | 60 | 41 | 1.116 | 0.556 | 1.099 | |
| (0.845–2.018) | (0.814–2.008) | (0.775–1.606) | (0.754–1.601) | ||||||||||
| WNT4 | 0.113 | ||||||||||||
| Low | 106 | 39 | NA | 1 | 1 | 63 | 35 | 1 | 1 | ||||
| High | 106 | 43 | NA | 1.229 | 0.352 | 1.116 | 0.624 | 53 | 53 | 0.828 | 0.313 | 0.742 | |
| (0.796–1.896) | (0.720–1.730) | (0.575–1.194) | (0.512–1.074) | ||||||||||
| WNT5A | 0.663 | ||||||||||||
| Low | 106 | 45 | NA | 1 | 1 | 62 | 36 | 1 | 1 | ||||
| High | 106 | 37 | NA | 0.847 | 0.456 | 0.915 | 0.690 | 54 | 51 | 0.875 | 0.473 | 0.922 | |
| (0.549–1.309) | (0.591–1.417) | (0.607–1.260) | (0.638–1.331) | ||||||||||
| WNT5B | 0.641 | ||||||||||||
| Low | 106 | 40 | NA | 1 | 1 | 64 | 46 | 1 | 1 | ||||
| High | 106 | 42 | NA | 1.068 | 0.766 | 1.328 | 0.215 | 52 | 43 | 0.803 | 0.240 | 0.915 | |
| (0.693–1.647) | (0.848–2.078) | (0.557–1.158) | (0.630–1.329) | ||||||||||
| WNT6 | 0.033 | ||||||||||||
| Low | 106 | 48 | 60 | 1 | 1 | 66 | 30 | 1 | 1 | ||||
| High | 106 | 34 | NA | 0.662 | 0.066 | 0.756 | 0.222 | 50 | 57 | 0.644 | 0.019 | 0.665 | |
| (0.426–1.027) | (0.483–1.184) | (0.446–0.931) | (0.457–0.968) | ||||||||||
| WNT7A | 0.180 | ||||||||||||
| Low | 106 | 43 | NA | 1 | 1 | 62 | 41 | 1 | 1 | ||||
| High | 106 | 39 | NA | 0.776 | 0.253 | 0.764 | 0.229 | 54 | 48 | 0.757 | 0.136 | 0.778 | |
| (0.503–1.198) | (0.492–1.185) | (0.526–1.091) | (0.539–1.123) | ||||||||||
| WNT7B | 0.522 | ||||||||||||
| Low | 106 | 36 | NA | 1 | 1 | 53 | 54 | 1 | 1 | ||||
| High | 106 | 46 | 60 | 1.421 | 0.115 | 1.057 | 0.812 | 63 | 30 | 1.390 | 0.078 | 1.132 | |
| (0.918–2.200) | (0.670–1.667) | (0.963–2.004) | (0.775–1.655) | ||||||||||
| WNT8B | 0.721 | ||||||||||||
| Low | 106 | 38 | NA | 1 | 1 | 57 | 48 | 1 | 1 | ||||
| High | 106 | 44 | NA | 1.192 | 0.428 | 0.904 | 0.654 | 59 | 37 | 1.103 | 0.597 | 0.935 | |
| (0.772–1.840) | (0.580–1.407) | (0.766–1.588) | (0.646–1.354) | ||||||||||
| WNT10B | 0.079 | ||||||||||||
| Low | 106 | 46 | 60 | 1 | 1 | 64 | 30 | 1 | 1 | ||||
| High | 106 | 36 | NA | 0.704 | 0.115 | 0.691 | 0.101 | 52 | 57 | 0.703 | 0.060 | 0.717 | |
| (0.455–1.090) | (0.445–1.0750) | (0.487–1.014) | (0.495–1.039) | ||||||||||
| WNT11 | 0.371 | ||||||||||||
| Low | 106 | 47 | 60 | 1 | 1 | 63 | 35 | 1 | 1 | ||||
| High | 106 | 35 | NA | 0.685 | 0.090 | 0.740 | 0.186 | 53 | 54 | 0.744 | 0.113 | 0.843 | |
| (0.442–1.061) | (0.474–1.156) | (0.515–1.073) | (0.581–1.225) | ||||||||||
| WNT16 | 0.349 | ||||||||||||
| Low | 106 | 40 | NA | 1 | 1 | 60 | 37 | 1 | 1 | ||||
| High | 106 | 42 | NA | 0.974 | 0.906 | 0.950 | 0.819 | 56 | 46 | 0.835 | 0.332 | 0.838 | |
| (0.632–1.503) | (0.611–1.477) | (0.580–1.202) | (0.580–1.212) | ||||||||||
Adjusted for cirrhosis, tumor size and BCLC stage. Bold print indicates statistical significance. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; OS, overall survival; RFS, recurrence-free survival; MST, median survival time; MRT, median recurrence time; HR, hazard ratio; CI, confidence interval; NA, not available; BCLC, Barcelona Clinic Liver Cancer.
Figure 7.
Kaplan-Meier survival curves for WNT genes in HBV-related HCC of GES14520. OS stratified by (A) WNT1; (B) WNT2; (C) WNT2B; (D) WNT3; (E) WNT4; (F) WNT5A; (G) WNT5B; (H) WNT6; (I) WNT7A; (J) WNT7B; (K) WNT8B; (L) WNT10B; (M) WNT11; and (N) WNT16. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; OS, overall survival.
Figure 8.
Kaplan-Meier survival curves for WNT genes in HBV-related HCC of GES14520. RFS stratified by (A) WNT1; (B) WNT2; (C) WNT2B; (D) WNT3; (E) WNT4; (F) WNT5A; (G) WNT5B; (H) WNT6; (I) WNT7A; (J) WNT7B; (K) WNT8B; (L) WNT10B; (M) WNT11; and (N) WNT16. HBV, hepatitis B virus; HCC, hepatocellular carcinoma; RFS, recurrence-free survival.
Joint effects analysis of WNT family genes
As shown above, the multivariate Cox regression analysis and Kaplan-Meier survival analysis demonstrated that only WNT1 and WNT6 were significantly associated with RFS. Joint effects survival analysis for WNT1 and WNT6 was performed following adjustment for cirrhosis, tumor size and BCLC stage. The results suggested that group 4 (high WNT1 expression and high WNT6 expression) had the longest RFS, whereas group 1 (low WNT1 expression and low WNT6 expression) had the shortest RFS (Table II and Fig. 9). Therefore, patients with a high expression of both WNT1 and WNT6 are expected to have a longer RFS.
Table II.
Joint effects analysis for the combination of WNT1 and WNT6.
| Group | WNT1 expression | WNT6 expression | Patients (n=212) | MRT (months) | Crude HR (95% CI) | Crude P-value | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|
| 1 | Low | Low | 56 | 18 | NA | 0.001 | NA | P<0.001 |
| 2 | Low | High | 50 | 49 | 0.524 (0.318–0.862) | 0.011 | 0.457 (0.276–0.755) | 0.002 |
| 3 | High | Low | 50 | 51 | 0.478 (0.290–0.787) | 0.004 | 0.413 (0.250–0.683) | 0.001 |
| 4 | High | High | 56 | NA | 0.401 (0.243–0.662) | P<0.001 | 0.405 (0.240–0.684) | 0.001 |
Adjusted for cirrhosis, tumor size and BCLC stage. MRT, median recurrence time; HR, hazard ratio; CI, confidence interval; NA, not available; BCLC, Barcelona Clinic Liver Cancer.
Figure 9.
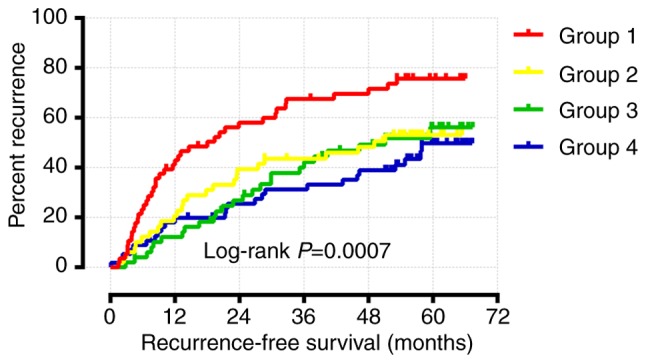
Joint effects analysis for the combination of WNT1 and WNT6.
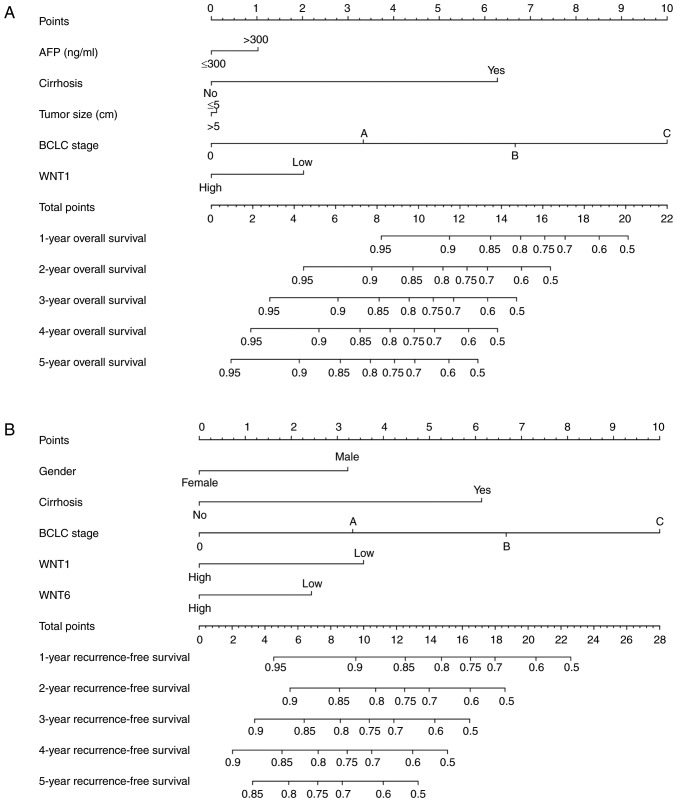
Prognostic nomogram for survival prediction
The prognostic risk factors that may predict the outcome of survival, including sex, serum AFP level, cirrhosis, BCLC stage, tumor size and WNT family gene expression, were selected to construct the nomogram, which can provide an individualized prognosis prediction. For the 212 patients with HBV-related HCC, nomogram analysis was performed for the probabilities of 1-, 2-, 3-, 4- and 5-year OS (Fig. 10A) and RFS (Fig. 10B). As shown in the nomogram, the expression level of WNT1 and WNT6 contributed to a certain extent to the patients' clinical prognosis outcome.
Figure 10.
Prognostic nomogram for survival prediction. (A) Nomogram for overall survival; (B) nomogram for recurrence-free survival. AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer.
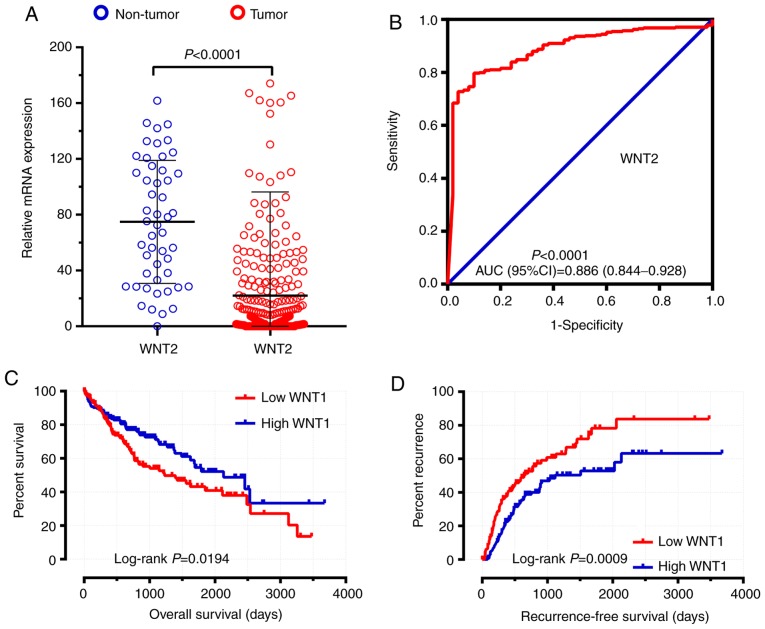
TCGA validation cohort
A total of 374 tumor tissues and 50 adjacent non-tumor tissues were included in the present study. Among those, 370 HCC patients with prognostic information were included in the survival analysis. The expression distribution of the WNT family genes between HCC tumor tissues and adjacent non-tumor tissues were calculated by DESeq and are shown in Table III. WNT2 was shown to be significantly differentially expressed between HCC and adjacent non-tumor tissues in the TCGA cohort (Table III, Fig. 11A and B). The clinical characteristics of HCC patients in the TCGA cohort are shown in Table SIV. The survival analysis results of the WNT family genes are shown in Table IV. WNT1 was found to be associated with both the OS and RFS of HCC in the TCGA cohort (Table IV, Fig. 11C and D).
Table III.
Expression distribution of WNT family genes between tumor tissue and adjacent non-tumor tissue in the TCGA validation cohort.
| Gene | Log2 (fold change) | P-value | FDR |
|---|---|---|---|
| WNT1 | 1.84421242 | 0.946083391 | 0.999996922 |
| WNT2 | −1.769264955 | 0.002852902 | 0.022026272 |
| WNT2B | 1.360412766 | 0.373384363 | 0.65436372 |
| WNT3 | 0.066523683 | 0.810682922 | 0.928938592 |
| WNT4 | 1.309792732 | 0.026270116 | 0.120328643 |
| WNT5A | 0.650962704 | 0.073901092 | 0.249424709 |
| WNT5B | −0.101243468 | 0.644473376 | 0.842930166 |
| WNT6 | 2.792018889 | 0.42971413 | 0.703074675 |
| WNT7A | 0.13572947 | 1 | 1 |
| WNT7B | 1.071320247 | 0.324934599 | 0.610626696 |
| WNT8B | 1.918325871 | 0.070391904 | 0.241649148 |
| WNT10B | 1.121445938 | 0.298549628 | 0.582917358 |
| WNT11 | −1.50622528 | 0.000105087 | 0.001433939 |
| WNT16 | −0.077860553 | 0.822321775 | 0.934493001 |
TCGA, The Cancer Genome Atlas; FDR, false discovery rate.
Figure 11.
Diagnostic and prognostic values of WNT genes in TCGA validation cohort. (A) the relative expression of WNT2 in HCC tumor tissues and non-tumor tissues; (B) ROC curves of WNT2; (C) Kaplan-Meier survival curves of WNT1 for OS; (D) Kaplan-Meier survival curves of WNT1 for RFS. TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; ROC, receiver operating characteristics; AUC, area under the ROC curve; OS, overall survival; RFS, recurrence-free survival.
Table IV.
Survival analysis results of the WNT family genes in the TCGA validation cohort.
| RFS | OS | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | P-value | HR | Low 95% CI | High 95% CI | P-value | HR | Low 95% CI | High 95% CI |
| WNT1 | 0.004872 | 0.5585569 | 0.37237755 | 0.83782122 | 0.038128301 | 0.67054664 | 0.459578646 | 0.978358776 |
| WNT2 | 0.165437 | 0.74242449 | 0.48738676 | 1.13091729 | 0.463552265 | 1.15543359 | 0.78518841 | 1.700262972 |
| WNT2B | 0.172666 | 0.75604408 | 0.5058112 | 1.13007116 | 0.949318195 | 0.9878679 | 0.678014074 | 1.439325557 |
| WNT3 | 0.111201 | 1.39315603 | 0.92641981 | 2.09503695 | 0.23053025 | 0.7955497 | 0.547382364 | 1.156228935 |
| WNT4 | 0.038296 | 0.65324005 | 0.43663377 | 0.97730085 | 0.308986119 | 0.82048053 | 0.560425379 | 1.201209516 |
| WNT5A | 0.25247 | 0.79158048 | 0.53045358 | 1.18125258 | 0.823749524 | 0.95871339 | 0.661532981 | 1.389396128 |
| WNT5B | 0.421213 | 0.84734859 | 0.56593122 | 1.26870474 | 0.701474468 | 0.92888443 | 0.637015977 | 1.354481386 |
| WNT6 | 0.571806 | 0.88958796 | 0.5929931 | 1.33452943 | 0.640628505 | 1.09289911 | 0.752662101 | 1.586938493 |
| WNT7A | 0.000396 | 0.47050498 | 0.31004378 | 0.71401184 | 0.668569853 | 0.92232626 | 0.636972232 | 1.335514626 |
| WNT7B | 0.156162 | 0.73979971 | 0.4877709 | 1.12205056 | 0.874887781 | 1.0304464 | 0.709391742 | 1.496803142 |
| WNT8B | 0.36875 | 0.83403019 | 0.56144901 | 1.23894839 | 0.230696394 | 0.79551932 | 0.547254355 | 1.156411068 |
| WNT10B | 0.052198 | 0.6701836 | 0.44743955 | 1.00381395 | 0.23391077 | 0.79061532 | 0.536982043 | 1.16404746 |
| WNT11 | 0.012024 | 0.59538341 | 0.397232 | 0.89237879 | 0.084433724 | 0.71556062 | 0.489264738 | 1.046523402 |
| WNT16 | 0.039333 | 0.65216604 | 0.43430059 | 0.97932296 | 0.457153569 | 1.15299749 | 0.792227421 | 1.678057559 |
TCGA, The Cancer Genome Atlas; RFS, recurrence-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
Discussion
The aim of the present study was to investigate the diagnostic and prognostic values of WNT family gene expression in HBV-related HCC established on public databases and a series of bioinformatics analyses. The results suggested that WNT2 may serve as potential diagnostic biomarker for HBV-related HCC. In addition, we demonstrated that the expression level of WNT1 was significantly associated with clinical prognostic outcome, with patients with a higher expression level of WNT1 expected to have a better prognostic outcome. Therefore, it may be concluded that WNT1 may serve as potential prognostic biomarker for patients with HBV-related HCC.
Previous studies confirmed that the WNT signaling pathway plays a crucial role in numerous physiological and pathological processes, including the regeneration of hair and skin (27), the repair of liver and lung after injury (28,29), hematopoiesis (30,31) and neurogenesis (32). Furthermore, other studies have demonstrated that the aberrant regulation of WNT signaling may contribute to various diseases, including cancer (6,33–35), osteoporosis (36,37), fibrosis (38–40), autoimmune diseases (41–43), neurological diseases (44,45), and disorders of endocrine function (46–48). The WNT signaling pathway has been shown to either promote or inhibit cancer biological progression in a cancer stage- and type-specific manner (6). WNT family genes, as the most important component of the WNT signaling pathway, participate in the initiation and progression of various cancers, such as esophageal carcinoma (16), gastric cancer (49), pancreatic cancer (50), prostate cancer (51), ovarian cancer (52,53), and leukemia (54). Numerous studies have reported that WNT family genes may regulate cell proliferation (50,54), differentiation, epithelial-to-mesenchymal transition (7,12) and WNT signaling (9,16). The conclusions of those studies were consistent with the results of gene function enrichment analysis in DAVID.
Early discoveries confirmed that WNT family genes may serve as potential diagnostic biomarkers for certain types of cancer. Sin et al selected RNA sequencing as a discovery method for specific RNA markers in bladder cancer, and found that WNT5A detection was a valuable complementary strategy in cystoscopy that may reduce unnecessary diagnostic procedures for bladder cancer (55). Jiang et al had reported that WNT6 may serve as a diagnostic biomarker for osteosarcoma, with an AUC of 0.854, a specificity of 88.4% and a sensitivity of 77.8% (56). Based on these previous studies, it may be hypothesized that WNTs may also predict HCC. To test this hypothesis and evaluate the diagnostic value of WNT genes, an ROC curve was constructed, and the analysis suggested that WNT2 may serve as potential diagnostic biomarker for patients with HBV-related HCC.
In addition, we also investigated the prognostic prediction ability of WNT family genes. The results demonstrated that the expression level of WNT1 was associated with OS and RFS, with patients exhibiting a higher expression level of WNT1 having a better prognostic outcome. Therefore, WNT1 may serve as potential prognostic biomarker for HBV-related HCC. It has been reported that WNT family genes may predict the prognostic outcome in several types of cancer. As previously reported, WNT1 expression may be one of the mechanisms underlying WNT/β-catenin signaling pathway activation in non-small cell lung cancer, and aberrant WNT1 expression level was found to be a predictor of adverse prognosis (57). Shi et al reported that the WNT2B genetic variant may be a biomarker for the outcome of patients with cutaneous melanoma (58). Jiang et al observed that high expression of WNT6 was a predictor of poor survival of osteosarcoma (56). Numerous studies have demonstrated that the expression level of WNT5A is associated with prognostic outcome and may serve as a prognostic biomarker in hepatocellular carcinoma (59), gallbladder carcinoma (60) and medulloblastoma (61). Based on these early discoveries, a prognostic predictive function for WNT family genes in HBV-related HCC has been confirmed in the present study.
We herein explored the diagnostic and prognostic value of WNT family gene expression in HBV-related HCC by collecting data from public databases and performing a series of bioinformatics analyses, with the aim of identifying more sensitive biomarkers and design a novel strategy for HCC diagnosis and treatment. There were certain limitations to the present study that must be addressed. First, the data were obtained from a public database and the sample size was limited; therefore, a larger population and multi-centered clinical studies are required to increase the credibility of our conclusions. Second, complete clinical parameters must be included to better evaluate the association between WNT family genes and HCC prognosis. Third, further functional validation and clinical trials are required to reveal the underlying molecular mechanism. Finally, although we explored the diagnostic and prognostic value at the mRNA level, the protein level was not investigated in the present study. Therefore, a comprehensive research design is required to check the consistency between mRNA and protein expression.
In conclusion, the findings of the present study demonstrated that WNT2 may serve as diagnostic biomarker and WNT1 may serve as prognostic biomarker for patients with HBV-related HCC in the GSE14520 cohort. Furthermore, through verification of the TCGA cohort, the diagnostic value of WNT2 and the prognostic value of WNT1 may be further validated and generalized to HCC patients. Therefore, our results require further confirmation.
Supplementary Material
Acknowledgements
The authors would like to acknowledge the laboratory equipment and platform support sponsored by the Key Laboratory of Early Prevention and Treatment for Regional High-Incidence Tumors (Guangxi Medical University; Ministry of Education, Nanning, China). The authors would also like to acknowledge the helpful comments on this article received from our reviewers. In addition, we would also like to thank the contributors of GSE14520 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14520) and The Cancer Genome Atlas (https://cancergenome.nih.gov/).
Funding
The present study was sponsored in part by the 2018 Innovation Project of Guangxi Graduate Education (grant nos. JGY2018037 and YCBZ2018036), the Guangxi Key Laboratory for the Prevention and Control of Viral Hepatitis (grant no. GXCDCKL201902), the National Natural Science Foundation of China (grant no. 81802874), the Natural Science Foundation of the Guangxi Province of China (grant no. 2018GXNSFBA138013), the Key laboratory of High-Incidence-Tumor Prevention & Treatment (Guangxi Medical University), Ministry of Education (grant no. GKE2018-01), and the Guangxi Key R & D Program (grant no. GKEAB18221019).
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QH and XY designed this study. XW, XL, CH, TY, CY, GL, BH, KH, GZ, ZL, XZ, HS, LS, YG, XS, TP and XY conducted this study and analyzed the data. QH wrote the manuscript and XY revised the manuscript. All the authors have read and approved the final version of the manuscript for publication and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753–770. doi: 10.1016/j.bpg.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Fan JH, Wang JB, Jiang Y, Xiang W, Liang H, Wei WQ, Qiao YL, Boffetta P. Attributable causes of liver cancer mortality and incidence in China. Asian Pac J Cancer Prev. 2013;14:7251–7256. doi: 10.7314/APJCP.2013.14.12.7251. [DOI] [PubMed] [Google Scholar]
- 4.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190–1200. doi: 10.1002/hep.27969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balogh J, Victor D, III, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP., Jr Hepatocellular carcinoma: A review. J Hepatocell Carcinoma. 2016;3:41–53. doi: 10.2147/JHC.S61146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 7.Cheng R, Sun B, Liu Z, Zhao X, Qi L, Li Y, Gu Q. Wnt5a suppresses colon cancer by inhibiting cell proliferation and epithelial-mesenchymal transition. J Cell Physiol. 2014;229:1908–1917. doi: 10.1002/jcp.24566. [DOI] [PubMed] [Google Scholar]
- 8.Shiah SG, Hsiao JR, Chang WM, Chen YW, Jin YT, Wong TY, Huang JS, Tsai ST, Hsu YM, Chou ST, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014;74:7560–7572. doi: 10.1158/0008-5472.CAN-14-0978. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Jia C, Jia C, Jin X, Gu X. MicroRNA-374a inhibits aggressive tumor biological behavior in bladder carcinoma by suppressing Wnt/β-catenin signaling. Cell Physiol Biochem. 2018;48:815–826. doi: 10.1159/000491911. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, Yasui W, Kikuchi A. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29:2036–2046. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Sun Z, Su L, Wang F, Jiang Y, Yu D, Zhang F, Sun Z, Liang W. miRNA-185 serves as a prognostic factor and suppresses migration and invasion through Wnt1 in colon cancer. Eur J Pharmacol. 2018;825:75–84. doi: 10.1016/j.ejphar.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Bo H, Zhang S, Gao L, Chen Y, Zhang J, Chang X, Zhu M. Upregulation of Wnt5a promotes epithelial-to-mesenchymal transition and metastasis of pancreatic cancer cells. BMC Cancer. 2013;13:496. doi: 10.1186/1471-2407-13-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Fan L, Xia X, Rao Y, Ma Q, Yang J, Lu Y, Wang C, Huang X. Silencing Wnt2B by siRNA interference inhibits metastasis and enhances chemotherapy sensitivity in ovarian cancer. Int J Gynecol Cancer. 2012;22:755–761. doi: 10.1097/IGC.0b013e3182540284. [DOI] [PubMed] [Google Scholar]
- 14.Webster MR, Xu M, Kinzler KA, Kaur A, Appleton J, O'Connell MP, Marchbank K, Valiga A, Dang VM, Perego M, et al. Wnt5A promotes an adaptive, senescent-like stress response, while continuing to drive invasion in melanoma cells. Pigment Cell Melanoma Res. 2015;28:184–195. doi: 10.1111/pcmr.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen PH, Liu AJ, Ho KH, Chiu YT, Anne Lin ZH, Lee YT, Shih CM, Chen KC. microRNA-199a/b-5p enhance imatinib efficacy via repressing WNT2 signaling-mediated protective autophagy in imatinib-resistant chronic myeloid leukemia cells. Chem Biol Interact. 2018;291:144–151. doi: 10.1016/j.cbi.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Fu L, Zhang C, Zhang LY, Dong SS, Lu LH, Chen J, Dai Y, Li Y, Kong KL, Kwong DL, Guan XY. Wnt2 secreted by tumour fibroblasts promotes tumour progression in oesophageal cancer by activation of the Wnt/β-catenin signalling pathway. Gut. 2011;60:1635–1643. doi: 10.1136/gut.2011.241638. [DOI] [PubMed] [Google Scholar]
- 17.Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX, Wang XW. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessler S, Long EL, Budhu A, Chen Y, Zhao X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al. Integrative genomic identification of genes on 8p associated with hepatocellular carcinoma progression and patient survival. Gastroenterology. 2012;142:957–966.e12. doi: 10.1053/j.gastro.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao X, Liu X, Yang C, Wang X, Yu T, Han C, Huang K, Zhu G, Su H, Qin W, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma. J Cancer. 2018;9:2357–2373. doi: 10.7150/jca.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 23.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaul YD, Yuan B, Thiru P, Nutter-Upham A, McCallum S, Lanzkron C, Bell GW, Sabatini DM. MERAV: A tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 2016;44:D560–566. doi: 10.1093/nar/gkv1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso L, Fuchs E. Stem cells in the skin: Waste not, Wnt not. Genes Dev. 2003;17:1189–1200. doi: 10.1101/gad.1086903. [DOI] [PubMed] [Google Scholar]
- 28.Monga SP. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int J Biochem Cell Biol. 2011;43:1021–1029. doi: 10.1016/j.biocel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beers MF, Morrisey EE. The three R's of lung health and disease: Repair, remodeling, and regeneration. J Clin Invest. 2011;121:2065–2073. doi: 10.1172/JCI45961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemeth MJ, Mak KK, Yang Y, Bodine DM. Beta-Catenin expression in the bone marrow microenvironment is required for long-term maintenance of primitive hematopoietic cells. Stem Cells. 2009;27:1109–1119. doi: 10.1002/stem.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malhotra S, Kincade PW. Wnt-related molecules and signaling pathway equilibrium in hematopoiesis. Cell Stem Cell. 2009;4:27–36. doi: 10.1016/j.stem.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inestrosa NC, Arenas E. Emerging roles of Wnts in the adult nervous system. Nat Rev Neurosci. 2010;11:77–86. doi: 10.1038/nrn2755. [DOI] [PubMed] [Google Scholar]
- 33.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31:2737–2746. doi: 10.1038/emboj.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herr P, Hausmann G, Basler K. WNT secretion and signalling in human disease. Trends Mol Med. 2012;18:483–493. doi: 10.1016/j.molmed.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Kikuchi A, Yamamoto H. Tumor formation due to abnormalities in the beta-catenin-independent pathway of Wnt signaling. Cancer Sci. 2008;99:202–208. doi: 10.1111/j.1349-7006.2007.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 37.Glass DA, II, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Königshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson WR, Jr, Chi EY, Ye X, Nguyen C, Tien YT, Zhou B, Borok Z, Knight DA, Kahn M. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci USA. 2010;107:14309–14314. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhmetshina A, Palumbo K, Dees C, Bergmann C, Venalis P, Zerr P, Horn A, Kireva T, Beyer C, Zwerina J, et al. Activation of canonical Wnt signalling is required for TGF-β-mediated fibrosis. Nat Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen M, Lauterbach K, El-Gabalawy H, Firestein GS, Corr M, Carson DA. Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci USA. 2000;97:2791–2796. doi: 10.1073/pnas.050574297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura Y, Nawata M, Wakitani S. Expression profiles and functional analyses of Wnt-related genes in human joint disorders. Am J Pathol. 2005;167:97–105. doi: 10.1016/S0002-9440(10)62957-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keerthivasan S, Aghajani K, Dose M, Molinero L, Khan MW, Venkateswaran V, Weber C, Emmanuel AO, Sun T, Bentrem DJ, et al. β-catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci Transl Med. 2014;6:225ra228. doi: 10.1126/scitranslmed.3007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalkman HO. A review of the evidence for the canonical Wnt pathway in autism spectrum disorders. Mol Autism. 2012;3:10. doi: 10.1186/2040-2392-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Sáez K, Henríquez JP, Zhao A, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García-Jiménez C. Wnt and incretin connections. Vitam Horm. 2010;84:355–387. doi: 10.1016/B978-0-12-381517-0.00014-X. [DOI] [PubMed] [Google Scholar]
- 47.Welters HJ, Kulkarni RN. Wnt signaling: Relevance to beta-cell biology and diabetes. Trends Endocrinol Metab. 2008;19:349–355. doi: 10.1016/j.tem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 49.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, Yasui W, Kikuchi A. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz AL, Malgor R, Dickerson E, Weeraratna AT, Slominski A, Wortsman J, Harii N, Kohn AD, Moon RT, Schwartz FL, et al. Phenylmethimazole decreases Toll-like receptor 3 and noncanonical Wnt5a expression in pancreatic cancer and melanoma together with tumor cell growth and migration. Clin Cancer Res. 2009;15:4114–4122. doi: 10.1158/1078-0432.CCR-09-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Placencio V, Iturregui JM, Uwamariya C, Sharif-Afshar AR, Koyama T, Hayward SW, Bhowmick NA. Prostate tumor progression is mediated by a paracrine TGF-beta/Wnt3a signaling axis. Oncogene. 2008;27:7118–7130. doi: 10.1038/onc.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshioka S, King ML, Ran S, Okuda H, MacLean JA, II, McAsey ME, Sugino N, Brard L, Watabe K, Hayashi K. WNT7A regulates tumor growth and progression in ovarian cancer through the WNT/beta-catenin pathway. Mol Cancer Res. 2012;10:469–482. doi: 10.1158/1541-7786.MCR-11-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitler BG, Nicodemus JP, Li H, Cai Q, Wu H, Hua X, Li T, Birrer MJ, Godwin AK, Cairns P, Zhang R. Wnt5a suppresses epithelial ovarian cancer by promoting cellular senescence. Cancer Res. 2011;71:6184–6194. doi: 10.1158/0008-5472.CAN-11-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochoa-Hernández AB, Ramos-Solano M, Meza-Canales ID, García-Castro B, Rosales-Reynoso MA, Rosales-Aviña JA, Barrera-Chairez E, Ortíz-Lazareno PC, Hernández-Flores G, Bravo-Cuellar A, et al. Peripheral T-lymphocytes express WNT7A and its restoration in leukemia-derived lymphoblasts inhibits cell proliferation. BMC Cancer. 2012;12:60. doi: 10.1186/1471-2407-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sin MLY, Mach KE, Sinha R, Wu F, Trivedi DR, Altobelli E, Jensen KC, Sahoo D, Lu Y, Liao JC. Deep sequencing of urinary RNAs for bladder cancer molecular diagnostics. Clin Cancer Res. 2017;23:3700–3710. doi: 10.1158/1078-0432.CCR-16-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang K, Li S, Li L, Wang X, Gu Y, Jin Z. WNT6 is an effective marker for osteosarcoma diagnosis and prognosis. Medicine (Baltimore) 2018;97:e13011. doi: 10.1097/MD.0000000000013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X, Sun PL, Li JZ, Jheon S, Lee CT, Chung JH. Aberrant Wnt1/β-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. J Thorac Oncol. 2011;6:716–724. doi: 10.1097/JTO.0b013e31820c5189. [DOI] [PubMed] [Google Scholar]
- 58.Shi Q, Liu H, Han P, Li C, Wang Y, Wu W, Zhu D, Amos CI, Fang S, Lee JE, et al. Genetic variants in WNT2B and BTRC predict melanoma survival. J Invest Dermatol. 2017;137:1749–1756. doi: 10.1016/j.jid.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geng M, Cao YC, Chen YJ, Jiang H, Bi LQ, Liu XH. Loss of Wnt5a and Ror2 protein in hepatocellular carcinoma associated with poor prognosis. World J Gastroenterol. 2012;18:1328–1338. doi: 10.3748/wjg.v18.i12.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu ZC, Xiong L, Wang LX, Miao XY, Liu ZR, Li DQ, Zou Q, Liu KJ, Zhao H, Yang ZL. Comparative study of ROR2 and WNT5a expression in squamous/adenosquamous carcinoma and adenocarcinoma of the gallbladder. World J Gastroenterol. 2017;23:2601–2612. doi: 10.3748/wjg.v23.i14.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee SE, Lim SD, Kang SY, Suh SB, Suh YL. Prognostic significance of Ror2 and Wnt5a expression in medulloblastoma. Brain Pathol. 2013;23:445–453. doi: 10.1111/bpa.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.