Abstract
Background
We report the performance of a gene expression profile test to classify the recurrence risk of cutaneous melanoma tumors of the head and neck as low‐risk Class 1 or high‐risk Class 2.
Methods
Of note, 157 primary head and neck cutaneous melanoma tumors were identified. Survival analyses were performed using Kaplan‐Meier and Cox methods.
Results
Gene expression profile class and node status stratified tumors into significantly different 5‐year survival groups by Kaplan‐Meier method (P < .0001 for all end points), and both were independent predictors of recurrence in multivariate analysis. Overall, 74% of distant metastases and 88% of melanoma‐specific deaths had Class 2 risk.
Conclusion
The gene expression profile test identifies cases at increased risk for metastasis and death independent of a clinically or pathologically negative nodal status, suggesting that incorporation of this molecular tool could improve clinical management of patients with head and neck cutaneous melanoma, especially in those with a negative sentinel lymph node biopsy.
Keywords: cutaneous melanoma, gene expression profiling, metastasis, prognosis, staging
1. INTRODUCTION
In the absence of effective adjuvant treatment for early stage cutaneous melanoma, initial detection and accurate staging are critical for optimal disease management. Previous studies have identified several indicators of overall and disease‐free survival, including patient age, Breslow thickness, and ulceration status.1, 2 The strongest prognostic factor is the status of the sentinel lymph node (SLN), as determined by the SLN biopsy procedure.1, 3, 4 SLN biopsy has been adopted as standard of care and is recommended as part of the National Comprehensive Cancer Network (NCCN) guidelines for accurate staging of melanoma patients,5 and the prognostic value of the procedure has been validated by the Multicenter Selective Lymphadenectomy Trial‐1 (MSLT‐I).3, 6
However, cutaneous melanoma tumors of the head and neck have shown higher rates of recurrence in SLN‐negative nodal basins compared to other anatomical regions,7, 8, 9, 10 as well as low rates of SLN positivity compared to lesions of the trunk or extremities.11, 12 SLN biopsy in patients with melanoma of the head and neck poses several unique challenges, as surgeons must navigate the complex lymphatic drainage system of this region while preserving critical neurovascular structures.13 The close proximity of the nodal basin to the primary tumor site may also cause difficulty in locating the SLN during lymphoscintigraphy,14, 15 and additional imaging methods may be necessary to accurately visualize the area.16 Given that the results of MSLT‐I show that 2 of 3 patients with cutaneous melanoma who metastasize and die from their disease are SLN negative,3, 6 there is an unmet clinical need for improved risk assessment and staging accuracy, particularly for patients with cutaneous melanoma with tumors in the head and neck region. Thus, additional prognostic tools are ever more critical. This need could be addressed by a prognostic molecular assay that determines the risk of recurrence in patients with cutaneous melanoma.
We have previously described a gene expression profile (GEP) test that utilizes primary tumor biology to determine a patient's risk of metastasis within 5 years as either low (Class 1) or high (Class 2), with further stratification of patients near the cutoff between Class 1 and 2 (eg, Class 1A, 1B, 2A, and 2B). The test has been validated in 3 multicenter studies17, 18, 19 and was recently shown to improve identification of high‐risk stage I and II melanomas when used in combination with American Joint Committee on Cancer (AJCC) staging criteria.20 The GEP is a noninvasive test, and a Class 2 result is highly associated with systemic spread of disease, making it a potentially valuable tool when used in combination with the SLN biopsy procedure. Here we report a subgroup analysis from prior validation studies using GEP classification to prognosticate risk in a cohort of patients with cutaneous melanoma of the head and neck.
2. PATIENTS AND METHODS
2.1. Sample and clinical data collection
A total of 690 cutaneous melanoma cases followed for ≥5 years or to first documented recurrence or metastatic event were collected from 2 previously reported validation studies and 1 performance study following Institutional Review Board approval.17, 18, 21 From this sample set, 157 patients with primary stage I, II, or III cutaneous melanoma tumors of the head and neck were identified from 16 US centers with original date of diagnosis between 1999 and 2011. A total of 110 patients had a documented SLN biopsy procedure. Patients younger than 18 years or those diagnosed with another malignant tumor type were excluded. Clinical parameters were reported by the contributing centers. For analysis, recurrence was defined as any local or distant disease recurrence, not including positive SLN status assessed during staging. Distant metastasis was defined as a recurrence outside the nodal basin of the primary tumor or to a different organ site, and also did not include positive SLN status at the time of diagnosis.
2.2. Gene expression profile class assignment
Gene expression profile (GEP) class assignment was performed using the commercially available 31‐GEP test (DecisionDx‐Melanoma) available from Castle Biosciences, Inc. (Friendswood, Texas), the development and validation of which has been previously described.17, 18, 19 This clinical test reports a class probability assignment of either Class 1 (low risk) or Class 2 (high risk), as well as a reduced confidence assignment with “A” reflecting a better and “B” reflecting worse outcomes resulting in assignments of Class 1A, 1B, 2A, and 2B. The melanoma cases included in this study are exclusive of the training set used in the test's algorithm.18 The cases in this study have been previously analyzed with the 31‐GEP test in broader cohorts but have not been analyzed as a head and neck subgroup.
2.3. Study endpoints
Primary endpoints for the study were recurrence‐free survival (RFS), defined as the time to any event including regional or distant metastasis but exclusive of a positive SLN biopsy result, distant metastasis‐free survival (DMFS), defined as the time to any distant metastatic event beyond the regional nodal basin, overall survival (OS), defined as survival until death from any cause, and melanoma‐specific survival (MSS), defined as survival until death documented as resulting from melanoma. The secondary endpoint was the analysis of the 31‐GEP predicted outcome in combination with node status to determine prognostic value added by the GEP. Patients with either clinically or pathologically negative nodes were recorded as nodenegative.
2.4. Statistical analysis
Survival curves were estimated applying the Kaplan‐Meier method; univariate and multivariate analyses were carried out using Cox regression, characterized with 95% confidence intervals (CIs) on the hazard ratio scale. For proportional hazards analysis, Breslow thickness and mitotic rate were measured as a continuous variable, while all other factors were dichotomized. In all cases the assumptions of proportional hazards were not violated. AJCC Stage I‐IIA vs IIB‐III were assessed based upon differences in clinical management recommendations from the NCCN.5 Statistical tests were performed using R version 3.3.0 (University of Auckland, New Zealand), with P < .05 considered statistically significant.
3. RESULTS
3.1. Cohort demographics
A total of 157 patients with cutaneous melanoma with tumors in the head and neck region who met the study inclusion and exclusion criteria were identified. Clinical characteristics of the cohort are shown in Table 1. The median age was 65 years (range 25‐89 years) and median Breslow thickness was 1.6 mm (range 0.2‐15.0 mm). Ulceration was present in 48 cases (31%) and 84 cases had a mitotic rate ≥1 mm2 (54%). The median time to recurrence was 1.4 years; for patients without a metastatic event, the median length of follow‐up was 7.1 years.
Table 1.
Patient demographics
| Clinical characteristics | Overall (n = 157) | Class 1 (n = 79) | Class 2 (n = 78) | P‐value |
|---|---|---|---|---|
| Median age (range), years | 65 (25‐89) | 62 (25‐89) | 66 (25‐85) | .27 |
| Recurrence/distant metastasis | 73/65 | 19/17 | 54/48 | <.001 |
| Median time to recurrence/follow‐up for non‐metastatic, years | 1.4/7.1 | 1.8/7.5 | 1.2/6.0 | … |
| AJCC stage | … | … | … | <.001 |
| I | 64 (41%) | 52 (81%) | 12 (19%) | … |
| II | 53 (32%) | 19 (37%) | 32 (63%) | … |
| III | 40 (27%) | 8 (19%) | 34 (81%) | … |
| Breslow thickness | … | … | … | <.001 |
| Median (range), mm | 1.6 (0.2‐15.0) | 1.0 (0.2‐6.0) | 2.5 (0.6‐15.0) | … |
| ≤1 mm | 44 (28%) | 39 (89%) | 5 (11%) | … |
| >1 mm | 112 (71%) | 40 (36%) | 72 (64%) | … |
| Not assessed | 1 (1%) | 0 (0%) | 1 (100%) | … |
| Mitotic index | … | … | … | .002 |
| <1/mm2 | 23 (14%) | 17 (74%) | 6 (26%) | … |
| ≥1/mm2 | 84 (54%) | 32 (38%) | 52 (62%) | … |
| Not assessed | 50 (32%) | 30 (60%) | 20 (40%) | … |
| Ulceration | … | … | … | <.001 |
| Absent | 92 (59%) | 63 (68%) | 29 (32%) | … |
| Present | 48 (31%) | 7 (15%) | 41 (85%) | … |
| Not assessed | 17 (10%) | 9 (53%) | 8 (47%) | … |
| Node status | … | … | … | <.001 |
| Negative | 118 (75%) | 72 (61%) | 46 (39%) | … |
| Positive | 39 (25%) | 7 (18%) | 32 (82%) | … |
| Growth pattern | … | … | … | <.001 |
| Superficial spreading | 40 (25%) | 27 (68%) | 13 (32%) | … |
| Nodular | 37 (24%) | 11 (30%) | 26 (70%) | … |
| Desmoplastic | 10 (6%) | 2 (20%) | 8 (80%) | … |
| Lentigo maligna | 11 (7%) | 10 (91%) | 1 (9%) | … |
| Other/not reported | 59 (38%) | 29 (49%) | 30 (51%) | … |
| Site | … | … | … | <.001 |
| Mid‐face | 52 (33%) | 34 (65%) | 18 (35%) | … |
| Lateral face | 40 (25%) | 21 (53%) | 19 (47%) | … |
| Scalp | 49 (32%) | 13 (27%) | 36 (73%) | … |
| Neck | 16 (10%) | 11 (69%) | 5 (31%) | … |
Abbreviations: AJCC, American Joint Committee on Cancer.
P‐values indicate statistical differences between Class 1 and Class 2.
3.2. Survival outcomes stratified by gene expression profile and nodal predictions of risk
Seventy‐nine patients (50%) had a low‐risk Class 1 result using 31‐GEP molecular classification, with 60 patients classified as Class 1A and 19 patients as Class 1B. Of the 78 (50%) called high‐risk Class 2, 19 were classified as Class 2A and 59 as Class 2B. Kaplan‐Meier analysis resulted in 5‐year RFS, DMFS, OS and MSS rates for Class 1A patients of 80%, 83%, 97%, and 98%, respectively, compared to 25%, 33%, 43%, and 61%, respectively, for those with a Class 2B result (P < .0001 for all comparisons; Figure 1). Similar trends were observed in the cohort of patients who underwent SLN biopsy (n = 110; data not shown).
Figure 1.
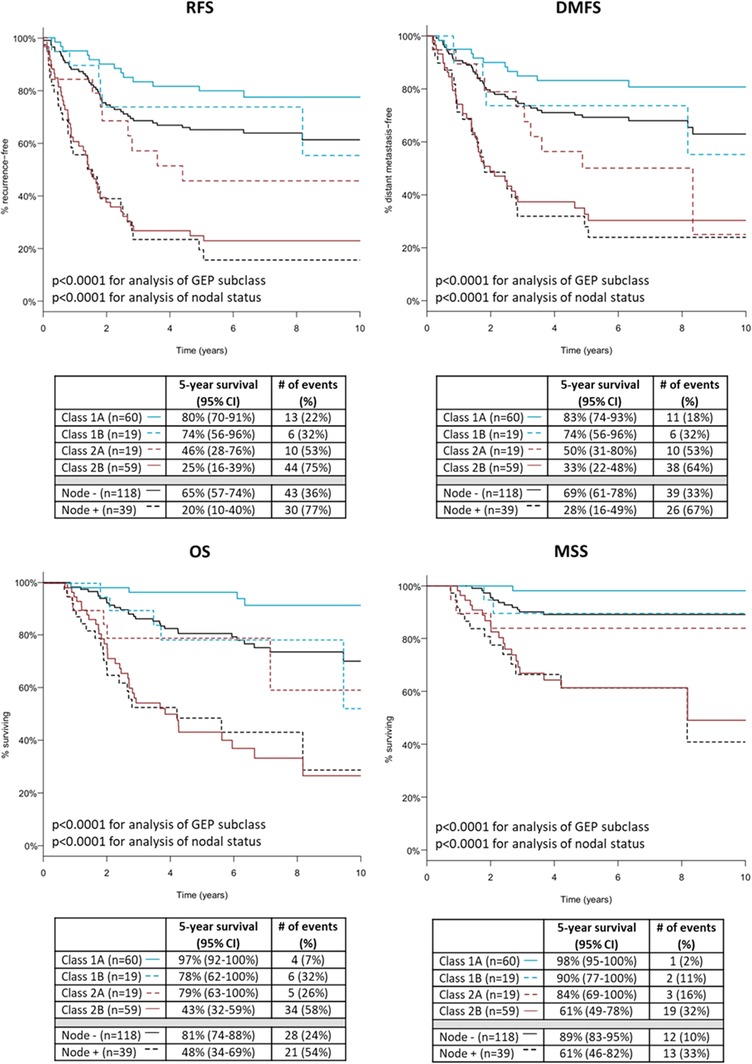
Kaplan‐Meier analysis of recurrence‐free (RFS), distant metastasis‐free (DMFS), overall survival (OS), and melanoma‐specific survival (MSS) by gene expression profile (GEP) class. RFS, DMFS, OS, and MSS rates for patients with cutaneous melanoma located in the head and neck (n = 157) using molecular GEP subclassification or nodal status in Kaplan‐Meier analysis. Tables below curves show the number of patients with each GEP class or a sentinel lymph node status, 5‐year survival rates for the outcome with 95% confidence intervals, and number of events with percentages experiencing the event. P‐values determined by log rank test for either all comparisons of GEP class or nodal status [Color figure can be viewed at wileyonlinelibrary.com]
Of the Class 2B patients, 75% (44 of 59) experienced recurrence, 64% (38 of 59) had a distant metastasis, 58% (34 of 59) died from any cause, and 32% (19 of 59) were documented to have died from their disease. By comparison, 22% (13 of 60) Class 1A patients recurred, 18% (11 of 60) developed distant metastases, 7% (4 of 60) died from any cause, and 2% (1 of 60) was documented to have died from melanoma.
Of the 157 patients in the cohort, 118 had a clinically or pathologically negative node, while 39 patients were SLN positive. Five‐year Kaplan‐Meier outcomes for RFS, DMFS, OS, and MSS in node‐negative cases were 65%, 69%, 81%, and 89%, respectively, compared to 20%, 28%, 48%, and 61%, respectively, for SLN‐positive cases (P < .0001 for all; Figure 1). For SLN‐positive patients, 77% (30 of 39) patients experienced recurrence, 67% (26 of 39) developed distant metastasis, 54% (21 of 39) died from any cause, and 33% (13 of 39) were documented to have died from melanoma. In node‐negative patients, 36% (43 of 118) had a recurrence, 33% (39 of 118) developed distant metastasis, 24% (28 of 118) died from any cause, and 10% (12 of 118) were documented to have died from melanoma.
3.3. Cox regression analysis of risk with gene expression profile and clinical factors
Since prognostic staging has traditionally been performed according to AJCC guidelines, the clinicopathologic features recommended by AJCC staging guidelines (version 7) were used in regression analyses with 31‐GEP, including Breslow thickness, mitotic rate, ulceration, and node status. In univariate Cox proportional hazard models, Breslow thickness, ulceration, node positivity, and GEP Class 2 results were all significant predictors of recurrence, distant metastasis, all‐cause death, and melanoma‐specific death (P ≤ .01 for all; Table 2). Mitotic rate was not a significant predictor of any endpoint. In multivariate analysis, Breslow thickness was an independent predictor of all survival endpoints (P ≤ .03 for all), and node positivity and molecular Class 2 were independent predictors of recurrence (P = .02 and .01, respectively; Table 2). Molecular Class 2 result was also a significant predictor of distant metastasis (P = .04). When comparing GEP Class 2 to AJCC stage IIB and above in multivariate analysis, both classifications were significant predictors of recurrence, distant metastasis, and all‐cause death (P ≤ .006 for all; Table 3). Only GEP Class 2 was significant for melanoma‐specific death (P = .005; Table 3).
Table 2.
Cox proportional hazard models evaluating gene expression profile classification along with standard clinicopathologic factors
| Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | ||
| RFS | Breslow | 1.2 | 1.1‐1.2 | <.0001 | 1.2 | 1.1‐1.5 | .02 |
| Mitotic rate | 1.0 | 1.0‐1.1 | .1 | 1.0 | 0.9‐1.0 | .1 | |
| Ulceration present | 2.5 | 1.5‐4.1 | .0002 | 1.2 | 0.6‐2.5 | .5 | |
| SLN‐positive | 3.8 | 2.4‐6.1 | <.0001 | 2.2 | 1.1‐4.1 | .02 | |
| GEP Class 2 | 4.8 | 2.8‐8.2 | <.0001 | 3.0 | 1.3‐7.1 | .01 | |
| DMFS | Breslow | 1.2 | 1.1‐1.3 | <.0001 | 1.3 | 1.1‐1.6 | .008 |
| Mitotic rate | 1.0 | 1.0‐1.1 | .5 | 0.9 | 0.9‐1.0 | .03 | |
| Ulceration present | 2.8 | 1.7‐4.8 | <.0001 | 2.0 | 0.9‐4.2 | .07 | |
| SLN‐positive | 3.4 | 2.1‐5.7 | <.0001 | 1.7 | 0.8‐3.4 | .1 | |
| GEP Class 2 | 4.5 | 2.5‐7.8 | <.0001 | 2.5 | 1.0‐6.3 | .04 | |
| OS | Breslow | 1.2 | 1.1‐1.3 | .0001 | 1.4 | 1.0‐1.7 | .01 |
| Mitotic rate | 1.0 | 1.0‐1.1 | .06 | 1.0 | 0.9‐1.0 | .5 | |
| Ulceration present | 4.6 | 2.4‐8.6 | <.0001 | 1.9 | 0.8‐4.7 | .1 | |
| SLN‐positive | 3.9 | 2.2‐6.9 | <.0001 | 1.5 | 0.7‐3.4 | .3 | |
| GEP Class 2 | 6.7 | 3.3‐13.5 | <.0001 | 2.9 | 1.0‐9.0 | .06 | |
| MSS | Breslow | 1.2 | 1.1‐1.3 | .0008 | 1.4 | 1.0‐1.9 | .03 |
| Mitotic rate | 1.0 | 0.9‐1.1 | .7 | 0.9 | 0.8‐1.0 | .1 | |
| Ulceration present | 3.1 | 1.3‐7.6 | .01 | 0.9 | 0.3‐3.1 | .9 | |
| SLN‐positive | 4.9 | 2.2‐10.9 | <.0001 | 2.6 | 0.8‐8.6 | .1 | |
| GEP Class 2 | 10.5 | 3.1‐35.4 | .0001 | 5.4 | 1.0‐30.7 | .06 | |
Abbreviations: CI, confidence interval; DMFS, distant metastasis‐free survival; HR, hazard ratio; MSS, melanoma‐specific survival; OS, overall survival; RFS, recurrence‐free survival; SLN, sentinel lymph node.
Table 3.
Cox multivariate analysis evaluating high risk classification by American Joint Committee on Cancer (AJCC) stage (stage IIB and above) along with gene expression profile class
| HR | 95% CI | P‐value | ||
|---|---|---|---|---|
| RFS | ≥AJCC stage IIB | 2.9 | 0.2‐0.6 | .0003 |
| GEP Class 2 | 2.8 | 1.5‐5.1 | .0009 | |
| DMFS | ≥ AJCC stage IIB | 2.4 | 0.2‐0.8 | .006 |
| GEP Class 2 | 2.8 | 1.5‐5.4 | .002 | |
| OS | ≥ AJCC stage IIB | 2.8 | 0.2‐0.7 | .005 |
| GEP Class 2 | 4.1 | 1.9‐9.1 | .0004 | |
| MSS | ≥ AJCC stage IIB | 2.3 | 0.2‐1.1 | .1 |
| GEP Class 2 | 6.8 | 1.8‐25.4 | .005 |
Abbreviations: DMFS, distant metastasis‐free survival; GEP, gene expression profile; MSS, melanoma‐specific survival; OS, overall survival; RFS, recurrence‐free survival.
3.4. Combining gene expression profile risk classification with node status
Binary GEP result (Class 1 or 2) was combined with node status to create four stratifications of risk using Kaplan‐Meier analysis (Figure 2). Comparing Class 1/node‐negative patients with Class 2/node‐negative patients, the 5‐year RFS was 83% vs 37%, DMFS was 85% vs 45%, OS was 91% vs 62%, and MSS was 96% vs 78%. For Class 1/SLN‐positive cases compared to Class 2/SLN‐positive cases, RFS rates were 29% vs 19%, DMFS was 43% vs 23%, OS was 100% vs 36%, and melanoma‐specific survival was 100% vs 50%.
Figure 2.
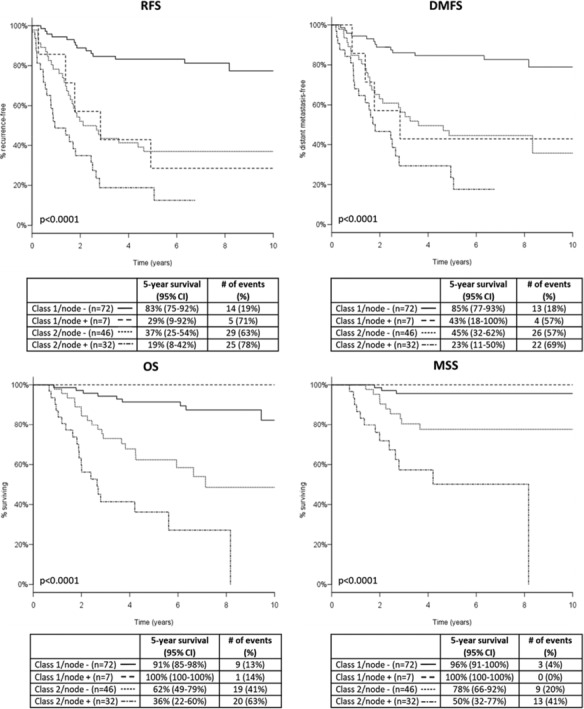
Kaplan‐Meier analysis of recurrence‐free (RFS), distant metastasis‐free (DMFS), overall survival (OS), and melanoma‐specific survival (MSS) outcomes by combined gene expression profile (GEP) and nodal risk classification. RFS, DMFS, OS, and MSS rates for patients with cutaneous melanoma located in the head and neck (n = 157) using molecular GEP class and clinical nodal status in Kaplan‐Meier analysis. Tables below curves show the number of patients with each GEP class and a sentinel lymph node status combination, 5‐year survival rates for the outcome with 95% confidence intervals, and number of events with percentages experiencing the event. P‐values determined by log rank test for all comparisons
3.5. Accuracy of risk prediction according to gene expression profile and nodal status
As shown in Table 4, molecular class resulted in sensitivities for prediction of recurrence, distant metastasis, death from any cause, and melanoma‐specific death of 74%, 74%, 80%, and 88%, respectively, compared to sensitivities for node status of 41%, 40%, 43%, and 52%, respectively. The negative predictive values of molecular class in predicting recurrence, distant metastasis, all‐cause death, and death due to melanoma were 76%, 78%, 87%, and 96%, respectively, compared to negative predictive values of 64%, 67%, 76%, and 90%, respectively, for node status. Combining the prognostic outcomes of both GEP and SLN status increased the accuracy of identifying high‐risk cases, with sensitivities for recurrence, distant metastasis, all‐cause death, and death due to melanoma of 81%, 80%, 82%, and 88%, respectively, and negative predictive values of 81%, 82%, 88%, and 96%, respectively.
Table 4.
Accuracy values of node status and gene expression profile alone and in combination for prediction of recurrence, distant metastasis, all‐cause death, and melanoma‐specific death
| GEP (95% CI) | Node (95% CI) | GEP + node (95% CI) | ||
|---|---|---|---|---|
| RFS | Sensitivity | 74% (62%‐84%) | 41% (30%‐53%) | 81% (70%‐89%) |
| Specificity | 71% (61%‐81%) | 89% (81%‐95%) | 69% (58%‐79%) | |
| PPV | 69% (58%‐79%) | 77% (61%‐89%) | 69% (62%‐76%) | |
| NPV | 76% (65%‐85%) | 64% (54%‐74%) | 81% (72%‐87%) | |
| DMFS | Sensitivity | 74% (61%‐84%) | 40% (28%‐53%) | 80% (68%‐89%) |
| Specificity | 67% (57%‐77%) | 86% (77%‐92%) | 64% (53%‐74%) | |
| PPV | 62% (50%‐72%) | 67% (50%‐81%) | 61% (54%‐68%) | |
| NPV | 78% (68%‐87%) | 67% (58%‐75%) | 82% (73%‐88%) | |
| OS | Sensitivity | 80% (66%‐90%) | 43% (29%‐58%) | 82% (68%‐91%) |
| Specificity | 64% (54%‐73%) | 83% (75%‐90%) | 58% (48%‐68%) | |
| PPV | 50% (38%‐62%) | 54% (37%‐70%) | 47% (41%‐53%) | |
| NPV | 87% (78%‐94%) | 76% (68%‐84%) | 88% (79%‐93%) | |
| MSS | Sensitivity | 88% (69%‐97%) | 52% (31%‐72%) | 88% (69%‐97%) |
| Specificity | 58% (49%‐66%) | 80% (72%‐87%) | 52% (43%‐61%) | |
| PPV | 28% (19%‐40%) | 33% (19%‐50%) | 26% (22%‐31%) | |
| NPV | 96% (89%‐99%) | 90% (83%‐95%) | 96% (89%‐99%) |
Abbreviations: CI, confidence interval; DMFS, distant metastasis‐free survival; GEP, gene expression profile; MSS, melanoma‐specific survival; NPV, negative predictive value; OS, overall survival; PPV, positive predictive value; RFS, recurrence‐free survival.
4. DISCUSSION
SLN biopsy is the standard for prognostication in cutaneous melanoma, although it is recognized that its sensitivity in patients with head and neck cutaneous melanoma is lower than for other anatomic sites due to a higher rate of false negatives and lower rate of true positives.9, 22, 23, 24 In addition, damage to cranial nerves and critical vascular structures is a risk.25 Detection and identification of the SLN itself is complicated by the generally short distances between primary lesion and nodal basin, and due to the complexities of the lymphatic drainage patterns in this region, clinically predicted pathways are often discordant from those identified using preoperative lymphoscintigraphy.14, 15 A trained multidisciplinary team of physicians who treat melanoma of the head and neck is required for accurate technical performance of the procedure in this group of patients with cutaneous melanoma.26
Given the higher rate of false negatives with SLN biopsy in patients with head and neck melanoma, a negative biopsy derives less confidence in a favorable patient outcome. Based on MSLT‐II, fewer completion lymph node dissections may be performed in the future and their added prognostic information will thus be mitigated.16 Despite improved survival seen in head and neck cases from MSLT‐II, these data may need to be interpreted with caution, making additional methods of prognostication even more vital in this population. Molecular classification can improve the identification of high‐risk tumors beyond clinical staging factors and could have a role in identifying patients with high risk of recurrence who had a negative node.
As shown in several recent reports, the prognostic information provided by the 31‐GEP, used in conjunction with node status, further identified high‐risk cases.17, 18, 21 Results from this study show that the 31‐GEP test identified 74% of patients who developed distant metastases and 88% of patients who died from their disease as high‐risk Class 2, and that the 5‐year risk of recurrence is more than doubled for the Class 2/node‐negative group compared to the Class 1/node‐negative group (63% vs 17%). Survival rates for Class 2 cases align closely with those of node‐positive cases (Figure 1), suggesting that prognoses for these two groups are similar and that patients should then be followed more intensively. The results suggest that the GEP is an independent predictor of recurrence and distant metastasis, and this subset analysis showed greater accuracy in detecting cases at high risk for recurrence, distant metastasis, death from any cause, and melanoma‐specific death compared to node status. However, the highest sensitivities were achieved when both methods of prognostication were combined. Similarly, the addition of molecular classification to node‐negative status improved identification of true low‐risk cases, with 5‐year RFS and DMFS rates of 83% and 85%, respectively, for Class 1/node‐negative patients compared to 65% and 69%, respectively, when considering node‐negative status alone.
Cutaneous melanoma is a heterogeneous disease believed to develop through different pathways depending on anatomical site.27 BRAF mutations arise more frequently in cutaneous melanoma tumors located on the trunk compared to those in the head and neck region, so a patient who develops distant metastasis following diagnosis of a cutaneous melanoma on the head or neck may have fewer options for cure.28, 29 The location‐specific etiology of the disease may result in a more aggressive biology in lesions of the head and neck, as these cells may have greater proliferative potential due in part to chronic sun exposure.30 Based on the results of this study, the 31‐GEP is able to identify high‐risk tumor biology to provide accurate and independent prognostic information in addition to the standard clinicopathologic features used in staging.
Intensive follow‐up and surveillance are critical for early detection of metastasis. GEP testing allows patients with intrinsic high‐risk tumor biology to be promptly identified at diagnosis and subsequently managed with high‐intensity surveillance to identify metastasis as early as possible. Multiple studies reveal that cross‐sectional imaging reveals melanoma progression well before it becomes symptomatic.31, 32, 33 More importantly, early detection of metastasis is becoming clinically important as treatment with modern therapeutic agents shows greater benefit when disease burden is low.34, 35
This study is limited by sample size, which could impact the multivariate analysis given the low numbers of MSS events, but we addressed this with a model including only GEP class and AJCC stage. Additionally, this cohort has some features that are more aggressive than the clinical population. Despite these limitations, the results indicate that molecular classification may enhance the selection of patients to undergo aggressive imaging and physical examination, leading to better resource utilization and the earliest possible detection of recurrence and metastases.
In conclusion, the early detection of distant metastases is of particular importance for patients with cutaneous melanoma in the head and neck region, as surgical resection can be limited by both technical and cosmetic concerns and regional treatment options may be limited due to anatomic site.36, 37 Furthermore, regional head and neck melanoma is often well controlled by operation (the head and neck was the only site for which completion node dissection offered improved OS in the MSLT‐II trial).38 Thus, the addition of the 31‐GEP test to standard staging offers the opportunity to complement node status and identify more patients at risk for distant metastasis and death who could potentially benefit from more aggressive surveillance and earlier therapeutic intervention,34, 35, 36 at a point when these treatments are most effective.
CONFLICT OF INTEREST
R.W.C. and K.R.C. are employees and stock option holders of Castle Biosciences, Inc. J.S.Z. and P.G. are consultants for Castle Biosciences, Inc. B.R.G., P.G., and J.V. are on the Speaker Bureau for Castle Biosciences, Inc.
ACKNOWLEDGEMENTS
The authors wish to thank the following centers for their contributions to the study: Dr. Laura Ferris, University of Pittsburgh Medical Center, Dr. Stephen Lyle, University of Massachusetts Medical School, Dr. Daniel Rosen, Baylor College of Medicine, Dr. Gilchrist Jackson, Kelsey‐Seybold Clinic, Dr. Anthony Greisinger, Kelsey Research Foundation, Drs. David Lawson, Maria Russell, and Keith Delman, Winship Cancer Institute of Emory University, Dr. Lewis Kaminester, Dermatology Palm Beach, Dr. Lee Cranmer, University of Arizona Cancer Center, Dr. Rene Gonzalez, University of Colorado Cancer Center, and Dr. Martin Fleming, The University of Tennessee Health Science Center.
Gastman BR, Zager JS, Messina JL, et al. Performance of a 31‐gene expression profile test in cutaneous melanomas of the head and neck. Head & Neck. 2019;41:871–879. 10.1002/hed.25473
Data from this manuscript was presented at the American Society of Clinical Oncology annual meeting, June 3, 2017, Chicago, Illinois.
Funding information This study was sponsored by Castle Biosciences, Inc. which provided funding for tissue and clinical data retrieval to contributing centers.
REFERENCES
- 1. Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27(36):6199‐6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McMasters KM. The sunbelt melanoma trial. Ann Surg Oncol. 2001;8(suppl 9):41S‐43S. [PubMed] [Google Scholar]
- 3. Morton DL, Thompson JF, Cochran AJ, et al. Sentinel‐node biopsy or nodal observation in melanoma. N Engl J Med. 2006;355(13):1307‐1317. [DOI] [PubMed] [Google Scholar]
- 4. Leong SP. Sentinel lymph node mapping and selective lymphadenectomy: the standard of care for melanoma. Curr Treat Options Oncol. 2004;5(3):185‐194. [DOI] [PubMed] [Google Scholar]
- 5. Coit DG, Thompson JA, Algazi A, et al. Melanoma, version 2.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2016;14(4):450‐473. [DOI] [PubMed] [Google Scholar]
- 6. Morton DL, Thompson JF, Cochran AJ, et al. Final trial report of sentinel‐node biopsy versus nodal observation in melanoma. N Engl J Med. 2014;370(7):599‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saltman BE, Ganly I, Patel SG, et al. Prognostic implication of sentinel lymph node biopsy in cutaneous head and neck melanoma. Head Neck. 2010;32(12):1686‐1692. [DOI] [PubMed] [Google Scholar]
- 8. Lee DY, Huynh KT, Teng A, et al. Predictors and survival impact of false‐negative sentinel nodes in melanoma. Ann Surg Oncol. 2016;23(3):1012‐1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlson GW, Page AJ, Cohen C, et al. Regional recurrence after negative sentinel lymph node biopsy for melanoma. Ann Surg. 2008;248(3):378‐386. [DOI] [PubMed] [Google Scholar]
- 10. Gomez‐Rivera F, Santillan A, McMurphey AB, et al. Sentinel node biopsy in patients with cutaneous melanoma of the head and neck: recurrence and survival study. Head Neck. 2008;30(10):1284‐1294. [DOI] [PubMed] [Google Scholar]
- 11. Fadaki N, Li R, Parrett B, et al. Is head and neck melanoma different from trunk and extremity melanomas with respect to sentinel lymph node status and clinical outcome? Ann Surg Oncol. 2013;20(9):3089‐3097. [DOI] [PubMed] [Google Scholar]
- 12. Penicaud M, Cammilleri S, Giorgi R, et al. Sentinel lymph node biopsy in head and neck melanoma: a single institution analysis. Rev Laryngol Otol Rhinol (Bord). 2014;135(3):115‐120. [PubMed] [Google Scholar]
- 13. Kaveh AH, Seminara NM, Barnes MA, et al. Aberrant lymphatic drainage and risk for melanoma recurrence after negative sentinel node biopsy in middle‐aged and older men. Head Neck. 2016;38(suppl 1):E754‐E760. [DOI] [PubMed] [Google Scholar]
- 14. O'Brien CJ, Uren RF, Thompson JF, et al. Prediction of potential metastatic sites in cutaneous head and neck melanoma using lymphoscintigraphy. Am J Surg. 1995;170(5):461‐466. [DOI] [PubMed] [Google Scholar]
- 15. Leong SP, Achtem TA, Habib FA, et al. Discordancy between clinical predictions vs lymphoscintigraphic and intraoperative mapping of sentinel lymph node drainage of primary melanoma. Arch Dermatol. 1999;135(12):1472‐1476. [DOI] [PubMed] [Google Scholar]
- 16. Doepker MP, Yamamoto M, Applebaum MA, et al. Comparison of single‐photon emission computed tomography‐computed tomography (SPECT/CT) and conventional planar lymphoscintigraphy for sentinel node localization in patients with cutaneous malignancies. Ann Surg Oncol. 2017;24(2):355‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerami P, Cook RW, Russell MC, et al. Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol. 2015;72(5):780‐785. e783. [DOI] [PubMed] [Google Scholar]
- 18. Gerami P, Cook RW, Wilkinson J, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res. 2015;21(1):175‐183. [DOI] [PubMed] [Google Scholar]
- 19. Zager JS, Gastman BR, Leachman S, et al. Performance of a prognostic 31‐gene expression profile in an independent cohort of 523 cutaneous melanoma patients. BMC Cancer. 2018;18(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferris LK, Farberg AS, Middlebrook B, et al. Identification of high‐risk cutaneous melanoma tumors is improved when combining the online American joint committee on cancer individualized melanoma patient outcome prediction tool with a 31‐gene expression profile‐based classification. J Am Acad Dermatol. 2017;76:818‐825.e3. [DOI] [PubMed] [Google Scholar]
- 21. Zager JS, Gastman BR, Messina J, et al. Performance of a 31‐gene expression profile in a previously unreported cohort of 334 cutaneous melanoma patients. J Clin Oncol. 2016;34(suppl 15):9581. [Google Scholar]
- 22. Nowecki ZI, Rutkowski P, Nasierowska‐Guttmejer A, Ruka W. Survival analysis and clinicopathological factors associated with false‐negative sentinel lymph node biopsy findings in patients with cutaneous melanoma. Ann Surg Oncol. 2006;13(12):1655‐1663. [DOI] [PubMed] [Google Scholar]
- 23. Scoggins CR, Martin RC, Ross MI, et al. Factors associated with false‐negative sentinel lymph node biopsy in melanoma patients. Ann Surg Oncol. 2010;17(3):709‐717. [DOI] [PubMed] [Google Scholar]
- 24. Jones EL, Jones TS, Pearlman NW, et al. Long‐term follow‐up and survival of patients following a recurrence of melanoma after a negative sentinel lymph node biopsy result. JAMA Surg. 2013;148(5):456‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmalbach CE, Nussenbaum B, Rees RS, Schwartz J, Johnson TM, Bradford CR. Reliability of sentinel lymph node mapping with biopsy for head and neck cutaneous melanoma. Arch Otolaryngol Head Neck Surg. 2003;129(1):61‐65. [DOI] [PubMed] [Google Scholar]
- 26. Giudice G, Leuzzi S, Robusto F, et al. Sentinel lymph node biopsy in head and neck melanoma*. G Chir. 2014;35(5–6):149‐155. [PMC free article] [PubMed] [Google Scholar]
- 27. Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001;12(1):69‐82. [DOI] [PubMed] [Google Scholar]
- 28. Bauer J, Buttner P, Murali R, et al. BRAF mutations in cutaneous melanoma are independently associated with age, anatomic site of the primary tumor, and the degree of solar elastosis at the primary tumor site. Pigment Cell Melanoma Res. 2011;24(2):345‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deichmann M, Krahl D, Thome M, Wust K, Hassanzadeh J, Helmke B. The oncogenic B‐raf V599E mutation occurs more frequently in melanomas at sun‐protected body sites. Int J Oncol. 2006;29(1):139‐145. [PubMed] [Google Scholar]
- 30. Kvaskoff M, Pandeya N, Green AC, et al. Site‐specific determinants of cutaneous melanoma: a case‐case comparison of patients with tumors arising on the head or trunk. Cancer Epidemiol Biomarkers Prev. 2013;22(12):2222‐2231. [DOI] [PubMed] [Google Scholar]
- 31. Livingstone E, Krajewski C, Eigentler TK, et al. Prospective evaluation of follow‐up in melanoma patients in Germany ‐ results of a multicentre and longitudinal study. Eur J Cancer. 2015;51(5):653‐667. [DOI] [PubMed] [Google Scholar]
- 32. Podlipnik S, Carrera C, Sanchez M, et al. Performance of diagnostic tests in an intensive follow‐up protocol for patients with American joint committee on cancer (AJCC) stage IIB, IIC, and III localized primary melanoma: a prospective cohort study. J Am Acad Dermatol. 2016;75(3):516‐524. [DOI] [PubMed] [Google Scholar]
- 33. Leon‐Ferre RA, Kottschade LA, Block MS, et al. Association between the use of surveillance PET/CT and the detection of potentially salvageable occult recurrences among patients with resected high‐risk melanoma. Melanoma Res. 2017;27:335‐341. [DOI] [PubMed] [Google Scholar]
- 34. Kaufman HAT, Nemunaitis JJ, Chesne JA, et al. Tumor size and clinical outcomes in melanoma patients (MEL pts) treated with talimogene laherparepvec (T‐VEC). J Clin Oncol. 2015;33(suppl 15):9074. [Google Scholar]
- 35. Ribas A, Hamid O, Daud A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315(15):1600‐1609. [DOI] [PubMed] [Google Scholar]
- 36. Andtbacka RH, Agarwala SS, Ollila DW, et al. Cutaneous head and neck melanoma in OPTiM, a randomized phase 3 trial of talimogene laherparepvec versus granulocyte‐macrophage colony‐stimulating factor for the treatment of unresected stage IIIB/IIIC/IV melanoma. Head Neck. 2016;38(12):1752‐1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hayes AJ, Clark MA, Harries M, Thomas JM. Management of in‐transit metastases from cutaneous malignant melanoma. Br J Surg. 2004;91(6):673‐682. [DOI] [PubMed] [Google Scholar]
- 38. Faries MB, Thompson JF, Cochran AJ, et al. Completion dissection or observation for sentinel‐node metastasis in melanoma. N Engl J Med. 2017;376(23):2211‐2222. [DOI] [PMC free article] [PubMed] [Google Scholar]


