Abstract
Up-regulation of human prion protein (PrP) in patients with pancreatic ductal adenocarcinoma (PDAC) is associated with a poor prognosis. However, the underlying molecular mechanism of PrP-mediated tumorigenesis is not completely understood. In this study, we found that PDAC cell lines can be divided into either PrP high expresser or PrP low expresser. In addition to filamin A (FLNA), PrP interacts with Notch1, forming a PrP/FLNA/Notch1 complex. Silencing PrP in high-expresser cells decreases Notch1 expression and Notch1 signaling. These cells exhibited decreased proliferation, xenograft growth, and tumor invasion but show increased tumor apoptosis. These phenotypes were rescued by ectopically expressed and activated Notch1. By contrast, overexpression of PrP in low expressers increases Notch1 expression and signaling, enhances proliferation, and increases tumor invasion and xenograft growth that can be blocked by a Notch inhibitor. Our data further suggest that PrP increases Notch1 stability likely through suppression of Notch proteosome degradation. Additionally, we found that targeting PrP combined with anti-Notch is much more effective than singularly targeted therapy in retarding PDAC growth. Finally, we show that coexpression of PrP and Notch1 confers an even poorer prognosis than PrP expression alone. Taken together, our results have unraveled a novel molecular pathway driven by interactions between PrP and Notch1 in the progression of PDAC, supporting a critical tumor-promoting role of Notch1 in PrP-expressing PDAC tumors.
Nearly 90% of pancreatic cancers are pancreatic ductal adenocarcinomas (PDAC). PDAC carries one of the most dismal prognoses of all of the solid tumors, with mean survival time of <6 months and an overall 5-year survival rate <5%.1 Traditional cytotoxic chemotherapy provides only limited benefit to patients with PDAC. Oncogenic Kras signaling is essential for both progression and maintenance of PDAC.2, 3, 4 However, all of the clinical trials targeting Kras have failed. Thus, identifying novel molecules and pathways, which may serve as potential therapeutic targets to curb pancreatic cancer cell growth and/or metastasis, is urgently needed.
Notch is a heterodimeric transmembrane protein. Notch activation is initiated by binding of Notch ligand to Notch receptor, which results in cleavages by disintegrin, metalloprotease, and γ-secretase.5 This proteolytic process mediates the release of Notch intracellular domain (ICN), which then enters into the nucleus and to regulate transcription of Notch target genes such as the HES family genes.5 Data indicate that aberrant Notch activation is implicated in the initiation and progression as well as the aggressiveness of PDAC,6, 7, 8, 9, 10, 11 and dysregulated Notch pathway is one of the common molecular signatures of pancreatic cancer.12 Thus, targeting Notch pathway may represent a novel strategy for pancreatic cancer treatment.13 However, results from clinical trials targeting Notch have so far shown little if any success. The limited antitumor response observed in anti-Notch therapy may be due to complex interactions between Notch and other signaling pathways.
In this study, we report that Notch1 signaling is potentiated through cross talks with the cellular prion protein (PrP) in pancreatic cancer. PrP is a glycosylphosphatidylinositol (GPI)-anchored membrane protein mainly known for its role in a group of fatal neurodegenerative diseases.14 Human PrP is up-regulated in gastric, breast, and pancreatic cancers.15 However, not much is known about the role PrP plays in tumor biology or in PDAC carcinogenesis. Our previous studies revealed that PrP is not expressed in normal pancreatic tissue, chronic pancreatitis, pancreatic intraepithelial neoplasia 1 (PanIN1) or PanIN2, but is expressed weakly in PanIN3.16 In comparison, 41% of PADCs show focal or diffuse labeling for PrP, and its expression correlates with poorer prognosis. A correlation with poorer PDAC prognosis was later confirmed by analysis of additional 142 PDAC cases derived from three cancer registries.17
Interestingly, the PrP detected in PDAC cells is a pro-PrP as defined by retaining the C-terminal GPI anchor peptide signal sequences (PSSs). The GPI-PSS is normally removed in the endoplasmic reticulum during PrP maturation.16, 18 The GPI-PSS of pro-PrP transverses the membrane and interacts with filamin A (FLNA), a cytolinker protein that links cell surface receptors to the cytoskeleton.19, 20 Binding of PrP with FLNA perturbs the normal functions of FLNA and therefore confers a growth advantage contributing to the invasiveness of PDAC.16
Here, we show that, in addition to FLNA, PrP also forms a complex with Notch1 in human PDAC cells. The PrP-Notch1 complex regulates Notch1 stability and enhances Notch activation, resulting in enhanced tumor proliferation and invasion in vitro, enhanced xenograft growth in vivo, while suppressing apoptosis. In addition, we show that expression of Notch1 in PrP+ human PDAC biopsies is associated with even worse survival than PrP+-only tumors compared with PrP− cases, supporting a critical tumor-promoting role of Notch1 in PrP-expressing PDAC tumors.
Materials and Methods
Cell Culture, shRNA Knockdown, and Overexpression
All cell lines were obtained from AATC (Manassas, VA) with certified characterization and cultured according to the instructions. PrP knockdown was performed in PDAC cells transfected with four shRNA against PRNP (Sigma-Aldrich MISSION ShRNA library, St. Louis, MO) by Lipofetamine 2000 (Invitrogen, Carlsbad, CA). PDAC cell lines were transfected with pLKO.1-puro Vector as control. Two shPrP clones (NM_000311.2-1250s1c1 and NM_000311.3-803s21c1) showed >70% efficiency and were used in this study. Two shRNA clones against human NOTCH1 (TRCN0000003359 and TRCN0000003362) were used as described.21 Stabilized cell lines were used for proliferation, apoptosis, or tumor invasion assay and xenograft experiments. For PrP overexpression, PrP cDNA was subcloned into the cytomegalovirus-3Flag-1A vector (Clontech, Mountain View, CA) to generate cytomegalovirus-3Flag-PrP-1A, and transfected into Capan-1 cells by Lipofectamine 2000.22 To express activated Notch1, enhanced green fluorescent protein-N1IC plasmid was transfected into PrP knock-down PDAC cells. Green fluorescent protein-positive cells were sorted by FACSAria (BD Bioscience, San Diego, CA).
Western Blot Analysis, Immunoprecipitation, Quantification, and Stability Analysis
Antibodies used in Western blot analysis include anti-PrP (8H4),23 anti-Notch1 (Cell Signaling Technology, Danvers, MA; D1E11), anti–cleaved and activated intracellular form of Notch1 (NICD; Cell Signaling Technology; Val1744), anti-Notch2 (Millipore Corporation, Billerica, MA; 07-1234), anti-Notch3 (Santa Cruz Technology, Santa Cruz, CA; sc-5593), anti-Hes1 (Abcam, Cambridge, MA; ab108937), and anti-FLNA (Millipore; MAB1678). Two percent of total lysates before immunoprecipitation served as input, and rabbit or mouse IgG was used as negative control for co-immunoprecipitation. Proteins were then transferred and blotted with antibody as indicated. Horseradish peroxidase-conjugated secondary antibody and chemiluminescence blotting system (Pierce, Rockford, IL) were used for detection of signal. Photoshop CS5.5 Web Premium software (Adobe Systems, Mountain View, CA) was used to measure the expression level of protein normalized to β-actin. Cycloheximide (CHX; 10 μmol/L), proteasome inhibitor MG132 (10 μmol/L), and lysosome inhibitor chloroquine (CLQ; 25 μmol/L) were used in cell culture.
Cell Proliferation, Apoptosis, Tumor Migration, and Invasion Assay
For MTT assay, 5 × 103 cells in triplicates were plated in 96-well plates according to the manufacturer's instruction (Promega, Madison, WI). Apoptosis analysis and invasion analysis were performed using a terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay (ApopTag Peroxidase In Situ Apoptosis Detection Kit; Millipore Corporation) and BD Biocoat Tumor Invasion System, respectively. In brief, 25 × 103 cells were plated with serum-free medium to the apical chambers of the Matrigel-coated 24-well plate and incubated for 22 hours, followed by labeling of the invading cells by calcein AM fluorescent dye (BD Bioscience) and measured by the PerkinElmer (Waltham, MA) fluorescent plate reader. Noncoated BD Falcon FluoroBlok multiwell insert plates were used for cell migration assay. Fetal bovine serum (5%) was added to the basal chamber as chemoattractant. Cells migrated to the bottom membrane are fixed and counterstained with DAPI for enumeration.
RNA Isolation and RT-PCR
Total RNA was prepared with TRIzol reagent (Invitrogen). One microgram of RNA was used to synthesize cDNA by reverse transcription (iScript cDNA Synthesis Kit II; Bio-Rad, Hercules, CA). Quantitative PCR was performed using SYBR Green Master Mix (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase was used to normalize mRNA. Primer sequences used are as follows: PRNP forward, 5′-CGAGCTTCTCCTCTCCTCA-3′; reverse, 5′-ACAAAGAGAACCAGCATCCA-3′. All other primer sequences were used as described.24
Xenograft and DBZ Treatment
The Institutional Animal Care and Use Committee approved all aspects of the animal research described in this study. Athymic nude mice obtained from Case Athymic Mouse Core (Cleveland, OH) were injected subcutaneously in the back on either side of 3 to 10 × 106 cells. Dibenzazepine (DBZ; Selleck Chemicals, Houston, TX) treatment (i.p. injection) was started when tumors reach 0.5 mm3, starting on day 3 in mice receiving BxPC-3 cells or on day 7 in mice receiving Panc-1 or Capan-1 cells. DBZ (10 μmol/kg body weight) was given for 3 days of a 7-day cycle. Animals were monitored up to 8 weeks. Tumor diameters were measured with digital calipers, and volume was calculated by the following formula: volume (mm3) = (width)2 × length/2. Tumors were dissected and fixed in 10% formalin for pathology evaluation.
Immunostaining and Confocal Microscopy
PDAC cell lines were cultured in poly-d-lysine–coated glass bottom Petri dishes (MatTek Corporation, Ashland, MA) overnight. Mouse anti-PrP (8H4; dilution 1:100) and rabbit anti-Notch1 (D1E11; dilution 1:100) were applied to fixed/permealized cells. Alexa 488 goat–anti-rabbit (dilution 1:1000; Invitrogen) and cyanine 3 anti-mouse (dilution 1:250; Jackson ImmunoResearch, West Grove, PA) were used as secondary antibodies. Samples were analyzed on a LSM 510 Meta confocal microscope (Carl Zeiss Inc., Thornwood, NY).
Human PDAC Collection, Immunohistochemistry, and Survival Analysis
All studies have been approved by the University Hospital Case Medical Center Institutional Review Board for Human Investigation (University Hospital Institutional Review Board no. 08-05-29). Paraffin-embedded blocks of 67 surgically resected, primary infiltrating PDACs (stage IIA and IIB) were collected from the Pathology Archives (2001 to 2006). Tissue slides were processed using a BenchMark Ultra automated immunostainer (Ventana Medical Systems, Tucson, AZ). Slidehs were deparaffinized, antigen retrieved, and incubated at 37°C with the primary anti-PrP (8H4), or rabbit anti-Notch1 (D1E11). Detection was performed with Ultraview-diaminobenzidine (Ventana Medical Systems) and subsequently counterstained. The cytoplasmic and membrane intensity and distribution of Notch1 was graded as positive (>5% neoplastic cells stained strongly positive) or negative (<5% neoplastic cells stained). Notch1 expression comparison in PrP+ versus PrP− tissues was done using the Fisher's exact test (two-tailed). Kaplan-Meier method was used to determine overall survival with respect to PrP and Notch1 expression. The log-rank test was used to generate P values (P < 0.05 was considered significant).
Results
PrP Forms Complex with Notch1 in Pancreatic Cancer Cells
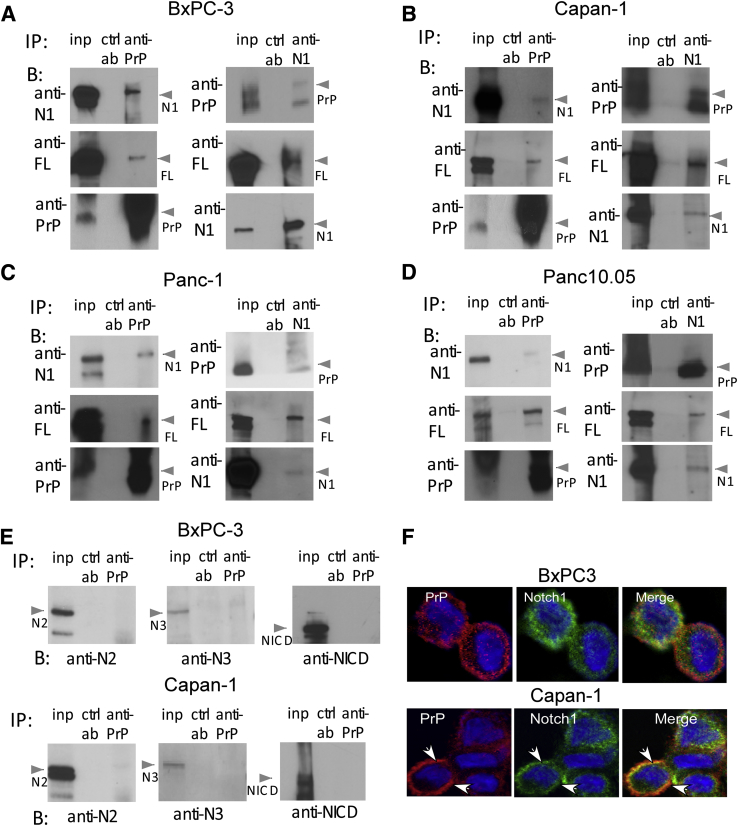
Both Notch signaling and elevated PrP expression mediate cell proliferation and are associated with adverse clinical features of pancreatic cancer. Hence, we asked if Notch signaling cross-interacts with PrP in pancreatic cancer cells. We immunoprecipitated four PDAC cell lines (BxPC-3, Capan-1, Panc10.05, and Panc-1) with anti-PrP monoclonal antibody (Mab) and then immunoblotted with either an anti-FLNA Mab or an anti-Notch1 Mab. We found that FLNA and Notch1 were copurified with PrP in BxPC-3 (Figure 1A), Capan-1 cells (Figure 1B), as well as in Panc-1 and Pan10.05 cells (Figure 1, C and D). Similarly, when cells were immunoprecipitated with anti-Notch1 Mab, PrP and FLNA were copurified with Notch1 (Figure 1, A–D). Notch2 and Notch3 have been found implicated in pancreatic cancer carcinogenesis,8, 9 but neither was found to be associated with PrP in BxPC-3 and Capan-1 cells (Figure 1E) and the other two cells (data not shown). However, the low level of Notch3 expression precludes a definitive conclusion whether there is an active interaction between PrP and Notch3. Consistently, Notch1 and PrP colocalized on the cell membranes of BxPC-3 and Capan-1 cells (Figure 1F) and the other two cell lines as well (Figure 2B) by immunofluorescence. However, NICD was not found to form complex with PrP in either BXPC-3 or Capan-1 cells (Figure 1E). The lack of interaction between PrP and NICD was further confirmed by cell fractionation in which we found that PrP was mainly present in the membrane fractions of PDAC cells, but was not detected in the nucleus fraction where NICD was located (data not shown). Collectively, these results show that Notch1, PrP, and FLNA interact with each other in PDAC cells, forming complexes mainly on cell membranes.
Figure 1.
PrP binds with Notch1 and filamin A in pancreatic cancer cells. A–D: Cell lysate of BxPC-3 (A), Capan-1 (B), Panc-1 (C), and Panc10.05 (D) were IP with anti-PrP or anti-N1, shown at the top, and blotted with other antibodies, shown in the left of each panel, as indicated. In each panel, the first column is total inp, second is the ctrl ab, and the third column the IP ab. E: Anti-N2, anti-N3, and NICD were used in blotting lysates of BxPC-3 cells and Capan-1 cells IP with anti-PrP. F: IF staining of N1 and PrP shows colocalization of PrP (8H4; red) with N1 (D1E11; green) mainly on membranes of BxPC-3 and Capan-1 cells, as indicated by the arrows. n = 3 independent experiments. Anti-N1, anti-Notch1; antu-N2, anti-Notch2; anti-N3, anti-Notch3; B, blot; ctrl ab, control IP antibody; FL, filamin A; IF, Immunofluorescence; inp, input protein; IP, immunoprecipitated; N1, Notch1; NICD, anti-activated form of Notch1; PrP, prion protein.
Figure 2.
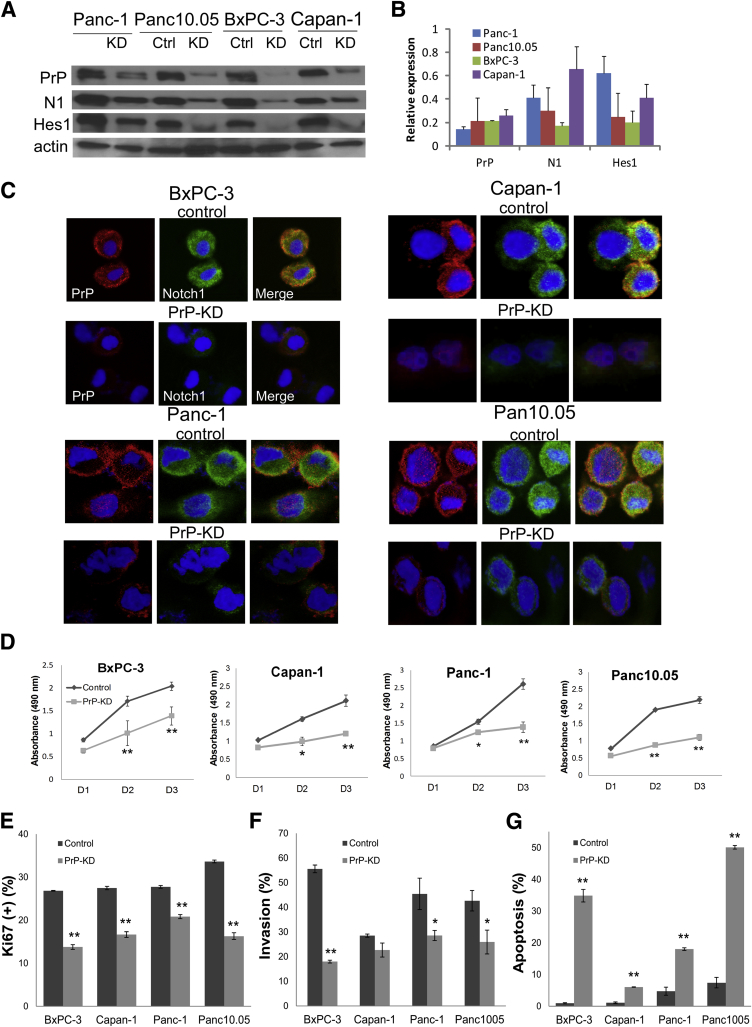
Silencing PrP suppresses Notch1 signaling, PDAC growth, invasion, but increases apoptosis. A: Immunoblots of PrP (8H4), Notch1 (D1E11), and Hes1 (ab108937) from cell lysates of four PDAC cells transfected with control shRNA or PRNP shRNA (results shown with shPrP clone NM_000311.2-1250s1c1). Actin is used as loading control. B: Expression of PrP, Notch1, and Hes1 after PrP silencing was quantified and normalized to the β-actin in each cell line examined in which the level of expression in parental cells was set as 1. C: If staining of Notch1 and PrP of four cells transfected with control shRNA or PRNP shRNA. D: Cell proliferation measured by MTT assay. Absorbance at 490 nm was recorded after 24 hours (D1), 48 hours (D2), and 72 hours (D3), respectively. E–G: PrP silencing in PDAC cells decreases cell proliferation by Ki-67 labeling (E) and cell invasion by Biocoat Invasion assay (F) but induces cell apoptosis by TUNEL assay (G). G: Apoptotic cells were counted. Data are expressed as means ± SEM (B) or as means ± SD (D, F, and G) n = 3 independent experiments (A and C); n = 5 biological replicates (D and F); n = 6 to approximately 10 biological replicates (E and G). t-test was performed (D–G). P values that were significantly different are shown as indicated. *P < 0.05, **P < 0.01. Ctrl, control; KD, knocked down; N1, Notch1; PDAC, pancreatic ductal adenocarcinoma; PrP, prion protein; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling.
Silencing PrP Suppresses Notch1 Signaling, Tumor Growth, and Invasion but Increases Tumor Cell Apoptosis
Next, we asked whether PrP regulates Notch1 expression and signaling in PDAC. First, we examined the effect of PrP down-regulation (PrP knockdown, PrP-KD) on Notch1 in four PDAC cells. All four PrP-KD cell lines have decreased levels of Notch1 with strongest effect seen in BxPC-3 cells (Figure 2, A and B), whereas PrP silencing has no significant effect on Notch1 mRNA expression (data not shown). Consistently, Hes1, a downstream signaling target of Notch, was also suppressed on PrP silencing in all four cell lines (Figure 2A). Expression level of Notch1, Hes1, and PrP after PrP silencing was compared after normalization with β-actin in each cell line examined in which the level of expression in parental cells was set as 1 (Figure 2B). Therefore, PrP not only modulated the expression level of Notch1, it also influenced the signaling activation of Notch1. Down-regulation of Notch1 in PrP-KD cells was also confirmed by immunofluorescent staining in PDAC cell lines. In all four cells, the immunoreactivity of both PrP and Notch1 was greatly diminished compared with those in the control parental cells (Figure 2C).
We then investigated whether the interaction between PrP and Notch1 was reciprocal. It appeared that knocking down Notch1 (N1-KD) did not affect the expression of PrP (data not shown).
We reported earlier that down-regulation of PrP in PDAC cell lines reduced their proliferation.16 With the use of a more quantitative MTT proliferation assay, we found that PrP silencing (PrP-KD) decreased growth of all four PDAC cell lines (Figure 2D). PrP silencing also decreased Ki-67 indexes by 48.7%, 39.3%, 24.9%, and 51.5% for BxPC-3, Capan-1, Panc-1, and Panc10.05 cells, respectively (Figure 2E). Furthermore, the in vitro invasion of all four PrP-KD cell lines were also decreased (Figure 2F). In contrast, PrP silencing increased apoptosis by 36.7-, 5.5-, 3.8-, and 6.8-fold, respectively, of the four cell lines (Figure 2G). Collectively, these findings reveal that down-regulation of PrP in PDAC cells resulted in a decrease in tumor proliferation and invasion, but an increase in their apoptosis, while simultaneously suppressing Notch1 signaling.
Notch1 Functions Downstream of PrP
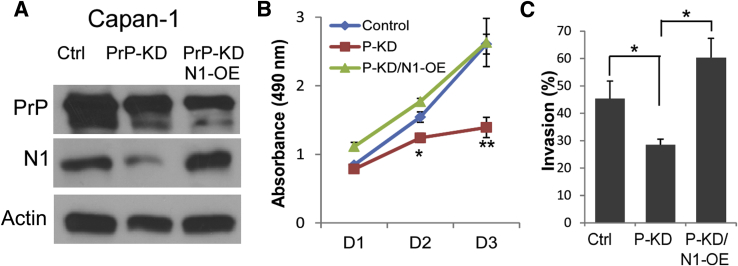
To further investigate if Notch1 is a downstream effector of PrP in PDAC cells, we asked whether overexpression of activated Notch1 could restore PrP silencing-mediated suppression of tumor phenotypes. To test this, an activated form of Notch1 (ICN1) was expressed in Panc-1 PrP-KD cells. As expected, expression of ICN1 restored expression of activated Notch1 but had no discernible effect on PrP expression (Figure 3A). Enforced expression of ICN1 in PrP-KD PDAC cells also restored down-regulated tumor proliferation (Figure 3B) and invasion (Figure 3C) to the levels comparable with the control cells. Hence, Notch1 was indeed a downstream effector of PrP-mediated tumor proliferation and invasion.
Figure 3.
Enforced Notch1 expression rescues suppressed cell proliferation and invasion mediated by PrP silencing. A: Capan-1 cells were transfected with ICN1 (pEGFP-N1IC), and immunoblotted to detect PrP and Notch1. B and C: The effect of enforced Notch1 expression in PrP-silenced cells (P-KD/N1-OE) on cell proliferation (B) and invasion (C) was compared with those in PrP-silenced cells (P-KD) and Ctrl cells. Data are expressed as means ± SD. n = 3 independent experiments (A); n = 5 biological replicates (B and C). t-test was performed (B and C). P values that are significantly different are shown as indicated. ∗P < 0.05, ∗∗P < 0.01. Ctrl, control; D1, 24 hours; D2, 48 hours; D3, 72 hours; ICN1, control plasmid or plasmid expressing activated Notch1; N1-OE, overexpression of Notch1; pEGFP, phosphorylated enhanced green fluorescent protein; P-KD, Prp knockdown; PrP, prion protein.
Notch Blockade Collaborates with Silencing PrP to Suppress Tumor Growth in Xenografts
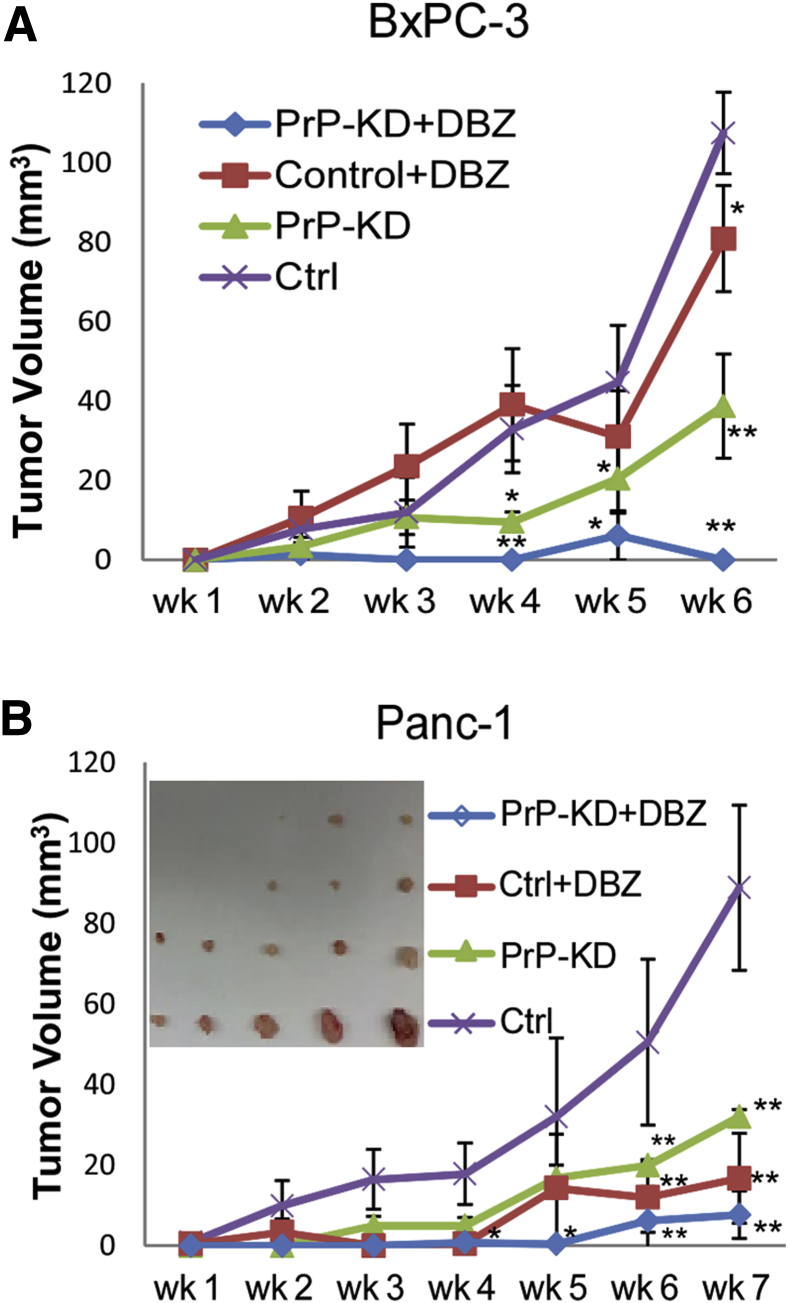
Next, we examined the effect of targeting PrP and blocking Notch signaling on tumor growth in vivo. We treated xenografts of two cell lines, BxPC-3 (Figure 4A) or Panc-1 cells (Figure 4B), and their PrP-KD derivatives, with phosphate-buffered saline or a γ-secretase inhibitor of Notch, DBZ. We did not test Panc10.05 cells in xenografts because they require human insulin for optimal growth. As expected, the growth of PrP-KD BxPC-3 cells (Figure 4A) and PrP-KD Panc-1 cells (Figure 4B) were significantly retarded compared with their respective controls (Figure 4). Tumor volume was not decreased in DBZ-treated, control BxPC-3 xenografts (Figure 4A), but it was significantly decreased in similarly treated, control Panc-1 xenografts (Figure 4B). This disparity most likely reflects the fact that BxPC-3 cells express a higher level of Notch1; therefore, they may require a higher concentration of DBZ to elicit the same effect. However, tumor growth was suppressed to a much greater extent in both Panc-1 and BxPC-3 PrP-KD xenografts that were also treated with DBZ (Figure 4). Collectively, these findings suggest that PrP knockdown elicits additional antitumor effect when combined with Notch inhibition.
Figure 4.
PrP silencing and blocking Notch1 activation collaborates to suppress cancer cell growth in xenografts. A and B: BxPC-3 (A) and Panc-1 (B) control or PrP-KD cells were injected ipsilaterally on the back of nude mice. Paired animals received either PBS (control or PrP-KD) or DBZ (control + DBZ or PrP-KD + DBZ) for 3 consecutive days every 7-day cycle. Tumor volumes were measured weekly and at the end of experiments (6 weeks for BxPC-3 and 7 weeks for Panc-1). B: Shown are Panc-1 tumor sizes at week 7 (inset). t-test was performed (A and B). Data are expressed as tumor volume ± SD. n = 5 in each group. ∗P < 0.05, ∗∗P < 0.01. Ctrl, control; DBZ, dibenzazepine; KD, knock down; PBS, phosphate-buffered saline; PrP, prion protein.
Overexpression of PrP Enhances Notch1 Signaling and Promotes Tumor Cell Growth and Invasion
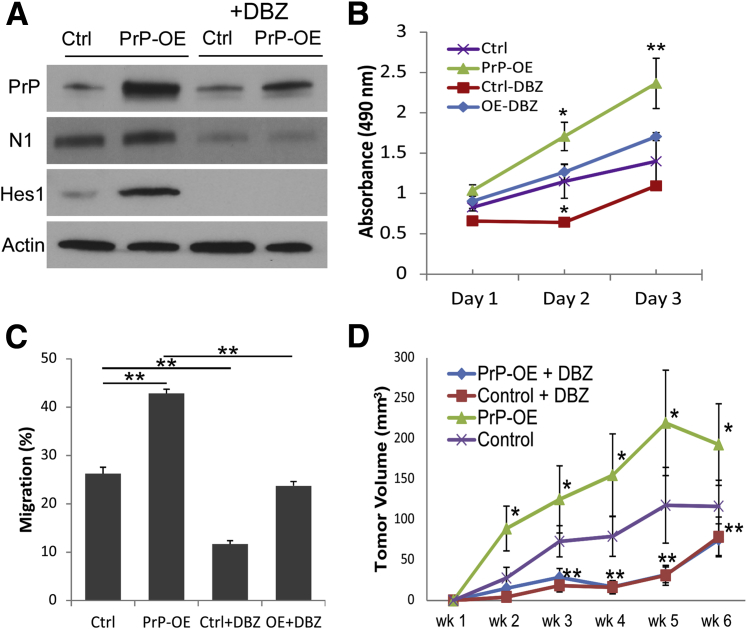
The level of PrP expression appeared to correlate with Notch1 expression and Notch1 signaling. We investigated whether forced expression of PrP in low PrP-expressing Capan-1 cells could induce an opposite effect on tumor cells and also on Notch1 signaling as seen with PrP silencing. Indeed, overexpression of PrP (PrP-OE) in Capan-1 cells increased the level of Notch1 and Hes1 expression (Figure 5A, left two panels). Accordingly, PrP-OE Capan-1 cells also exhibited increased proliferation (Figure 5B) and migration (Figure 5C), compared with control Capan-1 cells. We did not overexpress PrP in other PDAC cells because these cells expressed a high level of endogenous PrP.
Figure 5.
Overexpression of PrP enhances Notch1 signaling and cancer cell growth and invasion. Capan-1 cells were transfected with control or PrP-expressing plasmid. A: Expression of PrP, Notch1, and Hes1 was assessed by immunoblotting. N1 and Hes1 expression was also assessed after DBZ treatment (20 μmol/L for 48 hours) (right two columns). B and C: PrP-OE increases cell growth (B) and migration (C), although DBZ treatment decelerates Capan-1 proliferation and migration. D: Control or PrP-OE Capan-1 cells were injected ipsilaterally on the back of nude mice. Paired animals received either PBS (control or PrP-OE) or DBZ (control + DBZ or PrP-OE + DBZ) for 3 consecutive days every 7 days. Tumors were dissected at the end of 6 weeks. Tumor volumes were measured weekly and at the end of experiments. Data are expressed as means ± SD. t-test was performed (B–D). n = 3 independent experiments (A); n = 6 mice (B and C); n = 8 biological replicates (B and C); n = 5 mice in each group (D). ∗P < 0.05, ∗∗P < 0.01. Ctrl, control; DBZ, dibenzazepine; N1, Notch1; OE, overexpression; PBS, phosphate-buffered saline; PrP, prion protein.
To investigate whether PrP overexpression promoted cell proliferation and tumor migration via Notch signaling, we treated Capan-1 cells with DBZ or vehicle in vitro (20 μmol/L, 48 hours). DBZ treatment decreased both endogenous and up-regulated levels of Notch1 and Hes1 in Capan-1 cells (Figure 5A). DBZ treatment mildly decreased proliferation of control Capan-1 cells but inhibited cell migration by 50%. The increased proliferation and migration of PrP-OE Capan-1 cells were both brought down to the levels seen in control Capan-1 cells after DBZ treatment (Figure 5, B and C).
To test the effect of PrP overexpression on tumor growth in vivo, we injected parental or PrP-OE Capan-1 cells to either flank of nude mice. Consistent with in vitro assays, xenografts excised at the end of 6 weeks showed that PrP-OE Capan-1 tumors have a close to 50% increase of tumor volume (192.9 ± 50.2 mm3) compared with controls (116.5 ± 32.3 mm3) (P < 0.05). Further, growth comparison of parental and PrP-OE tumors that were treated with either DBZ or vehicles indicated that increased growth of PrP-OE tumors over parental tumors was completely blocked by DBZ (Figure 5D).
Collectively, these in vitro and in vivo findings support that enhanced cell proliferation and migration induced by PrP overexpression are mainly mediated through Notch signaling activation and that Notch1 is a main downstream effector of PrP in PDAC cells.
Down-Regulation of PrP Enhances Notch1 Degradation
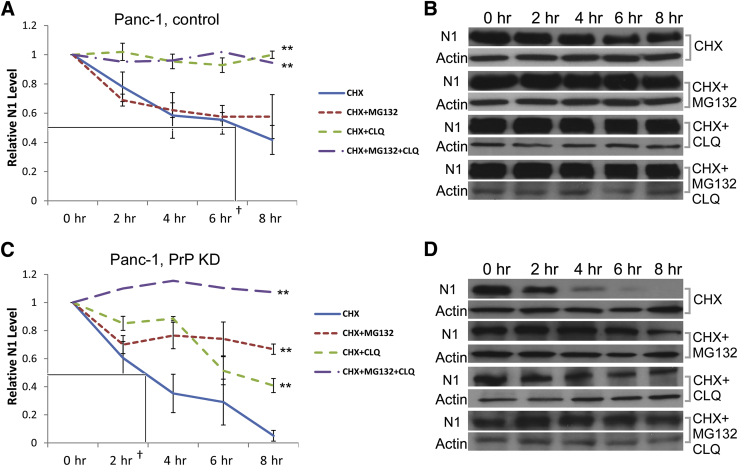
To investigate the mechanism of enhanced Notch1 signaling mediated by PrP/Notch1 complex, we examined the effect of PrP silencing on the half-life of Notch1. Membrane-tethered Notch1 is mainly internalized and degraded in the lysosomes by the E3 protein ubiquitin ligase Itch.25, 26 Itch can also mediate Notch1 proteasome degradation.27 To investigate the half-life of Notch1, we cultured the cells in the presence of CHX to prevent synthesis of new Notch1. We then prepared the cell lysates at various time points after CHX treatment and immunoblotted the cell lysate with anti-Notch1 antibody. We found that in PrP-KD Panc-1 cells, the half-life of Notch1 was significantly shortened from about 7.4 hours in control cells to about 3 hours in PrP-KD cells (Figure 6, A and C). In contrast, ICN1 displayed only marginal changes in its half-life (data not shown).
Figure 6.
Down-regulation of PrP enhances degradation of Notch1. Panc-1 parental cells (control; A and B) or PrP-KD cells (C and D) were cultured for up to 8 hours in the presence of CHX (10 μmol/L), CHX plus proteasome inhibitor MG132 (10 μmol/L; CHX + MG132, CHX plus lysosome inhibitor CLQ (25 μmol/L; CHX + CLQ), or CHX plus MG132 as well as CLQ (CHX + MG132 + CLQ). Cell lysates were prepared at indicated time points and blotted with anti-N1 and anti-actin (B and D). Notch1 was normalized to actin and expressed as relative level compared with that in the beginning of cell culture (A and C). Boxed areas in A and C indicate the half-life of CHX. Data are expressed as means ± SD (A and C). t-test was performed (A and C). n = 3 biological replicates. ∗∗P < 0.01. †Half-life of Notch1. CHX, cyclohexamide; CLQ, chloroquine; KD, knocked down; N1, Notch1; PrP, prion protein.
Protein half-life is controlled either by the proteasome or lysosome pathway or both. Next, we asked if either or both pathways are responsible for Notch1 degradation in PDAC cells, and if there is altered degradation of Notch1 in PDAC cells as a result of change in PrP expression level. We treated control and PrP-KD Panc-1 cells with proteasome inhibitor, MG132 (10 μmol/L), or lysosome inhibitor, CLQ (25 μmol/L), individually, or in combination, all in the presence of CHX. We found that control Panc-1 cells cultured in the presence of either CHX alone or CLQ combined with MG132 showed similar partial block of Notch1 degradation, whereas cells cultured in the presence of CHX combined with CLQ displayed a complete block of Notch1 degradation (Figure 6, A and B), suggesting that Notch1 in Panc-1 cells was degraded mainly through the lysosome pathway but not much through the proteasome pathway. In contrast, in PrP-KD Panc-1 cells, either MG132 or CLQ in the presence of CHX partially blocked Notch1 degradation, whereas combined MG132 and CLQ treatment completely blocked Notch1 degradation (Figure 6, C and D), suggesting that Notch1 is degraded through both the proteasome and lysosome pathway in PrP-KD Panc-1 cells. A similar pattern was observed in other PDAC cells before and after PrP silencing (data not shown). These findings indicate that in PDAC cells, a high level of PrP prolonged Notch1 half-life likely through the suppression of proteasome degradation of Notch1.
Notch1 Activity in PrP-Expressing Human PDACs Confers Even Poorer Prognosis
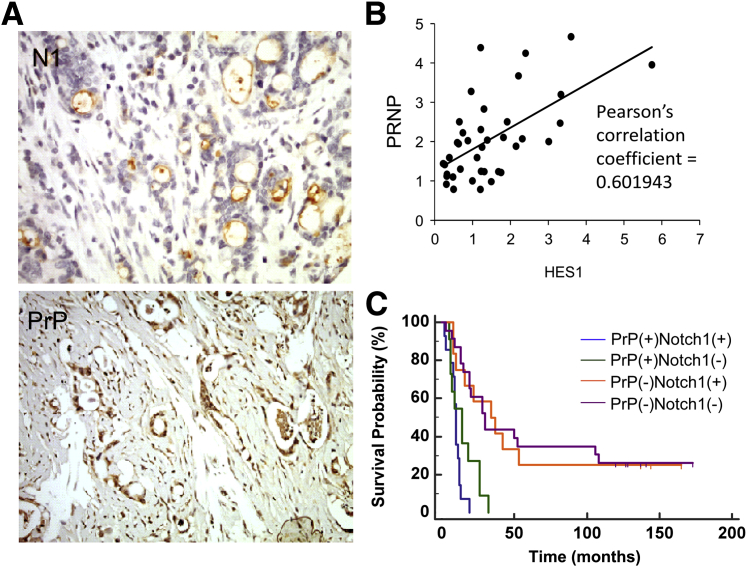
To extend our results from cell lines to humans, we examined the expression pattern of PrP and Notch1 in human PDAC specimens (Figure 7A). Notch1 expression was mainly found on cell membrane and the cytoplasm using the antibody that recognizes the transmembrane/intracellular fragment, whereas antibody recognizing Hes1 showed variable staining or no staining in the nucleus (data not shown). In a series of 67 resected, localized PDAC samples, we found that, consistent with our previous reports, 42% (28 of 67) of cases express a high level of PrP,23 whereas 45% (30 of 67) have Notch1 activity (defined by strong Notch1 staining). A correlation between PrP expression and Notch1 activity was also found, because 17 of 28 cases (61%) of PrP+ tumor also showed Notch1 activity, whereas only 13 of 39 cases (33%) of PrP− tumors showed Notch1 expression (Table 1) (P < 0.05). Because there is no clear explanation as to why certain tumors show strong activity of both Notch1 and Hes1 whereas others show only Notch1 staining, we further examined RNA expression of PRNP and HES1 and revealed a modest correlation between PrP and Notch activation (Pearson's correlation coefficient = 0.60) (Figure 7B). These findings support our PDAC cell line data that PrP expression in PDAC tissues appears to promote Notch signaling activation.
Figure 7.
PrP expression correlates with Notch1 expression and the co-expression of these two confers worse prognosis in human PDAC samples. A: PDAC specimens were stained for N1 (top) and anti-PrP (bottom). B: PrP and HES1 mRNA levels were normalized to those expressed by normal pancreatic tissues, respectively. Pearson correlation was calculated. C: Kaplan-Meier survival analysis was performed on PrP+Notch1+, PrP+Notch1−, PrP−Notch1+, and PrP−Notch1− tumors. Spikes indicated three surviving patients in the PrP−Notch1+ group (surviving months: 165, 144 and 137), and six in the PrP−Notch1− group (surviving months: 173, 144, 137, 128, 127, and 120) as of August 2015. n = 67 PDAC specimens (A); n = 14 PrP+Notch1+ tumors (B); n = 11 PrP+Notch1− tumors (B); n = 13 PrP−Notch1+ tumors (B); n = 23 PrP−Notch1− tumors (B). Original magnification: ×20 (A). N1, anti-Notch1; PDAC, pancreatic ductal adenocarcinoma; PRNP, human prion protein; PrP, prion protein.
Table 1.
PrP and Notch1 Immunoreactivity in Human PDAC
| Expression | Notch1+, n (%) | Notch1−, n (%) | Total, n (%) |
|---|---|---|---|
| PrP+ | 17 | 11 | 28 (42) |
| PrP− | 13 | 26 | 39 (58) |
| Total | 30 (45) | 37 (55) | 67 |
Fisher's exact test (two-tailed) was performed, P < 0.05.
PDAC, pancreatic ductal adenocarcinoma; PrP, prion protein.
Finally, we performed Kaplan-Meier survival analysis among four groups of specimens based on the pattern of PrP and Notch1 immunoreactivity. Of these 67 cases, 25 PrP+ and 35 PrP− cases have clinical follow-up information (Figure 7C). Among the 25 PrP+ cases, median survival was 10 months for Notch1+ tumors (n = 14) versus 14 months for Notch1− tumors (n = 11) [hazard ratio (HR) = 1.9; 95% CI, 0.84– 4.1; log-rank test P = 0.07]. Of 35 PrP− cases, median survival was 34 months for Notch1+ tumors (n = 13) versus 30 months for Notch1− tumors (n = 23) (HR = 1.0; 95% CI, 0.5–2.4; log-rank test P = 0.91) (Table 2). Therefore, although Notch1 immunoreactivity in localized PrP+ tumors was associated with a trend of decreased survival, although not reaching statistical significance, its expression in PrP− cases had no effect on prognosis. Of note, all nine surviving cases are PrP−, with three in the Notch1+ group and six in the Notch1− group. The risk of death was 4.3 to approximately 4.5 times higher for PrP+Notch1+) than PrP−Notch1+ (HR = 4.3; 95% CI, 1.5–12.1; P = 0.0003) or PrP−Notch1− tumors (HR = 4.5; 95% CI, 1.7–11.8; P < 0.0001), and 3.0 to approximately 3.1 times higher for PrP+Notch1− than PrP−Notch1+ (P = 0.005) or PrP−Notch1− tumors (P = 0.001).
Table 2.
Survival Analysis
| Group | Medium survival, months | 95% CI |
|---|---|---|
| PrP+Notch1+ | 10 | 8–12 |
| PrP+Notch1− | 14 | 6–26 |
| PrP−Notch1+ | 34 | 16–53 |
| PrP−Notch1− | 30 | 19–106 |
PrP, prion protein.
Discussion
In this study, we provide novel findings that PrP and Notch1 forms an interacting network to promote pancreatic cancer survival and invasion. First, we found that in PDAC cell lines, PrP, FLNA, and Notch 1 exist as a complex and that PrP and Notch1 colocalizes on the cell membrane. Second, we found Notch1 is a downstream effector of PrP, because increased expression of PrP up-regulates the expression and activation of Notch1, resulting in enhanced tumor cell growth and invasion but suppressed apoptosis. In contrast, PrP down-regulation has the opposite effect. These antitumor effects mediated by PrP down-regulation can be rescued by activated Notch1. Mechanistically, we found that binding of PrP prolongs the half-life of Notch1 by blocking proteasome degradation of Notch1. Third, we show that compared with the PDAC xenograft that is singularly targeted, the growth of PDAC xenograft is more effectively retarded when both PrP and Notch1 are targeted. Finally, we confirm these findings in human PDAC specimens, showing that most PrP-expressing PDAC tumors also show Notch1 expression, and coexpression of Notch1 and PrP confers even poorer prognosis than expression of PrP alone. These findings have unraveled an important pathway identifying Notch1 as an important effector downstream of PrP mediating PDAC tumor growth, invasion, and survival in a subset of patients with PDAC.
NOTCH1 mutation is frequently found in head and neck squamous cell carcinoma, lymphoma and in T-cell acute lymphoblastic leukemia but less frequently seen in human pancreatic cancer.28, 29, 30, 31 Whether Notch signaling is important in PDAC carcinogenesis is unsettled. Expression of activated Notch1 in a mouse model of PDAC increased PanIN formation,32 suggesting an oncogenic role for Notch in PDAC initiation. A γ-secretase inhibitor also inhibited the progression of PDAC in a transgenic mouse model.33 However, more recent work reveals a tumor suppressive role for Notch signaling in the context of PanIN development. Loss of Notch1 function may even promote PDAC development.34 Furthermore, different Notch receptors may have opposing roles in pancreatic cancer biology.8 Recent studies revealed that pancreatic cancer could be divided into different subtypes based on their molecular signatures.12 Therefore, whether Notch functions as a tumor suppressive or an oncogenic promoter may be cellular context, and, depending on Notch interacting, partners in a receptor-specific manner.
To the best of our knowledge, our findings in this study show for the first time that Notch1 forms a partner with PrP in human pancreatic cancer. Despite its relative small size, a plethora of more than 40 PrP-interacting partners have been reported. These include proteins, lipids, nucleic acids, glycosyaminoglycans, and divalent cations, such as copper and zinc.23 In infectious prion disease, up-regulation and activation of Notch1 during prion infection causes dendritic atrophy, and a γ-secretase inhibitor can ameliorate the pathology.35, 36 However, there is no evidence suggesting a direct interaction between PrP and Notch1. Note that the pro-PrP expressed in PDAC, unlike normal PrP, retains its GPI-PSS, thus enabling it to bind FLNA. It is likely the interaction between PrP and Notch1 is also due to a gain-of-function of pro-PrP in PDAC cells. This fatal attraction has significant biological significances because it promotes tumor proliferation and invasion but reduces tumor apoptosis.
Although we have demonstrated a physical interaction between Notch1, PrP, and FLNA, the detail biochemical pathways subsequent to this interaction remain uncharacterized. Notch activation is known to promote cancer cell proliferation and invasion and to suppress tumor apoptosis, some of these effects have been shown to be mediated through Notch activation of NF-κB in pancreatic cancer37 and Akt in other cancers.38, 39 Further, overexpression of Notch1 can lead to up-regulation of tumor-initiating cell markers.40 Similarly, PrP signaling displays antiapoptotic activity.41, 42 Overexpression of PrP also contributes to cancer metastasis by promoting cancer adhesiveness and invasiveness.43 Likely a complex formed by these three molecules, PrP, FLNA, and Notch1, will offer further advantage for PDAC progression and survival. This hypothesis is supported by our findings that combined anti-Notch treatment with silencing PrP shows more effective antitumor growth than anti-Notch treatment or silencing of PrP alone.
Binding of PrP with Notch1 prolongs Notch1 half-life. We show that down-regulation of PrP in PDAC cells enhances the proteosome degradation of Notch1 that is otherwise blunted in unmodified PDAC cells. Future studies are needed to elucidate if PrP suppresses Notch1 ubiquitination and degradation by modulating Itch.27 Furthermore, dysregulation of other E3 ligases or deubiquitinating enzymes, or alterations of Notch endosomal trafficking, could be linked to Notch1 stability dysregulation.44 Binding of PrP may also cause Notch1 aggregation, modulate Notch1 cleavage, or enhance binding of its ligands resulting in activation.
The relevance of our findings in clinical setting is supported in our study that a significant portion of localized PrP+ human PDAC biopsies also show Notch1 immunoreactivity and that PrP transcript expression is modestly correlated with Hes1 expression. In addition, a survival analysis demonstrates that coexpression of PrP with Notch1 in PDAC confers even poorer prognosis than expression of PrP only, although the differences in survival probability between these two groups are not statistically significant. In comparison, expression of Notch1 in PrP− specimens has no effect on patient survival. These findings support an additive tumor-promoting effect mediated by the PrP-Notch1 complex observed in PDAC cell lines. Additional clinical materials are required to confirm that adding Notch1 expression will have additional prognostic value in localized as well as in more advanced PDAC. We did not observe significant pathologic differences between PrP+ and PrP− groups. The underlying molecular signature of PrP-expressing PDAC and its effect on clinical courses warrants further investigation.
Conclusions
We have provided compelling evidence to suggest that PrP, FLNA, and Notch1 are partners in crime in enabling PDAC aggressiveness. These findings provide critical mechanistic insights to the biological significance of PrP-Notch1 cross talk in promoting PDAC progression and survival. Elucidating the molecular events driving PrP, FLNA, and Notch1 interacting network will reveal whether targeting this pathway will have diagnostic, prognostic, or therapeutic potential to benefit patients with PDAC.
Footnotes
Supported by GI spore pilot grant 1P50CA150964-01A1 (W.X.; Sanford Markowitz principal investigator), Department of Pathology Case Western Reserve University faculty startup fund (W.X. and L.Z.), NIH grant HL103827 (L.Z.) and American Cancer Society grant LIB-125064 (L.Z.).
Disclosures: None declared.
W.X. and L.Z. contributed equally.
References
- 1.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 2.Eser S., Schnieke A., Schneider G., Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins M.A., Brisset J.C., Zhang Y., Bednar F., Pierre J., Heist K.A., Galban C.J., Galban S., di Magliano M.P. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012;7:e49707. doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong B., Qia C., Erkan M., Kleeff J., Michalski C.W. Overview on how oncogenic Kras promotes pancreatic carcinogenesis by inducing low intracellular ROS levels. Front Physiol. 2013;4:246. doi: 10.3389/fphys.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 6.Buchler P., Gazdhar A., Schubert M., Giese N., Reber H.A., Hines O.J., Giese T., Ceyhan G.O., Muller M., Buchler M.W., Friess H. The Notch signaling pathway is related to neurovascular progression of pancreatic cancer. Ann Surg. 2005;242:791–800. doi: 10.1097/01.sla.0000189115.94847.f1. discussion 800–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto Y., Maitra A., Ghosh B., Zechner U., Argani P., Iacobuzio-Donahue C.A., Sriuranpong V., Iso T., Meszoely I.M., Wolfe M.S., Hruban R.H., Ball D.W., Schmid R.M., Leach S.D. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 8.Mazur P.K., Einwachter H., Lee M., Sipos B., Nakhai H., Rad R., Zimber-Strobl U., Strobl L.J., Radtke F., Kloppel G., Schmid R.M., Siveke J.T. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2010;107:13438–13443. doi: 10.1073/pnas.1002423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucas H., Mann C.D., Sutton C.D., Garcea G., Neal C.P., Berry D.P., Manson M.M. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol. 2008;97:63–68. doi: 10.1002/jso.20894. [DOI] [PubMed] [Google Scholar]
- 10.Lobry C., Oh P., Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH what you think. J Exp Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu H., Zhou L., Awadallah A., Xin W. Significance of Notch1-signaling pathway in human pancreatic development and carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:242–247. doi: 10.1097/PAI.0b013e3182655ab7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Ahmad A., Li Y., Azmi A.S., Miele L., Sarkar F.H. Targeting notch to eradicate pancreatic cancer stem cells for cancer therapy. Anticancer Res. 2011;31:1105–1113. [PubMed] [Google Scholar]
- 14.Harris D.A. Cellular biology of prion diseases. Clin Microbiol Rev. 1999;12:429–444. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antony H., Wiegmans A.P., Wei M.Q., Chernoff Y.O., Khanna K.K., Munn A.L. Potential roles for prions and protein-only inheritance in cancer. Cancer Metastasis Rev. 2012;31:1–19. doi: 10.1007/s10555-011-9325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C., Yu S., Nakamura F., Yin S., Xu J., Petrolla A.A., Singh N., Tartakoff A., Abbott D.W., Xin W., Sy M.S. Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J Clin Invest. 2009;119:2725–2736. doi: 10.1172/JCI39542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sy M.S., Altekruse S.F., Li C., Lynch C.F., Goodman M.T., Hernandez B.Y., Zhou L., Saber M.S., Hewitt S.M., Xin W. Association of prion protein expression with pancreatic adenocarcinoma survival in the SEER residual tissue repository. Cancer Biomark. 2011;10:251–258. doi: 10.3233/CBM-2012-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretzschmar H.A., Stowring L.E., Westaway D., Stubblebine W.H., Prusiner S.B., Dearmond S.J. Molecular cloning of a human prion protein cDNA. DNA. 1986;5:315–324. doi: 10.1089/dna.1986.5.315. [DOI] [PubMed] [Google Scholar]
- 19.Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., Shapiro S.S. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y., Walsh C.A. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K., Wong P., Zhang L., Jacobs B., Borden E.C., Aster J.C., Bedogni B. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene. 2012;31:4609–4618. doi: 10.1038/onc.2011.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin H.M., Minter L.M., Cho O.H., Gottipati S., Fauq A.H., Golde T.E., Sonenshein G.E., Osborne B.A. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Xin W., Sy M.S. Binding of pro-prion to filamin A: by design or an unfortunate blunder. Oncogene. 2010;29:5329–5345. doi: 10.1038/onc.2010.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinnix C.C., Lee J.T., Liu Z.J., McDaid R., Balint K., Beverly L.J., Brafford P.A., Xiao M., Himes B., Zabierowski S.E., Yashiro-Ohtani Y., Nathanson K.L., Bengston A., Pollock P.M., Weeraratna A.T., Nickoloff B.J., Pear W.S., Capobianco A.J., Herlyn M. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res. 2009;69:5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu L., Joazeiro C., Fang N., Wang H.Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y.C. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 26.Chastagner P., Israel A., Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M.Y., Mo J.S., Ann E.J., Yoon J.H., Park H.S. Dual regulation of notch1 signaling pathway by adaptor protein fe65. J Biol Chem. 2012;287:4690–4701. doi: 10.1074/jbc.M111.289637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 30.Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agrawal N., Frederick M.J., Pickering C.R., Bettegowda C., Chang K., Li R.J., Fakhry C., Xie T.X., Zhang J., Wang J., Zhang N., El-Naggar A.K., Jasser S.A., Weinstein J.N., Trevino L., Drummond J.A., Muzny D.M., Wu Y., Wood L.D., Hruban R.H., Westra W.H., Koch W.M., Califano J.A., Gibbs R.A., Sidransky D., Vogelstein B., Velculescu V.E., Papadopoulos N., Wheeler D.A., Kinzler K.W., Myers J.N. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De La O.J., Emerson L.L., Goodman J.L., Froebe S.C., Illum B.E., Curtis A.B., Murtaugh L.C. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plentz R., Park J.S., Rhim A.D., Abravanel D., Hezel A.F., Sharma S.V., Gurumurthy S., Deshpande V., Kenific C., Settleman J., Majumder P.K., Stanger B.Z., Bardeesy N. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2009;136:1741–1749.e6. doi: 10.1053/j.gastro.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanlon L., Avila J.L., Demarest R.M., Troutman S., Allen M., Ratti F., Rustgi A.K., Stanger B.Z., Radtke F., Adsay V., Long F., Capobianco A.J., Kissil J.L. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010;70:4280–4286. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishikura N., Clever J.L., Bouzamondo-Bernstein E., Samayoa E., Prusiner S.B., Huang E.J., DeArmond S.J. Notch-1 activation and dendritic atrophy in prion disease. Proc Natl Acad Sci U S A. 2005;102:886–891. doi: 10.1073/pnas.0408612101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spilman P., Lessard P., Sattavat M., Bush C., Tousseyn T., Huang E.J., Giles K., Golde T., Das P., Fauq A., Prusiner S.B., Dearmond S.J. A gamma-secretase inhibitor and quinacrine reduce prions and prevent dendritic degeneration in murine brains. Proc Natl Acad Sci U S A. 2008;105:10595–10600. doi: 10.1073/pnas.0803671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Banerjee S., Li Y., Rahman K.M., Zhang Y., Sarkar F.H. Down-regulation of notch-1 inhibits invasion by inactivation of nuclear factor-kappaB, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Res. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 38.Zhao N., Guo Y., Zhang M., Lin L., Zheng Z. Akt-mTOR signaling is involved in Notch-1-mediated glioma cell survival and proliferation. Oncol Rep. 2010;23:1443–1447. doi: 10.3892/or_00000782. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z., Li Y., Banerjee S., Kong D., Ahmad A., Nogueira V., Hay N., Sarkar F.H. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 40.Bao B., Wang Z., Ali S., Kong D., Li Y., Ahmad A., Banerjee S., Azmi A.S., Miele L., Sarkar F.H. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307:26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.McEwan J.F., Windsor M.L., Cullis-Hill S.D. Antibodies to prion protein inhibit human colon cancer cell growth. Tumour Biol. 2009;30:141–147. doi: 10.1159/000225243. [DOI] [PubMed] [Google Scholar]
- 43.Pan Y., Zhao L., Liang J., Liu J., Shi Y., Liu N., Zhang G., Jin H., Gao J., Xie H., Wang J., Liu Z., Fan D. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–1888. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- 44.Le Bras S., Loyer N., Le Borgne R. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic. 2011;12:149–161. doi: 10.1111/j.1600-0854.2010.01126.x. [DOI] [PubMed] [Google Scholar]