Abstract
BACKGROUND
Elucidation of the genetic factors underlying chronic liver disease may reveal new therapeutic targets.
METHODS
We used exome sequence data and electronic health records from 46,544 participants in the DiscovEHR human genetics study to identify genetic variants associated with serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST). Variants that were replicated in three additional cohorts (12,527 persons) were evaluated for association with clinical diagnoses of chronic liver disease in DiscovEHR study participants and two independent cohorts (total of 37,173 persons) and with histopathological severity of liver disease in 2391 human liver samples.
RESULTS
A splice variant (rs72613567:TA) in HSD17B13, encoding the hepatic lipid droplet protein hydroxysteroid 17-beta dehydrogenase 13, was found to be associated with reduced levels of ALT (P=4.20×10−12) and AST (P=6.2×10−10). Among DiscovEHR study participants, this variant was found to be associated with a reduced risk of alcoholic liver disease (by 42% [95% confidence interval {CI}, 20 to 58] among heterozygotes and by 53% [95% CI, 3 to 77] among homozygotes), nonalcoholic liver disease (by 17% [95% CI, 8 to 25] among heterozygotes and by 30% [95% CI, 13 to 43] among homozygotes), alcoholic cirrhosis (by 42% [95% CI, 14 to 61] among heterozygotes and by 73% [95% CI, 15 to 91] among homozygotes), and nonalcoholic cirrhosis (by 26% [95% CI, 7 to 40] among heterozygotes and by 49% [95% CI, 15 to 69] among homozygotes). Associations were confirmed in two independent cohorts. The rs72613567:TA variant was associated with a reduced risk of nonalcoholic steatohepatitis, but not steatosis, in human liver samples. The rs72613567:TA variant mitigated liver injury associated with the risk-increasing PNPLA3 p.I148M allele and resulted in an unstable and truncated protein with reduced enzymatic activity.
CONCLUSIONS
A loss-of-function variant in HSD17B13 is associated with a reduced risk of chronic liver disease and of progression from steatosis to steatohepatitis. (Funded by Regeneron Pharmaceuticals and others.)
CHRONIC LIVER DISEASE AND CIRRHOSIS are leading causes of illness and death, accounting for 38,170 deaths (1.5% of total deaths) in 2014 in the United States.1 The most common causes of cirrhosis are alcoholic liver disease, chronic hepatitis C, and nonalcoholic fatty liver disease. The prevalence of nonalcoholic fatty liver disease is between 19% and 46%2–4 and is rising over time5 in conjunction with increasing rates of obesity, its primary risk factor.6
Genomewide association studies have identified sequence variations associated with an increased risk of chronic liver disease. The most robustly validated association is with a common missense variant in PNPLA3, encoding patatin-like phospholipase domain-containing 3 protein. This variant (rs738409, p.I148M) is associated with increased hepatic triglyceride levels7 and an increased risk of nonalcoholic steatohepatitis8,9 and cirrhosis.10 A missense variant in TM6SF2, encoding transmembrane 6 superfamily member 2, is associated with nonalcoholic fatty liver disease.11–13 The mechanisms underlying these associations have yet to be elucidated,14–17 and much of the genetic risk of chronic liver disease remains unexplained.
In this study, we used exome sequencing to identify variants associated with serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), markers of hepatocyte injury, using data from DiscovEHR, a cohort study that links exome sequence data to electronic health records (EHRs), and three additional studies. We then evaluated the associations between implicated genetic variants and chronic liver disease and the histopathological severity of liver disease.
METHODS
STUDY DESIGN AND PARTICIPANTS
We carried out tests of association using genomic DNA samples and data from six cohorts, including two DiscovEHR study populations from the MyCode Community Health Initiative of Geisinger Health System (GHS).18 The GHS discovery cohort consisted of 46,544 persons of European descent who were recruited from outpatient primary care and specialty clinics, and the GHS bariatric-surgery cohort consisted of 2644 additional persons of European descent who underwent bariatric surgery. Associations with ALT and AST levels were replicated in 1357 persons of European ancestry from the Dallas Heart Study (DHS) and 8526 persons of European ancestry from the Penn Medicine BioBank. Associations with chronic liver disease were evaluated in a total of 37,173 persons: 31,938 DiscovEHR study participants, and in replication studies, 517 case patients from the Dallas Liver Study (DLS) and 4279 controls from the DHS, as well as 205 case patients and 234 controls from the Dallas Pediatric Liver Study (DPLS). Full study descriptions and clinical phenotype and disease definitions are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org. The studies were approved by the institutional review boards at each participating site.
SAMPLE PREPARATION, SEQUENCING, AND GENOTYPING
DNA sample preparation and whole-exome sequencing for the participants in DiscovEHR, DHS, and Penn Medicine BioBank were performed at the Regeneron Genetics Center, as described elsewhere18 and in the Supplementary Appendix. HSD17B13 rs72613567 was genotyped by TaqMan assay (and verified by Sanger sequencing in five persons of each genotype: homozygous for risk or reference variant and heterozygous) in the DLS and DPLS cohorts.
EXOMEWIDE ASSOCIATION ANALYSIS OF LIVER ENZYMES AND PHENOTYPES OF CHRONIC LIVER DISEASE
We used linear mixed models to test biallelic variants meeting quality-control criteria for association with aminotransferase levels, as described in the Supplementary Appendix. For variants with exomewide significant associations (P<1×10−7) in the GHS discovery cohort, we performed association analyses and a meta-analysis, as described in the Supplementary Appendix, of the replication studies. For the test of replication, we used a Bonferroni significance threshold determined by the number of variants tested. We also carried out a metaanalysis of the discovery and replication studies.
We subsequently tested aminotransferase-associated variants for associations with phenotypes of chronic liver disease. (See the Methods section in the Supplementary Appendix.) We used a Bonferroni significance threshold determined by the number of variants and broad categories of chronic liver disease that were tested. We further tested replicated novel variants for association with histopathologically defined liver phenotypes from the GHS bariatric-surgery cohort. We also performed a phenomewide study of associations of replicated novel variants with 405 quantitative clinical measurements and 3168 clinical diagnoses, as described in the Supplementary Appendix.
RNA SEQUENCING STUDIES
RNA samples from human liver biopsies were prepared, pooled, and sequenced with the use of 75-bp paired-end sequencing on an Illumina HiSeq 2500 system, version 4. (For details on the preparation of RNA samples, see the Supplementary Appendix.)
IDENTIFICATION AND VALIDATION OF NOVEL HSD17B13 TRANSCRIPTS
Reads were mapped, and HSD17B13 transcripts were identified. Custom gene models were built to incorporate novel transcripts of HSD17B13, and transcript quantification was estimated by read alignment to the custom gene model. Novel transcripts were validated with the use of reverse-transcriptase–polymerase-chain-reaction assay and PacBio long-read sequencing. (See the Methods section in the Supplementary Appendix.)
SUBCELLULAR LOCALIZATION OF HSD17B13 ISOFORMS
HepG2 cells were infected with lentivirus carrying the HSD17B13 A and D transcripts, stable cell lines were selected, and HSD17B13 isoforms and lipid droplets were visualized with the use of immunofluorescence, as described in the Supplementary Appendix. Lipid-droplet isolation and characterization of subcellular localization of HSD17B13 were performed as described in the Supplementary Appendix.
IN VITRO AND CELLULAR CHARACTERIZATION OF HSD17B13 ENZYMATIC ACTIVITY
Recombinant human HSD17B13 protein was purified from Escherichia coli transformed with plasmid DNA harboring HSD17B13 transcript A or transcript D. Enzymatic activity was determined through luciferase-based measurement of NADH production, as described in the Supplementary Appendix. HEK293 cells overexpressing HSD17B13 transcript A, transcript D, or green fluorescent protein were used to investigate the activity of HSD17B13 against estradiol in a cell-based assay.
RESULTS
GENETIC ASSOCIATIONS WITH AMINOTRANSFERASE LEVELS
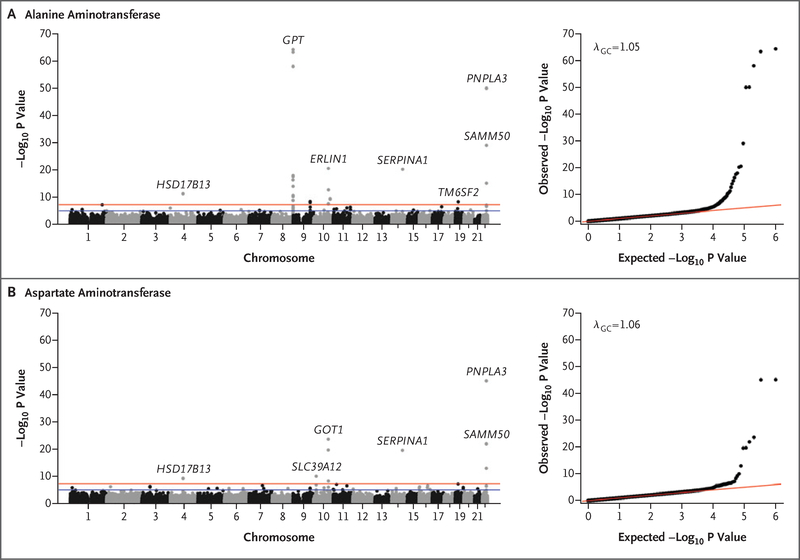
We tested 502,219 single genetic variants for association with serum levels of ALT or AST in 46,544 persons of European descent from the DiscovEHR study. (Basic demographic and clinical characteristics of the participants are shown in Table S1 in the Supplementary Appendix.) A total of 35 variants in 19 genes were found to be associated with ALT or AST levels at P<1.0×10−7 (Fig. 1, and Table S2 in the Supplementary Appendix). We performed replication studies in three cohorts of persons of European descent (Table S1 in the Supplementary Appendix). In a meta-analysis of the replication cohorts (12,527 persons), 13 variants in 9 genes were significantly associated with ALT or AST levels (Bonferroni significance threshold of P<1.43×10−3 for 35 variants tested) (Table S3 in the Supplementary Appendix). These included variants in genes previously reported to be associated with liver disease: PNPLA3,7 TM6SF2,11 SERPINA1,19 SAMM50,20 and ERLIN1.21 We also identified variants in genes not previously reported to be associated with liver disease: GPT and GOT1, the genes encoding ALT and AST, respectively, and SLC39A12, encoding solute carrier family 39 member 12.
Figure 1. Association of Single-Nucleotide Variants with Aminotransferase Levels in the GHS Discovery Cohort.
Each panel includes a Manhattan plot (left) and quantile–quantile plot (right). Variants that are indicated by gene name, including HSD17B13, remained significantly associated with levels of alanine aminotransferase or aspartate aminotransferase in a replication meta-analysis of three separate cohorts of persons of European ancestry (Table S3 in the Supplementary Appendix). GHS denotes Geisinger Health System.
We identified a reproducible association between a variant in HSD17B13, encoding hydroxysteroid 17-beta dehydrogenase 13, an uncharacterized member of the hydroxysteroid 17-beta dehydrogenase family, and decreased levels of ALT (P=4.2×10−12 in the discovery cohort and P=1.7×10−4 in the replication cohorts) and AST (P = 6.2×10−10 in the discovery cohort and P=1.7×10−4 in the replication cohorts) (Table S3 in the Supplementary Appendix). The associated variant, rs72613567, is an insertion of an adenine adjacent to the donor splice site of exon 6 (rs72613567:TA allele); it had an allele frequency of 26.0% in the GHS discovery cohort. Previously, Chambers et al. identified a nearby locus at 4q22 (rs6834314) that was associated with ALT levels22; an association of aminotransferase levels with rs72613567 has not heretofore been reported. HSD17B13 is 30 kb upstream of HSD17B11, another member of the same gene family. We did not observe exomewide significant associations between coding or splice variants in HSD17B11 and aminotransferase levels in the dis-covery cohort (Fig. S1 in the Supplementary Appendix) or in the meta-analysis of the discovery cohort and three replication cohorts. Furthermore, linkage disequilibrium of rs72613567 with variants in HSD17B11 was modest across all ancestry groups (r2<0.4 by Pearson’s correlation of genotypic allele counts with all ascertained variants in HSD17B11) (Fig. S2 in the Supplementary Appendix).
ASSOCIATION OF EXONIC VARIANTS WITH CHRONIC LIVER DISEASE
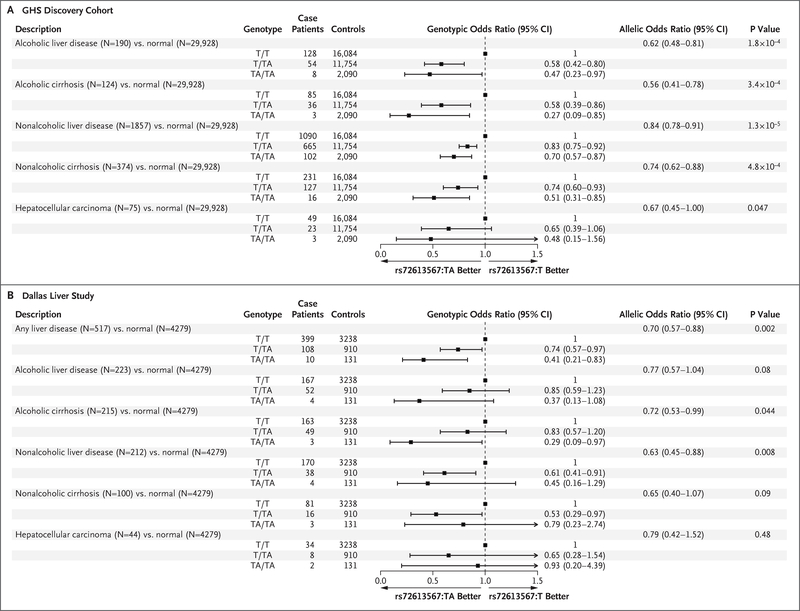
Next, we analyzed the relationship between the 13 aminotransferase-associated variants and alcoholic and nonalcoholic (nonviral) chronic liver diseases. Using a Bonferroni significance threshold of P<1.92×10−3 for the 13 variants and two broad categories of liver disease tested, we found significant associations between 6 variants in five genes (HSD17B13, SERPINA1, TM6SF2, PNPLA3, and SAMM50) and chronic liver disease (Table S4 in the Supplementary Appendix). In the discovery cohort, HSD17B13 rs72613567:TA was associated with lower odds of all categories of liver disease in an allele dose-dependent manner (Fig. 2A). This allele was associated with a reduced risk of alcoholic liver disease (by 42% [95% confidence interval {CI}, 20 to 58] among heterozygotes and by 53% [95% CI, 3 to 77] among homozygotes) and nonalcoholic liver disease (by 17% [95% CI, 8 to 25] among heterozygotes and by 30% [95% CI, 13 to 43] among homozygotes), as well as with a reduced risk of alcoholic cirrhosis (by 42% [95% CI, 14 to 61] among heterozygotes and by 73% [95% CI, 15 to 91] among homozygotes) and nonalcoholic cirrhosis (by 26% [95% CI, 7 to 40] among heterozygotes and by 49% [95% CI, 15 to 69] among homozygotes). HSD17B13 rs72613567:TA was also nominally associated with lower odds of hepatocellular carcinoma (P=0.047).
Figure 2. Associations of HSD17B13 rs72613567:TA with Phenotypes of Alcoholic and Nonalcoholic Liver Disease in the GHS Discovery Cohort and in the Dallas Liver Study.
In the GHS discovery cohort (Panel A), allelic odds ratios were calculated with the use of logistic regression, with adjustment for age, age squared, sex, body-mass index (BMI), and the first four principal components of ancestry. Genotypic odds ratios for reference allele homozygotes (T/T), heterozygotes (T/TA), and alternate allele homozygotes (TA/TA) are also shown. All reported P values correspond to the allelic model. In the Dallas Liver Study (Panel B), allelic odds ratios were calculated with the use of logistic regression, with adjustment for age, age squared, sex, BMI, and patient-reported ethnic group.
We tested these associations in the multiethnic DLS and DPLS cohorts (Table S5 in the Supplementary Appendix). In the DLS cohort, rs72613567:TA was associated with lower odds of any liver disease in an allele dose-dependent manner (Fig. 2B). Similar effects were observed across subtypes of liver disease, including alcoholic and nonalcoholic cirrhosis. In the DPLS cohort, the TA allele was associated with lower odds of pediatric liver disease (allelic odds ratio, 0.61; 95% CI, 0.37 to 0.99; P=0.046).
GENETIC INTERACTION BETWEEN PNPLA3 AND HSD17B13 VARIANTS
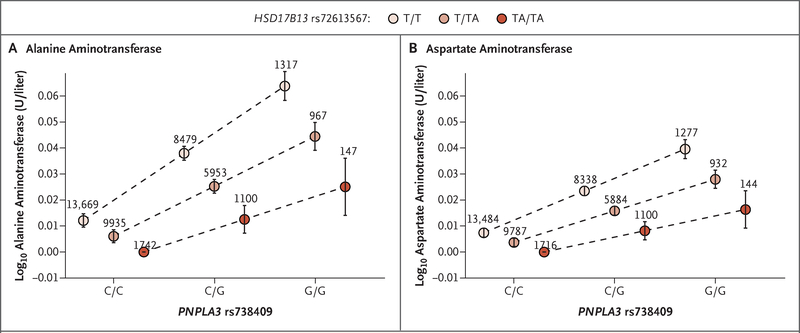
We wondered whether the implicated HSD17B13 allele modifies the risk of liver injury associated with PNPLA3 rs738409 (p.I148M) and so tested for interaction between the two variants in association with ALT level, AST level, and chronic liver disease in DiscovEHR. We observed nominally significant interactions in association analyses with ALT level (P=0.002 for interaction) and AST level (P=0.004 for interaction), such that each rs72613567:TA allele mitigated increases in aminotransferase levels associated with each PNPLA3 148M allele (Fig. 3, and Figs. S3 and S4 in the Supplementary Appendix). These associations were primarily observed in obese persons (Table S6 in the Supplementary Appendix). RNA sequencing- based expression analysis revealed that HSD17B13 rs72613567:TA was associated with decreased PNPLA3 messenger RNA (mRNA) expression in an allele dose-dependent manner (Fig. S5 in the Supplementary Appendix).
Figure 3. Association of HSD17B13 rs72613567 with Aminotransferase Levels in Persons with Each PNPLA3 p.I148M Genotype.
Effect estimates were calculated with the use of linear regression, with adjustment for age, age squared, sex, BMI, and the first four principal components of ancestry. PNPLA3 rs738409 genotypes on the x axis indicate reference allele homozygotes (C/C), heterozygotes (C/G), and alternate allele homozygotes (G/G). The rs738409:G allele corresponds to the p.I148M amino acid change. The P values for interaction between HSD17B13 rs72613567:TA and PNPLA3 rs738409:G (p.I148M) in association analyses of levels of alanine aminotransferase and aspartate aminotransferase were P=0.002 and P=0.004, respectively. The numbers above the circles indicate the number of persons with each genotype combination. I bars indicate 95% confidence intervals.
HSD17B13 AND LIVER PATHOLOGY
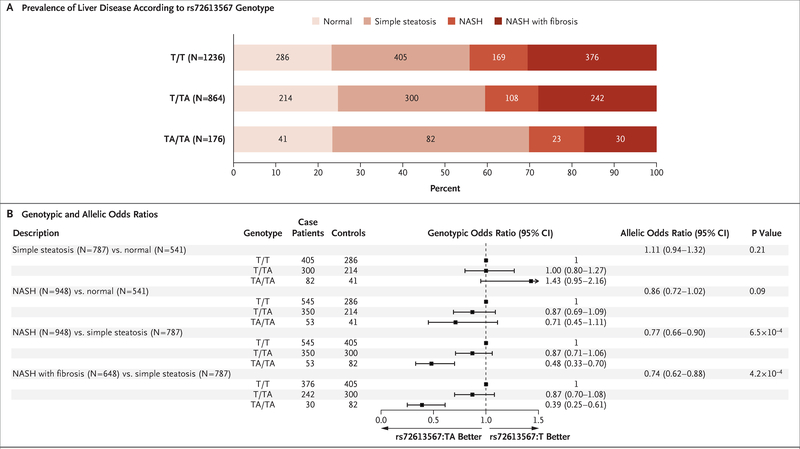
To understand the association between HSD17B13 rs72613567:TA and histopathological progression of simple steatosis to nonalcoholic steatohepatitis, we performed tests of association in 2391 persons with liver biopsies from the GHS bariatric-surgery cohort. The prevalence of normal liver did not appear to differ according to genotype (P=0.59 by chi-square test for trend in proportions), but the prevalence of nonalcoholic steatohepatitis decreased (P=7.3×10−4) and that of simple steatosis increased (P=0.002) with each rs72613567:TA allele (Fig. 4A). Among persons with steatosis, rs72613567:TA was associated with lower odds of nonalcoholic steatohepatitis (by 13% [95% CI, −6 to 29] among heterozygotes and by 52% [95% CI, 30 to 67] among homozygotes) and fibrosis (by 13% [95% CI, −8 to 30] among heterozygotes and by 61% [95% CI, 39 to 75] among homozygotes), as compared with simple steatosis (Fig. 4B).
Figure 4. Associations of HSD17B13 rs72613567:TA with Liver Pathology in Patients Undergoing Bariatric Surgery.
Panel A shows the prevalence of histopathologically characterized liver disease according to HSD17B13 rs72613567 genotype in 2391 persons with liver biopsies from the GHS bariatric-surgery cohort. Panel B shows associations of HSD17B13 rs72613567:TA with liver pathology in the GHS bariatric-surgery cohort, according to logistic regression with adjustment for age, age squared, sex, BMI, and the first four principal components of ancestry. NASH denotes nonalcoholic steatohepatitis
HSD17B13 AND CLINICAL QUANTITATIVE TRAITS AND DIAGNOSES
To more comprehensively examine the clinical consequences of the HSD17B13 splice variant, we performed a phenomewide study of associations of HSD17B13 rs72613567:TA. Using Bonferroni significance thresholds of P = 1.23×10−4 and P = 1.58×10−5 for associations with 405 clinical measurements and 3168 clinical diagnoses, respectively, from EHRs, we identified significant associations of HSD17B13 rs72613567:TA with higher platelet counts, in addition to the associations with liver aminotransferase levels (Table S7 in the Supplementary Appendix). There were no significant associations with clinical diagnoses other than chronic liver disease (Table S8 in the Supplementary Appendix).
EFFECT OF VARIANT ON HSD17B13 MRNA AND PROTEIN
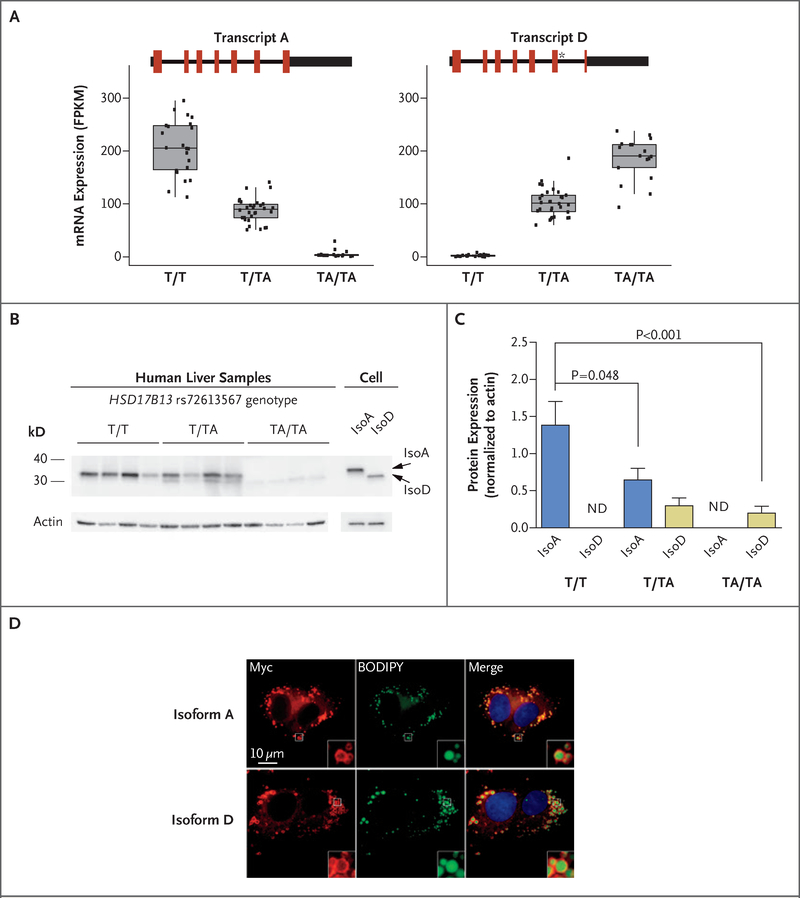
We next used RNA sequencing to examine the effect of HSD17B13 rs72613567:TA on transcript expression in human liver samples. We observed, in addition to two known HSD17B13 transcripts (A and B), two novel transcripts (Figs. S6 through S8 in the Supplementary Appendix): transcript C, lacking exon 6, and transcript D, containing an insertion of a guanine nucleotide at the 3′ end of exon 6, which is predicted to result in premature truncation of the protein. Levels of transcripts A and B decreased, whereas those of transcripts C and D increased, with each rs72613567:TA allele (Fig. 5A, and Fig. S6 in the Supplementary Appendix). Transcript A, encoding the full-length protein, was the predominant transcript in reference allele homozygotes (T/T), whereas transcript D, encoding the prematurely truncated protein, was the predominant transcript in alternate allele homozygotes (TA/TA). In tissue from human liver biopsies, isoform D protein was minimally present in heterozygotes and TA/TA homozygotes, and isoform A protein abundance was reduced in an allele dose-dependent manner (Fig. 5B and 5C). Overexpression of isoforms A or D in HEK293 or HepG2 cells indicated further reduction in abundance of isoform D protein, which suggests instability of the D protein isoform (Figs. S9 and S10 in the Supplementary Appendix). These data are consistent with HSD17B13 rs72613567 altering mRNA splicing, resulting in the synthesis of a truncated protein with substantially reduced abundance in human liver.
Figure 5. Expression and Subcellular Localization of a Novel HSD17B13 Transcript.
Panel A shows the expression of HSD17B13 ranscripts A and D in rs72613567 reference allele homozygotes (T/T), heterozygotes (T/TA), and alternate allele homozygotes (TA/TA). Coding regions are indicated in red, untranslated regions as thick black lines, and introns as thin black lines. The asterisk in transcript D indicates the A insertion from rs72613567. Box plots show the median and interquartile range (IQR) of fragments per kilobase of transcript per 1 million mapped reads (FPKM). The length of the whiskers is 1.5 times the IQR. Dots represent individual messenger RNA expression levels. Panel B shows the results of Western blot analysis of human liver and HEK293 cell samples. Human liver samples were from T/T, T/TA, and TA/TA carriers of the HSD17B13 rs72613567 splice variant. Cell samples were from HEK293 cells overexpressing nontagged HSD17B13 transcripts A and D. Panel C shows levels of HSD17B13 isoform A (isoA) and isoform D (IsoD) protein in human liver samples. ND denotes not determined. Panel D shows localization of HSD17B13 isoforms A and D. HepG2 cells stably overexpressing HSD17B13 transcripts A or D were labeled with boron-dipyrromethene (BODIPY) to show lipid droplets and anti-Myc to show HSD17B13 localization. All figures are magnified to the same extent. Insets represent 4× amplification of the original images.
EXPRESSION OF HSD17B13 IN HUMAN LIVER CELLS
HSD17B13 is expressed primarily in the liver,23 where it localizes to lipid droplets,24 which is consistent with a role in the pathogenesis of fatty liver disease. We evaluated HSD17B13 expression and localization in stable human liver cell lines expressing HSD17B13 transcript A or D. HSD17B13 isoforms A and D were mainly detected on membranes surrounding lipid droplets (Fig. 5D, and Figs. S10 and S11 in the Supplementary Appendix). No significant differences in intracellular triglyceride content were observed with oleic acid treatment of cell lines overexpressing HSD17B13 isoforms A or D (Fig. S10 in the Supplementary Appendix).
EFFECT OF RS72613567:TA ON HSD17B13 ACTIVITY
We evaluated the enzymatic activity of isoforms A and D in vitro using recombinant protein and nicotinamide adenosine dinucleotide as cofactor. We tested 265 unique putative substrates (Table S9 in the Supplementary Appendix) and identified steroid substrates and bioactive lipids (e.g., leukotriene B4) as enzymatic substrates of HSD17B13. Isoform D showed much less activity toward estradiol in vitro and in cell-based enzymatic con-version assays than isoform A (Fig. S12 in the Supplementary Appendix).
DISCUSSION
By linking large-scale exome sequencing to EHR- derived clinical phenotypes, we identified a novel association of a splice variant in HSD17B13 with decreased serum aminotransferase levels, as well as with a reduced risk of nonalcoholic and alcoholic forms of liver disease. These associations were observed consistently, in an allele dose-dependent manner, in four independent cohorts and across several categories of liver disease, including advanced cirrhotic forms of liver disease and hepatocellular carcinoma. The HSD17B13 rs72613567:TA allele was not associated with simple steatosis but was associated with a reduced risk of nonalcoholic steatohepatitis and fibrosis, findings that suggest that this variant allele protects against progression to more clinically advanced stages of chronic liver disease. The HSD17B13 rs72613567:TA allele also mitigated the risk of liver injury in persons who were genetically predisposed to steatotic liver disease by the PNPLA3 p.I148M variant and was associated with reduced PNPLA3 mRNA expression. The 434K allele of PNPLA3 has been reported to mitigate the effect of the PNPLA3 148M allele on chronic liver disease by reducing hepatic PNPLA3 mRNA and protein expression,25 thus providing precedent for modulation of PNPLA3 148M allele expression as a mechanism for modifying the risk of liver disease. This finding suggests an important subpopulation for therapeutic inhibition of HSD17B13 — persons heterozygous or homozygous for the PNPLA3 148M allele. In a phenomewide association study, HSD17B13 rs72613567:TA was significantly associated only with chronic liver disease and related clinical measurements (hepatic aminotransferase levels and platelet counts), findings that suggest that the clinical effects of the variant allele may be specific to chronic liver disease.
Other members of the hydroxysteroid 17-beta dehydrogenase family are involved in the metabolism of sex steroids and fatty acids,26 but little is known about the function of HSD17B13. Overexpression of human HSD17B13 was shown previously to increase lipogenesis in mouse liver and to increase the number and size of lipid droplets in cultured hepatocytes.24 An increase in hepatic expression of HSD17B13 protein has been observed in patients with fatty liver disease.24,27 Our data suggest that both HSD17B13 isoforms (encoded by the A and D transcripts) are expressed on the lipid droplet membrane but do not appear to affect intracellular neutral fat content, a finding consistent with a lack of observed association between HSD17B13 rs72613567:TA and simple steatosis in humans. Although we are unaware of physiologic substrates of HSD17B13, we found that the isoform encoded by the protective allele is catalytically defective against estradiol. We do not know whether any of the substrates that we tested are critical for liver disease, but HSD17B13 has enzymatic activity against several bioactive lipid species (e.g., leukotriene B4) that have been implicated in lipid-mediated inflammation.28 A genetic variant near MBOAT7, encoding an acyltransferase involved in remodeling of bioactive phospholipids, is associated with an increased risk of chronic liver disease, a finding that provides suggestive evidence for modulation of bioactive lipids as an important factor in the pathogenesis of chronic liver disease.29,30
The associations were observed in Hispanic Americans and Americans of European descent who had an elevated body-mass index; we do not know whether these findings are generalizable to other populations. HSD17B13 is in close proximity to HSD17B11, a member of the same gene family but with broader expression across tissues.26 We did not observe an association between variants in HSD17B11 and serum aminotransferase levels, but perhaps rs72613567 is in linkage disequilibrium with a functional variant in HSD17B11 that we did not capture in our sequence analysis. These limitations notwithstanding, our data support a role for HSD17B13 in the progression of liver disease from steatosis to later stages of nonalcoholic steatohepatitis, fibrosis, and cirrhosis.
Supplementary Material
Acknowledgments
Supported by Regeneron Pharmaceuticals. The Geisinger Health System bariatric-surgery biobank is partly supported by a grant (P30DK072488, to Drs. Carey, Still, and Mirshahi) that was awarded to the Mid-Atlantic Nutrition Obesity Research Center by the National Institutes of Health (NIH). The Dallas Heart Study is supported by the National Center for Advancing Translational Sciences of the NIH under award number UL1TR001105. Dr. Kozlitina is partially supported by this award (UL1TR001105) and by a Regeneron–University of Texas Southwestern sponsored-research agreement. The Penn Medicine BioBank is funded by the Perelman School of Medicine at the University of Pennsylvania and by a gift from the Smilow family. Dr. Carey is an author on behalf of the Geisinger–Regeneron DiscovEHR Collaboration.
We thank the participants from the MyCode Community Health Initiative for their permission to use their health and genomics information as part of the DiscovEHR study, and Weixing Shi of the Geisinger Health System for his contribution to the study.
APPENDIX
The authors’ full names and academic degrees are as follows: Noura S. Abul-Husn, M.D., Ph.D., Xiping Cheng, M.D., Ph.D., Alexander H. Li, Ph.D., Yurong Xin, Ph.D., Claudia Schumann, Ph.D., Panayiotis Stevis, Ph.D., Yashu Liu, Ph.D., Julia Kozlitina, Ph.D., Stefan Stender, M.D., Ph.D., G. Craig Wood, M.S., Ann N. Stepanchick, Ph.D., Matthew D. Still, Shane McCarthy, Ph.D., Colm O’Dushlaine, Ph.D., Jonathan S. Packer, B.S., Suganthi Balasubramanian, Ph.D., Nehal Gosalia, Ph.D., David Esopi, M.S., Sun Y. Kim, B.A., Semanti Mukherjee, Ph.D., Alexander E. Lopez, M.S., Erin D. Fuller, B.S., John Penn, M.S., Xin Chu, Ph.D., Jonathan Z. Luo, B.S., Uyenlinh L. Mirshahi, Ph.D., David J. Carey, Ph.D., Christopher D. Still, D.O., Michael D. Feldman, M.D., Ph.D., Aeron Small, B.A., Scott M. Damrauer, M.D., Daniel J. Rader, M.D., Brian Zambrowicz, Ph.D., William Olson, Ph.D., Andrew J. Murphy, Ph.D., Ingrid B. Borecki, Ph.D., Alan R. Shuldiner, M.D., Jeffrey G. Reid, Ph.D., John D. Overton, Ph.D., George D. Yancopoulos, M.D., Ph.D., Helen H. Hobbs, M.D., Jonathan C. Cohen, Ph.D., Omri Gottesman, M.D., Tanya M. Teslovich, Ph.D., Aris Baras, M.D., Tooraj Mirshahi, Ph.D., Jesper Gromada, Ph.D., D.M.Sc., and Frederick E. Dewey, M.D.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
Contributor Information
N.S. Abul-Husn, Regeneron Genetics Center, Tarrytown, NY
X. Cheng, Regeneron Pharmaceuticals, Tarrytown, NY
A.H. Li, Regeneron Genetics Center, Tarrytown, NY
Y. Xin, Regeneron Pharmaceuticals, Tarrytown, NY
C. Schurmann, Regeneron Genetics Center, Tarrytown, NY
P. Stevis, Regeneron Pharmaceuticals, Tarrytown, NY
Y. Liu, Regeneron Pharmaceuticals, Tarrytown, NY
J. Kozlitina, University of Texas Southwestern Medical Center at Dallas, Dallas
S. Stender, University of Texas Southwestern Medical Center at Dallas, Dallas
G.C. Wood, Geisinger Health System, Danville, Pennsylvania
A.N. Stepanchick, Geisinger Health System, Danville, Pennsylvania
M.D. Still, Geisinger Health System, Danville, Pennsylvania
S. McCarthy, Regeneron Genetics Center, Tarrytown, NY
C. O’Dushlaine, Regeneron Genetics Center Tarrytown, NY
J.S. Packer, Regeneron Genetics Center, Tarrytown, NY
S. Balasubramanian, Regeneron Genetics Center Tarrytown, NY
N. Gosalia, Regeneron Genetics Center, Tarrytown, NY
D. Esopi, Regeneron Pharmaceuticals, Tarrytown, NY
S.Y. Kim, Regeneron Pharmaceuticals, Tarrytown, NY
S. Mukherjee, Regeneron Genetics Center, Tarrytown, NY
A.E. Lopez, Regeneron Genetics Center Tarrytown, NY
E.D. Fuller, Regeneron Genetics Center, Tarrytown, NY
J. Penn, Regeneron Genetics Center, Tarrytown, NY
X. Chu, Geisinger Health System, Danville, Pennsylvania
J.Z. Luo, Geisinger Health System, Danville, Pennsylvania
U.L. Mirshahi, Geisinger Health System, Danville, Pennsylvania
D.J. Carey, Geisinger Health System, Danville, Pennsylvania
C.D. Still, Geisinger Health System, Danville, Pennsylvania
M.D. Feldman, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania
A. Small, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania
S.M. Damrauer, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania
D.J. Rader, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania
B. Zambrowicz, Regeneron Pharmaceuticals, Tarrytown, NY
W. Olson, Regeneron Pharmaceuticals, Tarrytown, NY
A.J. Murphy, Regeneron Pharmaceuticals, Tarrytown, NY
I.B. Borecki, Regeneron Genetics, Center Tarrytown, NY
A.R. Shuldiner, Regeneron Genetics Center, Tarrytown, NY
J.G. Reid, Regeneron Genetics Center, Tarrytown, NY
J.D. Overton, Regeneron Genetics Center, Tarrytown, NY
G.D. Yancopoulos, Regeneron Pharmaceuticals, Tarrytown, NY
H.H. Hobbs, University of Texas Southwestern Medical Center at Dallas, Dallas
J.C. Cohen, University of Texas Southwestern Medical Center at Dallas, Dallas
O. Gottesman, Regeneron Genetics Center, Tarrytown, NY
T.M. Teslovich, Regeneron Genetics Center, Tarrytown, NY
A. Baras, Regeneron Genetics Center, Tarrytown, NY
T. Mirshahi, Geisinger Health System, Danville, Pennsylvania
J. Gromada, Regeneron Pharmaceuticals, Tarrytown, NY
F.E. Dewey, Regeneron Genetics Center, Tarrytown, NY
REFERENCES
- 1.Kochanek KD, Murphy SL, Xu J, Tejada-Vera B. Deaths: final data for 2014. Natl Vital Stat Rep 2016;65:1–122. [PubMed] [Google Scholar]
- 2.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 2004;40:1387–95. [DOI] [PubMed] [Google Scholar]
- 3.Lazo M, Hernaez R, Eberhardt MS, et al. Prevalence of nonalcoholic fatty liver disease in the United States: the Third National Health and Nutrition Examination Survey, 1988–1994. Am J Epidemiol 2013;178:38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams CD, Stengel J, Asike MI, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology 2011;140:124–31. [DOI] [PubMed] [Google Scholar]
- 5.Younossi ZM, Stepanova M, Afendy M, et al. Changes in the prevalence of the most common causes of chronic liver diseases in the United States from 1988 to 2008. Clin Gastroenterol Hepatol 2011;9(6): 524–530.e1. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science 2011;332:1519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010;52: 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res 2009;50:2111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trépo E, Romeo S, Zucman-Rossi J, Nahon P. PNPLA3 gene in liver diseases. J Hepatol 2016;65:399–412. [DOI] [PubMed] [Google Scholar]
- 11.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu YL, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with nonalcoholic fatty liver disease. Nat Commun 2014;5:4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sookoian S, Castaño GO, Scian R, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology 2015;61:515–25. [DOI] [PubMed] [Google Scholar]
- 14.Smagris E, Gilyard S, BasuRay S, Cohen JC, Hobbs HH. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J Biol Chem 2016;291:10659–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A 2014; 111:8913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem 2011;286:37085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pirazzi C, Adiels M, Burza MA, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276–82. [DOI] [PubMed] [Google Scholar]
- 18.Dewey FE, Murray MF, Overton JD, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 2016;354:aaf6814. [DOI] [PubMed] [Google Scholar]
- 19.Brantly M, Nukiwa T, Crystal RG. Molecular basis of alpha-1-antitrypsin deficiency. Am J Med 1988;84:6A:13–31. [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto T, Kitamoto A, Yoneda M, et al. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet 2013;132:783–92. [DOI] [PubMed] [Google Scholar]
- 21.Feitosa MF, Wojczynski MK, North KE, et al. The ERLIN1-CHUK-CWF19L1 gene cluster influences liver fat deposition and hepatic inflammation in the NHLBI Family Heart Study. Atherosclerosis 2013; 228:175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43: 1131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Huang C, Li D, et al. Molecular cloning and expression analysis of a new gene for short-chain dehydrogenase/reductase 9. Acta Biochim Pol 2007;54:213–8. [PubMed] [Google Scholar]
- 24.Su W, Wang Y, Jia X, et al. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc Natl Acad Sci U S A 2014;111:11437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donati B, Motta BM, Pingitore P, et al. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology 2016;63:787–98. [DOI] [PubMed] [Google Scholar]
- 26.Moeller G, Adamski J. Integrated view on 17beta-hydroxysteroid dehydrogenases. Mol Cell Endocrinol 2009;301:7–19. [DOI] [PubMed] [Google Scholar]
- 27.Kampf C, Mardinoglu A, Fagerberg L, et al. The human liver-specific proteome defined by transcriptomics and antibodybased profiling. FASEB J 2014;28:2901–14. [DOI] [PubMed] [Google Scholar]
- 28.Li P, Oh DY, Bandyopadhyay G, et al. LTB4 promotes insulin resistance in obese mice by acting on macrophages, hepatocytes and myocytes. Nat Med 2015; 21:239–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buch S, Stickel F, Trepo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443–8. [DOI] [PubMed] [Google Scholar]
- 30.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology 2016;150(5):1219–1230.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.