Abstract
The current study aimed to identify the potential clinical significance and molecular mechanisms of kinesin (KIF) family member genes in lung adenocarcinoma (LUAD) using genome-wide RNA sequencing (RNA-seq) datasets derived from The Cancer Genome Atlas (TCGA) database. Clinical parameters and RNA-seq data of patients with LUAD from the TCGA database enabled the assessment of the clinical significance of KIF genes, while the potential mechanisms of their interactions in LUAD were investigated by gene set enrichment analysis (GSEA). A gene signature with potential prognostic value was constructed via a stepwise multivariable Cox analysis. In total, 23 KIF genes were identified to be differentially expressed genes (DEGs) between the LUAD tumor and adjacent non-cancerous tissues. Of these, 8 differentially expressed KIF genes were strongly found to be strongly associated with the overall survival of patients with LUAD. Three of these genes were found to be able to be grouped as a potential prognostic gene signature. Patients with higher risk scores calculated using this gene signature were found to have a markedly higher risk of mortality (adjusted P=0.003; adjusted HR, 1.576; 95% CI, 1.166–2.129). Time-dependent receiver operating characteristic analysis indicated that this prognostic signature was able to accurately predict patient prognosis with an area under curve of 0.636, 0.643,0.665, 0.670 and 0.593 for the 1-, 2-, 3-, 4- and 5-year survival, respectively. This prognostic gene signature was identified as an independent risk factor for LUAD and was able to more accurately predict prognosis in comparison to other known clinical parameters, as shown via comprehensive survival analysis. GSEA enrichment revealed that that KIF14, KIF18B and KIF20A mediated basic cell physiology through the regulation of the cell cycle, DNA replication, and DNA repair biological processes and pathways. On the whole, the findings of this study identified 23 KIF genes that were DEGs between LUAD tumor and adjacent non-cancerous tissues. In total, 8 of these genes had the potential to function as prognostic and diagnostic biomarkers in patients with LUAD.
Keywords: genome-wide, kinesin, lung adenocarcinoma, prognosis, The Cancer Genome Atlas
Introduction
Lung cancer is the primary contributor towards cancer mortality and morbidity in the developed world, including in countries such as China. The latest global cancer statistics report an estimated 2,093,876 new cases and 1,761,007 deaths due to lung cancer worldwide in 2018 (1). These statistics are reflected in China, where there were 733,300 new lung cancer cases and 610,200 deaths due to lung cancer in 2015 (2). Lung cancer presents as either non-small cell (NSCLC) or small cell lung cancer, with the latter further classified into lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). In recent years, an increased number of cases of LUAD has been observed, which has surpassed the incidence of LUSC. LUAD is mostly associated with genetic factors, environmental and other external factors, including smoking. Genetic factors are able to function as more objective biomarkers for the diagnosis, treatment and prognosis of lung cancer.
The kinesin (KIF) family member genes are mainly found in eukaryotic cells, primarily in microtubules. In vitro experiments have demonstrated that the transport of proteins is unidirectional, moving along the negative pole of microtubule towards the positive pole. Therefore, the KIF family genes control mass protein transfer both intracellularly and extracellularly, including functions, such as transporting organelles and material vesicles, and participating in cell mitosis (3–5).
The use of whole-genome sequencing data combined with bioinformatics analysis is an effective method with which to explore prospective molecular mechanisms. The Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov) is an open-source project using large-scale genomic sequencing to map the genomes of 33 types of human cancer (6,7), including complete RNA sequencing (RNA-seq) data for LUAD. Numerous studies have reported that KIF family member genes are dysregulated in multiple types of cancer, and can be used as diagnostic and prognostic biomarkers for cancers (5,8–10). In our previous study, we analyzed genome-wide breast cancer RNA-seq dataset from the TCGA database and found that multiple genes belonging to the KIF family could be used as biomarkers for the diagnosis and prognosis of breast cancer (11). Therefore, we concluded that some of the KIF family genes may also be used as diagnostic and prognostic biomarkers of LUAD. In addition, previous studies have also reported that some of the KIF family genes may be used as prognostic indicators of LUAD (12–14). However, the comprehensive systemic analysis of KIF family genes in LUAD has not yet been reported, at least to the best of our knowledge, and thus the potential underlying molecular mechanism still require further investigation. In order to fill this gap in knowledge, the present study aimed to elucidate the potential molecular mechanisms of KIF family member genes, and to determine their prognostic value in LUAD.
Materials and methods
Data source and pre-processing
Clinical data, as well as the complete RNA sequencing (RNA-seq) library of TCGA LUAD cohort were derived from the TCGA database (https://portal.gdc.cancer.gov/projects/TCGA-LUAD) (6,7,15). Raw RNA-seq was normalized using the R platform of the DESeq package (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html), allowing the identification of the differentially expressed genes (DEGs) of KIF family members between LUAD tumor and adjacent non-cancerous tissues (16). This study does not contain any experiments using human participants or animals performed by any of the authors. Since all datasets included in tge current study were downloaded from the TCGA database and data acquisition and application are consistent with the publication guidelines of TCGA, additional approval by an ethics committee is thus not necessary.
Prognostic KIF gene screening
The inclusion criteria and exclusion criteria of the patients with LUAD for survival analysis were as follows: Inclusion criteria: i) LUAD tumor tissues RNA sequencing data set were available; ii) overall survival (OS) time was available and not zero. Exclusion criteria: i) Patient tumor tissues were not subjected RNA sequencing; ii) the OS time was zero or unavailable. Survival analysis was performed using the normalized mRNA gene expression dataset of KIF-related genes and clinical outcome parameters. The subjects were grouped as having either a low or high-expression based on the median expression value of each gene. The prognostic values of KIF family member genes were evaluated via multivariate Cox proportional hazards regression analysis using the R platform of the survival package (https://cran.r-project.org/web/packages/survival/index.html). The group that had a low KIF gene expression was used as the reference group, with all data adjusted for tumor stage. A P-value <0.05 was considered to indicate a statistically significant difference, with the respective gene designated as a prognostic KIF genes.
Construction of a prognostic gene signature based on KIF gene expression
A prognostic gene signature was constructed based on the linear combination of gene expression levels multiplied by a regression coefficient (β), which was derived from multivariate Cox proportional hazards regression analysis. The prognostic KIF family member genes were inserted into the multivariate Cox regression model using overall survival as the dependent variable. The risk score formula of the prognosis signature was as follows (17–22): Risk score = expression of KIF1 × β1 KIF1 + expression of KIF2 × β2 KIF2 + … expression of KIFn × βn KIFn. Patients were classified as having low or high risks based on the median value of risk scores. A time-dependent receiver operating characteristic (ROC) curve was drawn by the R platform of the survivalROC package (https://cran.r-project.org/web/packages/survivalROC/index.html) in order to evaluate the predictive accuracy of KIF genes expression based prognostic signature for the prognosis of LUAD (23).
Comprehensive survival analysis of mRNA expression-based prognostic signature
The association between LUAD clinical features and the contrasted prognostic signature was investigated using stratified and joint effects survival analysis. A nomogram was generated to evaluate the individualized prognosis risk score based on clinical characteristics and KIF gene expression-based prognostic signature.
Gene set enrichment analysis (GSEA)
To further assess the biological pathways that underlie prognostic KIF genes in LUAD OS, GSEA (http://software.broadinstitute.org/gsea/index.jsp) was performed (24,25). GSEA uncovered the potential mechanisms of prognostic-KIF genes using the Molecular Signatures Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb/index.jsp) c2(c2.all.v6.2.symbols.gmt) and c5(c5.all.v6.2.symbols.gmt) (26). The results of GSEA that had a false discovery rate (FDR) <0.25, |Normalized Enrichment Score (NES)| >1 and a nominal P-value <0.05 were considered to indicate a statistically significant difference.
Statistical analysis
SPSS version 20.0 software (IBM Corp.) and R3.3.1 (https://www.r-project.org). were used to compute all statistical analyses. The diagnostic receiver operating characteristic (ROC) curves of KIF genes between tumor and adjacent non-cancerous tissues were analyzed and plotted by SPSS version 20.0. The independent samples t-test was used to compare the mRNA expression levels of tumor and adjacent normal tissues. The co-expression correlation between KIF family member genes was assessed by Pearson's correlation coefficient. Survival analyses were assessed using the Kaplan-Meier method and Cox proportional hazard regression model. Clinical parameters with a log-rank test P-value <0.05 in LUAD OS were subjected to further multivariate Cox proportional hazards regression model for adjustment. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Study cohort
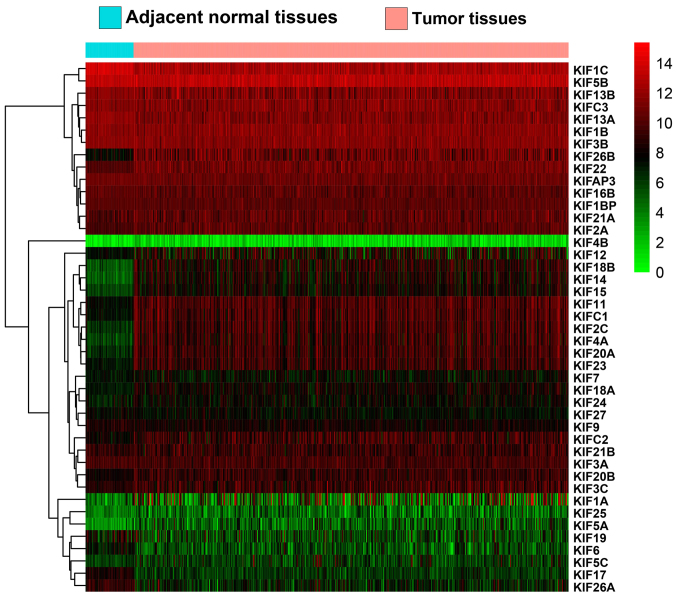
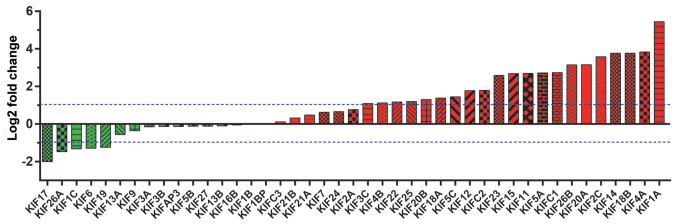
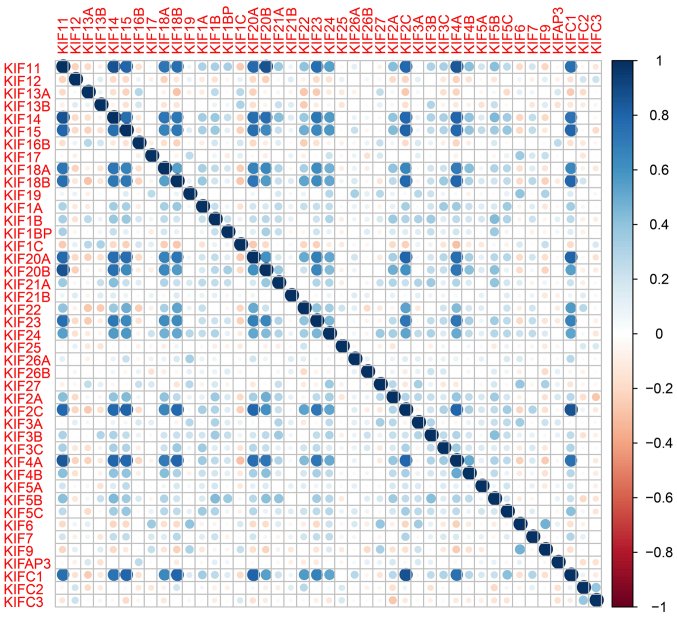
A total of 515 patients that contributed 535 tumor tissues and 59 adjacent non-cancerous tissues were extracted from the TCGA database LUAD project. In total, 500 patients with LUAD had complete clinical outcome parameters and RNA-seq data, and these were included into further survival analysis. Univariate survival analysis of the clinical parameters in LUAD OS suggested that tumor stage was significantly associated with LUAD OS (Table I). Expression heatmaps and differential expression fold changes are shown in Figs. 1 and 2, respectively. In total, 25 KIF genes were found to be significantly dysregulated between the LUAD tumor and adjacent non-cancerous tissues, of these, 23 KIF genes were identified as DEGs based on the following criteria: |log2 Fold Change(FC)| ≥1, P-value <0.05 and FDR <0.05. In total, 5 DEGs were found to be downregulated in the LUAD tumor tissues, whereas the others were upregulated (Table II). Further analysis of the co-expressed KIF genes in the tumor tissues revealed that a majority of KIF genes existed in complex co-expression associations (Fig. 3 and Table SI).
Table I.
Clinical parameters of patients with LUAD from TCGA.
| Variables | Patients (n=500) | MST (days) | Crude HR (95% CI) | Log-rank P-value |
|---|---|---|---|---|
| Age (years)a | 0.386 | |||
| ≤65 | 215 | 1,499 | 1 | |
| >65 | 264 | 1,454 | 1.143 (0.845–1.546) | |
| Sex | 0.754 | |||
| Female | 270 | 1,454 | 1 | |
| Male | 230 | 1,528 | 1.048 (0.783–1.403) | |
| Tumor stageb | <0.0001 | |||
| Stage I | 268 | 2,620 | 1 | |
| Stage II | 119 | 1,209 | 2.473 (1.719–3.559) | |
| Stage III | 80 | 879 | 3.495 (2.383–5.126) | |
| Stage IV | 25 | 826 | 3.819 (2.201–6.629) | |
| Tumor stageb | <0.0001 | |||
| Stage I+II | 387 | 1,632 | 1 | |
| Stage III+IV | 105 | 826 | 2.585 (1.894–3.528) |
Information of age was unavailable for 21 patients
Information of tumor stage was unavailable for 8 patients. TCGA, The Cancer Genome Atlas; MST, median survival time; LUAD, lung adenocarcinoma; HR, hazard ratio; CI, confidence interval.
Figure 1.
Heatmap of KIF family genes in LUAD tumor and adjacent normal tissues. KIF, kinesin; LUAD, lung adenocarcinoma.
Figure 2.
Fold change of KIF family genes between LUAD tumor and adjacent normal tissues. KIF, kinesin; LUAD, lung adenocarcinoma.
Table II.
Differential expression analysis and survival analysis of KIF family genes in patients with LUAD.
| Differential expression analysis | Survival analysis | ||||||
|---|---|---|---|---|---|---|---|
| Genes | log2 Fold change | P-value | FDR | HRa | Low 95% CI | High 95% CI | P-valueb |
| KIF14 | 3.761199 | <0.0001 | <0.0001 | 1.686098 | 1.246964 | 2.279878 | 0.000689 |
| KIF18A | 1.373027 | 0.002178 | 0.00919 | 1.610726 | 1.190265 | 2.179715 | 0.002012 |
| KIF23 | 2.582725 | <0.0001 | <0.0001 | 1.51088 | 1.117299 | 2.043104 | 0.007355 |
| KIF20A | 3.14896 | <0.0001 | <0.0001 | 1.478097 | 1.092943 | 1.998979 | 0.011181 |
| KIF11 | 2.689239 | <0.0001 | <0.0001 | 1.437782 | 1.064161 | 1.94258 | 0.01803 |
| KIF20B | 1.301735 | <0.0001 | 0.000271 | 1.43011 | 1.059873 | 1.929678 | 0.019265 |
| KIF18B | 3.766396 | <0.0001 | <0.0001 | 1.421903 | 1.052975 | 1.920092 | 0.021631 |
| KIF4A | 3.830272 | <0.0001 | <0.0001 | 1.405441 | 1.04261 | 1.894539 | 0.025494 |
| KIF1B | 0.008518 | 0.925271 | 0.972874 | 0.749876 | 0.558315 | 1.007161 | 0.055805 |
| KIF27 | −0.11005 | 0.603581 | 0.758646 | 0.763263 | 0.567524 | 1.02651 | 0.073954 |
| KIFC1 | 2.734491 | <0.0001 | <0.0001 | 1.307037 | 0.968747 | 1.763459 | 0.079741 |
| KIF4B | 1.122955 | 0.585909 | 0.74532 | 1.282052 | 0.954614 | 1.721803 | 0.098684 |
| KIFC2 | 1.792038 | <0.0001 | <0.0001 | 0.79411 | 0.590222 | 1.068429 | 0.127819 |
| KIF2C | 3.573061 | <0.0001 | <0.0001 | 1.25987 | 0.93572 | 1.696312 | 0.127964 |
| KIFAP3 | −0.1271 | 0.49553 | 0.668045 | 0.820523 | 0.610544 | 1.102719 | 0.189646 |
| KIF16B | −0.04692 | 0.831639 | 0.914865 | 0.822771 | 0.611296 | 1.107406 | 0.198118 |
| KIF1BP | 0.032733 | 0.883941 | 0.946259 | 1.196995 | 0.890981 | 1.608112 | 0.232603 |
| KIF13A | −0.5605 | 0.00104 | 0.00482 | 0.838206 | 0.624352 | 1.125311 | 0.240241 |
| KIF5A | 2.712286 | 0.007712 | 0.026645 | 0.850435 | 0.633679 | 1.141335 | 0.280467 |
| KIF17 | −1.99907 | <0.0001 | 0.000566 | 0.854866 | 0.637723 | 1.145947 | 0.29427 |
| KIF21B | 0.316249 | 0.296082 | 0.472991 | 0.863796 | 0.641166 | 1.163729 | 0.335624 |
| KIF3C | 1.08788 | 0.000112 | 0.000682 | 1.155325 | 0.860101 | 1.551884 | 0.337567 |
| KIF21A | 0.471146 | 0.078692 | 0.175791 | 0.869257 | 0.647914 | 1.166216 | 0.35006 |
| KIF5B | −0.11057 | 0.509291 | 0.680306 | 1.149144 | 0.857356 | 1.540238 | 0.352275 |
| KIF26A | −1.46358 | 0.000133 | 0.000793 | 0.877007 | 0.652637 | 1.178514 | 0.38403 |
| KIF12 | 1.77177 | <0.0001 | <0.0001 | 0.878517 | 0.6541 | 1.179931 | 0.389461 |
| KIF24 | 0.658401 | 0.275437 | 0.450236 | 1.132135 | 0.843729 | 1.519126 | 0.408083 |
| KIF15 | 2.685009 | <0.0001 | <0.0001 | 1.133063 | 0.842076 | 1.524602 | 0.40941 |
| KIF25 | 1.190456 | 0.336607 | 0.51667 | 0.918596 | 0.685346 | 1.23123 | 0.569947 |
| KIF7 | 0.621628 | 0.265307 | 0.439337 | 1.083857 | 0.807299 | 1.455156 | 0.592125 |
| KIF6 | −1.28474 | 0.013328 | 0.042003 | 0.924712 | 0.688201 | 1.242503 | 0.603526 |
| KIF13B | −0.09452 | 0.4315 | 0.610314 | 1.07943 | 0.804877 | 1.447637 | 0.609761 |
| KIF19 | −1.23658 | 0.000403 | 0.002133 | 0.931921 | 0.694829 | 1.249914 | 0.637848 |
| KIF5C | 1.444426 | 0.067916 | 0.156645 | 0.937923 | 0.697936 | 1.260429 | 0.670823 |
| KIF26B | 3.141348 | <0.0001 | <0.0001 | 0.938876 | 0.698727 | 1.261564 | 0.675623 |
| KIF1A | 5.445563 | <0.0001 | <0.0001 | 1.064054 | 0.791266 | 1.430885 | 0.681207 |
| KIF2A | 0.760907 | 0.001016 | 0.004726 | 1.062085 | 0.791335 | 1.42547 | 0.688285 |
| KIF22 | 1.166602 | <0.0001 | <0.0001 | 1.057282 | 0.787208 | 1.420012 | 0.711293 |
| KIFC3 | 0.108768 | 0.504868 | 0.675814 | 1.056382 | 0.787667 | 1.41677 | 0.714182 |
| KIF3B | −0.13168 | 0.409013 | 0.588436 | 0.954955 | 0.712533 | 1.279855 | 0.757712 |
| KIF3A | −0.14052 | 0.500277 | 0.671849 | 0.955389 | 0.71235 | 1.281346 | 0.760587 |
| KIF1C | −1.31875 | <0.0001 | <0.0001 | 0.990563 | 0.737947 | 1.329656 | 0.949668 |
| KIF9 | −0.34716 | 0.253979 | 0.425275 | 0.992495 | 0.739871 | 1.331377 | 0.959913 |
Low expression group is the reference group
adjusted for tumor stage in the Cox proportional hazard regression model. KIF, kinesin; LUAD, lung adenocarcinoma; FDR, false discovery rate; HR, hazard ratio; CI, confidence interval.
Figure 3.
Co-expression heatmap of KIF family genes in LUAD tumor tissues. KIF, kinesin; LUAD, lung adenocarcinoma.
Prognostic KIF gene screening
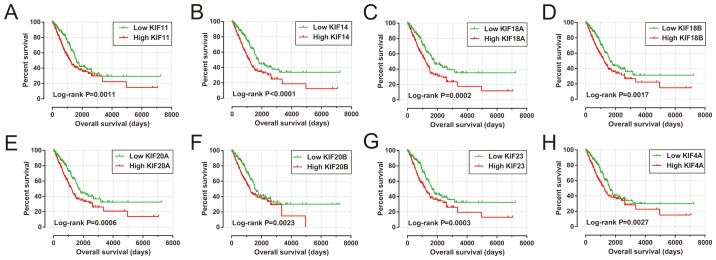
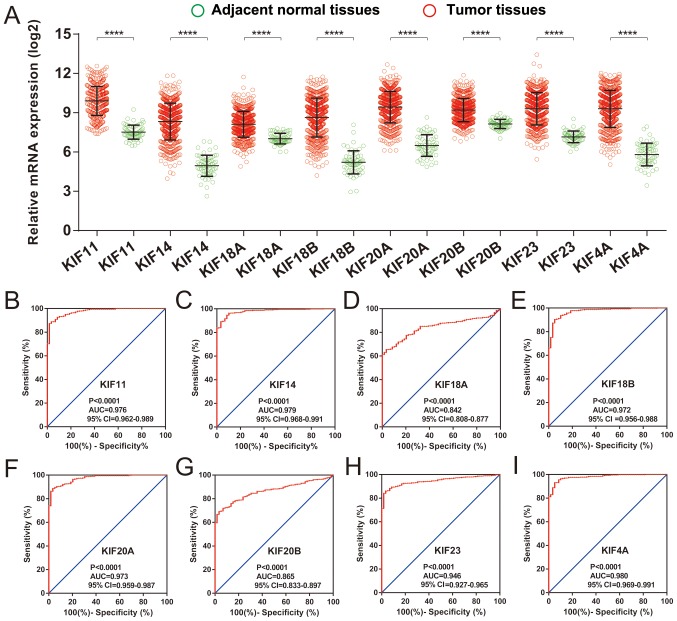
Survival analysis of KIF genes in the present study cohort based on multivariate Cox proportional hazards regression model demonstrated a total of 8 KIF genes that were significantly associated with LUAD OS (Table II and Fig. 4). The upregulation of these 8 prognostic KIF genes was associated with significantly higher mortality risks in the patients with LUAD. In addition, we also observed that these 8 prognostic KIF genes were notably upregulated in the LUAD tumor tissues (Fig. 5A), and ROC curve analysis also substantiated that these 8 prognostic KIF genes may serve as potential diagnostic biomarkers for LUAD (Fig. 5B-I).
Figure 4.
The Kaplan-Meier curves of 8 prognostic KIF family genes in LUAD. The order of the Kaplan-Meier curves of the 8 prognostic KIF family genes was as follows: (A) KIF11; (B) KIF14; (C) KIF18A; (D) KIF18B; (E) KIF20A; (F) KIF20B; (G) KIF23; (H) KIF4A. KIF, kinesin; LUAD, lung adenocarcinoma.
Figure 5.
Differential expression distribution and diagnostic ROC curves of 8 prognostic KIF family genes in LUAD tumor and adjacent normal tissues. (A) Differential expression distribution of 8 prognostic KIF family genes between LUAD tumor and healthy adjacent tissues; the order of ROC curves for the 8 prognostic KIF family genes was as follows: (B) KIF11; (C) KIF14; (D) KIF18A; (E) KIF18B; (F) KIF20A; (G) KIF20B; (H) KIF23; (I) KIF4A. KIF, kinesin; LUAD, lung adenocarcinoma; ROC: receiver operating characteristic.
Construction of a prognostic gene signature
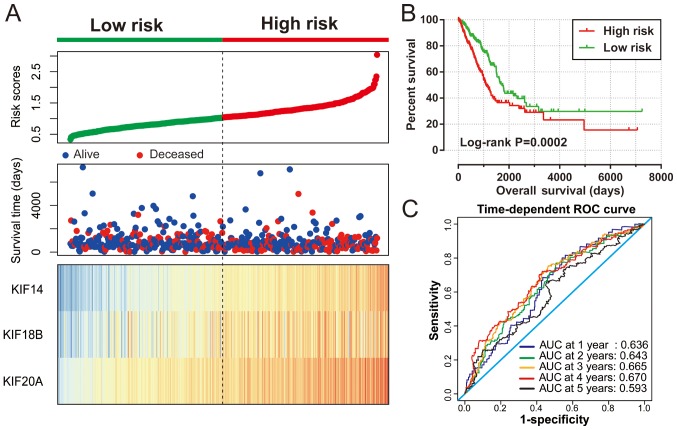
The 8 KIF genes that were significantly associated with LUAD OS on single gene survival analysis were subjected to screening for potential prognostic gene signature combination using the ‘step’ function. The most significant KIF candidate gene combinations of these 8 KIF genes were further screened for prognostic signature construction. Finally, KIF14, KIF18B and KIF20A were used for the prognostic signature construction based on the following formula: Expression of KIF14 × (0.2437) + expression of KIF18B × (−0.1541) + expression of KIF20A × (0.1926). Survival analysis revealed that patients with high risk scores were more likely to have an increased risk of death (log-rank P=0.0002, adjusted P=0.003; adjusted HR, 1.576; 95% CI, 1.166–2.129; Fig. 6A and B) and a poorer clinical outcome (median survival time, high risk vs. low risk: 1,081 vs. 1,725 days). The predictive accuracy of this prognostic signature was determined using time-dependent ROC curve analysis, with the results suggesting that the constructed signature was able to accurately predict the 1-, 2-, 3-, 4- and 5-year patient survival, based on the respective area under curves 0.636, 0.643,0.665, 0.670 and 0.593 (Fig. 6C), respectively. We also noted that the expression levels of the KIF14, KIF18B and KIF20A genes exhibited a strongly and positive correlation with each other (Pearson's correlation coefficient r=0.713 for KIF14 and KIF18B; r=0.760 for KIF14 and KIF20A; r=0.722 for KIF20A and KIF18B; Table SI).
Figure 6.
Prognostic risk score model analyses of 3 KIF family genes in patients with LUAD. (A) From top to bottom are the risk score, distribution of patient survival status, and the expression heat maps of 3 prognostic KIF family genes between the low- and high-risk groups. (B) Kaplan-Meier curves of the high and low risk groups. (C) ROC curves used for survival prediction in LUAD patients based on risk score. KIF, kinesin; LUAD, lung adenocarcinoma; ROC: receiver operating characteristic.
Stratified and joint effects analysis
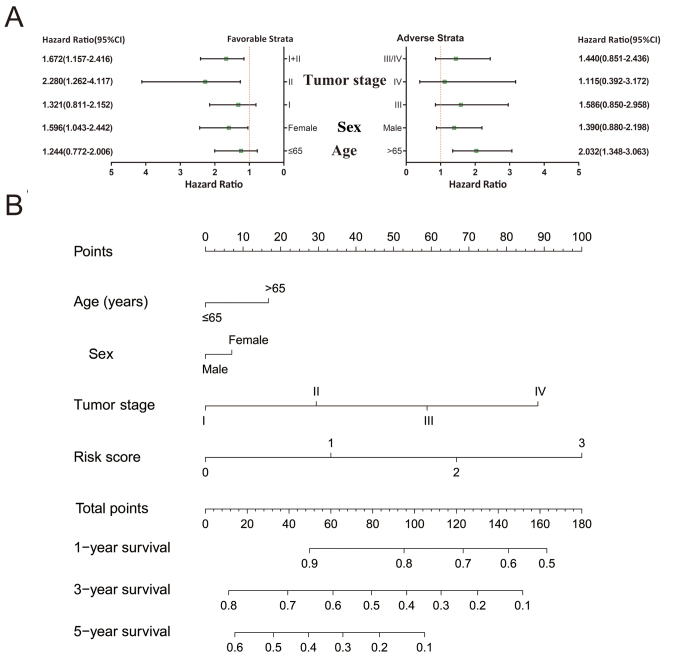
A comprehensive analysis of the nomogram and stratified and joint effects survival analysis was used to further investigate the association between clinical parameters and the prognostic gene signature. Patients that had stage I and stage I+II disease, were of the female sex and were >65 years of age were more likely to succumb to the disease if they also had higher risk scores (Fig. 7A). A nomogram constructed of the risk scores and clinical LUAD parameters demonstrated that the KIF gene expression-based prognostic signature was more accurate compared to other parameters (Fig. 7B).
Figure 7.
Stratified analysis and nomogram for the risk score and clinical features. (A) Stratified analysis of the risk score in LUAD OS. (B) Nomogram for predicting the 1-, 3-, and 5-year OS using the risk scores and clinical features in LUAD. LUAD, lung adenocarcinoma; OS, overall survival.
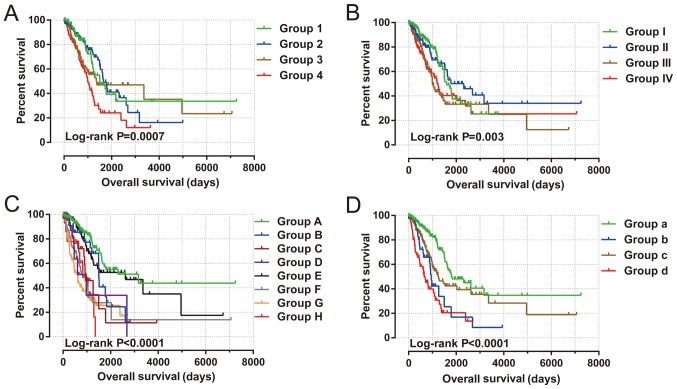
Joint effects survival analysis between the KIF gene expression-based clinical parameters and prognostic gene signatures indicated that the constructed signature was able to accurately predict the OS of patients with LUAD, particularly when combined with clinical parameters (Fig. 8 and Table III).
Figure 8.
Joint effects analysis of risk score and clinical features in patients with LUAD. Joint effects analysis of risk score stratified by the following clinical features in patients with LUAD: (A) Age: Group 1 is low risk and age ≤65 combination, group 2 is low risk and age >65 combination, group 3 is high risk and age ≤65 combination, and group 4 is high risk and age >65 combination. (B) Sex: Group I is low risk and female combination, group II is low risk and male combination, group III is high risk and female combination, and group IV is high risk and male combination. (C) Tumor stage: Group A is low risk and stage I combination, group B is low risk and stage II combination, group C is low risk and stage III combination, group D is low risk and stage IV combination, group E is high risk and stage I combination, group F is high risk and stage II combination, group G is high risk and stage III combination, and group H is high risk and stage IV combination. (D) Tumor stage stratified by early stage and advanced stage: Group a is low risk and stage I+II combination, group b is low risk and stage III+IV combination, group c is high risk and stage I+II combination, and group d is high risk and stage III+IV combination. LUAD, lung adenocarcinoma.
Table III.
Joint effects survival analysis of clinical parameters and the risk score in LUAD patients from TCGA.
| Group | Risk score | Variables | Events/total (n=500) | MST (days) | Crude HR (95% CI) | Crude P | Adjusted HR (95% CI) | Adjusted P-valuea |
|---|---|---|---|---|---|---|---|---|
| Age (years) | ||||||||
| 1 | Low risk | ≤65 | 29/95 | 1,501 | 1 | 1 | ||
| 2 | Low risk | >65 | 43/144 | 1653 | 0.958 (0.598–1.536) | 0.86 | 0.984 (0.607–1.595) | 0.948 |
| 3 | High risk | ≤65 | 45/120 | 1,357 | 1.362 (0.853–2.173) | 0.195 | 1.223 (0.760–1.967) | 0.406 |
| 4 | High risk | >65 | 56/120 | 999 | 2.021 (1.288–3.171) | 0.002 | 1.937 (1.226–3.060) | 0.005 |
| Sex | ||||||||
| I | Low risk | Female | 43/151 | 1,600 | 1 | 1 | ||
| II | Low risk | Male | 32/99 | 2,318 | 1.011 (0.638–1.601) | 0.963 | 0.935 (0.582–1.503) | 0.781 |
| III | High risk | Female | 53/119 | 999 | 1.763 (1.177–2.640) | 0.006 | 1.644 (1.088–2.4830 | 0.018 |
| IV | High risk | Male | 54/131 | 1,235 | 1.747 (1.169–2.609) | 0.006 | 1.434 (0.952–2.159) | 0.084 |
| Tumor stage | ||||||||
| A | Low risk | Stage I | 34/154 | 3,169 | 1 | 1 | ||
| B | Low risk | Stage II | 17/48 | 1,501 | 1.751 (0.977–3.140) | 0.06 | 1.751 (0.977–3.140) | 0.06 |
| C | Low risk | Stage III | 15/32 | 952 | 3.025 (1.643–5.571) | 0.0004 | 3.025 (1.643–5.571) | 0.0004 |
| D | Low risk | Stage IV | 7/11 | 976 | 3.957 (1.749–8.952) | 0.001 | 3.957 (1.749–8.952) | 0.001 |
| E | High risk | Stage I | 31/114 | 2,620 | 1.317 (0.809–2.144) | 0.268 | 1.317 (0.809–2.144) | 0.268 |
| F | High risk | Stage II | 37/71 | 864 | 3.883 (2.421–6.227) | <0.0001 | 3.883 (2.421–6.227) | <0.0001 |
| G | High risk | Stage III | 30/48 | 593 | 4.697 (2.868–7.694) | <0.0001 | 4.697 (2.868–7.694) | <0.0001 |
| H | High risk | Stage IV | 9/14 | 826 | 4.712 (2.244–9.892) | <0.0001 | 4.712 (2.244–9.892) | <0.0001 |
| Tumor stage | ||||||||
| a | Low risk | Stage I+II | 51/202 | 1,798 | 1 | 1 | ||
| b | Low risk | Stage III+IV | 22/43 | 952 | 2.767 (1.674–4.574) | <0.0001 | 3.948 (2.024–7.699) | <0.0001 |
| c | High risk | Stage I+II | 68/185 | 1,258 | 1.750 (1.216–2.520) | 0.003 | 1.644 (1.139–2.373) | 0.008 |
| d | High risk | Stage III+IV | 39/62 | 656 | 3.974 (2.612–6.047) | <0.0001 | 5.700 (3.061–10.613) | <0.0001 |
Adjusted for tumor stage in the Cox proportional hazard regression model. TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; MST, median survival time; HR, hazard ratio; CI, confidence interval.
GSEA
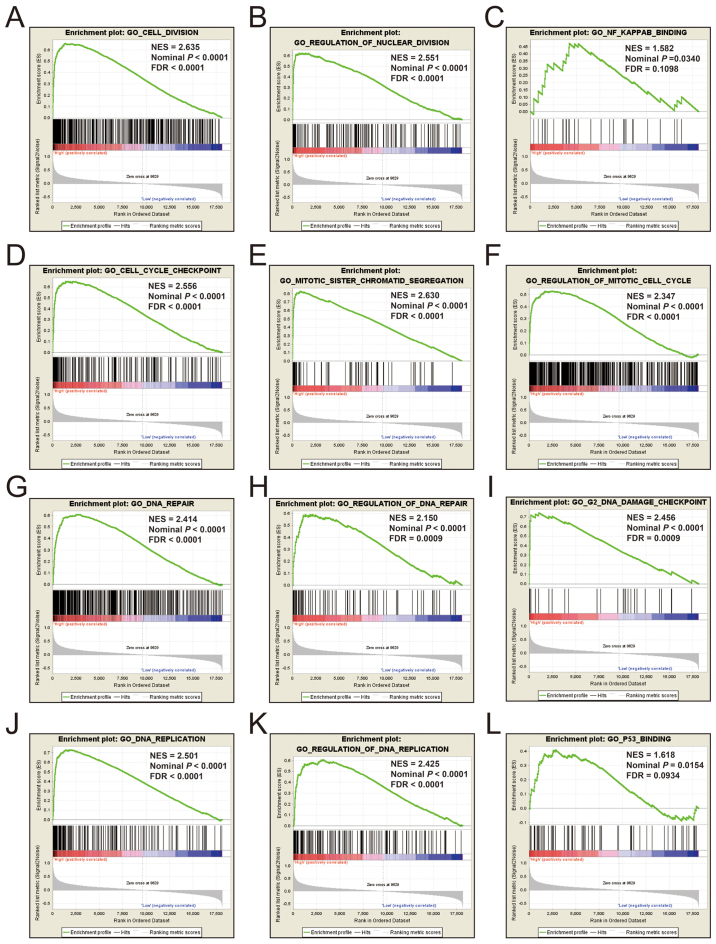
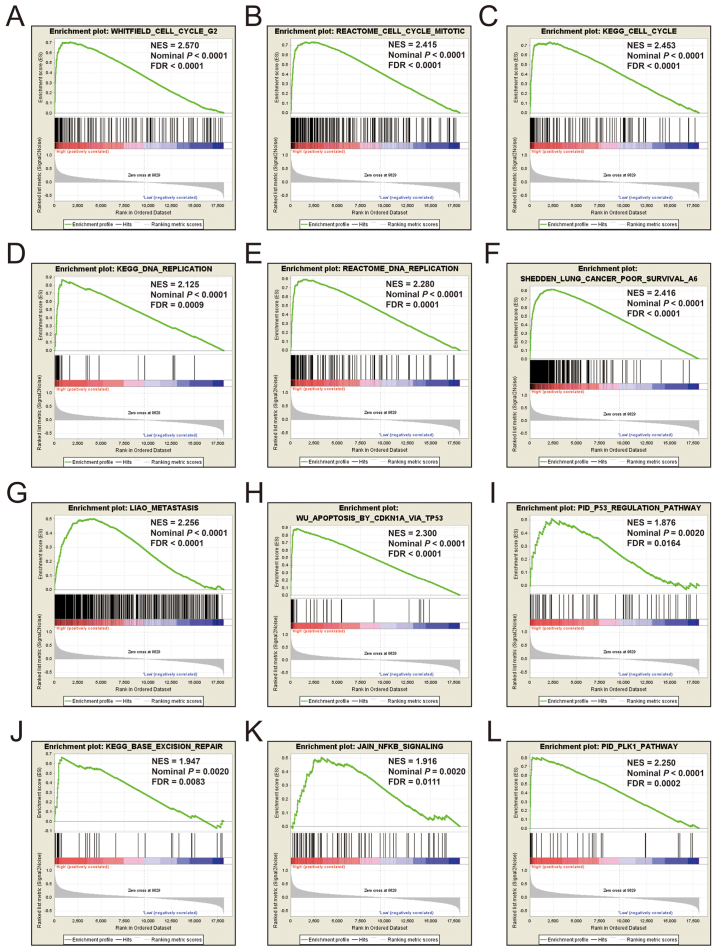
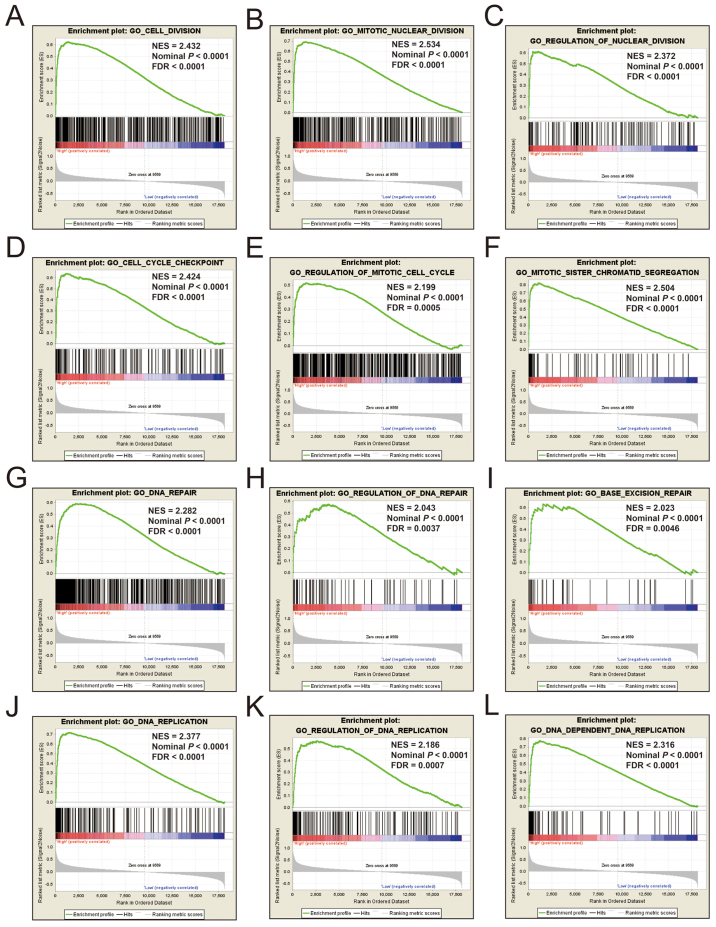
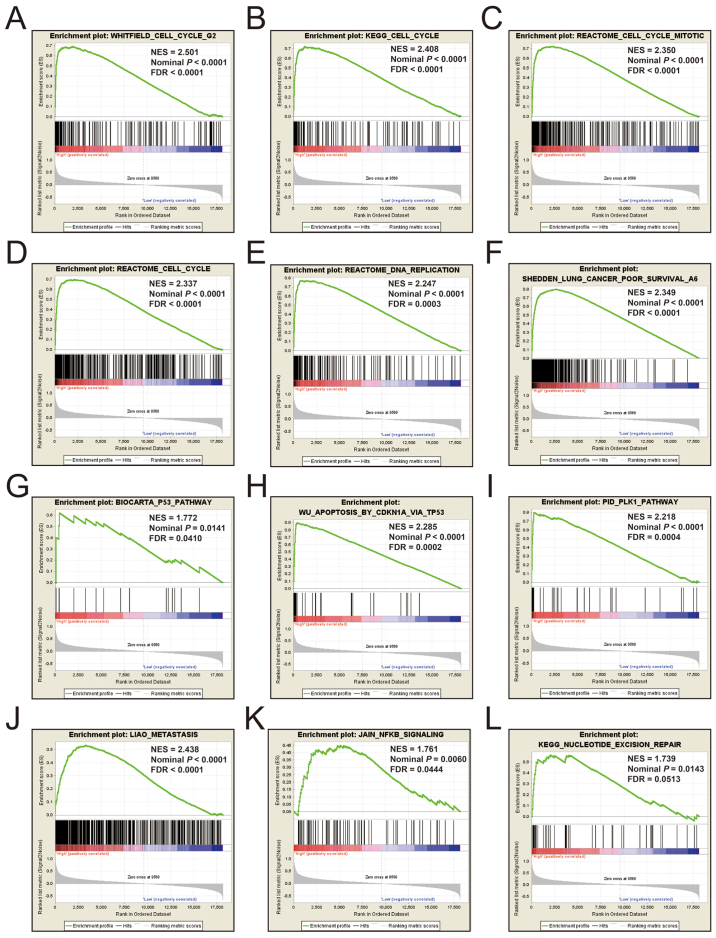
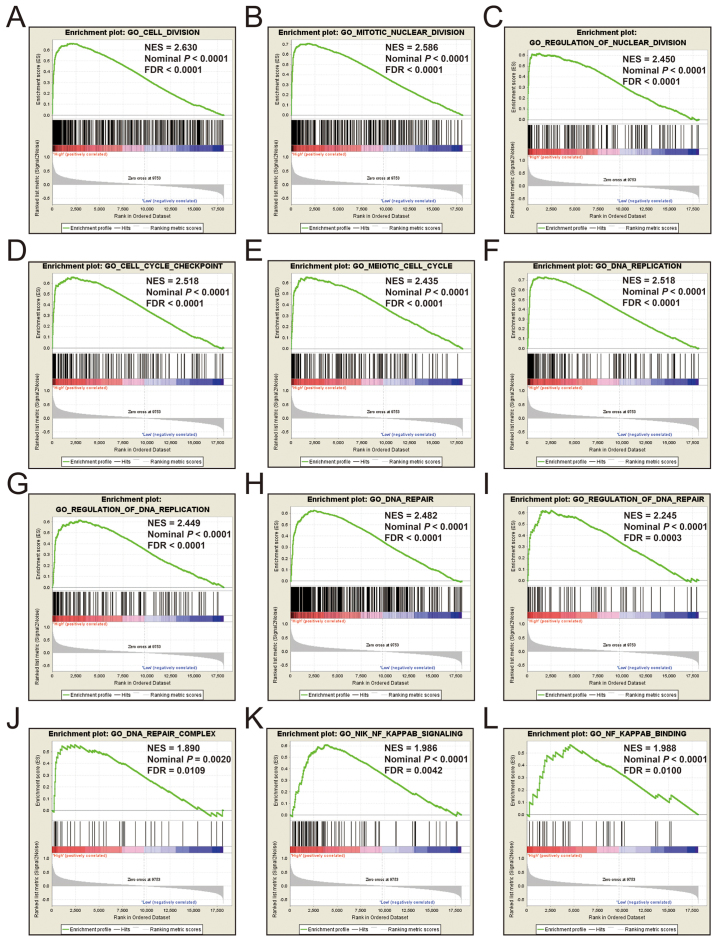
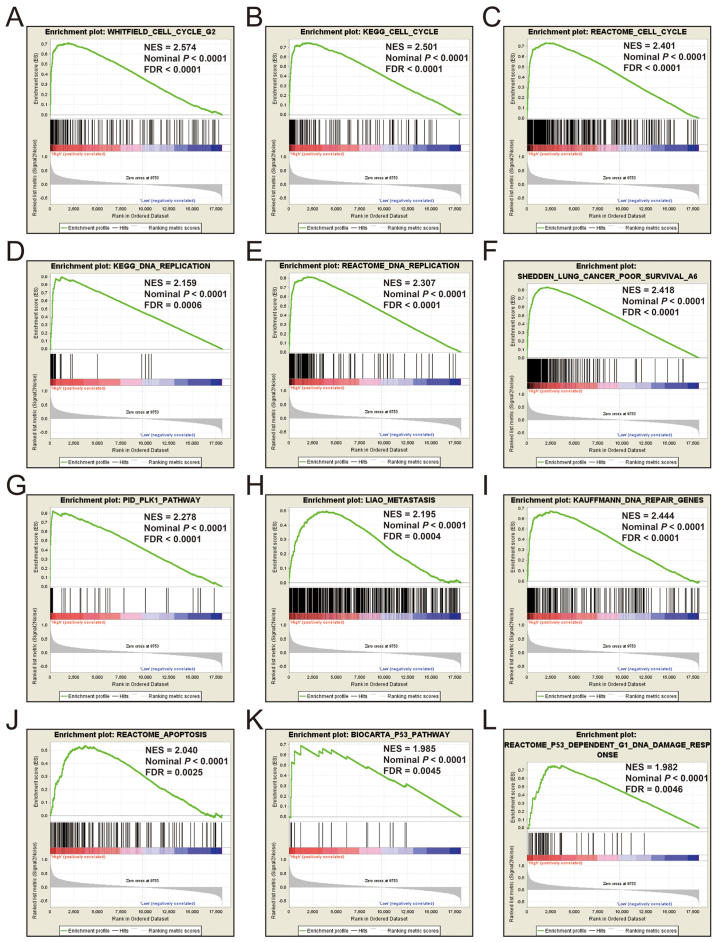
Additional exploration of the biological pathways of the selected KIF genes in relation to LUAD was carried out using a single gene GSEA. An enrichment of c5 suggested that a high expression of KIF14 was involved in DNA repair, DNA replication, cell cycle, tumor protein p53 (TP53) binding and mitotic sister chromatid separation biological processes (Fig. 9 and Table SII). Whereas, an enrichment of c2 indicated that a high expression of KIF14 influenced the cell cycle, DNA replication, lung cancer poor survival, metastasis, base excision repair, the PLK1 pathway, nuclear factor-κB (NF-κB) and the TP53 pathway (Fig. 10 and Table SIII). c5 enrichment suggested that a high expression of KIF18B was also involved in cell division, the cell cycle, DNA replication and DNA repair (Fig. 11 and Table SIV), whereas c2 enrichment suggested that a high expression of KIF18b was involved in the cell cycle, DNA replication, lung cancer poor survival, metastasis, base excision repair, the PLK1 pathway, NF-κB and TP53 pathway (Fig. 12 and Table SV). Similar results were also found for KIF20A, where c5 enrichment suggested that a high expression of KIF20A was involved in cell division, cell cycle, DNA replication, DNA repair and the NF-κB pathway (Fig. 13 and Table SVI), whereas c2 enrichment suggested that high expression of KIF20A was involved in cell cycle, DNA replication, lung cancer poor survival, apoptosis, metastasis, DNA repair, the PLK1 and TP53 pathway (Fig. 14 and Table SVII). It is evident that the potential mechanisms of KIF14, KIF18B and KIF20A are likely mediated through their influence on cell cycle regulation, DNA replication and DNA repair. Furthermore, all the c2 enrichment analyses of KIF14, KIF18B and KIF20A were enriched in the gene set of SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6, which indicated that the upregulated expression levels of the genes of this gene set in patients with lung cancer were predictors of a poor survival outcome.
Figure 9.
(A-L) GSEA results of c5 reference gene set of high KIF14 expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Figure 10.
(A-L) GSEA results of c2 reference gene set of high KIF14 expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Figure 11.
(A-L) GSEA results of c5 reference gene set of high KIF18B expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Figure 12.
(A-L) GSEA results of c2 reference gene set of high KIF18B expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Figure 13.
(A-L) GSEA results of c5 reference gene set of high KIF20A expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Figure 14.
(A-L) GSEA results of c2 reference gene set of high KIF20A expression group. ES, enrichment score; NES, normalized enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; KIF, kinesin.
Discussion
KIF family member genes encoded proteins are required for numerous processes, including intracellular transport, chromosome segregation, mitotic spindle formation and cytokinesis, and multiple family member genes have been reported to be dysregulated in various types of cancer (5,8,12,27–29). The prognostic and diagnostic capabilities of KIF family member genes have been demonstrated in various types of cancer. In a previous study, immumohistochemical staining suggested that KIF3A expression was significantly higher in breast cancer (BC) tumor tissues than healthy adjacent tissues (8). Previous studies have demonstrated that an increased KIF2A expression is a predictor of an unfavorable clinical outcome in patients with LUAD and diffuse large B cell lymphoma (12,30). An increased KIF4A expression has also been shown to be strongly associated with a poorer prognosis of patients with BC (9), prostate cancer (PCa) (10) and hepatocellular carcinoma (HCC) (31,32). Similar prognostic values of KIF11 in oral cancer (33) and BC (34) have also been reported. Other members of the KIF gene family have also exhibited similar prognostic values in other types of cancer, such as KIF26B in ovarian cancer (OC) (35) and KIF20B in HCC (36). In the present study, we observed that 26 KIF family member genes were differentially expressed in LUAD and healthy adjacent tissues and identified as DEGs, with 5 DEGs were downregulated and 18 DEGs were upregulated. In total, 8 of these DEGs were identified as diagnostic and prognostic genes for LUAD, which was consistent with the findings of the above-mentioned studies. Three of these genes were used to construct a potential prognostic signature for LUAD.
For 3 KIF genes of the prognostic signature that were identified in the present study, previous studies have observed that KIF14 is notably upregulated in tumor tissues of OC (37,38), pancreatic carcinoma (39), cervical cancer (40), BC (41), PCa (28), glioma (42) and gastric cancer (GC) (43). The prognostic analysis of previous studies has demonstrated that an elevated KIF14 expression confers unfavorable clinical outcomes in patients with OC (37,38), pancreatic carcinoma (39), cervical cancer (40), BC (41), PCa (28), glioma (42), medulloblastoma (44), lung cancer (45), HCC (46) and GC (43). The function of KIF14 may serve as an oncogene in cancers, and inhibiting KIF14 has been shown to inhibit the growth of PCa and LUAD cell lines (13,28), to suppress the proliferation of medulloblastoma and NSCLC (44,45), to decrease cancer cell migration and induce apoptosis in HCC (46), as well as to inhibit tumor metastasis in GC (43), PCa (28) and LUAD (13). In the present study, our results of KIF14 in LUAD were also consistent with those of these previous studies. Our results suggest that KIF14 may be adopted as diagnostic and prognostic indicator for LUAD. Similar with the KIF14, we also identified that KIF18B was upregulated in LUAD tumor tissues, suggesting its utility as a diagnostic and prognostic biomarker in patients with LUAD. Wu et al demonstrated that KIF18B expression was increased in cervical cancer tumor tissues with an advanced tumor grade and stage. This gene may also function as a cervical cancer oncogene, as the downregulation of KIF18B has been shown to inhibit cervical cancer migration, invasion and cell in vitro (47). Itzel et al identified KIF18B as a novel oncogene that drives carcinogenesis in HCC (48). Our results were very consistent with the results of these previous studies. To the best of our knowledge, this study was the first to suggest that KIF18B may serve as potential diagnostic and prognostic indicator for LUAD.
Another of our candidate prognostic signature gene, KIF20A, has also been reported to be strongly expressed in nasopharyngeal carcinoma (NPC) (49), NSCLC (14,50,51), HCC (52), cervical cancer (53), glioma (54), OC (55) and clear cell renal cell carcinoma (ccRCC) (56). Furthermore, a high expression of KIF20A in these types of cancer has also been shown to be associated with an increased risk of an unfavorable prognosis (14,49–53,55,56). In addition, previous studies have also demonstrated that a high KIF20A expression is associated with a poor clinical outcome in patients with melanoma (57) and ovarian clear cell carcinoma cells (58). Previous studies have also observed that KIF20A is significantly related to tumor progression, and advanced stage tumor tissues exhibit an inceased KIF20A expression level (55,56). Functional experiment assessment in cancers infers that KIF20A may play a carcinogenic role in cancer, and cancer cell proliferation can be regulated by the overexpression or inhibition of KIF20A (14,52,54,55,58).
In the present study, we also identified the prospective molecular mechanisms using GSEA. KIF family genes play critical roles in chromosome segregation, mitotic spindle formation and cytokinesis. GSEA analysis further verified that KIF14, KIF18B and KIF20A were significant participators in cell cycle regulation, thereby influencing the clinical outcome of patients with LUAD. Previous studies have demonstrated that KIF14 functions to regulate cell apoptosis and proliferation, cytokinesis and cell division (46,59,60). Xu et al demonstrated that inhibiting KIF14 in HCC cell lines can influence the cell cycle and cytokinesis biological process (29). The overexpression of KIF14 in colorectal cancer (CRC) has been shown to promote cell proliferation and accelerate cell cycle progression (61). A similar oncogenic function of KIF20A in the cell cycle and proliferation has also been reported in pan-cancers (14,55,58,62). Itzel et al observed that the overexpression of KIF18B increased the proliferation of HCC cells (48). Based on literature reviewing and prospective molecular mechanism analysis from the current study, it can be concluded that the one of the molecular mechanisms of KIF family genes is the involvement in the prognosis of LUAD mainly by affecting cell cycle-related biological processes and pathways.
Among one of the limitations of this study is that clinical information derived from TCGA was not comprehensive, barring a complete assessment of risk profiles. The results of the current study were also based on a single cohort and lack additional validation cohorts, with verification in larger sample sizes across differing cohorts needed to further verify the findings. Furthermore, the results of this study were derived from RNA sequencing data from the TCGA LUAD cohort and were not validated in additional cohorts by RT-PCR and immunohistochemistry in both the mRNA and protein level. Nevertheless, the resultant 3 KIF gene-signature developed in this study was proven to be a more accurate prognosticator in contrast to other clinical data. These results lay the foundation for further studies into the mechanistic functions of KIF genes as regards the prognosis of patients with LUAD, allowing for further development targeted LUAD therapy.
In conclusion, in this study, using an integrated assessment of KIF family member genes RNA-seq dataset and clinical data of LUAD derived from the TCGA database, we systematically evaluated the differential expression and prognostic values of KIF family member genes, and found that 23 KIF genes were DEGs between LUAD tumor and adjacent normal tissues. In total, 8 of these were found to be potential prognostic and diagnostic biomarkers in patients with LUAD. In addition, we also developed a novel 3 KIF gene-expression-based signature, including KIF14, KIF18B and KIF20A, which may aid in the prognosis of patients with LUAD.
Supplementary Material
Acknowledgements
The authors would like to thank the contributors of The Cancer Genome Atlas (https://cancergenome.nih.gov/) for sharing the LUAD dataset on open access. In addition, we would also like to acknowledge the helpful comments on this manuscript received from our reviewers.
Funding
This study was supported in part by the self-raised Scientific Research Fund of the Ministry of Health of Guangxi Province (Z2016318 and Z2016364), The Basic Ability Improvement Project for Middle-aged and Young Teachers in Colleges and Universities in Guangxi (2018KY0110).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request. All raw data of LUAD, which were included in the current study, can be downloaded from TCGA (https://portal.gdc.cancer.gov/).
Authors' contributions
LZ and PW designed this study; LZ, GZ, XW, XL, RH, CH, PH, JZ and PW conducted and further performed the study, processed and analyzed the data, as well as interpret the results. LZ wrote this manuscript, and PW guided the writing. All authors have read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The current study does not contain any studies with human participants or animals performed by any of the authors. Since all LUAD datasets included in this manuscript were obtained from The Cancer Genome Atlas, therefore, additional approval by an Ethics Committee was not necessary. In addition, the procedures of this manuscript were in accordance with the Helsinki declaration of 1964 and its later amendments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 4.Lu W, Gelfand VI. Moonlighting motors: kinesin, dynein, and cell polarity. Trends Cell Biol. 2017;27:505–514. doi: 10.1016/j.tcb.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12:527–539. doi: 10.1038/nrc3310. [DOI] [PubMed] [Google Scholar]
- 6.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 19A. 2015:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia P, Chu S, Liu G, Chen G, Yi T, Feng S, Zhou H. High expression of KIF3A is a potential new parameter for the diagnosis and prognosis of breast cancer. Biomed Rep. 2018;8:343–349. doi: 10.3892/br.2018.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue D, Cheng P, Han M, Liu X, Xue L, Ye C, Wang K, Huang J. An integrated bioinformatical analysis to evaluate the role of KIF4A as a prognostic biomarker for breast cancer. OncoTargets Ther. 2018;11:4755–4768. doi: 10.2147/OTT.S164730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao H, Chen X, Cai Q, Shang Z, Niu Y. Increased KIF4A expression is a potential prognostic factor in prostate cancer. Oncol Lett. 2018;15:7941–7947. doi: 10.3892/ol.2018.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song X, Zhang T, Wang X, Liao X, Han C, Yang C, Su K, Cao W, Gong Y, Chen Z, et al. Distinct diagnostic and prognostic values of kinesin family member genes expression in patients with breast cancer. Med Sci Monit. 2018;24:9442–9464. doi: 10.12659/MSM.913401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie T, Li X, Ye F, Lu C, Huang H, Wang F, Cao X, Zhong C. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem Biophys Res Commun. 2018;497:65–72. doi: 10.1016/j.bbrc.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Hung PF, Hong TM, Hsu YC, Chen HY, Chang YL, Wu CT, Chang GC, Jou YS, Pan SH, Yang PC. The motor protein KIF14 inhibits tumor growth and cancer metastasis in lung adenocarcinoma. PLoS One. 2013;8:e61664. doi: 10.1371/journal.pone.0061664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Zhou LL, Li X, Ni J, Chen P, Ma R, Wu J, Feng J. Overexpression of KIF20A confers malignant phenotype of lung adenocarcinoma by promoting cell proliferation and inhibiting apoptosis. Cancer Med. 2018;7:4678–4689. doi: 10.1002/cam4.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao X, Zhu G, Huang R, Yang C, Wang X, Huang K, Yu T, Han C, Su H, Peng T. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi: 10.2147/CMAR.S161334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao X, Liu X, Yang C, Wang X, Yu T, Han C, Huang K, Zhu G, Su H, Qin W, et al. Distinct diagnostic and prognostic values of minichromosome maintenance gene expression in patients with hepatocellular carcinoma. J Cancer. 2018;9:2357–2373. doi: 10.7150/jca.25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao X, Wang X, Huang K, Yang C, Yu T, Han C, Zhu G, Su H, Huang R, Peng T. Genome-scale analysis to identify prognostic microRNA biomarkers in patients with early stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. Cancer Manag Res. 2018;10:2537–2551. doi: 10.2147/CMAR.S168351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao X, Yang C, Huang R, Han C, Yu T, Huang K, Liu X, Yu L, Zhu G, Su H, et al. Identification of potential prognostic long non-coding RNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cell Physiol Biochem. 2018;48:1854–1869. doi: 10.1159/000492507. [DOI] [PubMed] [Google Scholar]
- 21.Huang R, Liao X, Li Q. Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. OncoTargets Ther. 2017;10:5243–5254. doi: 10.2147/OTT.S147717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao X, Huang K, Huang R, Liu X, Han C, Yu L, Yu T, Yang C, Wang X, Peng T. Genome-scale analysis to identify prognostic markers in patients with early-stage pancreatic ductal adenocarcinoma after pancreaticoduodenectomy. OncoTargets Ther. 2017;10:4493–4506. doi: 10.2147/OTT.S142557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng N, Xu YZ, Xi QH, Jiang HY, Wang CY, Zhang Y, Ye Q. Overexpression of KIF2A is suppressed by miR-206 and associated with poor prognosis in ovarian cancer. Cell Physiol Biochem. 2018;50:810–822. doi: 10.1159/000494467. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Yuan Y, Liang P, Zhang Z, Guo X, Xia L, Zhao Y, Shu XS, Sun S, Ying Y, et al. Overexpression of a novel candidate oncogene KIF14 correlates with tumor progression and poor prognosis in prostate cancer. Oncotarget. 2017;8:45459–45469. doi: 10.18632/oncotarget.17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Choe C, Shin SH, Park SW, Kim HS, Jung SH, Yim SH, Kim TM, Chung YJ. Silencing of KIF14 interferes with cell cycle progression and cytokinesis by blocking the p27(Kip1) ubiquitination pathway in hepatocellular carcinoma. Exp Mol Med. 2014;46:e97. doi: 10.1038/emm.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, You X, Liu H, Xu M, Dang Q, Yang L, Huang J, Shi W. High KIF2A expression predicts unfavorable prognosis in diffuse large B cell lymphoma. Ann Hematol. 2017;96:1485–1491. doi: 10.1007/s00277-017-3047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, Deng M, Huang Y. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis. 2018;9:141. doi: 10.1038/s41419-017-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou G, Dong C, Dong Z, Liu G, Xu H, Chen L, Liu L, Wang H, Zhou W. Upregulate KIF4A enhances proliferation, invasion of hepatocellular carcinoma and indicates poor prognosis across human cancer types. Sci Rep. 2017;7:4148. doi: 10.1038/s41598-017-04176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daigo K, Takano A, Thang PM, Yoshitake Y, Shinohara M, Tohnai I, Murakami Y, Maegawa J, Daigo Y. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer. Int J Oncol. 2018;52:155–165. doi: 10.3892/ijo.2017.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pei YY, Li GC, Ran J, Wei FX. Kinesin family member 11 contributes to the progression and prognosis of human breast cancer. Oncol Lett. 2017;14:6618–6626. doi: 10.3892/ol.2017.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Zhang L, Xie L. Upregulation of KIF26B, cell migration and proliferation of human ovarian cancer cell lines in vitro, and patient outcomes from human bioinformatic analysis. Med Sci Monit. 2018;24:3863–3872. doi: 10.12659/MSM.907889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Li Y, Zhang X, Liu XY, Peng A, Chen Y, Meng L, Chen H, Zhang Y, Miao X, et al. Inhibition of kinesin family member 20B sensitizes hepatocellular carcinoma cell to microtubule-targeting agents by blocking cytokinesis. Cancer Sci. 2018;109:3450–3460. doi: 10.1111/cas.13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu HL, Deng SZ, Li C, Tian ZN, Song XQ, Yao GD, Geng JS. High expression of KIF14 is associated with poor prognosis in patients with epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21:239–245. [PubMed] [Google Scholar]
- 38.Thériault BL, Pajovic S, Bernardini MQ, Shaw PA, Gallie BL. Kinesin family member 14: An independent prognostic marker and potential therapeutic target for ovarian cancer. Int J Cancer. 2012;130:1844–1854. doi: 10.1002/ijc.26189. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Z, Cheng Y, Jiang Y, Liu S, Zhang M, Liu J, Zhao Q. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci. 2018;14:124–136. doi: 10.7150/ijbs.22619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Shi Y, Li J, Cui W, Yang B. Up-regulation of KIF14 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in cervical cancer. Biosci Rep. 2016;36:e00315. doi: 10.1042/BSR20150314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corson TW, Gallie BL. KIF14 mRNA expression is a predictor of grade and outcome in breast cancer. Int J Cancer. 2006;119:1088–1094. doi: 10.1002/ijc.21954. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Wang L, Li D, Deng J, Zhao Z, He S, Zhang Y, Tu Y. Kinesin family member 14 is a candidate prognostic marker for outcome of glioma patients. Cancer Epidemiol. 2013;37:79–84. doi: 10.1016/j.canep.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Yang Z, Li C, Yan C, Li J, Yan M, Liu B, Zhu Z, Wu Y, Gu Q. KIF14 promotes tumor progression and metastasis and is an independent predictor of poor prognosis in human gastric cancer. Biochim Biophys Acta Mol Basis Dis 1865. 2019:181–192. doi: 10.1016/j.bbadis.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 44.Li KK, Qi Y, Xia T, Chan AK, Zhang ZY, Aibaidula A, Zhang R, Zhou L, Yao Y, Ng HK. The kinesin KIF14 is overexpressed in medulloblastoma and downregulation of KIF14 suppressed tumor proliferation and induced apoptosis. Lab Invest. 2017;97:946–961. doi: 10.1038/labinvest.2017.48. [DOI] [PubMed] [Google Scholar]
- 45.Corson TW, Zhu CQ, Lau SK, Shepherd FA, Tsao MS, Gallie BL. KIF14 messenger RNA expression is independently prognostic for outcome in lung cancer. Clin Cancer Res. 2007;13:3229–3234. doi: 10.1158/1078-0432.CCR-07-0393. [DOI] [PubMed] [Google Scholar]
- 46.Yang T, Zhang XB, Zheng ZM. Suppression of KIF14 expression inhibits hepatocellular carcinoma progression and predicts favorable outcome. Cancer Sci. 2013;104:552–557. doi: 10.1111/cas.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, Wang A, Zhu B, Huang J, Lu E, Xu H, Xia W, Dong G, Jiang F, Xu L. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. OncoTargets Ther. 2018;11:1707–1720. doi: 10.2147/OTT.S157440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itzel T, Scholz P, Maass T, Krupp M, Marquardt JU, Strand S, Becker D, Staib F, Binder H, Roessler S, et al. Translating bioinformatics in oncology: Guilt-by-profiling analysis and identification of KIF18B and CDCA3 as novel driver genes in carcinogenesis. Bioinformatics. 2015;31:216–224. doi: 10.1093/bioinformatics/btu586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu SL, Lin HX, Qiu F, Zhang WJ, Niu CH, Wen W, Sun XQ, Ye LP, Wu XQ, Lin CY, et al. Overexpression of kinesin family member 20A correlates with disease progression and poor prognosis in human nasopharyngeal cancer: A retrospective analysis of 105 patients. PLoS One. 2017;12:e0169280. doi: 10.1371/journal.pone.0169280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G, Chen Q, Xiao J, Zhang H, Wang Z, Lin X. Identification of genes and analysis of prognostic values in nonsmoking females with non-small cell lung carcinoma by bioinformatics analyses. Cancer Manag Res. 2018;10:4287–4295. doi: 10.2147/CMAR.S174409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ni M, Liu X, Wu J, Zhang D, Tian J, Wang T, Liu S, Meng Z, Wang K, Duan X, et al. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of non-small cell lung cancer via integrated bioinformatics analysis. Front Genet. 2018;9:469. doi: 10.3389/fgene.2018.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi C, Huang D, Lu N, Chen D, Zhang M, Yan Y, Deng L, Lu Q, Lu H, Luo S. Aberrantly activated Gli2-KIF20A axis is crucial for growth of hepatocellular carcinoma and predicts poor prognosis. Oncotarget. 2016;7:26206–26219. doi: 10.18632/oncotarget.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, He W, Shi Y, Gu H, Li M, Liu Z, Feng Y, Zheng N, Xie C, Zhang Y. High expression of KIF20A is associated with poor overall survival and tumor progression in early-stage cervical squamous cell carcinoma. PLoS One. 2016;11:e0167449. doi: 10.1371/journal.pone.0167449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito K, Ohta S, Kawakami Y, Yoshida K, Toda M. Functional analysis of KIF20A, a potential immunotherapeutic target for glioma. J Neurooncol. 2017;132:63–74. doi: 10.1007/s11060-016-2360-1. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Zhang W, Sun X, Chen J, Li Y, Niu C, Xu B, Zhang Y. Overexpression of kinesin family member 20A is associated with unfavorable clinical outcome and tumor progression in epithelial ovarian cancer. Cancer Manag Res. 2018;10:3433–3450. doi: 10.2147/CMAR.S169214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan L, Chen L, Qian K, Qian G, Wu CL, Wang X, Xiao Y. Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC) Genom Data. 2017;14:132–140. doi: 10.1016/j.gdata.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita J, Fukushima S, Jinnin M, Honda N, Makino K, Sakai K, Masuguchi S, Inoue Y, Ihn H. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012;92:593–597. doi: 10.2340/00015555-1416. [DOI] [PubMed] [Google Scholar]
- 58.Kawai Y, Shibata K, Sakata J, Suzuki S, Utsumi F, Niimi K, Sekiya R, Senga T, Kikkawa F, Kajiyama H. KIF20A expression as a prognostic indicator and its possible involvement in the proliferation of ovarian clear-cell carcinoma cells. Oncol Rep. 2018;40:195–205. doi: 10.3892/or.2018.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carleton M, Mao M, Biery M, Warrener P, Kim S, Buser C, Marshall CG, Fernandes C, Annis J, Linsley PS. RNA interference-mediated silencing of mitotic kinesin KIF14 disrupts cell cycle progression and induces cytokinesis failure. Mol Cell Biol. 2006;26:3853–3863. doi: 10.1128/MCB.26.10.3853-3863.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singel SM, Cornelius C, Zaganjor E, Batten K, Sarode VR, Buckley DL, Peng Y, John GB, Li HC, Sadeghi N, et al. KIF14 promotes AKT phosphorylation and contributes to chemoresistance in triple-negative breast cancer. Neoplasia. 2014;16:247–256. doi: 10.1016/j.neo.2014.03.008. 256 e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang ZZ, Yang J, Jiang BH, Di JB, Gao P, Peng L, Su XQ. KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int J Oncol. 2018;53:1939–1952. doi: 10.3892/ijo.2018.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiu G, Sui X, Wang Y, Zhang Z. FOXM1 regulates radiosensitivity of lung cancer cell partly by upregulating KIF20A. Eur J Pharmacol. 2018;833:79–85. doi: 10.1016/j.ejphar.2018.04.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request. All raw data of LUAD, which were included in the current study, can be downloaded from TCGA (https://portal.gdc.cancer.gov/).