Abstract
Spatial information is crucial to interrogate developmental and tissue-level regulation of gene expression; this information is usually lost when mRNA levels from the entire tissue are measured using reverse transcriptase PCR, microarray or high-throughput sequencing. In contrast, single-molecule fluorescent in situ hybridization (smFISH) preserves the spatial information of the cellular mRNA content with subcellular resolution across tissue. We describe here an smFISH protocol that allows the quantification of single mRNAs in Drosophila embryos using commercially available smFISH probes, a wide-field epifluorescence, confocal or structured illumination microscope (SIM), a super-resolution imaging approach, and a spot-detection algorithm. Fixed Drosophila embryos are hybridized in solution with a mix of fluorescently-labeled probes, mounted onto a coverslip and imaged in three dimensions (3D). Single, fluorescently labeled mRNAs are then localized within tissue and counted using spot-detection software to generate quantitative, spatially-resolved gene expression datasets. With minimum guidance, a graduate student can successfully implement this protocol. The smFISH procedure described here can be completed in 4-5 days.
Keywords: smFISH, single molecule mRNA, mRNA localization, spot detection, SIM, super-resolution microscopy
INTRODUCTION
In situ hybridization (ISH) allows qualitative mRNA detection in intact tissue and has been a tool of choice to study the distribution of mRNAs in biological samples for nearly 40 years. However, after the development of single-molecule fluorescent in situ hybridization (smFISH) with its subsequent optimization that allowed single mRNA counting in 19981, the interest in detecting and quantifying all aspects of mRNA biology in individual cells with single mRNA sensitivity has rapidly increased. This mounting interest was further augmented in 2008 by the development of the smFISH protocol that utilized a mix of short, singly-labeled DNA probes to detect individual mRNAs in single cells2 accompanied by the subsequent launch of custom-designed, commercially available and affordable Stellaris® RNA probes. The year 2015 alone recorded 133 publications that used these commercially-available smFISH probes to detect mRNAs3 and strikingly, nearly half characterized mRNA expression quantitatively rather than qualitatively. These publications demonstrate that the broader scientific community is not only interested in whether the mRNA is present in the cell but rather how much of it is present and where in the cell it is located. Indeed, several groups have developed algorithms to quantify various aspects of mRNA biology with single molecule sensitivity in the fly4-8. While detection and counting of single smFISH-labeled mRNA molecules using a spot detection algorithm is a critical part of quantitative smFISH, to date a step-by-step protocol describing this part of quantitative smFISH has been absent in the Drosophila literature. As the field is rapidly evolving, we propose here an up-to-date protocol, combined with a thoroughly tested software and with cutting-edge microscopy that will be easy to use by a broad audience.
Overview of the procedure
The smFISH labeling and counting protocol described here is composed of four parts; embryo fixation and collection, embryo hybridization with smFISH probes, imaging and single mRNA detection and counting.
Drosophila embryos are collected, dechorionated and fixed with paraformaldehyde, PBS and paraformaldehyde-saturated heptane. Fixative is then removed and methanol added. Embryos are vigorously shaken to remove embryo’s vitelline membrane, washed with methanol and stored overnight at 4°C in methanol.
The next day embryos are rehydrated, post-fixed with paraformaldehyde, followed by several PBS washes containing Tween-20 detergent (PBT). Embryos are then treated with Proteinase K to allow probe penetration into deeper embryo volume, followed by an additional fixation with paraformaldehyde and afterwards extensively washed in PBT. During a pre-hybridization step, embryos are incubated in deionized formamide and SSC. Embryos are then resuspended in the hybridization mix containing smFISH probes, deionized formamide, probe competitor, dextran sulfate, bovine serum albumin (BSA), saline-sodium citrate buffer (SSC), vanadyl ribonucleoside complex VRC and ddH2O, and incubated overnight at 37°C. The next day, the hybridization mix is removed, embryos are extensively washed, first with pre-hybridization solution and afterwards with PBS. Embryos are then mounted on a coverslip and imaged. Afterwards, 3D images are deconvolved and single mRNAs are detected and counted using a spot detection algorithm.
Development of the protocol
Our groups have previously published biological applications of the labeling of mRNAs using commercially-available smFISH probes in Drosophila5, the development of the instant structured illumination microscopy (iSIM) super-resolution system9,10 as well as the application of Airlocalize, the algorithm that enables detection and counting of smFISH-hybridized mRNAs5,11,12. The current protocol covers all steps in enough detail to guide non-specialist users, with an emphasis on single-molecule counting, arguably the most complex step.
Our fluorescent staining protocol of mRNAs in Drosophila is robust, reproducible and easy to implement. The protocol detailing embryo fixation and collection followed by embryo hybridization with smFISH probes is a modification of protocols by Lécuyer et al.13, Lionnet et al12., Femino et al.1 and Raj et al.2. To achieve single-mRNA detection sensitivity in Drosophila embryos, we utilize multiple short, singly-labeled commercially-available smFISH probes2,5 to fluorescently label mRNAs in situ rather than a single, long and heavily fluorescently labeled in vitro-transcribed RNA probe traditionally used in whole-mount in situs to detect transcripts13. In a whole-mount in situ approach, a single antisense RNA probe is in vitro transcribed from a DNA template in the presence of digoxigenin-UTP (DIG-UTP), which is then hybridized to the target mRNA. DIG-modified FISH probes are not readily visualized per se; rather, a subsequent detection of digoxigenins with antibodies coupled to enzymes such as alkaline phosphatase, which react with chromogenic substrates, is necessary to reveal the spatial distribution of target mRNAs14. However, the chromogenic dyes produced by alkaline phosphatase diffuse away from the target mRNA thereby reducing the spatial resolution of the technique13-15. This shortcoming was mitigated by the use of horseradish peroxidase-mediated deposition of tyramides radicals labeled with fluorescent dyes such as Cyanine-3, which covalently react with the proteins at the site of horseradish peroxidase activity. The fluorescent signal becomes localized and the spatial resolution of the technique increases13,15. Whole-mount in situ approaches have been widely adopted to visualize mRNAs particularly in the fly because of their strong staining potential; every 20th to 25th nucleotide in the RNA FISH probe is a DIG-UTP whose detection is further enzymatically amplified. However, all FISH probes also non-specifically bind to the cellular background16-18 (see below and also section “Experimental design, smFISH probes”) and without the ability to distinguish between a bona fide hybridization of the probe to the target mRNA versus the cellular background, whole-mount in situs can only provide qualitative rather than quantitative information. Our protocol utilizes multiple short, singly-labeled smFISH probes2,5 that bind to different positions on the target mRNA to fluorescently label mRNAs in situ and thus readily distinguishes between specific binding to target mRNA and non-specific sticking of the probe to cellular background (Fig. 4b,c). The commercial availability of the probe and relative ease of their synthesis makes it possible to adapt our protocol to quantify various aspects of mRNA biology in Drosophila with subcellular resolution5-8,19,20.
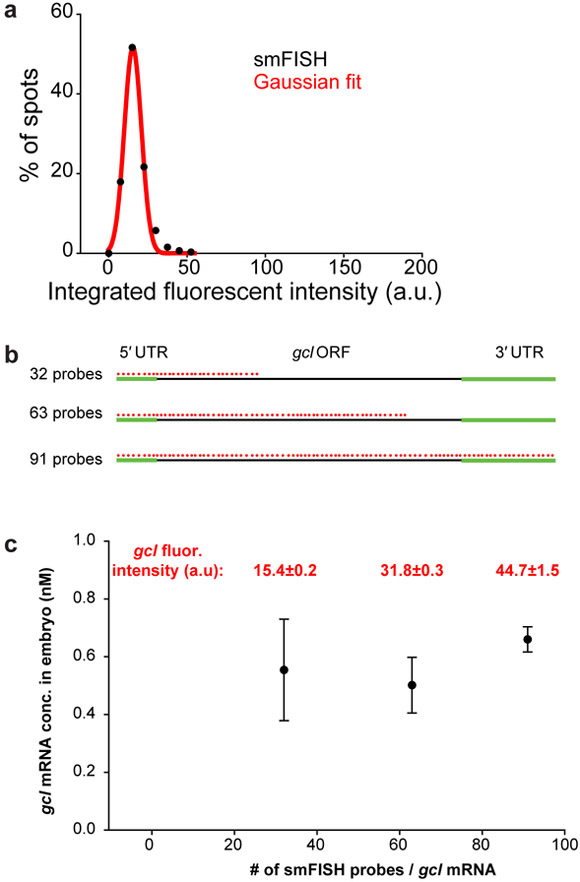
Figure 4. Determining the fluorescent intensity of a single mRNA molecule.
(a) To determine the average integrated fluorescent intensity of a single smFISH-labeled gcl mRNA detected in Fig. 3 g and h, the integrated fluorescence intensities of all detected spots in h are plotted as a histogram (black circles) and fitted to a Gaussian curve (red line). The peak of the Gaussian fit determines the average integrated fluorescence intensity of a single gcl mRNA, here hybridized with 32 singly-labeled Quasar 670 probes, which, with given imaging and microscope parameters was 15.4 ± 0.2a.u. (R2=0.99). (b,c) Adding more smFISH probes to the probe mix increases the integrated fluorescence intensity of a single mRNA but does not change the number of detected gcl mRNAs significantly. gcl was labeled with 32 (gcl 1st third probe mix), 63 (gcl 1st third and 2nd third smFISH probe mix) or 91 (gcl 1st third, 2nd third and 3rd smFISH probe mix) Quasar 670 smFISH probes (b) and an average integrated fluorescent intensity of a single gcl mRNA determined ventrally in the early embryo for each probe mix, as described above. The average integrated fluorescence intensity of a single gcl mRNA, hybridized with 32 singly-labeled Quasar 670 probes was 15.4 ± 0.2 a.u. (R2=0.99) (see Fig. 3), with 63 singly-labeled Quasar 670 probes was 31.8 ± 0.3 a.u. (R2=0.99) and with 91 singly-labeled Quasar 670 probes was 44.7 ± 1.5 a.u. (R2=0.99) (gcl fluorescent intensity reported in red above the graph). (c) Thus, integrated fluorescent intensity of a labeled mRNA scales linearly with the number of smFISH probes hybridizing to the mRNA, yet the number of detected gcl mRNAs, reported as nM concentration of gcl mRNA per embryo, does not change.
Stained Drosophila embryos are imaged in 3D to allow quantification of uniformly distributed and localized mRNAs in a volume5,12. Our smFISH-staining protocol is compatible with a range of imaging modalities, including widefield and confocal microscopy as well as structured illumination microscopy (SIM), a super-resolution method. Here we describe the application of a variation of SIM called instant SIM (iSIM) 9,10. Unlike conventional SIM, iSIM offers very rapid imaging (no loss in speed relative to widefield epifluorescence microscopy), enables superior imaging in thick samples due to better background rejection, and therefore provides superior detection sensitivity as well as localization precision during fluorescent mRNA detection. When coupled with smFISH, iSIM allows a highly detailed quantification of the spatial distribution of transcripts within a complex protein environment, such as an mRNA-bound germ granule5, otherwise unattainable with widefield epifluorescence or confocal microscopy.
3D images are analyzed with a custom spot-detection algorithm, Airlocalize5,12, to detect and count single, smFISH-labeled mRNAs. Individual, fluorescently-labeled transcripts can be detected with sub-pixel resolution in 2D images as well as 3D stacks. Owing to a local affine baseline subtraction step, the intensity measurements are robust even in the presence of high fluorescence background, as is often encountered in tissue. By providing a comprehensive guide to single molecule detection and counting, Airlocalize can be implemented to quantify the cellular mRNA content in any context5,11,12. While we showcase the Drosophila embryo as an example sample in this protocol, the mRNA staining with smFISH probes, imaging and image analysis techniques can be implemented to quantify cellular mRNAs in a variety of tissues and organisms.
Application of the method
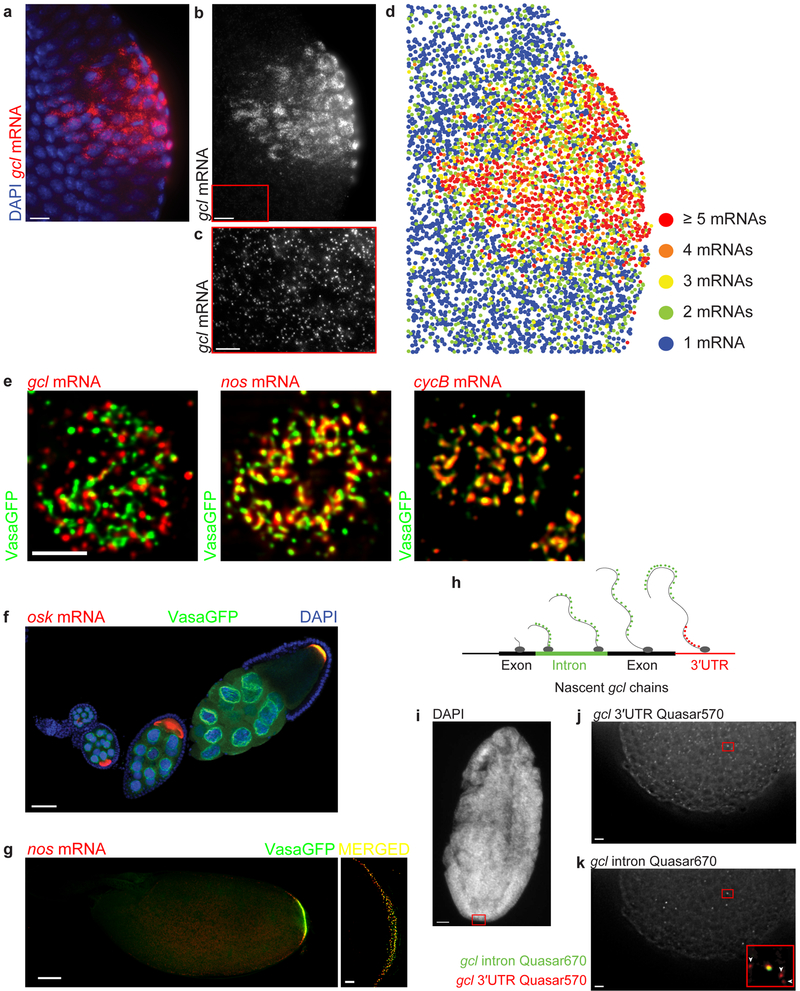
smFISH has been implemented to characterize mRNA transcription2,4,7,21-30, splicing31-33, nuclear export33,34, localization5,6,8,16,20,28,35-40 and mRNA decay34,41-43 in single cells or in developing organisms using a variety of imaging approaches. It has also been used to quantify expression and localization of long non-coding mRNAs44,45, to visualize genomic loci in fixed cells46 and to identify sites of active mRNA translation11. The mRNA staining protocol described here is compatible with a range of imaging modalities, including conventional widefield epifluorescence and confocal microscopy as well as structured illumination microscopy (SIM)5,11,12, a super-resolution imaging method. Our image analysis protocol can be used in diverse cell types, tissues and organisms5,11,12 to quantify uniformly distributed5,11,12, localized5,12 and translating mRNAs11 (Fig. 5a-g). By using smFISH probes hybridizing to introns of nascent mRNAs, our smFISH protocol also enables quantification of transcriptional dynamics by direct visualization of active transcription sites within individual nuclei throughout Drosophila development (Fig. 5h-k).
Figure 5. smFISH enables spatial and quantitative characterization of gene expression in the fly tissue.
(a-c) Images of gcl (red) localized at the posterior pole of an embryo forming crescents surrounding the nuclei (labeled with DAPI, blue) of the newly formed primordial germ cells during the 13th nuclear cycle of the embryonic development. The inset marked with a red box in b highlights un-localized, somatic gcl mRNA, further magnified and contrast adjusted in c. (d) Heat map of gcl mRNA demonstrates that outside of the posterior pole, the majority of gcl transcripts are found as single mRNAs. Only approximately 3% of maternally-deposited gcl mRNA is localized to germ granules found at posterior pole of an embryo5 (Fig. 2c, 5e) where it groups within a diffraction limited volume and forms homotypic clusters composed of multiple gcl mRNA molecules5. To create a heat map, the average integrated fluorescent intensity of a single gcl was first determined (see Fig. 3, 4a) after which this value was used to calibrate the intensities of clustered gcl spots at the posterior pole. These had higher fluorescent intensities and therefore contained multiple gcl mRNAs in a diffraction-limited volume. During spot detection, Airlocalize determines the position of each single mRNA and mRNA cluster in 2D and 3D with sub-pixel resolution. These coordinates are then used to plot the spatial distribution individual mRNAs and clusters in the embryo. In all panels, three consecutive Z planes (z = 400 nm) were maximally projected and subsequently analyzed. All images were acquired with a widefield microscope in 3D and subsequently deconvolved using Huygens. (e) iSIM super-resolution imaging reveals a detailed spatial organization of mRNA-bound germ granules. Germ granule-localized gcl, nos and cycB mRNAs group to form homotypic mRNA clusters that inhabit different positions within germ granules. gcl clusters are located at the germ granule periphery while cycB clusters are located in the center of the granule. nos mRNA clusters are located midway between gcl and cycB clusters. Shown are images of granules found in the 10th nuclear cycle of early embryonic development. Images were acquired in 3D and afterwards deconvolved using Huygens. (f,g) Images demonstrating accumulation of oskar (osk) and nanos (nos) mRNA (red), respectively at the posterior pole of a developing oocyte through oogenesis. During late oogenesis, Oskar protein recruits Vasa protein (green) to the posterior pole to form germ plasm. Germ plasm was visualized with a Vasa transgene, tagged with a green fluorescent protein (GFP)5. First 10 oogenic stages are shown in f. In g, an oocyte found in late oogenic stage 14 is shown. Nuclei in f were stained with the DAPI stain (blue). Images in f and g (left panel in g) were acquired with a laser scanning confocal microscope. An image of the germ plasm shown under “MERGED” was acquired with an iSIM. (h-k) Intronic smFISH probes reveal active sites of transcription and discriminate between spliced mature transcripts and unspliced nascent transcripts. (h) A schematic depicting detection of gcl mRNA transcription site using smFISH probes hybridizing to either gcl introns (green dots) or gcl 3′UTR (red dots). (i) An image of a DAPI stained embryo to visualize nuclei. (j,k) Before being spliced, smFISH-hybridized introns (green) co-localize with the smFISH probes hybridizing to gcl 3′UTR (red) at the site of transcription (inset in k, yellow spot). Mature gcl mRNAs do not bind intronic probes (inset in k, arrowheads). Scale bar in e is 2.5 µm, in c, g (right panel under merged) j and k is 5 µm, 10 µm in a and b and 50 µm in f, g (left panel) and h.
The analysis protocol relies on our freely available spot detection software that localizes individual, fluorescently-labeled transcripts with precision far better than the diffraction limit in 2D or 3D datasets. With a build-in “Batch mode” option, Airlocalize makes it possible to process a large series of images in sequence, automating the analysis process. The algorithm also allows the analysis of 3D regions of interest (ROI) of a known volume; this speeds up the analysis when the signal is restricted to specific regions of large datasets, or when one uses a representative region as a proxy for the concentration of homogeneously distributed mRNAs across the entire embryo5. The absolute number of localized mRNAs clustered in a diffraction-limited spot (e.g. germ granule-localized mRNAs5) can also be determined by measuring the cumulative fluorescent intensity of an individual mRNA cluster and normalizing it by the fluorescent intensity value of a single mRNA. Concurrently with the quantification of the absolute number of transcripts, Airlocalize also determines the spatial position of clusters with a sub-pixel resolution. Specifically for mRNA clusters localized in Drosophila germ granules and imaged with iSIM5, the precision with which we are able to determine the position of smFISH-labeled mRNA clusters in granules ranges between 9.1±2.9 nm and 14.7±1.6 nm. As single mRNA counting techniques are entering the mainstream, this approachable user interface combined with the potential to scale up the analysis to larger datasets constitutes a unique resource for the smFISH community.
Limitations
When quantifying uniformly dispersed, un-localized mRNAs, our quantitative smFISH performs exceptionally well unless the abundance of transcripts exceeds the concentration at which the individual transcripts are no longer discernible as diffraction-limited spots (see “Anticipated results”). For example, we have experimentally determined that in wild type (WT) Drosophila embryos and within the resolution of iSIM, this limit is reached at approximately 4nM concentration (~ 17 million copies per WT embryo)5. Additionally, smFISH is not a whole-transcriptome approach. Rather, the number of mRNAs that can be simultaneously imaged is limited by the number of spectrally-distinct dyes that covalently label the probes as well as by the number of filters, detectors and laser lines accommodated by the microscope (typically up to 3). This limit can be extended by using a defined color combination of spectrally distinct smFISH probes hybridizing to a specific target mRNA (spectral barcoding using smFISH or sequential hybridization)47-50, to characterize up to tens of distinct mRNAs simultaneously. Recently, a highly multiplexed smFISH imaging method called MERFISH (multiplexed error-robust fluorescence in situ hybridization) was developed that enables simultaneous visualization, counting and cellular localization of 140 different mRNAs28. This is a true high throughput approach that will find numerous applications in cultured cells as well as in tissues.
As with any resolution-enhancing implementation of SIM, achieving the full resolution gain (typically two-fold better than diffraction-limited) is dependent on appropriate deconvolution. Such deconvolution depends critically on the nature of the algorithm and point-spread-function (PSF, e.g. experimental or theoretical). We have obtained good results with an assumed Gaussian PSF and the Richardson-Lucy deconvolution algorithm51,52 (as well as the commercially supported Huygens software), as these choices describe the iSIM PSF sufficiently. However, we note that refractive index mismatch, mounting media, and the details of the microscope directly influence the PSF – and thus we urge the reader to verify that the assumed PSF indeed matches the experimentally measured PSF before attempting deconvolution. Suboptimal deconvolution can lead to image artifacts and degrade spatial resolution/localization precision of this method.
Finally, we note that optical aberrations and the working distance of the objective lens limit the effective imaging depth when using iSIM for this application. We recommend taking care to match the refractive index of oil, the NA of the objective, and the mounting medium to limit spherical aberration, which would otherwise lower the effective signal-to-noise and spatial resolution. When using a high-index mounting medium and high NA oil immersion lens, the working distance of the objective lens also limits the depth of smFISH – we conducted all experiments up to 40 µm from the coverslip when using an 1.45 NA oil immersion lens.
Comparison to alternative methods
An alternative to single mRNA detection using smFISH probes described here are molecular beacons. These are fluorescently labeled probes that fluoresce only upon hybridization to their target mRNA53,54. Molecular beacons are typically short and single-stranded DNA sequences that contain a stem-loop structure. The loop portion of the molecular beacon contains a sequence perfectly complementary to a target mRNA. The stem, which is only present when the molecular beacon is not bound to its target mRNA, is formed upon annealing of complementary sequences in the probe flanking the loop. A fluorophore and a quencher are covalently coupled to each end of the molecular beacon. In the absence of hybridization, the formation of the stem-loop brings the fluorophore and the quencher in close proximity thereby quenching the fluorescence of the fluorophore. This is a particular advantage of this approach since it eliminates fluorescence originating from non-specific probe binding and thereby increases the accuracy of detecting bona fide mRNA targets. When injected into live Drosophila oocytes, molecular beacons also enable quantification of mRNAs in live tissue54,55.
Since smFISH is carried out in fixed tissue, it typically only provides a snapshot of an mRNA life-cycle. To detect individual mRNAs with a detailed temporal and spatial resolution in living tissue one can genetically tag mRNAs such that they will bind fluorescent proteins to allow single molecule detection. Currently, two such systems are most commonly used: the MS2 and the PP7 system. They are based on the MS2- and PP7-bacteriophage derived nucleic sequences, respectively56,57. These sequences are repeated up to 24 times and typically inserted into the 3′ untranslated region (UTR) of a gene. Once the mRNA is transcribed, these sequences form stem-loops that enable binding of the MS2 or PP7 bacteriophage capsid proteins, tagged with a fluorescent protein such as GFP. Because a single transcript is tagged with multiple stem-loops, which will each bind a fluorescently-tagged capsid protein, the mature mRNA is strongly fluorescently labeled. This multiplication of fluorescent signal on a single mRNA enables single mRNA detection in single living cells as well as in live Drosophila tissue8,11,57-63. Development of additional tags such as U1A and λN further enables a possibility to visualize multiple mRNAs in live tissue concurrently11,56,57,64. For some mRNAs, genetic tagging of the 3′UTR of the gene with 24 MS2 or PP7 repeats seem not to affect the life-cycle of the mRNA12,57, while for others the insertion of repeats affects mRNA stability of the target mRNA65. Thus, before employing MS2- or PP7-like strategies to visualize mRNA, care must be taken to ascertain whether this genetic tagging affects the mRNAs life cycle.
Experimental design
smFISH probes
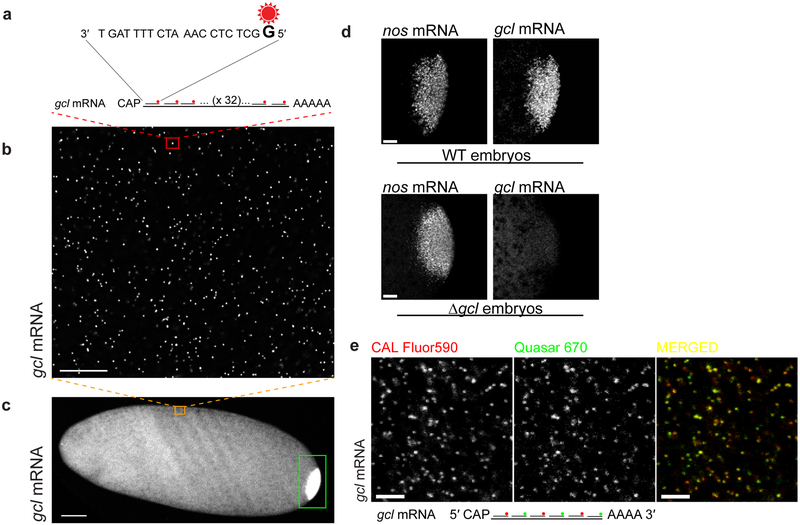
This protocol utilizes commercially available Stellaris® FISH probes to detect individual mRNA molecule in embryos. It follows a strategy introduced by Raj et al.2, which was later commercialized by Biosearch Technologies, Inc. In short, a typical smFISH probe is designed as a 20 nucleotide long DNA primer with a GC composition of approximately 50 per cent such that the minimal distance among neighboring probes is two nucleotides. Each probe is covalently conjugated to a single fluorophore via the 5′-most nucleotide and HPLC purified to remove free dye and unconjugated probes. This purification step is important as it ensures that the uncoupled dye does not contribute to the autofluorescent background during hybridization (non-specific sticking of unconjugated dye in the sample), while the unlabeled probes, which will also bind to the target mRNA but won’t fluoresce, do not reduce the overall fluorescence intensity of a smFISH-hybridized mRNA as well as increase the heterogeneity in the fluorescent intensity among individual mRNA. To improve the specificity of probe binding, stringent masking levels can be implemented to design organism specific probes and to avoid problematic RNA sequences (see www.biosearchtech.com/stellarisdesigner). Hybridization of target mRNAs with smFISH probes is carried out in the presence of 10% formamide and 2XSSC at 37ºC, conditions that are optimized to allow optimal hybridization of a 20 nucleotide long probe with a 50% GC composition. Changes in the length and GC composition of the probe, changes in the composition of the hybridization mix as well as in the hybridization temperature can affect the efficiency of smFISH probe binding to the target mRNA (see troubleshooting section “No or low smFISH design” and section “Parameters affecting single molecule detection, Image background”). smFISH probes are purchased as a mix where each probe mix contains up to 48 individual smFISH probes targeting different positions along the target mRNA (Fig. 2a-c). We most often use probes conjugated to CAL Fluor590, Quasar 570 or Quasar 670. Probes in each probe set are mixed in an equimolar ratio. Applying a mix of smFISH probes that anneal to different positions on the target mRNA strongly amplifies the signal-to-noise S/N ratio and therefore detection sensitivity during single-molecule counting. Typically, more probes per target mRNA are desirable to detect single mRNAs in the embryo however, one can reliably detect individual transcripts with less than 48 probes. We have successfully quantified single pgc mRNAs hybridized with 17 smFISH probes conjugated in the lab with Alexa488 dye and imaged with iSIM5. Similarly, gcl mRNAs hybridized with 32 Quasar 670-labeled smFISH probes, imaged on the widefield microscope followed by deconvolution, produced adequate amount of fluorescent signal to reliably quantify this mRNA in the early embryo (Fig. 4b,c).
Figure 2. Detection of single, smFISH-hybridized mRNAs in Drosophila.
(a-c) (a) Schematic of a germ-cell-less (gcl) mRNA hybridized with 32 fluorescently-labeled, commercially available smFISH probes. (b) gcl mRNA, hybridized with 32 smFISH probes, appears as a bright fluorescent spot (red box) against a uniform autofluorescent embryo background. (c) Uniformly dispersed, single gcl mRNAs located ventrally (orange box) in the Drosophila embryo. Nearly 3 per cent of gcl localize to embryo’s posterior5 (green box). (d) Hybridization of smFISH probes to their target RNA is specific; fluorescent signal cannot be detected if the target RNA is not expressed. In the absence of gcl mRNA expression (in embryos laid by mothers mutated for gcl gene expression (Δgcl embryos)), gcl smFISH signal cannot be detected, while the fluorescence of a control nanos (nos) mRNA, also enriched at the posterior pole, remains unaffected. (e) gcl hybridized with interchanging spectrally distinct CAL Fluor590 and Quasar 670 probes. The two fluorescent signals overlap indicating a bona fide detection of single mRNA molecules. Scale bar in b and e is 5 µm, in d is 10 µm and in c is 50 µm. Images in b and c were acquired with a widefield epifluorescence microscope and in d and e with a laser scanning confocal microscope.
iSIM
Since mRNAs hybridized with smFISH probes appear as diffraction limited spots, the ability to resolve features at or below the diffraction limit (‘super-resolution’) is helpful when attempting to count spots, particularly for spots in close (< 250 nm) proximity. Among the plethora of available super-resolution microscopy techniques, linear structured illumination microscopy66 (SIM) stands out due to its high speed, compatibility with standard fluorescent probes, and relatively low dose (102 – 106 lower than other methods, implying far less photobleaching). In SIM, higher spatial resolution (usually two-fold improvement over the diffraction limit) is obtained via multiplication of excitation and emission point spread functions. In practice, this is achieved by exciting the sample with a series of sharp illumination patterns, recording one or more diffraction limited images, and mathematically processing these images. In this protocol, we use a variant of SIM, ‘instant’ structured illumination microscopy (iSIM)9. iSIM enables much faster imaging (by a factor of 10–100) than conventional SIM because only a single image is recorded and most of the requisite image processing occurs during image formation (so less digital post-processing is needed). Additionally, and importantly for Drosophila samples, iSIM uses a pinhole array to block out-of-focus fluorescence, thus improving signal-to-noise and background rejection in these thick, densely labeled samples relative to conventional SIM.
Spot detection algorithm Airlocalize
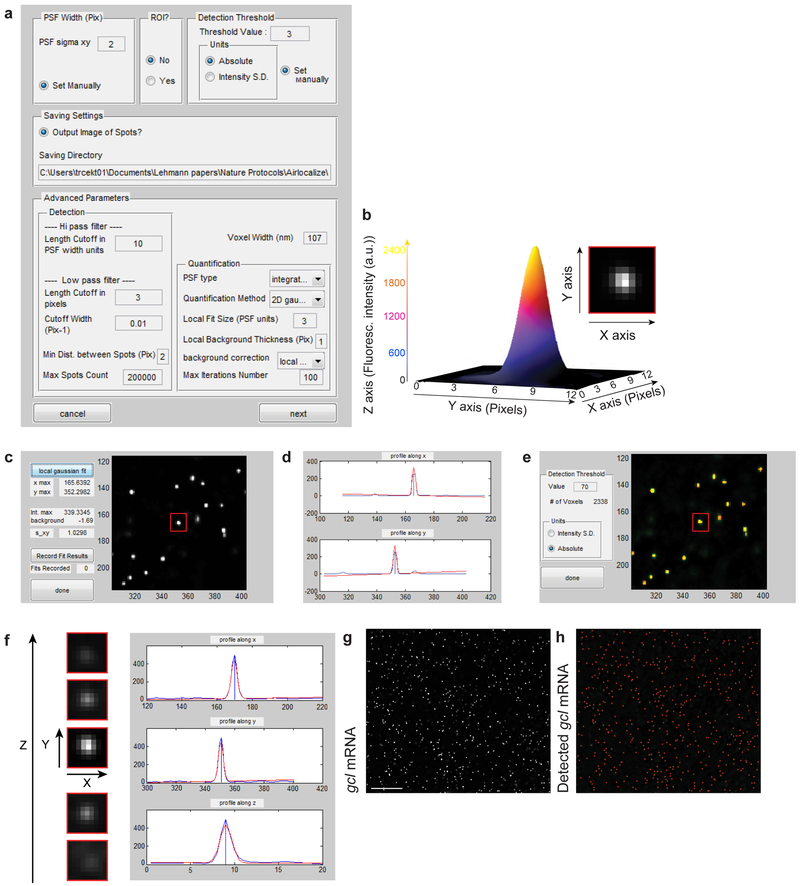
mRNAs hybridized with smFISH probes appear as bright fluorescent spots that are either uniformly dispersed through the embryo or localized within a sub-embryonic region. To automate the spot detection, an algorithm is employed that is capable of detecting individual spots and measuring the number, positions, and intensities of these fluorescently-labeled mRNAs in a 2D or 3D image. We use an algorithm called Airlocalize, which was developed in the MatLab programming language (MathWorks)12. An image of a fluorescently-labeled mRNA corresponds to a diffraction-limited image of a sub-resolution object (object with spatial extent less than the resolution of the light microscope). Thus, the size of an mRNA labeled with smFISH probes is determined by the optical properties of the microscope (known as the point spread function (PSF)) and not the physical size of the mRNA molecule. The spot detection algorithm uses a 3D Gaussian kernel to find the center and intensity of each spot (the kernel is calculated based on the average PSF shape)12. The robustness of spot identification and its localization against high autofluorescent background (for example created by neighboring fluorescent particles in a crowded environment) is enhanced by an affine local background subtraction prior to applying the Gaussian kernel. Detection and quantification of smFISH-labeled mRNAs proceeds through the following steps (Fig. 3): 1) spatial bandpass filter is implemented to identify candidate spots for further analysis, 2) local background surrounding the fluorescent spots is subtracted from the integral fluorescent intensity of a spot to remove any residual offset or unevenness in the image, 3) 2-dimensional (for 2D images) or 3-dimensional (for 3D images) Gaussian mask fitting of each fluorescent spot is implemented to find the center and the intensity of each spot, and 4) culling of spots based on duplication and/or an intensity threshold. Spot intensities and positions are written to a data file for further analysis (Fig. 3,4).
Figure 3. Workflow of the single mRNA molecule detection using Airlocalize.
(a) Defining the microscope parameters for single mRNA detection. (b) A heat map demonstrating the spatial distribution of the fluorescence intensity of a single smFISH-labeled gcl mRNA, reported in arbitrary units (a.u.). The inset demonstrates a single plane close-up of a gcl mRNA hybridized with 32 smFISH probes acquired on a widefield epifluorescence microscope. (c,d) Determining the two dimensional (2D) Intensity profile of a smFISH-labeled gcl mRNA. An example of a single fluorescently labeled mRNA (c, red box) and its Intensity profile are shown. Blue line in d indicates the profile of a fluorescent mRNA along the X and Y axis, red line is the Gaussian fit to the PSF. (e) Determining the fluorescent intensity threshold for mRNA detection. Green spots are smFISH labeled gcl mRNAs. Red spots are gcl mRNAs that have fluorescent intensity larger than the specified detection threshold intensity. Yellow indicates co-localization between smFISH-labeled gcl and thresholded gcl fluorescent signal. (f) Determining the Intensity profile of an mRNA in 3D. Blue line indicates the Intensity of a fluorescent mRNA along the X, Y and Z axis, red line is the Gaussian fit to the 3D profile. (g,h) Images showing smFISH-labeled gcl mRNAs found in the middle of the embryo (see Fig. 2b) before (g) and after (h) spot detection using Airlocalize. In h, detected single gcl mRNAs are marked as red spots. Scale bar in g is 5 µm.
Parameters affecting single molecule detection
a). Fluorescent intensity of labeled mRNAs
Typically, fluorescent intensity is the critical parameter for differentiating specific signal from background originating from non-specific probe binding and thus care must be taken in any algorithm to ensure the best possible recovery of the spot intensity from the image. A number of controls described in the “Experimental design” and “Anticipated results” sections can be used to differentiate between specific and non-specific fluorescent signal during the spot detection step.
Often, mRNAs are grouped to form a cluster in a smaller volume such that individual smFISH-labeled mRNAs cannot be individually resolved5. To determine the absolute number of mRNAs grouped within a smaller volume (clustered mRNAs), an integrated fluorescent intensity of a spot is first determined which is then calibrated by the fluorescent intensity value of a single mRNA located outside of the cluster to obtain the absolute number of mRNA molecules located within the cluster (Fig. 5d). When mRNAs labeled with smFISH-probes are particularly heavily clustered in a small volume (as is the case with germ granule localized mRNAs5), the abundance of fluorescent signal emanating from the cluster can saturate the dynamic range of the camera or the detector of the microscope. For example, a typical CCD camera mounted on a widefield epifluorescence microscope does not provide enough dynamic range to concurrently image single and heavily clustered transcripts such as nanos mRNA. Because nanos forms clusters that can contain more than 45 mRNAs packed in a volume comparable to the resolution limit of the widefield epifluorescence microscope, the multitude of smFISH probes within a cluster produces enough signal to saturate the camera. Such images are not quantifiable. This problem can be mitigated by reducing the number of probes in the probe mix, while making sure that the single mRNAs are labeled with enough probes that they are still observable as single mRNAs (see above, section “Experimental design, smFISH probes”). Alternatively, a detector or the camera with increased dynamic range can be selected to better match the sample. For example, the ~30000:1 dynamic range of the scientific cMOS camera mounted on our iSIM (see section “Equipment setup, Microscope) reliably captures the full fluorescence intensity profile of a single and of clustered nanos transcripts without signal saturation and without the need to reduce the number of smFISH probes during hybridization.
b). Image background
There are two main sources of background that deteriorate image quality, reduce the S/N ratio and single molecule sensitivity in the fly embryo. First is a very high autofluorescence originating from the embryo’s yolk, particularly in the green and red channels. Second is the fluorescence originating from non-specific smFISH probe hybridization to cellular background. To overcome autofluorescent background and to achieve single-mRNA detection sensitivity, our protocol utilizes multiple short, singly-labeled smFISH probes to fluorescently label mRNAs in situ (Fig. 2a) rather than a one long in vitro-transcribed RNA probe labeled with multiple fluorescent dyes to enhance detection capability traditionally used in whole-mount in situs to detect transcripts in Drosophila13. This probe iteration strongly amplifies the fluorescent signal of labeled mRNAs, which afterwards appear as bright fluorescent spots against a uniform autofluorescent background (Fig. 2b). We find that reducing the autofluorescent signal is particularly challenging and efforts to reduce it result only in minor changes in its intensity. When the autofluorescent signal is particularly high, multiple smFISH probe mixes can be designed to hybridize to 5′ and 3′ untranslated regions (UTRs) as well as the open reading frame (ORF) to further augment the fluorescent intensity of the smFISH-hybridized mRNA relative to the autofluorescent background.
smFISH probes also non-specifically hybridize to cellular background16,17. The length and the GC content of the smFISH probes we use (see Reagents setup) is designed such that all the probes in a probe mix optimally hybridize to the target mRNA at 37°C in the presence of 10% formamide and 2XSSC. Care must be taken to prepare a pre-hybridization solution with 10% formamide and 2XSSC and that the hybridization of the sample with the probe mix will proceed at 37°C. Lower formamide concentration and lower hybridization temperature will encourage non-specific probe binding and increase background fluorescence. Higher formamide concentration and higher hybridization temperature will prevent hybridization of probe to target mRNA. Exposed to air, deionized formamide also quickly hydrolytically breaks down to formic acid and ammonium formate, and acidifies. Acidified formamide quenches (inhibits) fluorescence of certain organic dyes such as Quasar 670 during hybridization. To prevent acidification, formamide can be aliquoted and stored at −80 °C until use. Monitor the pH of the stored deionized formamide using a pH strip. pH between 7.5 and 8 indicates good quality deionized formamide.
Control experiments
A series of control experiments can be performed to assay the specificity of smFISH probe hybridization to target mRNA. First, the hybridization specificity can be monitored by perturbing gene expression of a target mRNA and determining whether changes have occurred in the expression pattern of a target mRNA. For example, deletion of a gene will result in the specific loss in smFISH fluorescence of a target mRNA (Fig. 2d). Similarly, gene expression of a target mRNA can be reduced using fly lines expressing interfering mRNAs specifically designed to down-regulate a target mRNA (available through the Bloomington Stock Center). In this case, one should expect a reduction in the number of fluorescently-labeled target mRNAs in response to RNAi treatment.
A good way of determining whether the observed fluorescent signal originates from an mRNA hybridized with smFISH probes rather than from non-specific background accumulation of smFISH probes is to co-label a target mRNA with two sets of spectrally-distinct smFISH probe sets (Fig. 2e). Here, the two signals should co-localize and quantification of the number of mRNA molecules detected in both channels should give similar numbers. Additionally, several sets of probes with increasing number of probes per set can be used to label a target mRNA. In this case, the average integrated fluorescence intensity of a single mRNA should increase but the absolute number of detected mRNAs does not change (Fig. 4b,c).
Up to 200 different mRNAs are estimated to be localized to the posterior pole in an early Drosophila embryo67 where they instruct the formation of the fly abdomen68 and of germ cells69. We and others have characterized several maternally-deposited mRNAs for their ability to localize to the posterior pole and found that unlocalized mRNA counterparts found in the somatic regions of the embryo are not packaged into larger ribonucleoprotein particles (RNPs) but are mostly found as single mRNAs5,6. For these mRNAs, the average integrated fluorescent intensity of a single mRNA can be unambiguously determined by fitting the distribution of integrated fluorescent intensities of all identified fluorescent spots to a Gaussian curve and calculating the peak of the Gaussian fit (Fig. 4a). An exception is oskar (osk) mRNA. Approximately 50% of unlocalized osk mRNA is present in clusters containing more than one osk mRNA5,6,40,70 and the peak of the distribution of fluorescent intensity of spots containing a single osk mRNA is not easily identifiable. To troubleshoot quantification of clustered mRNAs such as osk, the mRNA expression levels determined for osk with smFISH can be compared with the relative mRNA expression levels determined by mRNA seq5 (freely available through FlyBase71). For example, we have previously quantified mRNA levels of ccr4, nanos (nos), germ-cell-less (gcl), polar granule component (pgc) and osk in early embryos laid by WT mothers and compared them to the relative mRNA levels determined for these mRNAs with mRNA-seq. The two measurements correlated exceptionally well (r=0.95) indicating that mRNA levels, including osk mRNA levels, determined by smFISH were in a good agreement with relative mRNA expression levels determined for these mRNAs by RNA-seq5. This control provided a strong indication that appropriate spot detection parameters were used during spot detection, which enabled identification of single smFISH-labeled osk mRNAs within a heterogeneous population of fluorescent spots containing multiple osk mRNA particles.
MATERIALS
Reagents
Wild type fruit flies (Bloomington Drosophila Stock Center, W1118)
Dry fine yeast (Lab Scientific Inc; FLY-8040–20F)
Apple juice plate (see reagent setup)
Clorox Ultra Germicidal Liquid Bleach (Fisher; 50371500)
Heptane (Fisher Chemical; O3008–4)
20% Paraformaldehyde (Electron Microscopy Sciences; 15713)
100 % Methanol (Fisher Chemical; A412–4)
Triton X-100 (Sigma; T8787–250 ml)
Tween-20 (Sigma; P2287–500 ml)
10XPBS (PBS - Phosphate-Buffered Saline (10X) pH 7.4) (Thermo Fisher Scientific; AM 9624)
Proteinase K (Sigma-Aldrich; 03 115879 001)
Glycine (Sigma-Aldrich; G8790–1kg)
20XSSC (Saline-Sodium Citrate buffer) (Thermo Fisher Scientific; AM 9770)
Deionized formamide (Amresco; 0606–100 ml)
Ribonucleoside Vanadyl Complex (VRC) (New England Biolabs; S1402S)
smFISH probe mix (Biosearch Technologies, Inc., commercially available and custom designed Stellaris® RNA FISH Probe Sets)
Sheared salmon sperm DNA (ThermoFisher Scientific; AM9680)
E.coli tRNA (Roche; 1010951001)
Bovine Serum Albumin (BSA)(Roche; 10711454001)
Dextran sulfate sodium salt (ACROS Organics; 433241000)
DAPI (4’,6-Diamidine-2’-phenylindole Dihydrochloride) (Roche; 10 236 276 001)
ProLong® Gold Antifade Reagent (Invitrogen (Molecular Probes); P36934)
Vectashield (Vector Labs Inc, H-1000)
Immersion oil (see Equipment setup, section Microscope)
Equipment:
25ºC incubator (ThermoFisher Scientific; Forma™ Enviromental Chambers)
Embryo collection cage (Large: 8.75cm x 14.8cm) (Flystuff.com; 59–101) (Fig. 1a). The cage accommodates a petri dish containing an apple juice plate, which is securely attached to the cage with a plastic cap. The top of the cage is covered with a stainless steel mesh to allow ample breathability while eliminating condensation. After each use, the cage must be cleaned with cold soapy water and must not clean with ethanol or other alcohols.
BD Falcon Petri Dishes, 60 × 15 mm, for small apple juice plates (Fisher Scientific; 25373–085)
Wooden tongue depressors (Fisher Scientific; 11–700-555)
Plastic scintillation vial, Wheaton™ 20mL HDPE Liquid Scintillation Vials with Polypropylene Caps (Fisher Scientific; 986700)
Nitex mesh (Dynamic Aqua-Supply Ltd.; NTX100)
Dumont Tweezers #5, 11cm, Straight, 0.1×0.06mm Tips, Dumostar; 500233)
Dissecting microscope (Zeiss; Stemi SV 11 equipped with a light source)
Squirt bottle (VWR; 16650–028)
Pyrex glass flask (Corning, Inc.; 1395–500)
Glass scintillation vial, wheaton glass 20 ml scintillation vial with poly sealed cap (Fisher; 03–341-25J)
Glass Pasteur Pipette (Fisher Scientific; 13–687-20C)
1.7 ml super clear microcentrifuge tubes (Crystalgen; L-2052)
Nanodrop (Nanodrop; The NanoDrop® ND-1000 UV/Vis Spectrophotometer)
pH-indicator strips pH 5.0–10.0 colorpHast (EMD; 9588)
Vortex (VWR; Standard Mini Vortex)
Falcon™ 15mL Conical Centrifuge Tubes (Fisher Scientific; 14–959-53A)
Tabletop centrifuge (Eppendorf, 5424)
37°C incubator (Fisher Scientific; Isotemp™ Standard lab incubator)
Microscope Slides (Fisher Scientific; 12–544-2)
Microscope Cover Slips 22X22–1.5 (Fisher Scientific; 12–544-D)
Wide-field epifluorescence, confocal or instant structured illumination microscope (iSIM), a super-resolution imaging system (see Equipment setup and Procedure Steps 27–28)
Huygens deconvolution algorithm (Scientific Volume Imaging)
Single molecule detection algorithm (Airlocalize© created in the laboratory, can be downloaded from the http://www.timotheelionnet.net/software/) (see Equipment setup and Procedure Steps 29–42)
SigmaPlot (Systat Software, Inc)
Figure 1. Collecting and fixing fly embryos for smFISH.
(a) Egg collection cage housing fruit flies and mounted with an apple juice plate containing yeast paste. (b) Apple juice plate with yeast paste and laid embryos. (c,d) Assembly of the egg collection basket.
Reagent Setup
CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare solutions used during smFISH hybridization.
Apple juice plates. This is a petri dish filled half way with agar mixed with apple juice. A dollop of yeast paste is placed in the middle of the apple juice plate to provide food for the flies while they lay eggs on the surface of the apple juice plate. For a detailed protocol on how to prepare apple juice plates, refer to “Drosophila. A laboratory Manual”72. Store at 4°C for one month.
Yeast paste. Dissolve 30 g of fine dry yeast powder in 20 ml of H2O. Stir frequently while incubating at R/T for several hours and adjusting with H2O until the yeast paste achieves the consistency similar to peanut butter. Store at 4°C for up to 2 months.
Fixative. To prepare 10 ml of fixative, combine 2 ml of 20% paraformaldehyde, 1 ml of 10XPBS and 7 ml of ddH2O. CRITICAL Prepare fresh. !CAUTION Paraformaldehyde is a toxic cross-linking agent. Wear protective gloves and handle in the fume hood. Discard according to the environmental health and safety instructions.
Saturated heptane Combine equal volumes of heptane and fixative (see above) in a glass flask. Close the flask and shake the mixture vigorously for 15 s. Let the solution settle into two phases. Repeat this mixing several times through the day and let settle over-night (O/N) to ensure that the heptane becomes saturated with paraformaldehyde. The saturated heptane is the upper phase in the 1:1 heptane: paraformaldehyde mixture. Wrap in aluminum foil and store at R/T for up to two months. !CAUTION Paraformaldehyde is a toxic cross-linking agent. Wear protective gloves and handle in the fume hood. Discard according to the environmental health and safety instructions.
Bleach solution. Prepare 50% bleach solution by mixing equal amounts of bleach and H2O. Store at R/T for up to two weeks.
PBT solution. To make 1000 ml of PBT, combine 100 ml of 10XPBS, 1 ml of Tween-20 and 899 ml of ddH2O. Mix thoroughly and keep at R/T for several months. CRITICAL Maintain RNase-free conditions and use RNase-free reagents to prepare PBT solution.
1XPBS solution. To make 1000 ml of 1XPBS, combine 100 ml of 10XPBS and 900 ml of ddH2O. Keep at R/T for several months. CRITICAL Maintain RNase-free conditions and use RNase-free reagents to prepare PBS solution.
3 μg/ml Proteinase K. To prepare 10 ml of working 3 μg/ml Proteinase K solution, combine 30 µl of 1 mg/ml Proteinase K with 10 ml of PBT. Stock 1 mg/ml Proteinase K solution is prepared by dissolving 10 mg of lyophilized powder in 10 ml of ddH2O and stored at −20°C for up to 12 months. Prepare working 3 μg/ml Proteinase K fresh. CRITICAL Maintain RNase-free conditions and use RNase-free reagents 3 μg/ml Proteinase K.
2 mg/ml glycine solution. Prepare 50 ml of 20 mg/ml glycine solution as a stock and keep at 4°C. Dilute 10 fold with ddH2O to obtain working 2 mg/ml glycine solution and store at R/T for several months. CRITICAL Maintain RNase-free conditions and use RNase-free reagents to prepare 2mg/ml glycine solution.
Pre-hybridization solution. 10 ml of pre-hybridization solution contains 1 ml of 100% deionized formamide (10% final), 1 ml of 20XSSC (2XSSC final) and 8 ml of ddH20. CRITICAL Prepare fresh. !CAUTION Formamide is toxic and should be handled with protective gloves in the hood and discarded according to the environmental health and safety instructions. CRITICAL Maintain RNase-free conditions and use RNase-free reagents to prepare the pre-hybridization mix.
Hybridization mix. To hybridize ~ 20 µl of embryos with a single smFISH probe mix, prepare 60 µl of hybridization mix. This mix will contain: 6 µl of 100% deionized formamide (10% final), 1 µl competitor DNA (see below), 2 µl of 40 ng/µl probe mix (see below), 30 µl of 20% dextran sulfate (10% final, see below), 5 µl BSA (2 mg/ml final), 5 µl 20XSSC (2XSSC final), 2.5 µl of 200 mM VRC (10 mM final) and 8.5 µl ddH2O. To hybridize multiple mRNA targets simultaneously using spectrally distinct probes, add 2 µl of 40 ng/µl probe mix for each target mRNA and adjust the total amount of ddH2O accordingly. CRITICAL Prepare fresh. Maintain RNase-free conditions and use RNase-free reagents to prepare the hybridization mix. !CAUTION Formamide is toxic and should be handled with protective gloves in the hood and discarded according to the environmental health and safety instructions.
smFISH probe mix. Commercially available Stellaris® FISH Probes were custom designed against gcl, nos and osk by utilizing the Stellaris® RNA FISH Probe Designer (Biosearch Technologies, Inc.) available online at www.biosearchtech.com/stellarisdesigner (Suppl. Table 1). Each smFISH probe mix is shipped as a dried set containing up to 48 individual smFISH probes hybridizing to different positions along the target mRNA (Fig. 2a-c). We most often use probes conjugated to CAL Fluor590, Quasar 570 or Quasar 670. Probes in each probe set are mixed in an equimolar ratio. Applying a mix of smFISH probes that anneal to different positions on the target mRNA strongly amplifies signal-to-noise (S/N) ratio and therefore detection sensitivity during single-molecule counting. The probe mix is resuspended in a TE buffer (see below) to final concentration of 40 ng/µl and stored at −20°C. CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare the smFISH probe mix.
TE buffer. To make 100 ml of TE buffer pH 8.0, combine 1 ml of 1 M Tris-HCl (pH 8.0) and 0.2 ml 0.5M EDTA and fill up with ddH2O to 100ml. Adjust pH to 8.0. Store at room temperature (R/T). CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare TE buffer.
Competitor DNA. To make 200 µl of 10 mg/ml of competitor DNA, combine 100 µl of 10 mg/ml sheared salmon sperm DNA with 100 µl of 10mg/ml E.coli tRNA. tRNA is prepared by dissolving 10 mg of powder in to 1ml of ddH2O. CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare Competitor DNA.
20% dextran sulfate. To prepare 50 ml of 20% dextran sulfate, combine 10 g of dextran sulfate with ddH2O to reach 50 ml. Store at 4°C for several months. CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare 20% dextran sulfate.
DAPI. Prepare a 0.5 µg/ml DAPI solution in 1XPBS. Stir for several hours at R/T in flask wrapped in aluminum foil. Store at 4°C for several months. CRITICAL Maintain DNase and RNase-free conditions and use DNase and RNase-free reagents to prepare DAPI.
Equipment Setup
Embryo collection basket.
To make an embryo collection basket, take a plastic scintillation vial and cut off the bottom of the vial (Fig. 1c,d). Remove the plastic cap and using a power tool or a flamed knife, cut a whole into the cap. Cut a circle the size of the plastic cap out of the nitex mesh and place it into the cap of the scintillation vial. Attach the cap with the mesh to the tube of the scintillation vial.
Microscope.
To acquire images with a widefield epifluorescence microscope, an API DeltaVision personal DV system equipped with the Olympus IX-71 inverted microscope, Nomarski differential interference contrast optics and Photometrics CoolSNAP HQ2 CCD camera with a pixel size of 6.45 µm (which meets the Nyquist criterion with a 60X magnification objective) was used. The microscope was guided by a SoftWorx suite. A xenon lamp and fiber optic module enabled a uniform illumination of the field. A high NA microscope objective such as Olympus PlanApo N 60x/1.42 oil objective is desirable to collect the maximum number of photons. This objective is optimized for imaging with # 1.5 microscope cover slip with a working distance of 0.15 mm. To minimize spherical aberration, it is important to use an immersion oil that closely matches the refractive index of the objective lens. We used an Olympus Type-F immersion oil (NA 1.516). The customized DeltaVision fluorescence filter set, which includes a DAPI filter, Tritc filter (for Quasar 570 and CAL Fluor590 detection), and Cy5 filter (for Quasar 670 detection), maximizes emission collection and minimizes the spectral overlap in case of multi-color smFISH.
To acquire images using a laser scanning confocal microscope, the Zeiss LSM780, AxioOberver microscope equipped with 32-channel GaAsp detector and two PMT detectors, a multiline 25mW argon laser, HeNe 633 laser, a 30 mW diode 405 laser, a DPSS 561–10 laser, a Plan-Apo40X/1.4 Oil DIC (microscope cover slip #1.5 and working distance of 0.13 mm) and EC Plan-Neofluar 10X/0.30 (microscope cover slip #1.5 and working distance of 5.2 mm) objectives was used. We used a Zeiss Immersol™ 518 F (NA 1.518) immersion oil. The microscope was guided by a Zeiss ZEN system.
To obtain super-resolution images of smFISH-labeled mRNAs, we employed the instant structured illumination microscope (iSIM)9,10, equipped with a 60X (although note that the full magnification from sample to cameras was ~117x, due to a 350 mm tube lens) NA1.45 oil objective (Olympus) (with a working distance of 0.10 mm) and accompanying oil (type DF, refractive index 1.515, Cargille), acquiring images in green or red channels with 488 nm and 561 nm excitation5,9 and filtering pump light with emission filters using a filter wheel (Sutter, FG-LB10-BIQ and FG-LB10-NW) and notch filters (Semrock, NF03–488E-25 and NF03–561E-25)9,10. These were placed immediately before the cameras to reject excitation laser light. Images were captured on a scientific complementary-metal-oxide-semiconductor (sCMOS) camera with 6.5 µm pixels (pco, edge 5.5) – the final imaging pixel size is 55.5 nm (=6.5 µm/117). Images are integrated from 5–2020 ms ‘sweeps’ of the galvanometric mirror that scans the structured illumination through the sample, for an average exposure time of 100–400 ms/image. Stacks were assembled by acquiring data with a piezoelectric stage (Applied Scientific Instrumentation, 200 µm axial travel) affixed to the microscope body – typical step sizes are 150 – 200 nm. We have previously fully described the alignment of the system10. Since its development in 2013, iSIM has become commercially available through VTi VisiTech International.
Deconvolution algorithm.
Microscopy images unavoidably suffer from artifacts like blurring, which arise due to the optics of the microscope, and noise, which is typically dominated by Poisson statistics arising from the recording of fluorescence emission. We use Huygens deconvolution software (version 16.05; Scientific Volume Imaging) for reducing these imaging artifacts. Specifically, we use the iterative Classic Maximum Likelihood Estimation (CMLE) algorithm in Huygens which is a restoration method that optimizes the likelihood of an estimate of an object given the measured image and the Point Spread Function (PSF). The object estimate is in the form of a regular 3D image. The likelihood in this procedure is computed by a Quality Criterion under the assumption that the photon noise is governed by Poisson statistics. For this reason it is well suited for increasing the S/N and effective resolution in an image, which facilitates the detection and counting of fluorescently labeled single mRNAs and importantly, markedly increases the precision with which individual molecules can be localized (Fig. 5a-d). Settings used with the CMLE deconvolution are 50 iterations, a quality threshold of 0.01, and a SNR value of 40. We typically use Huygens to deconvolve our data although a much simpler iterative Richardson-Lucy algorithm also worked well on our type of image data5.
Spot detection algorithm Airlocalize.
When imaged with an epifluorescent, confocal or structured illumination microscope, mRNA molecules hybridized with smFISH probes appear as bright fluorescent spots against a uniform auto-fluorescent background. A spot detection algorithm is employed to automate spot detection that detects and measures the number, positions, and intensities of individual fluorescently-labeled mRNAs in a 2D or 3D image. We use an algorithm called Airlocalize, which was developed in the MatLab programming language (MathWorks)12.
SigmaPlot.
SigmaPlot. Is a graphing and data analysis algorithm provided by the Systat Software, Inc. It is used to calculate the average fluorescent intensity of fluorescently labeled mRNAs by fitting the distribution of integrated fluorescent intensity of detected spots to a Gaussian curve, where the peak of the Gaussian curve determines the average fluorescent intensity of a single mRNA (Fig. 4a).
PROCEDURE
Collecting and fixing fly embryos for smFISH. TIMING up to 1 d.
-
1)
Place 200 to 400 flies with the 2:1 female-to-male ratio into an egg collection cage (Fig. 1a) mounted with an apple juice plate containing a dollop of yeast paste and pre-warmed to 25ºC (or the temperature specified by the experiment). ? TROUBLESHOOTING
-
2)
Place the egg collection bottles into a 25ºC incubator (or the temperature specified by the experiment) and allow flies to lay eggs onto the apple juice plate. ? TROUBLESHOOTING
-
3)
Remove the apple juice plate from the egg collection cage (Fig. 1b), scrape off the yeast paste with a wooden tongue depressor and remove trapped flies with the tweezers.
-
4)
Fill the apple juice plate half way with a bleach solution and incubate at R/T for up to 2 min to dechorionate the embryos. Immediately proceed to Step 5. CRITICAL STEP The potency of bleach can vary. Monitor dechorionation to prevent embryo damage by overexposure to bleach. ? TROUBLESHOOTING
-
5)
Pour the embryos with the bleach solution into the egg collection basket (Fig.1 c,d) and with a squirt bottle immediately wash the embryos with water. Wash the inner edges of the basket as well, so that all of the embryos assemble in the center of the nitex mesh. ? TROUBLESHOOTING
-
6)
Remove the mesh with dechorionated embryos from the egg collection basket and using tweezers place the mesh into the scintillation vial filled 5 ml of saturated heptane and 5 ml of fixative. !CAUTION Paraformaldehyde and heptane are toxic and should be handled with protective gloves in the fume hood and discarded according to the environmental and safety instructions.
-
7)
Shake the vial vigorously for 15 s. Let the vial stand at R/T for 20 min. The embryos will settle at the interface between the lower paraformaldehyde and upper heptane layer.
-
8)
Devitellinize the embryos by methanol cracking. Remove the bottom paraformaldehyde layer. Using a glass pipette, remove as much fixative as possible without removing the embryos. Add 5 ml of 100% methanol, cap the vial, shake vigorously for 15 s and let settle at R/T. Devitellinized embryos will settle at the bottom of the vial while vitellinized embryos will remain at the interphase between the two solutions. CRITICAL STEP Only devitellinized embryos will hybridize smFISH probes. !CAUTION Methanol is toxic when ingested or when in contact with skin or eyes. It is highly flammable. Handle with protective gloves in the fume hood and discard according to the environmental and safety instructions. ? TROUBLESHOOTING
-
9)
Using a glass Pasteur pipette or a cut-off pipette tip, collect devitellinized embryos and transfer them into a 1.7 ml tube. Wash three times with 100% methanol and store in 100% methanol at 4°C until further use. CRITICAL STEP. It is critical that a cut-off tip or a glass Pasteur pipette is used to preserve embryo’s integrity. !CAUTION Methanol is toxic when ingested or when in contact with skin or eyes. It is highly flammable. Handle with protective gloves in the fume hood and discard according to the environmental and safety instructions. ? TROUBLESHOOTING PAUSE POINT
Performing smFISH. TIMING 2 to 3 d
-
10)
Aliquot 20 µl of fixed embryos into a 1.7 ml tube. CRITICAL STEP Use a cut-off tip or a glass Pasteur pipette. ? TROUBLESHOOTING
-
11)
Wash the embryos with 1 ml of 100% methanol and incubate for 5 min at R/T. !CAUTION Methanol is toxic when ingested or when in contact with skin or eyes. It is highly flammable. Handle with protective gloves in fume hood and discard according to the environmental and safety instructions.
-
12)
Wash the embryos twice in a 1 ml of 1:1 mixture of 100% methanol:PBT, and incubate for 5 min at R/T each time. !CAUTION Methanol is toxic when ingested or when in contact with skin or eyes. It is highly flammable. Handle with protective gloves in the fume hood and discard according to the environmental and safety instructions.
-
13)
Wash the embryos twice in 1 ml of PBT and incubate at R/T for 5 min each time. PAUSE POINT. Once rehydrated, the embryos can be stored in PBT for several hours at R/T or several days at 4 °C before continuing with step 14.
-
14)
Postfix the embryos with 1 ml of fixative for 20 min at R/T. !CAUTION Paraformaldehyde is toxic and should be handled with protective gloves in the fume hood and discarded according to the environmental and safety instructions.
-
15)
Remove the fixative and wash 3 times with 1 ml of PBT at R/T, each time incubating for 2 min.
-
16)
Add 1 ml of Proteinase K and incubate at R/T for 13 minutes, inverting the tube frequently. Put the tube on ice and incubate for 1 h, inverting the tube every 15 min. CRITICAL STEP Proteinase K treatment is needed to ensure uniform smFISH probe penetration into tissue and to ensure efficient probe binding to the mRNA. It is particularly recommended when an mRNA is assumed to be tightly packed with proteins (masked)16 such that masking would preclude probe hybridization. The prolonged incubation on ice at low Proteinase K concentrations ensures slow and uniform penetration and activity of Proteinase K without over-digesting the tissue13. ? TROUBLESHOOTING
-
17)
Remove and inactivate Proteinase K by washing the embryos with 1 ml of glycine and incubating at R/T for 2 min. Repeat this step. CRITICAL STEP. Thoroughly inactivate and remove Proteinase K to prevent over-digestion of the sample. ? TROUBLESHOOTING
-
18)
Wash embryos twice with 1 ml of PBT, incubating each time for 2 min at R/T.
-
19)
Postfix the embryos with 1 ml of fixative for 20 min at R/T. !CAUTION Paraformaldehyde is toxic and should be handled with protective gloves in the fume hood and discarded according to the environmental and safety instructions.
-
20)
Wash the embryos five times in 1 ml of PBT at R/T, incubating the embryos each time for 2 min to remove all traces of the fixative.
-
21)
Completely remove the PBT and incubate the embryos at R/T for 10 min with 1 ml of pre-hybridization solution. !CAUTION Formamide is toxic and should be handled with protective gloves in the fume hood and discard according to the environmental and safety instructions. CRITICAL STEP. To ensure efficient probe hybridization and minimize non-specific probe binding, care must be taken to prepare a pre-hybridization solution with 10% formamide and 2XSSC and that the hybridization of the sample with the probe mix will proceed at 37°C.? TROUBLESHOOTING
-
22)
Remove the pre-hybridization solution and add 60 µl of hybridization solution containing the smFISH probe mix. Mix by gently flickering the tube with a finger and incubate protected from light at 37 °C O/N in an incubator. CRITICAL STEP Do not spin the sample with a centrifuge. This will disintegrate the embryos. It is preferable to incubate the embryos in an incubator to prevent excessive evaporation of water into the cap of the tube. This will ensure that the concentration of formamide does not change during hybridization. !CAUTION Formamide is toxic and should be handled with protective gloves in the hood and discarded according to the environmental health and safety instructions.? TROUBLESHOOTING
-
23)
Remove the hybridization solution and wash twice with 1 ml of pre-hybridization solution pre-warmed to 37°C, incubating each time for 15 min in a 37°C incubator. !CAUTION Formamide is toxic and should be handled with protective gloves in the hood and discard according to the environmental health and safety instructions.? TROUBLESHOOTING
-
24)
Remove the pre-hybridization solution and wash twice with 1 ml of 1X PBS, incubating each time for 1 h at R/T and protected from light. ? TROUBLESHOOTING
-
25)
Optional: to stain the nuclei, incubate with 1 ml of DAPI for 1 min at R/T and protected from light. Afterwards, remove the DAPI and incubate with 1 ml of 1X PBS for 5 min at RT. ? TROUBLESHOOTING
-
26)
Transfer the embryos onto the microscope slide using a pipette with a cut-off tip. Then remove as much PBS as possible without drying the embryos. Add a few drops of mounting medium on the embryos attached on the microscope slide and and cover with the microscope cover slip. Seal the edges of the microscope cover slip with nail polish or allow the mounting medium to cure following the manufacturer’s instructions. CRITICAL STEP Use a cut-off pipette tip or glass Pasteur pipette to transfer the embryos onto the microscope slide to prevent embryo disintegration. Do not dry the embryos as this increases autofluorescence. Certain commercially available mounting media, like Prolong Gold, need to cure for ~24 hours at RT in dark before the samples can be imaged. Water based mounting media like Vectashield do not need this step and samples can be imaged immediately after the microscope coverslip is sealed onto the microscope slides with a nail polish. After the mounting is completed, the slides can be stored at −20°C for several months before being imaged. ? TROUBLESHOOTING PAUSE POINT
Imaging smFISH-stained embryos TIMING ~1h-2h
-
27)
Embryos are imaged in 3D using a widefield epifluorescence microscope, laser scanning confocal microscope or iSIM (see Equipment Setup). Using a widefield epifluorescence microscope or a laser scanning confocal microscope, 3D images of embryos are acquired with a 200 – 400 nm Z step size, spanning 2.5–5.0 µm depth. When iSIM is used, 3D images of embryos are acquired with a finer Z step (due to the superior optical sectioning of this approach), typically with a 150 – 200 nm Z step size, spanning 2.5 – 5.0 µm depth. Whenever possible, exposure times, laser powers, number of scans and sweeps are chosen such that the range of fluorescent intensities of collected light occupies 90% of the dynamic range of the camera or the detector while also making sure to minimize bleaching of the sample. CRITICAL STEP Long exposure, high laser powers, multiple scans and sweeps can rapidly bleach the fluorescent signal. To increase the fluorescent intensity of a single mRNA during imaging, it is better to increase the number of probes hybridizing to an mRNA or choose probes coupled to a brighter and more photostable dye rather than increasing the exposure time or laser strength during imaging. ? TROUBLESHOOTING PAUSE POINT
Deconvolution of 3D image stacks TIMING ~1h-2h
-
28)
Deconvolve the 3D-acquired widefield epifluorescence or iSIM images. If images are acquired with a laser scanning confocal microscope, proceed to step 29. ? TROUBLESHOOTING PAUSE POINT
smFISH detection and counting using Airlocalize TIMING ~1 day (scales with proficiency of the user)
-
29)
Launch Airlocalize. CRITICAL STEP. Download the Airlocalize script together with the MCRInstaller, which allows one to run a MatLab algorithm without separately installing MatLab onto the computer.
-
30)
Select File Mode. Choose “2D single image file”. CRITICAL STEP As a proof of principle we provide here a step-by-step protocol of detection and counting of smFISH-labeled mRNAs in a single 2D image. However, a single 3D image file can be similarly analyzed. To further automate and speed up quantification, multiple 2D or 3D image files can be analyzed in a batch mode option. ? TROUBLESHOOTING
-
31)
Select Detection and Quantification parameters (Fig. 3a). Define the spatial characteristics of the point-spread-function (PSF), whether a region of interest (ROI) or an entire image is analyzed, and define how the detection threshold is determined. ? TROUBLESHOOTING
-
32)
Select the “Output Image of Spots” option if an image of detected smFISH-labeled mRNAs is desired (Fig. 3g,h).
-
33)
The Advanced Parameters section consists of software parameters, which typically do not need to be modified; the defaults are well-adapted to most single molecule detection applications. These parameters include: the maximum number of spots to consider in the image, the minimum distance allowed between spots (spots closer than this distance will be reduced to a single spot), the shape of the bandpass filter (High pass Length Cutoff, Low pass Length Cutoff, Cutoff Width), the PSF model used (PSF type, default: Gaussian Kernel Integrated over the pixel/voxel), the fitting algorithm (Quantification Method, default: Gaussian Mask), the size of the region around each spot used to compute position and intensity (Local Fit Size), the radius surrounding the fitting region used to estimate the background (Local background thickness), the algorithm used to compute the background value (default: local plane), the maximum number of iterations allowed for the mask to converge. Once all the parameters are defined, press “Next”.
-
34)
Define the shape of a 3D Gaussian mask. This mask will be used to quantify the integrated fluorescent intensity of smFISH-labeled mRNAs (Fig. 3b). Scroll over the image located in the left panel and choose a representative fluorescent spot of the smFISH-labeled mRNA. Find the center of the spot and left click to select the spot (a close-up of the image is located in the bottom right panel, which helps selecting the center of the fluorescent spot) (Fig. 3c). A 2D fluorescent profile of the spot will appear in the upper right two panels (Fig. 3d) demonstrating the distribution of the fluorescent intensity of the spot (Y axis; arbitrary units, a.u.) along the x (X axis in pixels; upper panel) and along the y (X axis in pixels; lower panel) in an image. ? TROUBLESHOOTING
-
35)
Click “local Gaussian fit” (Fig. 3c) to calculate the shape of the 3D Gaussian mask overlaying the fluorescent spot (Fig. 3d).
-
36)
Click “Record Fit Results”. CRITICAL STEP A good PSF is critical in making sure that the Gaussian mask will optimally fit the fluorescently-labeled mRNAs, calculate their integrated fluorescent intensity and determine the center of mass of the smFISH-labeled mRNA with a sub-pixel resolution. Therefore, it is important that the fit (red curve) deviates minimally from the measured data (blue curve) and thus appropriately describes the PSF of the fluorescent spot. ? TROUBLESHOOTING
-
37)
Repeat steps 34 to 36 five to 10 times to obtain an average 3D Gaussian mask, which will be used to detect and quantify the fluorescent intensity of smFISH-labeled mRNAs in subsequent steps. Click “Done”. ? TROUBLESHOOTING
-
38)
Define the fluorescent intensity threshold for detection of smFISH-labeled mRNAs (Fig 3e). The pixels whose fluorescent intensity is above the specified threshold are marked in red, pixels of the input image are marked in green and co-localized pixels are yellow. ? TROUBLESHOOTING
-
39)
Chose “Done”. Airlocalize provides three files important for further analysis and store them into the folder where the analyzed image originates from. The first is a TIF file corresponding to the “Output Image of Spots”. This is an image, in which the X,Y and Z positions of the centers of mass (the peak of fluorescent intensity) of all detected fluorescent spots are marked in red (Fig. 3g,h). The second is a localization (.loc or .loc3 for 3D images, ascii text) file that lists the spatial and quantitative information of all detected fluorescent spots. On this list, each spot is listed in a row, its position with a sub-pixel resolution specified (X,Y,Z in units of pixels in columns A B and C respectively) and its integrated fluorescence intensity (a.u.) encompassed by the 3D Gaussian mask provided (column D). The third, (.par, ascii text) file lists all the parameters specified during the analysis.
-
40)
Plot the distribution of integrated fluorescent intensities of all detected mRNAs as a histogram. To calculate the average fluorescent intensity of a single mRNA, fit the data (black dots) to a Gaussian function (red curve) (Fig. 4a) using a curve-fitting algorithm such as SigmaPlot. The peak of the Gaussian fit determines the average florescent intensity of a single smFISH-labeled mRNA. ? TROUBLESHOOTING
-
41)
To determine the absolute number of mRNAs within a cluster, the cumulative fluorescent intensity of each fluorescent cluster is normalized against the average fluorescent intensity of a single mRNA determined by the Gaussian fit (Fig. 5a-d). Clusters with fluorescent intensities ranging from 0.5≤X<1.5 are considered to contain 1 smFISH-labeled mRNA, those with fluorescent intensities ranging from 1.5≤X<2.5 are considered to contain 2 smFISH-labeled mRNAs, those with fluorescent intensities ranging from 2.5≤X<3.5 are considered to contain 3 smFISH-labeled mRNAs and so on (Fig. 5d). ? TROUBLESHOOTING
-
42)
When analyzing multiple images acquired in the same condition, it is crucial to ensure consistency of the detection parameters. The number of spots detected as well as their intensity can be sensitive to the choice of the PSF size and the threshold. To ensure consistent analysis, the PSF size and threshold can be set manually on an initial pilot image; the values can then be extracted from the resulting .par file (listed under “sigma_xy”, “sigma_z” for 3D images, and “thresh.level”) and typed in the corresponding dialog boxes (Fig. 3a) when analyzing subsequent images or launching a batch analysis. The “Set Manually” buttons need to be unchecked in this case.
? TROUBLESHOOTING
We have provided a list of solutions of how to troubleshoot detection and quantification of smFISH-hybridized mRNAs in Drosophila embryo in Table 1.
Table 1:
Troubleshooting table.
| STEP | PROBLEM | POSSIBLE REASON | SOLUTION |
|---|---|---|---|
| 2 | Flies lay few eggs. | a) Not enough flies in the cage. | a) This can be fly line dependent since some flies are not as fecund. Add more flies to the cage. |
| b) Apple juice plate are cold. | b) Apple juice plates must be pre-warmed to 25°C (or the temperature at which the eggs are collected) since the cold shock reduces egg-laying. | ||
| c) No surface for flies to lay eggs. | c) Place the yeast paste in the middle of the plate to allow the flies to lay eggs away from food or reduce the number of flies. Add filter paper to cage, so flies do not get stuck on plate. | ||
| d) Flies are too old. | d) Use flies that are at the peak of their fecundity (3 to 7 days old flies). | ||
| e) Flies are not acclimated to the apple juice plate. | e) Acclimate flies to apple juice plates by keeping them in an egg collection cage for one day before collecting eggs. | ||
| f) Flies are not well fed. | f) Feed the flies well with yeast paste to stimulate egg production. | ||
| g) Eggs are not collected at the appropriate time of the day. | g) Flies grow on a 12 h light cycle. Flies lay best towards dusk and worst at dawn. Collect later in the day or use an incubator with a controlled 12 h light cycle. | ||
| 4, 5, 9, 10, 16, 17, 22, 26, 27 | Embryos disintegrate or have impaired morphology. | a) Too much mechanical force applied when transferring the embryos. | a) Use only cut-off pipette tips or glass Pasteur pipettes to prevent shearing the embryos. Do not spin embryos with a centrifuge. This will disintegrate the embryos particularly after they have been treated with Proteinase K and formamide. |
| b) Bleach times are too long or the potency of bleach is too high. | b) As a rule of thumb, the embryos are dechorionated after they detach from an apple juice plate and start floating in the bleach solution. Monitor the efficiency of dechorionation under the dissecting microscope. When the dorsal appendages have dissolved in 80% of the embryos (~2 min), remove the bleach and wash the embryos with water. | ||
| c) Embryos are over-digested. | c) The duration of Proteinase K treatment is too long or the concentration of Proteinase K is too high. Adjust the time ond concentration of Proteinase K treatment. | ||
| d) Embryos are insufficiently fixed. | d) Increase the duration of fixation with fixative to 45 min. Autofluorescent background does not increase significantly with longer fixation times. When fixing for 20 min with 4% paraformaldehyde, heptane must first be saturated with 4% paraformaldehyde to ensure even and fast fixation. Saturated heptane must be prepared at least one day before and frequently shaken throughout the day to promote saturation. | ||
| 16, 17, 26, 27 | Oocytes disintegrate or have impaired morphology. | a) Oocytes are over-digested. | a) Oocytes can be susceptible to Proteinase K overdigestion and can lose their morphology during treatment. The duration and the potency of the Proteinase K treatment can be reduced or omitted during smFISH staining of oocytes. |
| b) Too much mechanical force applied when transferring the oocytes. | b) Use only cut-off pipette tips or glass Pasteur pipettes to prevent shearing the oocytes. Do not spin oocytes with a centrifuge as they will disintegrate. | ||
| 8, 9 | Embryos did not devitellinize efficiently. | a) Younger embryos did not devitellinize. | a) Younger embryos found in nuclear cell cycles one to 10 do not devitellinize efficiently using methanol cracking. Try hand-devitellinization instead. |
| b) Embryos of various stages poorly devitellinize. | b) It is important to remove as much fixative as possible since it inhibits devitellinization of the embryos. When fixing larger amounts of embryos, bigger vials with bigger methanol:heptanol volumes are preferred. By increasing the methanol surface, the embryos will crack their vitelline membrane more readily. Remove the devitellinized embryos from the bottom of the vial and add 5 ml of fresh 100% methanol and repeat methanol cracking. | ||
| 9, 10, 26 | Embryos stick to Pasteur pipettes and pipette tips. | Embryos are resuspeded in 1XPBS. | Wet the glass Pasteur pipette or cutoff pipette tip with methanol or PBT before pipetting the embryos. Alcohols and detergents prevent embryos from sticking to glass and plastic. If embryos are sticking too much, PBS can be replaced with PBT. |
| 8, 9, 16, 21, 22, 27, 28, 29-42 | No or low smFISH signal. | a) The number of smFISH probes is insufficient to detect single mRNAs. | a) Design more smFISH probes against the target mRNA. The probes can cover 5′ and 3′ untranslated regions (UTRs) as well |
| b) Imaging of mRNAs too deep in tissue | b) The smFISH signal might be visible at the surface of the embryo but not deeper in tissue. This occurs due to photon loss when emitted light scatters during image capture. | ||
| c) Inappropriate formamide concentration. | c) Formamide concentration above 10% prevents probe binding. Adjust formamide concentration to the final concentration of 10%. It is preferable to incubate the embryos in an incubator to prevent excessive evaporation of water into the cap of the tube. This will ensure that the concentration of formamide does not change during hybridization which could increase formamide concentration and prevent probe efficient hybridization. | ||
| d) Inappropriate hybridization temperature. | d) Temperature during hybridization is above 37°C, which prevents probe binding. | ||
| e) Inappropriate formamide pH. | e) pH of formamide is too acidic – formamide not deionized. When exposed to air, formamide becomes acidified and quenches (inhibits) fluorescence of certain organic dyes such as Quasar 670. Aliquot fresh formamide into 1.7 ml tubes and store at −80 °C until use. Over time monitor the quality of the stored deionized formamide using a pH strip. pH between 7.5 and 8 indicates high quality deionized formamide. | ||
| f) smFISH probes did not penetrate into the embryo. | f) It is necessary to incubate embryos in 100% methanol prior to hybridization for at least a couple of hours to ensure that smFISH probes will penetrate into the embryo during hybridization. If methanol is avoided, 1XPBS that contains 1% Triton X-100 must be used instead to perforate embryos. PBT containing 0.1% Tween-20 does not have sufficient detergent strength to perforate the embryos. Alternatively, embryos are not devitellinized. Repeat methanol cracking. | ||
| g) Dyes coupled to smFISH probes are not a bright or photostable. | g) Choose smFISH probes coupled to a bright and photostable dye. We most often use probes labeled with CAL Fluor590, Quasar 570 and Quasar 670. Quasar 670 is bright however, when imaged with a laser scanning confocal microscope, Quasar 670 does not excite well and bleaches rapidly. It is also sensitive to the pH of formamide (see above). Quasar 570 is not as bright as Quasar 670 or CAL Fluor 590. | ||
| h) Proteinase K treatment did not work. | h) Prepare fresh Proteinase K solution. | ||
| i) DNases and RNases contaminated the smFISH probe mix and/or sample and degraded probes and/or mRNAs in the sample. | i) Although we have not experienced signal loss due to smFISH probe or mRNA degradation during our smFISH staining (smFISH probes are DNA primers which are fairly resistant to degradation, while mRNAs in the embryos are fixed prior to hybridization and samples treated with VRC RNase inhibitor during hybridization and both treatments stabilize mRNAs during hybridization), care must be taken to use DNase and RNase free reagents and laboratory practices. | ||
| 9, 25, 26, 27, 28, 29-42 | Autofluorescence is too high. | a) Embryos have been stored too long. | a) Avoid using embryos that have been stored for too long as prolonged storage can increase autofluorescence. |
| b) Embryos became dried. | b) Do not dry the embryos during smFISH labeling as this can lead to increased autofluorescence. | ||
| c) Staining with DAPI was excessive. | c) Reduce the time and the concentration of DNA staining with DAPI. DAPI has a broad emission spectrum that can cause bleadthrough of signal into other channels when images of multiple channels are captured simultaneously. | ||
| d) Embryo’s autofluorescence is too high. | d) Use TrueBlack Lipofuscin Autofluorescence Quencher (Biotium) to reduce the autofluorescent background. | ||
| e) Embryos were stored in 1XPBS. | e) Store embryos in 100% methanol rather than 1XPBS or PBT. Methanol perforates the embryos and washes some autofluorescence away. | ||
| 24, 26, 27, 29-42 | Fluorescent signal bleaches. | a) Inappropriate mounting medium was used. | a) Use appropriate mounting medium that is an efficient scavenger of oxygen radicals, which cause bleaching of the fluorophores. We regularly use Vectashield and Prolong Gold. |
| b) Exposure time was too long and/or the power of the light or laser was too high, the number of sweeps was too high. | b) Reduce exposure time and/or the power of the light (widefield microscope), reduce laser power (confocal microscope, iSIM), reduce the number of scanned lines (confocal microscope), reduce the number of sweeps (iSIM) reduce number of optical Z sections (all microscopes). | ||
| c) Dyes coupled to smFISH probes are not a bright or photostable | c) Use a photostable fluorophore. CAL Fluor 590 is very photostable while Quasar 690 bleaches more rapidly, particularly when a laser scanning confocal microscope is used where a focused, high intensity laser line excites the fluorophores. To reduce the rate of photobleaching during imaging, add 1 mM Trolox antioxidant to the PBS washes (step 24). Curing mounting media might not diffuse deep into the embryo before they solidify to prevent photobleaching. By adding Trolox during PBS washes, the embryo is thoroughly perfused with the anti-bleaching reagent. | ||
| 22, 23, 24, 27, 29-42 | Background smFISH fluorescence is too high. | a) Too much smFISH probe used. | a) Reduce the amount of probe used. |
| b) Hybridization times were too long. | b) Implement shorter hybridization periods. | ||
| c) The frequency and the duration of washes was too short. | c) Increase the frequency and the duration of washes with the pre-hybridization solution and PBS. | ||
| d) The concentration of BSA or DNA competitor was too low. | d) Increase the concentration of BSA and competitor DNA. | ||
| e) Inappropriate formamide concentration. | e) Formamide concertation is below 10% and stimulates non-specific probe binding. | ||
| f) Inappropriate hybridization temperature. | f) Temperature during hybridization is below 37°C and stimulates nonspecific probe binding. | ||
| 27, 29-42 | Reduced resolution of iSIM. | a) Imaging too deep. | a) The optimal imaging depth of an iSIM microscope is sample dependent, but typically no better than 50-60 μm in transparent tissue (and in Drosophila tissue with a 60x 1.45 NA objective and imaging mRNA-bound germ granules located immediately beneath the surface of the embryo’s posterior pole, no more than 40 μm from the coverslip). Ensure that the working distance of the objective is sufficient to image at the chosen depth from the coverslip. |
| b) Wrong microscope cover slip. | b) It is important that the cover slip is # 1.5 (thickness 0.16 – 0.19 mm) | ||
| c) The refractive index of the mounting medium does not match the refractive index of glass, or objective. | c) We typically use Vectashield and Prolong Gold to mount our samples, it is therefore important to select an oil objective designed to work with similar refractive index oil. | ||
| 29-41 | Detection algorithm over/under counts. | a) Threshold parameters are not optimized. | a) Fluorescent intensities of the fluorescent mRNAs/probes change with the exposure time. For each imaged condition determine the threshold parameters separately. Threshold parameters will determine which fluorescent spots will be included as single mRNAs and optimizing threshold parameters is crucial to achieve successful single mRNA analysis. If the threshold is set too high, the algorithm will detect only the brightest fluorescent spots (those that contain multiple mRNAs in the same pixel), which will skew the center of the Gaussian fit towards higher fluorescent intensities. If it is set too low, pixels with a higher background fluorescence will also be detected skewing the Gaussian towards lower fluorescent intensities. It is crucial to use biological controls to confirm selectivity (using fluorescent probes that have no reactivity with any mRNA in the tissue, e.g. using a knock out sample Fig. 2d) and sensitivity (by comparing absolute counts to known abundance calculated by other means (see Anticipated results), or performing a double labeling experiment, Fig. 2e). Threshold can be set as absolute or as S.D. (standard deviation). Absolute Threshold (i.e. threshold in units of fluorescence counts) should be used when analyzing FISH data. It is crucial to analyze all images from the same dataset with the same threshold value to avoid biases. |
| b) Spots are difficult to observe when running Airlocalize. | b) Adjust minimal (min) and maximal (max) fluorescence intensity (Int.) settings located above the image panel. | ||
| 27 | Samples move during imaging. | A water based mounting medium was used. | Use a curing mounting medium. We use Prolong Gold. It crosslinks and fixes the embryos on the coverslip and the embryos do not move during the imaging. |
| 28 | Deconvolution artifacts. | a) Use of deconvolution. | a) 3D images acquired on a widefiled or iSIM are typically deconvolved before being analyzed to count single mRNAs. There are no problems intrinsic to widefield or iSIM deconvolution. 3D stacks acquired on a laser scanning confocal microscope are typically not deconvolved. If the fluorescent signals of smFISH-labeled mRNAs are strong enough, images can be analyzed without prior deconvolution. In this case, deconvolution only facilitates the detection and counting since it makes fluorescently labeled mRNAs brighter. However, due to an increase in contrast and signal assigned to the spot, properly applied deconvolution markedly increases the precision to which individual fluorescent molecules can be localized. Thus, when one wishes to determine where in a 3D space particular mRNAs lie with a sub-pixel resolution, deconvolution of image stacks prior to analysis is strongly advised. |
| b) Deconvolved images contain ringing artifacts (black regions in images or in axial reslices of volume) | b) Check that parameters of the experiment properly match the settings in the deconvolution software. Magnification and axial step size are particularly important. Also check that the axial step size is set to properly sample the axial extent of the PSF (ideally at Nyquist sampling or better). | ||
| c) Deconvolved images display degraded resolution relative to raw data. | c) Verify that the model of PSF used in the deconvolution software (e.g. Gaussian, theoretical, etc) well approximates the experimental PSF. If not, the model of the PSF needs to be revised for better agreement, or an experimental PSF should be used in the deconvolution software. If the latter option is pursued, ensure that the PSF is as noise-free as possible. | ||
| 31 | Analyzing an ROI with Airlocalize. | Image sets are too large to analyze as a whole. | To circumvent imaging and analyzing very large image data sets, a 3D ROI of a known volume can be used as a proxy for the concentration of unlocalized mRNAs across the entire embryo. Single mRNAs are then counted in the 3D image stack as described in the steps 29 through 41 and the concentration of mRNAs determined by extrapolating to the known embryo volume5. |
| 31 | Defining the PSF during spot detection. | Defining correct PSF for spot detection. | Visual aids (profile along x,y,z in Fig. 3d,f) can be checked to ensure that the software appropriately fits the intensity of the spots in the image. The program can fail to converge if the spots are extremely dim or the background very high. In this case, the PSF parameters can be computed using diffraction-limited beads imaged under identical conditions (objective, filter sets). |
TIMING
Step 1–9: Collecting and fixing fly embryos for smFISH: Up to 1 d
Step 10–26: Performing smFISH: 2 −3 d
Step 27: Imaging smFISH-stained embryos: 1–2 h
Step 28: Deconvolution of 3D image stacks: 1–2 h
Step 29–42: smFISH detection and counting using Airlocalize: 1 d (scales with the proficiency of the user)
ANTICIPATED RESULTS
Figures 2b,d,e, 3g,h 4b,c and 5 demonstrate anticipated results. Drosophila samples are stained, imaged, and afterwards 3D images are analyzed with Airlocalize to detect and count single smFISH-labeled mRNAs within a volume. Labeled mRNA molecules are observed as discrete bright fluorescent spots surrounded by a uniform autofluorescent background of lower intensity (Fig. 2b,e, 3g, 5c,d).The average integrated fluorescent intensity of a single smFISH-labeled mRNA scales linearly with the number of smFISH probes used in the hybridization mix (Fig. 4b,c), however the number of detected mRNAs per embryos does not change (Fig. 4b,c). This experiment provides a good control to determine whether the observed fluorescent signal originates from an mRNA hybridized with smFISH probes or from non-specific accumulation of smFISH probes. To bypass issues associated with analyzing large image data sets, a representative 3D ROI of a known volume can be analyzed that will serve as a proxy for the concentration of unlocalized mRNAs across the entire embryo. By determining the volume of an embryo73, the number of mRNA molecules, and hence the mRNA concentration, can be extrapolated from a 3D ROI onto the entire embryo5.
Because of its sensitivity, smFISH is a technique particularly suitable for detection and quantification of mRNAs expressed at lower concentrations. However, our smFISH protocol reaches its limit of detection at high mRNA concentrations, when the density of fluorescently-labeled mRNAs increases such that a spot detection algorithm can no longer reliably discern among the neighboring fluorescently-labeled mRNAs. An example of such mRNA is CycB. Using widefiled epifluorescence, confocal or structured illumination microscopy, CycB transcripts are not discernible as single molecules and the somatic regions of young WT embryos stain with uniform yet intense fluorescence5. We have experimentally determined that in WT Drosophila embryos and within the resolution of iSIM, quantification of mRNA levels with smFISH meets the limit when the concentration of the expressed mRNA target reaches approximately 4nM concentration (~ 17 million copies per WT embryo)5. Above this concentration, the algorithm begins undercounting the number of mRNA molecules expressed in the sample.
Once an average integrated fluorescent intensity of a single smFISH-labeled mRNA is determined (Fig. 4a), it is used to calibrate the fluorescent intensity of all fluorescent spots identified by the spot detection algorithm in an image and to determine the absolute number of mRNAs clustered in each fluorescent spot. mRNA clusters with fluorescent intensities ranging from 0.5≤X<1.5 are considered to contain 1 smFISH-labeled mRNA, those with fluorescent intensities ranging from 1.5≤X<2.5 are considered to contain 2 smFISH-labeled mRNAs, those with fluorescent intensities ranging from 2.5≤X<3.5 are considered to contain 3 smFISH-labeled mRNAs and so on (Fig. 5a-d)5.
A particular advantage of smFISH is that the position of a labeled mRNA in 2D and 3D can be determined with a sub-pixel resolution simply by measuring the center of mass of an mRNA hybridized with smFISH probes (Fig. 3b,c). The accuracy with which the center of mass can be measured is determined by the signal-to-noise ratio of an image. We have empirically determined that for mRNA clusters localized to Drosophila germ granules in an early embryo and imaged with iSIM5, the precision with which we are able to determine the position of smFISH-labeled mRNA clusters ranges between 9.1±2.9 nm and 14.7±1.6 nm. The sub-pixel position of each mRNA can be combined with the number of mRNAs located in each fluorescent spot to spatially quantify gene expression in the fly tissue in 3D. An example of such an analysis is provided in Fig. 5a-d, which shows the distribution of gcl mRNA at the posterior pole during the 13th nuclear cycle of the embryonic development in embryos laid by WT mothers. Approximately 3% of maternally-deposited gcl mRNA is localized to the posterior pole of an embryo where multiple gcl mRNAs group within a diffraction limited volume to form homotypic gcl mRNA clusters (Fig. 2c and 5a-e)5. The gcl mRNA heat map depicted in Fig. 5d demonstrates that outside of the posterior pole, the majority of gcl transcripts are found as single mRNAs and only organize into clusters once they are localized at the at the posterior pole, consistent with the literature5,6.
At the posterior pole, localized mRNAs interact with proteins assembled into membraneless organelles called germ granules5,6,67,74-76. smFISH can be combined with iSIM, a super-resolution microscope, to gain a detailed view of how localized mRNAs spatially organize within germ granules5. This approach revealed that in germ granules localized gcl, nos and cycB form homotypic mRNA clusters that are asymmetrically distributed within granules. Specifically, gcl clusters are located at the periphery of the granule, cycB clusters in the center of the granule while nos clusters occupy an intermediate space between gcl and cycB clusters5 (Fig. 5e). Such spatial sensitivity is not obtainable with conventional widefield epifluorescence or laser scanning confocal microscopy.
Aside from early embryogenesis, our staining, imaging and spot detection protocol can also be implemented to image mRNA localization during other Drosophila developmental stages such as oogenesis (Fig. 5f,g). Here, all steps 27 to 41 described for image capture and image analysis in the early embryo also apply to image and quantify fluorescently labeled mRNAs in the oocytes as well. Historically, detection of mRNAs during late fly oogenesis (stage 11 to 14) using whole-mount in situs has been challenging due to poor probe penetration into tissue during hybridization. Vitelline membrane, which forms by the end of oogenic stage 1077 reduces the accessibility of longer FISH probes used in whole-mount in situs in the oocyte and needs to be removed prior to staining15. This is typically achieved by rolling of fixed oocytes on a “rough” surface such as the frosted portion of the microscope slide15. This process however is laborious and can damage oocyte integrity if excessive mechanical force is applied during rolling. Short smFISH on the other hand easily penetrate oocyte tissue even without rolling and readily hybridize to target mRNAs during all oogenic stages6,8,19,20 (Fig. 5f,g). To prepare oocytes for smFISH, the ovaries of well-fed three to four day old female flies are dissected under a dissection microscope in cold 1XPBS. After several ovaries are collected, they are immediately resuspended in 1 ml of fixative containing 4% paraformaldehyde and 1XPBS dissolved in ddH20 and incubated for 20 min at RT. Unlike for embryos, saturated heptane is not needed to fix oocytes. Ovaries are then washed three to five times in 1XPBS, each time for 5 min at RT. Ovaries can then be further dissected to release individual ovarioles or oocytes from the ovary. Before smFISH can precede, ovaries, ovarioles or oocytes must be resuspended in 100% methanol and incubated overnight (or longer) to allow smFISH probe penetration into the oocytes6,8. Afterwards, the smFISH staining continues as described for embryos in this protocol starting with Step 11. However, oocytes are susceptible to Proteinase K over-digestion and can quickly lose their morphology during treatment. It is preferable therefore that the duration and the potency of the Proteinase K treatment in oocytes is reduced or the treatment is omitted during smFISH staining (omitting Steps 16 through 20).
The protocol described here can also be implemented to detect active sites of transcription in Drosophila embryos undergoing zygotic gene expression (Fig. 5h-k). It has been shown that active transcription sites are often bigger and brighter than single cytoplasmic transcripts or mature nuclear transcripts that have diffused away from the transcription site, indicating that multiple RNA II polymerases were actively transcribing a gene at the time of fixation and smFISH probe hybridization1,21,22,78,79. Therefore, several nascent mRNA chains are associated with an active site of transcription, which accommodate binding of multiple smFISH probes, thereby enhancing the fluorescent intensity of an active transcription site relative to a mature, single nucleoplasmic or cytoplasmic mRNA (Fig. 5h). Owning to the multitude of RNA II polymerases transcribing a gene at a given time, one can identify active sites of transcription simply by designing exonic mRNA probes and look for the brightest nuclear signal, as has been shown in Drosophila embryos, mammalian cells and tissue and in budding yeast4,7,21,22,41,50,79-82.
As the transcription proceeds, the intronic regions in the pre-mRNA are also transcribed and can be detected by designing smFISH probes hybridizing specifically to the intronic regions in the unspliced nascent mRNA. Because a typical Drosophila gene contains multiple introns and because an entire intron must be transcribed before it can be spliced, some proportion of the nascent mRNAs will contain introns and bind intronic smFISH probes. On the other hand, smFISH probes targeting mRNA exons will label both nascent as well as spliced transcripts. The latter are also smaller and have less fluorescence than active transcription sites (Fig. 5h,k, red dots)4,7,21,22,41,50,79,80,83. Bigger size, increased fluorescence and the ability to bind smFISH probes designed against the intronic pre-mRNA regions are suitable markers that distinguish active transcription sites from mature freely diffusing nuclear or cytoplasmic transcripts (Fig. 5h,k, green dots)1,31,32,50,83,84. It must be noted however that some transcripts are spliced once they diffuse away from the transcription site. These pre-mRNAs are easily identified since they retain their introns and thus their intronic and exonic smFISH signals co-localize in multiple discrete fluorescent spots of similar size and brightness in the nucleoplasm away from transcription site or in the cytoplasm31,32.
As the field is rapidly evolving, we put forward an up-to-date protocol, combined with thoroughly tested software and with cutting-edge microscopy that will be easy to use by a broad audience. Our spot detection step, arguably the most complex part of the protocol relies on a custom build and thoroughly tested algorithm with an approachable user interface combined with the potential to scale up the analysis to larger datasets constitutes a unique resource for the smFISH community. While we showcase the Drosophila embryo as an example in this protocol, the software and analysis techniques can be implemented to analysis diverse cell types, tissues and organisms. The ease of probe synthesis makes it possible to adapt the protocol to quantify transcription, translation or splicing events with subcellular resolution. We therefore expect that any lab interested in studying gene expression with modern, quantitative tools will find this protocol indispensable.
Supplementary Material
ACKNOWLEDGEMENTS
We thank NYULMC OCS Microscopy Core for providing the API DeltaVision personal DV microscope, particularly Yang Deng and Michael Cammer of the NYULMC OCS Microscopy Core for his help with the widefield epifluorescence microscope. We thank Vincent Schoonderwoert (Scientific Volume Imaging) for his help with Huygens deconvolution software. This work was supported by the Intramural Research Programs of the US National Institute of Biomedical Imaging and Bioengineering. TT is an HHMI fellow of the Jane Coffin Childs Memorial Fund. RL is an HHMI investigator. Funding for TL was provided by the Howard Hughes Medical Institute.
REFERENCES
- 1.Femino AM, Fay FS, Fogarty K & Singer RH Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A & Tyagi S Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5, 877–879, doi: 10.1038/nmeth.1253 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.biosearchtech.com/support/resources/stellaris-citation-center.
- 4.Little SC, Tikhonov M & Gregor T Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789–800, doi: 10.1016/j.cell.2013.07.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trcek T et al. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 6, 7962, doi: 10.1038/ncomms8962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little SC, Sinsimer KS, Lee JJ, Wieschaus EF & Gavis ER Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol 17, 558–568, doi: 10.1038/ncb3143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Sepulveda LA, Figard L, Sokac AM & Golding I Combining protein and mRNA quantification to decipher transcriptional regulation. Nat Methods 12, 739–742, doi: 10.1038/nmeth.3446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trovisco V et al. bicoid mRNA localises to the Drosophila oocyte anterior by random Dynein-mediated transport and anchoring. Elife 5, doi: 10.7554/eLife.17537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.York AG et al. Instant super-resolution imaging in live cells and embryos via analog image processing. Nat Methods 10, 1122–1126, doi: 10.1038/nmeth.2687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curd A et al. Construction of an instant structured illumination microscope. Methods 88, 37–47, doi: 10.1016/j.ymeth.2015.07.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead JM et al. Translation. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347, 1367–1671, doi: 10.1126/science.aaa3380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lionnet T et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nature methods 8, 165–170, doi: 10.1038/nmeth.1551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecuyer E, Necakov AS, Caceres L & Krause HM High-resolution fluorescent in situ hybridization of Drosophila embryos and tissues. CSH Protoc 2008, pdb prot5019, doi: 10.1101/pdb.prot5019 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Lecuyer E, Parthasarathy N & Krause HM Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol 420, 289–302, doi: 10.1007/978-1-59745-583-1_18 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Wilkie GS & Davis I Visualizing mRNA by in situ hybridization using ‘high resolution’ and sensitive tyramide signal amplification. Pages 94–97 (Academic Press, 1994). [Google Scholar]
- 16.Buxbaum AR, Wu B & Singer RH Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 343, 419–422, doi: 10.1126/science.1242939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trcek T et al. Single-mRNA counting using fluorescent in situ hybridization in budding yeast. Nat Protoc 7, 408–419, doi: 10.1038/nprot.2011.451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zenklusen D & Singer RH Analyzing Mrna Expression Using Single Mrna Resolution Fluorescent in Situ Hybridization. Method Enzymol 470, 641–659, doi: 10.1016/S0076-6879(10)70026-4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinsimer KS, Lee JJ, Thiberge SY & Gavis ER Germ plasm anchoring is a dynamic state that requires persistent trafficking. Cell Rep 5, 1169–1177, doi: 10.1016/j.celrep.2013.10.045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson A, Parton RM, Rabouille C, Weil TT & Davis I Localized Translation of gurken/TGF-alpha mRNA during Axis Specification Is Controlled by Access to Orb/CPEB on Processing Bodies. Cell Rep 14, 2451–2462, doi: 10.1016/j.celrep.2016.02.038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandhi SJ, Zenklusen D, Lionnet T & Singer RH Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol 18, 27–34, doi: 10.1038/nsmb.1934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenklusen D, Larson DR & Singer RH Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15, 1263–1271, doi: 10.1038/nsmb.1514 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nair G, Walton T, Murray JI & Raj A Gene transcription is coordinated with, but not dependent on, cell divisions during C. elegans embryonic fate specification. Development 140, 3385–3394, doi: 10.1242/dev.098012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castelnuovo M et al. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20, 851–858, doi: 10.1038/nsmb.2598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin JD & Cook PR Super-resolution measurement of distance between transcription sites using RNA FISH with intronic probes. Methods 98, 150–157, doi: 10.1016/j.ymeth.2015.11.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinner SO, Sepulveda LA, Xu H & Golding I Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat Protoc 8, 1100–1113, doi: 10.1038/nprot.2013.066 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itzkovitz S et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol 14, 106–114, doi: 10.1038/ncb2384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090, doi: 10.1126/science.aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniguchi Y et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538, doi: 10.1126/science.1188308 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battich N, Stoeger T & Pelkmans L Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nature methods 10, 1127–1133, doi: 10.1038/nmeth.2657 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Vargas DY et al. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147, 1054–1065, doi: 10.1016/j.cell.2011.10.024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong S et al. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol Cell 25, 559–573, doi: 10.1016/j.molcel.2007.01.012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyle NP & Ish-Horowicz D Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proc Natl Acad Sci U S A 110, E4316–4324, doi: 10.1073/pnas.1308811110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trcek T, Sato H, Singer RH & Maquat LE Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27, 541–551, doi: 10.1101/gad.209635.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batish M, van den Bogaard P, Kramer FR & Tyagi S Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A 109, 4645–4650, doi: 10.1073/pnas.1111226109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weil TT et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nature cell biology 14, 1305–1313, doi: 10.1038/ncb2627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colak D, Ji SJ, Porse BT & Jaffrey SR Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 153, 1252–1265, doi: 10.1016/j.cell.2013.04.056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park HY, Trcek T, Wells AL, Chao JA & Singer RH An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep 1, 179–184, doi: 10.1016/j.celrep.2011.12.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell PD, Chao JA, Singer RH & Marlow FL Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development 142, 1368–1374, doi: 10.1242/dev.118968 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Besse F, Lopez de Quinto S, Marchand V, Trucco A & Ephrussi A Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev 23, 195–207, doi: 10.1101/gad.505709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trcek T, Larson DR, Moldon A, Query CC & Singer RH Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell 147, 1484–1497, doi: 10.1016/j.cell.2011.11.051 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahar Halpern K & Itzkovitz S Single molecule approaches for quantifying transcription and degradation rates in intact mammalian tissues. Methods 98, 134–142, doi: 10.1016/j.ymeth.2015.11.015 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Messier V, Zenklusen D & Michnick SW A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell 153, 1080–1093, doi: 10.1016/j.cell.2013.04.035 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Cabili MN et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16, 20, doi: 10.1186/s13059-015-0586-4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Werven FJ et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150, 1170–1181, doi: 10.1016/j.cell.2012.06.049 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng W, Shi X, Tjian R, Lionnet T & Singer RH CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci U S A 112, 11870–11875, doi: 10.1073/pnas.1515692112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lubeck E & Cai L Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods 9, 743–748, doi: 10.1038/nmeth.2069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levsky JM, Shenoy SM, Pezo RC & Singer RH Single-cell gene expression profiling. Science 297, 836–840, doi: 10.1126/science.1072241 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M & Cai L Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 11, 360–361, doi: 10.1038/nmeth.2892 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levesque MJ & Raj A Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods 10, 246–248, doi: 10.1038/nmeth.2372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucy LB Iterative Technique for Rectification of Observed Distributions. Astron J 79, 745–754 (1974). [Google Scholar]
- 52.Richardson WH Bayesian-Based Iterative Method of Image Restoration. J Opt Soc Am 62, 55–59, doi:Doi 10.1364/Josa.62.000055 (1972). [DOI] [Google Scholar]
- 53.Tyagi S & Kramer FR Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol 14, 303–308, doi: 10.1038/nbt0396-303 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Bratu DP Molecular beacons light the way: Imaging native mRNAs in living cells. Discov Med 3, 44–47 (2003). [PubMed] [Google Scholar]
- 55.Mhlanga MM et al. In vivo colocalisation of oskar mRNA and trans-acting proteins revealed by quantitative imaging of the Drosophila oocyte. PLoS One 4, e6241, doi: 10.1371/journal.pone.0006241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chao JA, Patskovsky Y, Almo SC & Singer RH Structural basis for the coevolution of a viral RNA-protein complex. Nat Struct Mol Biol 15, 103–105, doi: 10.1038/nsmb1327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hocine S, Raymond P, Zenklusen D, Chao JA & Singer RH Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nature Methods 10, 119–121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abbaszadeh EK & Gavis ER Fixed and live visualization of RNAs in Drosophila oocytes and embryos. Methods 98, 34–41, doi: 10.1016/j.ymeth.2016.01.018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park HY, Buxbaum AR & Singer RH Single mRNA tracking in live cells. Methods Enzymol 472, 387–406, doi: 10.1016/S0076-6879(10)72003-6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buxbaum AR, Haimovich G & Singer RH In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16, 95–109, doi: 10.1038/nrm3918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forrest KM & Gavis ER Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13, 1159–1168 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Tyagi S Imaging intracellular RNA distribution and dynamics in living cells. Nature Methods 6, 331–338, doi: 10.1038/Nmeth.1321 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Bertrand E et al. Localization of ASH1 mRNA particles in living yeast. Molecular Cell 2, 437–445, doi:Doi 10.1016/S1097-2765(00)80143-4 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Daigle N & Ellenberg J lambda(N)-GFP: An RNA reporter system for live-cell imaging. Nature Methods 4, 633–636, doi: 10.1038/Nmeth1065 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Garcia JF & Parker R MS2 coat proteins bound to yeast mRNAs block 5 ‘ to 3 ‘ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. Rna 21, 1393–1395, doi: 10.1261/rna.051797.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gustafsson MG Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc 198, 82–87 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Frise E, Hammonds AS & Celniker SE Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol 6, 345, doi: 10.1038/msb.2009.102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gavis ER & Lehmann R Localization of nanos RNA controls embryonic polarity. Cell 71, 301–313 (1992). [DOI] [PubMed] [Google Scholar]
- 69.Arkov AL, Wang JY, Ramos A & Lehmann R The role of Tudor domains in germline development and polar granule architecture. Development 133, 4053–4062, doi: 10.1242/dev.02572 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Jambor H, Brunel C & Ephrussi A Dimerization of oskar 3’ UTRs promotes hitchhiking for RNA localization in the Drosophila oocyte. RNA 17, 2049–2057, doi: 10.1261/rna.2686411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.mod EC et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797, doi: 10.1126/science.1198374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashburner M Drosophila. (Cold Spring Harbor Laboratory, 1989). [Google Scholar]
- 73.Markow TA, Beall S & Matzkin LM Egg size, embryonic development time and ovoviviparity in Drosophila species. J Evol Biol 22, 430–434, doi: 10.1111/j.1420-9101.2008.01649.x (2009). [DOI] [PubMed] [Google Scholar]
- 74.Gao M & Arkov AL Next generation organelles: structure and role of germ granules in the germline. Mol Reprod Dev 80, 610–623, doi: 10.1002/mrd.22115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahowald AP Fine Structure of Pole Cells and Polar Granules in Drosophila Melanogaster. J Exp Zool 151, 201–215, doi:DOI 10.1002/jez.1401510302 (1962). [DOI] [Google Scholar]
- 76.Voronina E, Seydoux G, Sassone-Corsi P & Nagamori I RNA granules in germ cells. Cold Spring Harb Perspect Biol 3, doi: 10.1101/cshperspect.a002774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mahowald AP Ultrastructural observations on oogenesis in Drosophila. J Morphol 137, 29–48, doi: 10.1002/jmor.1051370103 (1972). [DOI] [PubMed] [Google Scholar]
- 78.Cho WK et al. RNA Polymerase II cluster dynamics predict mRNA output in living cells. Elife 5, doi: 10.7554/eLife.13617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raj A, Peskin CS, Tranchina D, Vargas DY & Tyagi S Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4, e309, doi: 10.1371/journal.pbio.0040309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagha M et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987, doi: 10.1016/j.cell.2013.04.045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levsky JM, Shenoy SM, Pezo RC & Singer RH Single-cell gene expression profiling. Science 297, 836–840, doi:DOI 10.1126/science.1072241 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Pezo RC et al. Single-cell transcription site activation predicts chemotherapy response in human colorectal tumors. Cancer Research 68, 4977–4982, doi: 10.1158/0008-5472.Can-07-6770 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bothma JP, Magliocco J & Levine M The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr Biol 21, 1571–1577, doi: 10.1016/j.cub.2011.08.019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Turris V, Nicholson P, Orozco RZ, Singer RH & Muhlemann O Cotranscriptional effect of a premature termination codon revealed by live-cell imaging. RNA 17, 2094–2107, doi: 10.1261/rna.02918111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.