Highlights
-
•
A novel HLA-A2 restricted phosphopeptide was identified from a tumor-associated antigen, TRAP1.
-
•
The phosphopeptide is immunogenic for CTL induction to lysis the cancer cell with overexpressed TRAP1.
-
•
Vaccination of novel phosphopeptide can suppress the tumor-growth rate in AAD transgenic mice.
Keywords: TRAP1, HLA-A2, Phosphopeptide, Cytotoxic T lymphocyte, Immunotherapy, Cancer vaccine
Abstract
The tumor necrosis factor receptor associated protein 1 (TRAP1) is a mitochondria chaperon protein that has been previously implicated as a target for cancer therapy due to its expression level is linked to tumor progression. In this study, an immunodominant phosphopeptide of TRAP1 was identified from an HLA-A2 gene transfected mouse cancer cell line using mass spectrometry, and a synthetic phosphopeptide was generated to evaluate the potency on cancer immunotherapy. In the transporter associated with antigen processing (TAP) deficient cell, the conjugated phosphate group plays a critical role to enhance the binding affinity of phosphopeptide with HLA-A2 molecule. On the basis of immunological assay, immunization of synthetic phosphopeptide could induce a high frequency of IFN-γ-secreting CD8+ T cells in HLA-A2 transgenic mice, and the stimulated cytotoxic T lymphocytes showed a high target specificity to lysis the epitope-pulsed splenocytes in vivo and the human lung cancer cell in vitro. In a tumor challenge assay, vaccination of the HLA-A2 restricted phosphopeptide appeared to suppress the tumor growth and prolong the survival period of tumor-bearing mice. These results suggest that novel phosphopeptide is naturally presented as a HLA-A2-restricted CTL epitope and capable of being a potential candidate for the development of therapeutic vaccine against high TRAP1-expressing cancers.
1. Introduction
TRAP1 is a homolog of the heat shock protein 90 (Hsp90) chaperon that was majorly found in mitochondria [1], [2]. Accumulated evidence has indicated TRAP1 is highly expressed in a wide range of carcinomas, including colorectal [3], non-small cell lung cancer [4] and breast cancer [5]. These highly expressed TRAP1 can present multiple functions via the interaction with cyclophilin D [6] and succinate dehydrogenase (SDH) tumor suppression factors, respectively, to suppress the cell apoptosis and promote the cell transformation [7], [8], [9]. On the other hand, downregulation of TRAP1 level was shown to accelerate cancer cell apoptosis and to inhibit the neoplastic transformation [4], which indicated that targeting TRAP1 is a potential strategy for cancer therapy. Previously, Hsp90 inhibitor and its derivatives were hypothesized as potential drug candidates to inactivate TRAP1 [10], [11] due to the sequence similarity with Hsp90 protein. However, their anti-tumor potency remained controversial since these molecules may lack the specificity to distinguish TRAP1 from other chaperone homologs, raising a concern of interfering the client protein expression and homeostasis in normal cells [12], [13]. Thus, a novel TRAP1-targeting strategy is required.
Major histocompatibility complex class I (MHC I) are expressed on the surface of nucleated cell to display a broad repertoire of peptides for recognition by CD8+ cytotoxic T lymphocytes (CTL) via the T cell receptors (TCRs). In the human immune system, MHC I molecules are referred to as human leukocyte antigens class I molecules (HLAs). Typically, these peptides are predominantly 9–12 amino acids in length, which are generated from the proteasome degradation of intracellular proteins and are then transported into the endoplasmic reticulum via the transporter associated with antigen processing (TAP) to form the complex with MHC I proteins (pMHC I) [14]. Basically, thousands of different peptides can be loaded on individual MHC I allele, and it was estimated that one peptide can be presented 1 to 4000 copies per cell [15], leading the exposed peptide-MHC I complex (pMHC I) as “nature’s gene chip” to display internal proteome of cell for scanning by the circulating CD8+ T cell, a process called the immune surveillance [16]. Mutation in gene product or changes in protein expression level associated with cell transformation or viral infection would alternate the peptide frame displayed on the HLA I proteins [17], which serves as a flag to probe the transformed cell for CTL recognition. Sykulev and colleagues have demonstrated that immune surveillance is highly sensitive, such that a single antigenic peptide-MHC complex can be readily sensed and discriminated by CD8+ T cell and trigger degradation [18]. Previously, study reports demonstrated that high abundant TRAP1 was phosphorylated via the ERK pathway to regulate the metabolic mechanism and to promote the tumor proliferation [19], [20]. In general, the highly expressed phosphoproteins would be ubiquitinated by E3 ligase for rapid proteasome degradation to regulate their activities for maintaining the cellular homeostasis [20]. Interestingly, such mechanism was found to elevate the extent of phosphopeptide presented on MHC I molecules [21], [22], [23]. By mass spectrometry analysis, results revealed that pMHC I complex presents the peptide with phosphorylation, and indicated that this post translational modification can enhance the affinity with HLA protein and lead to CTL priming [23]. Therefore, the MHC I-bound peptides derived from the tumor-associated TRAP1 in cancer cells could be tumor specific, and may be a potential vaccine candidates for development of cancer immunotherapy [24].
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jvacx.2019.100017.
TC1/AAD is an HLA-A2 gene-transfected mouse lung epithelial cancer cell line, which can express a chimeric class I molecule, AAD, consisting of α1 and α2 domains of the HLA-A*0201 for peptide binding specificity and α3 transmembrane domain of the mouse H2-Db to stabilize the molecule on mouse cell membrane [25], [26]. Such hybrid HLA-AAD molecule was designed to present the human epitope for murine T cell recognition. It should be noted that the TRAP1 from human and mouse are highly similar based on the sequence alignment from UniProt database with the respective access numbers of Q12931 and Q9CQN1 (supplemental Fig. S1). Therefore, it is reasonable to expect TC1/AAD cells to present the identical HLA-A2 restricted peptides of TRAP1 as those found in human cell line. In the study, MS coupling liquid chromatography (LCMS) was carried out to analyze the peptide pools immunoprecipitated from TC1/AAD cells [27], [28]. The mono HLA allele lead the determination of HLA-type of MS-identified sequence becomes straightforward. Through phosphoproteomic analysis, a novel HLA-A2-restricted phosphopeptide of TRAP1 was identified in TC1/AAD cells. The peptide contains a novel phosphorylation site that was first observed in the TRAP1 protein. Via the immunological characterization and tumor challenge assays, the TRAP1-derived HLA-A2 restricted phosphopeptide was examined as a potential agent for the development of cancer immunotherapeutic treatment.
Supplementary Fig. S1.
Sequence alignment of mouse (Q9CQN1, UniProt) and human (Q12931, UniProt) TRAP1 protein. The results showed that the sequence of KLISVETDI was conserved in protein from these two mammalian species.
2. Material and methods
2.1. Animals and cell lines
C57BL/6 wild-type mice were obtained from the National Animal Center. AAD (containing the alpha1 + alpha2 domains of HLA-A2 and the alpha3 domain of H-2Db) transgenic mice were purchased from The Jackson Laboratory (Sacramento, CA). All animal studies were approved by the Institutional Animal Care and Use Committee of the NHRI (NHRI-IACUC-105156). The mouse lung cancer epithelial TC-1 cells were kindly provided by Dr. T. C. Wu (Johns. Hopkins University, USA). The TC-1/AAD was derived from parental TC-1 cells expressing AAD from AAD transgenic mice. In brief, the DNA fragment encoding the AAD chimeric protein was amplified with a pair of primers (5′-CCCAAGCTTATGGCCGTCATGGCGCCCCGA-3′ and 5′-GCTCTAGATCACACTTTACAATCTGGGAG-3′) from splenocytes of HLA- AAD transgenic mice. The amplified DNA fragment was further cloned into pcDNA4/TO/myc-His plasmid (Thermo Fisher Scientific Inc., USA) to generate pcDNA4/TO/myc-His/AAD. The TC-1 cells were transfected with pcDNA4/TO/myc-His/AAD to generate the TC-1/AAD stable cell line. The human lung cancer H2981 cell line (HLA-A2 positive) was a gift from Dr. Y-P, Sher [29]. The cell lines were cultivated in complete RPMI medium (RPMI 1640, GIBCO) containing 10% heat-inactivated fetal bovine serum (HyClone), 100 units/mL of penicillin and 100 µg/mL of streptomycin (GIBCO), 1 mM sodium pyruvate, and 5 mM HEPES in the incubator at 37 °C with 5% CO2.
2.2. Western blot analysis
The harvested cells were washed with chilled PBS buffer and then lysed with RIPA buffer (0.1% in 100 mM PBS buffer, pH 7.8) containing a protease inhibitor tablet (cOmplete, Roche). AAD transgenic mouse lung tissue served as a control in the SDS page electrophoresis. The lung tissue was pulverized with ceramic beads (100 μm, EE-TEC Ltd., Taiwan) in an ice bath and then lysed with 10 mL RIPA buffer at 4 °C. The protein concentrations of the cell lysate and tissue control were quantified with a BCA kit (BCA Protein Assay Kit, Pierce). The BLUeye prestained protein ladder (GeneDireX, Canada) was used for SDS-PAGE. A total of 50 μg protein was separated by 4–12% SDS-PAGE (NuPAGE, ThermoFisher Scientific) and then transferred to a PVDF membrane. To reduce non-specific binding, washing with PBST buffer (1 × PBS buffer containing 0.05% Tween 20) was after every wash step. The blotting membrane was blocked with 5% non-fat milk in PBST at 4 °C overnight. Subsequently, the membrane was cut and separately incubated with diluted anti-TRAP1 (1 : 1000 in PBST, rabbit polyclonal, ab8227, Abcam) and anti-actin (1 : 10,000 in PBST, rabbit polyclonal, ab151239, Abcam). The membranes were then stained with horseradish peroxidase (HRP)-conjugated detection antibodies (1 : 10,000 in PBST, goat polyclonal, ab6721, Abcam) in enhanced chemiluminescence solution (Millipore).
2.3. Immunohistochemical staining
The lung non-small cell cancer tissue array was purchased from US Biomax (LC10012). The TRAP1 expression level in clinical specimens from patients with lung cancer was evaluated by immunohistochemical staining as previously described [30]. The specimens were obtained from the files of the Department of Pathology of China Medical University Hospital in compliance with protocols approved by the CMUH IRB (DMR99-IRB-131). A pair of tumor tissues and adjacent tissue were used to compare the staining intensity, and 44 paired specimens from a total of 45 cases were included in the analysis due to the loss of one core from the tissue array. The tissue sections were incubated with rabbit anti-human TRAP1 protein (LS-C31383; LifeSpan BioSciences) and horseradish peroxidase-conjugated Avidin-biotin complex (ABC) from the Vectastain Elite ABC Kit (Vector Laboratories) and AEC chromogen (Vector Laboratories). The TRAP1 staining intensity in all samples was scored by a pathologist.
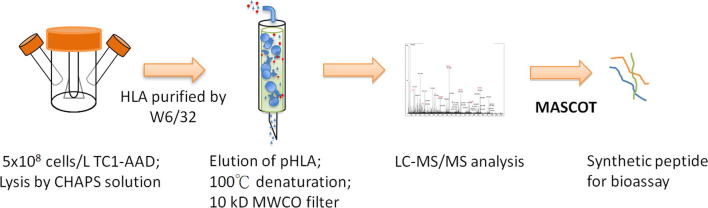
2.4. Isolation of HLA-AAD peptides
HLA-AAD-bound peptides were obtained by the immunoprecipitation as previously described [31] with slight modifications. The flow chart of CTL peptide identification is shown in supplemental Fig. S2. TC1/AAD cells were harvested from two spinner flasks (1 × 109 cells/liter), lysed with CHAPS solution (1% in chilled PBS buffer, pH 7.8) and then centrifuged at 2000 rpm for 10 min to remove the cell debris. The HLA-AAD-peptide complex was immunoprecipitated using an anti-HLA A2 antibody (BB7.2)-immobilized column. After heat denaturation (95 °C, 10 min) and a cooling step, the associated peptides were extracted sequentially using an ultracentrifuge column (10 kDa cutoff membrane, Millipore, Billerica, MA) and reverse-phase spin column (C18, Invitrogen) [31], [32]. The desalted analyte was lyophilized and then resuspended in 5% formic acid solution for LC-MS/MS analysis.
Supplementary Fig. S2.
Flow chart for identifying the HLA-A2 restricted CTL epitope from mouse TC1/AAD cancer cell line.
2.5. Identification of HLA-AAD-bound peptides by LCMS/MS
A Q Exactive mass spectrometer (Thermo Scientific, USA) coupled to an Ultimate 3000 RSLC system (Dionex) was utilized for HLA-peptide sequencing. The complex peptide mixture was separated using a reverse-phase column (C18, Acclaim pepmap RSLC, 75 µm × 150 mm, 2 µm pore size, Dionex). The mobile phases were prepared as follows: A: 0.1% FA in 5% acetonitrile (ACN) (V/V), and B: 0.1% FA in 95% ACN (V/V). A linear gradient was initiated from 1% B to 25% B in 40 min, and then increased to 80% B in 18 min to elute all the analytes in the column. Full MS scan was performed within the range of m/z 300–2000, and the ten most intense ions in the MS scan were subjected to fragmentation to acquire the MS/MS (tandem MS) spectra. The MS raw data was converted into peak list (PKL) using Proteome Discoverer 1.3. Sequence analysis was carried out using the MASCOT searching engine with self-constructed databases (mouse species, UniProt) with parameters that included the following: no enzyme, variable modification of oxidization (M) and phosphorylation (ST, Y), intensity ratio cutoff of 10%, tolerance of ±0.002 Da, mass tolerance of ±10 ppm. The MS spectra of the identified sequences were confirmed by further manual examination.
2.6. Prediction of binding motif of peptide
The MHC class I binding predictions were performed using IEDB analysis resource NetMHC (ver. 4.0) tool [33], [34], [35] with parameters including the following: human allele of HLA-A*02:01, mouse allele of H2-Db and peptide length of 8–12 amino acids.
2.7. T2 cell-based stabilization assay
The binding affinity between the peptide and HLA-A2 molecule was evaluated by the TAP-deficient T2-cell stabilization assay as described previously [36]. Briefly, 2 x105 T2 cells (ATCC, HLA-A2-positive) were incubated with 10 μM synthetic peptide (purity greater than 95%, UV 210 nm) in DMEM containing 0.1% FBS, 5 × 10−5 M β-mercaptoethanol, and 5 × 10−7 M β2-microglobumin at 25 °C in a 5% CO2 filling incubator overnight. Subsequently, the temperature was adjusted to 37 °C for 3 h incubation, stained with PE-conjugated anti-HLA-A2 antibody (BB7.2, BD Bioscience, USA), and then detected the mean fluorescence intensity (MFI) by flow cytometry (FACSCalibur, BD Bioscience, USA). Cell marked with isotype Ab (27–35, monoclonal anti-dansyl, BD) served as control in the T2 assay. The affinity was determined according to the mean fluorescence intensity (MFI) of peptide-pulsed T2 cells. In the assay, synthetic peptides of VYCKQQLLR (VYC, HLA-A11-restricted epitope of HPV16 E638−46) and YMLDLQPETT (YML, HLA-A2-restricted epitope of HPV16 E711−20) were applied as the negative and positive controls, respectively.
2.8. Peptide immunization and IFN-γ secretion assay
The peptide-specific T cell response was performed using the enzyme-linked immunospot (ELISPOT) assay as previously described with some modifications [37]. To enhance the peptide immunogenicity, an equal amount of the CD4 helper T cell epitope (AKFVAAWTLKAAA, PADRE) was mixed with the target peptide in incomplete Freund’s adjuvant (50% in PBS buffer) solution with the final peptide concentration of 1 mg/mL[38], [39]. Synthetic peptides of VYC and YML were applied as the negative and positive controls, respectively. Next, 50 µL of the prepared immunogen was injected into the mice via the footpad on Day 0 and 7. One week after the final immunization, a total of 5 × 105 lymph node cells were harvested and incubated with peptide (final: 5 μg/ml) in complete RPMI medium at 37 °C with 5% CO2 for 48 hrs. After incubation, the cells were washed with PBST buffer, and then 50 µL of biotin-conjugated anti-IFN-γ antibody (R46A2, eBioscience, CA) solution and streptavidin-conjugated HRP (eBioscience) were added to the plate in sequence. The spots were developed with 3-amine-9-ethyl carbazole solution (ACE, Sigma), and then the number of spots was counted using an ELISOT reader (Cellular Technology Ltd., Shaker Heights, OH).
2.9. Intracellular staining
Splenocytes (5 × 106) were harvested from peptide-immunized AAD transgenic mice and incubated with the corresponding immunogen (final concentration 10 μg/ml) overnight at 37 °C with 5% CO2. After the incubation period, the cells were washed once with chilled PBS and stained with FITC-conjugated monoclonal anti-mouse CD8 antibody (53–6.7, BD). The FITC-labeled cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% saponin/PBS for 5 mins, and then the supernatant was decanted. Intracellular cytokine staining was detected with PECy7-conjugated anti-mouse IFN-γ (R3-34, BD), and the number of stained cells was then determined by flow cytometry (BD Bioscience). The data analysis was performed using FCS Express software (De Novo Software).
2.10. In vivo killing assay
Splenocytes (5 × 107) were harvested from AAD transgenic mice and then incubated with either peptide (5 μg/ml) or PBS (untreated control) for 30 min at 37 °C. After incubation, carboxylfluorescein succinimidyl ester (CFSE, Molecular Probes, Eugene, OR) was used to label the peptide-pulsed spleen cells and untreated control at a concentration of 10 µM and 1 µM, respectively, for 15 min at 37 °C. Ice-cold complete RPMI medium was then added to stop the CFSE-labeling reaction. The non-peptide pulsed cells and the peptide-pulsed splenocytes were mixed at a ratio of 1:1, and a total of 2 × 107 CFSE-labeled cells were injected into the peptide-immunized AAD transgenic mice via the tail vein. At 18 h post-adoptive transfer, the spleen cells were harvested and analyzed by flow cytometry (BD Bioscience). The data analysis was performed using FCS Express software (De Novo Software).
2.11. 51Cr release assay
The CD8+ T cells were isolated from spleen of KLIpS-immunized mice using the CD8+ T Cell Isolation Kit (Miltenyl Biotech). H2981 cells (5 × 105/ml) were labeled with 100 μCi of 51Cr (Na251CrO4, PerkinElmer, MA) at 37 °C for 1 h as the target cells. The CD8+ T cells (5 × 105) was then mixed with 51Cr-labeled target cells (5 × 103) at a ratio of 100:1 (Effector : Target, E : T), and incubated at 37 °C for 18 h. After the incubation period, the supernatants were harvested to measure the radioactivity using a gamma counter. The percentage of specific lysis was calculated using the following formula: 100 × [(experimental release − spontaneous release)/(maximal release - spontaneous release)].
2.12. Tumor challenge assay
AAD transgenic mice aged 6–8 weeks were inoculated with 2 × 105 TC1/AAD cells for tumor development. One week after the tumor injection, the tumor-bearing mice (n = 5) were immunized twice with 50 µg peptide in IFA with PADRE on Day 7 and 14 by footpad injection. Tumor-bearing mice (n = 5) immunized with PBS buffer served as the untreated control group in the assay. Tumor size was monitored twice a week by palpation with calipers, and the tumor volume was calculated using the following formula: length × width × width/2. The tumor-bearing mice were sacrificed when the tumor size exceeded 2000 mm3.
2.13. Statistical analysis
Statistical significance was evaluated by the Student’s t-test (two-tailed) and ANOVA at the 5% level.
3. Results
3.1. Expression levels of TRAP1 in lung cancer cells and tumors
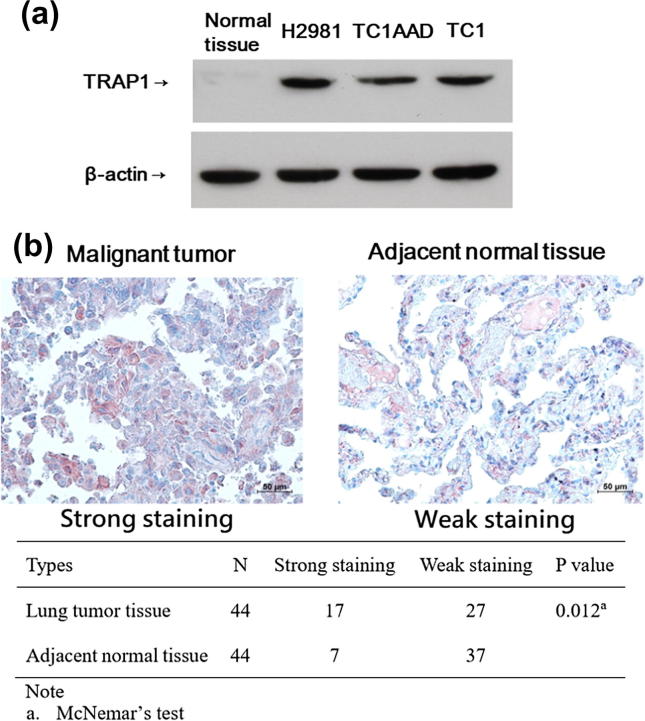
To assess the TRAP1 expression level in mouse and human lung cancer cell lines, we quantified this protein target using a western blot assay and compared its abundance to that in normal mouse lung cells. Consistent with another study [40], the expression level of TRAP1 was significantly higher in the tested lung cancer cell lines, but it was nearly undetectable in normal lung tissue, as shown in Fig. 1A. In order to validate the expression of TRAP1, immunohistochemical staining was carried out to assess the abundance of TRAP1 in human lung tumor tissues and their adjacent normal tissues. IHC results showed that positive TRAP1 expression was located in the cytoplasm as red color staining, as shown in Fig. 1B. The strong staining of TRAP1 in tumor tissue and normal tissue were 38.6% (17/44) and 15.9% (7/44), respectively, and the difference was statistically significant. Among these 7 normal tissue samples with strong staining of TRAP1, 5 of them were sampled from the adjacent tumor tissues with highly abundant of TRAP1, suggesting the high expression rate of TRAP1 could be found in normal tissues adjacent to the tumor tissue.
Fig. 1.
TRAP1 protein is highly expressed in cancer cell lines and tumor tissues. (A) 50 μg total protein of cell lysate was analyzed using Western blotting to quantify the relative abundance of TRAP1 protein in TC1/AAD, TC1, H2981, and normal mouse lung tissue. β-actin served as the loading internal control in the assay. (B) The IHC staining of TRAP1 protein expression in patient malignant lung tissue and adjacent normal tissue. The intensity of TRAP1 staining in all samples was scored by a pathologist and the IHC analysis result is summarized in the attached table.
3.2. Identification of HLA-A2-bound peptides by mass spectrometry
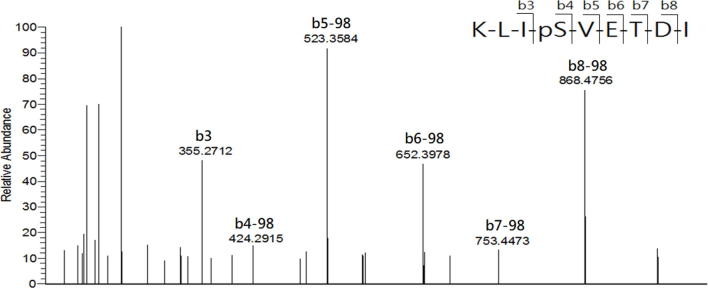
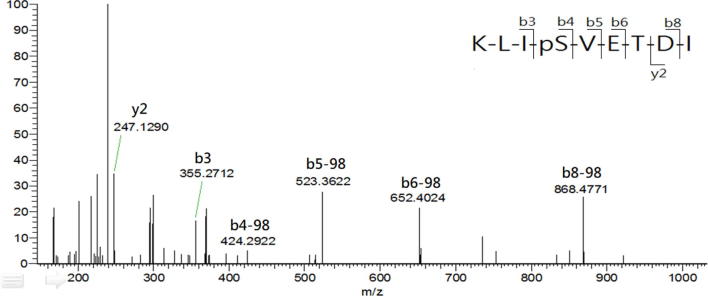
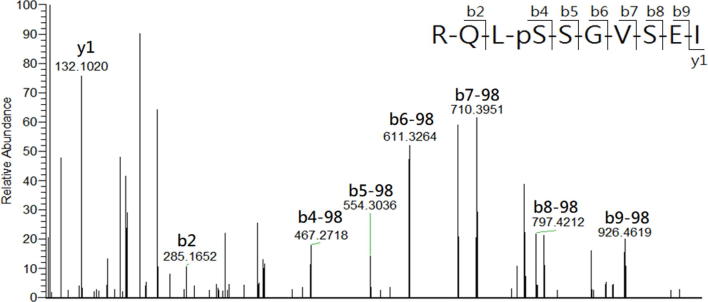
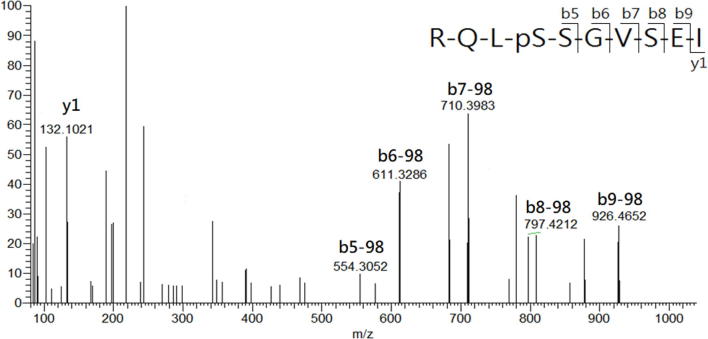
In the study, LCMS was conducted to analyze the HLA-AAD-bound peptides that immunoprecipitated from TC1/AAD cells. The MASCOT search resulted in the identification of a total of 12 significant HLA-A2-restricted peptides in 1 × 109 TC1/AAD cells, and their sequences were further confirmed by manual inspection of their corresponding tandem MS spectra (Table S1). By importing the sequences into the IEDB (www.IEDB.org) database, all the MS-identified peptides fit the binding motif of HLA-A2 (A*02:01) molecule but not the mouse MHC molecule (H2-Db). Among these MS-identified peptides, KLIpSVETDI (abbreviate as KLIpS, lowercase letter denotes phosphate group in the article) and RQLpSSGVSEI (ROLpS) were phosphopeptides derived from the cancer-associated protein of TRAP1_mouse (Q9CQN1, UniProt) and HSP27_mouse (P14602, UniProt), respectively. Additionally, synthetic references were prepared and their tandem MS spectra presented nearly similar b/y fragment patterns as those acquired from the immunoprecipitated phosphopeptides, which confirmed the sequence identities of the phosphopeptides (Supplementary Fig. S3A, Supplementary Fig. S3B, Supplementary Fig. S4A, Supplementary Fig. S4B). It is noteworthy that RQLpS is a known HLA-A2-restricted CTL epitope that has been detected in human ovarian carcinoma (cov413) [41] and melanoma cells (DM331) [27], [42]. Such Hsp27-derived HLA-A2 restricted phosphopeptide was first observed in a mouse lung cancer cell line, further supporting the MS sequencing results of the study. As the structure features indicated phosphate group of HLA-phosphopeptide is critical for T cell receptor (TCR) recognition, [43], the immunogenicity of TRAP1-derived phosphopeptide on CTL induction was analyzed in the following study.
Supplementary Table S1.
HLA-A2 bound-peptides identified from mouse TC1/AAD cancer cell.
Supplementary Fig. S3A.
Comparison of tandem MS spectra of the (A) synthetic reference and (B) immunoprecipitated KLIpS peptide. The corresponding b and y series are marked on top of significant ion with matched m/z.
Supplementary Fig. S3B.
Comparison of tandem MS spectra of the (A) synthetic reference and (B) immunoprecipitated KLIpS peptide. The corresponding b and y series are marked on top of significant ion with matched m/z.
Supplementary Fig. S4A.
Comparison of tandem MS spectra of the (A) synthetic reference and (B) immunoprecipitated RQLpS peptide. The corresponding b and y series are marked on top of significant ion with matched m/z.
Supplementary Fig. S4B.
Comparison of tandem MS spectra of the (A) synthetic reference and (B) immunoprecipitated RQLpS peptide. The corresponding b and y series are marked on top of significant ion with matched m/z.
3.3. HLA-peptide stabilization assay
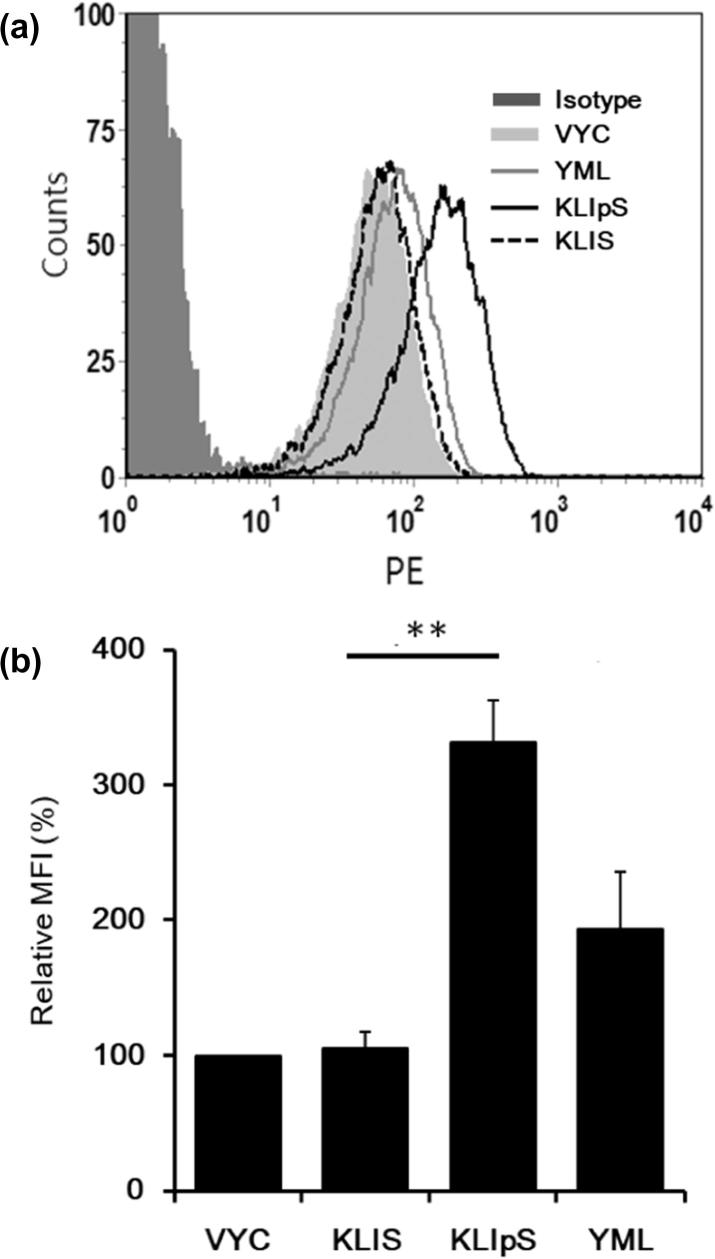
A stabilized peptide-HLA (pHLA) complex is essential for TCR recognition. The T2 cell stabilization assay was applied to assess the binding affinity of the phosphopeptide to HLA-A2 molecule. The T2 cell line is TAP-deficient but expresses peptide-unloaded HLA-A2 molecules on the cell membrane, which can be stabilized by docking with allele-restricted peptides. As shown in Fig. 2, the KLIpS was found to form a stabilized complex with the HLA-A2 molecule in the T2 cell as well as the positive control of YMLDLQPET peptide (HLA-A2 binder). In contrast, only a few or a less stabilized HLA-peptide complex was detected when T2 cell was cocultured with non-phosphorylated peptide (KLIS). The observed results indicated that such site-specific phosphate moiety of HLA-A2 phosphopeptide is critical for the peptide binding affinity with HLA-A2 molecule, and such result is consistent with the finding of other investigations [21].
Fig. 2.
The KLIpS could form a stable complex with HLA-A2 molecule. (A) 2 × 105 TAP-deficiency T2 cells, incubated with either KLIpS (solid line), KLIS (dashed line), YML (light line, HLA-A2 binder) or VYV peptide (light-shaded, HLA-A11 binder) as indicated, were stained with PE-labeled anti-HLA-A2 Ab (BB7.2), and then monitored the MFI by flow cytometry. Cells without peptide incubation were as negative control (NC, dot line). Cells stained with isotype Ab are marked in dark Gray-fill. 1 × 104 gating cells were analyzed. (B) The bar chart showed the relative MFI of peptide detected in T2 assays using the MFI of negative control as reference. Significant difference calculated with the Student’s t-test (two-tailed) and ANOVA is marked by asterisks. (**, p < 0.01) The error bar represented the standard deviation (SD) of triplicate experiments.
3.4. Immunization with KLIpS induces cytokine responses in transgenic mice
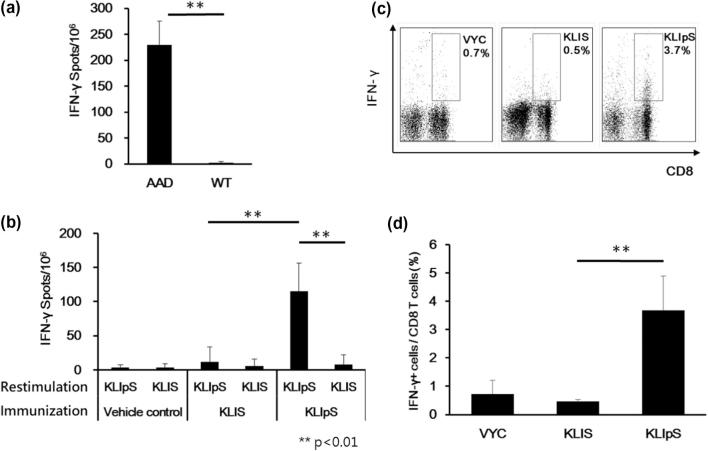
Based on the T2 assays result, we further evaluated the T cell-inducing ability of KLIpS in the animals of C57BL/6 wild type (WT) and AAD transgenic mice. ELISPOT result revealed that a high frequency of interferon gamma (IFN-γ)-secreting cells was detected only in the lymph nodes of peptide-immunized AAD transgenic mice, but not in the wild-type control, as shown in Fig. 3A. The mice-immunization results not only validated the binding motif of KLIpS, but also indicated that the cytokine response was mediated by the AAD molecule rather than other mouse MHC molecules. Subsequently, the AAD transgenic mice were immunized with peptides of KLIpS, KLIS (non-phosphorylated counterpart), YML and VYC to compare their efficacy for cytotoxic T cell induction. Upon peptide restimulation, ELISPOT assay results demonstrated that a significantly larger number of IFN-γ-secreting cells in the lymph nodes of AAD transgenic mice immunized with KLIpS (spots: 230 ± 45) or YML peptide (spots: 161 ± 81, positive control), but such cytokine response was not observed in the mice immunized with either KLIS or VYC peptide (Fig. S5). Moreover, a cross-restimulation assay was performed, and the robust cytokine response was restricted to KLIpS but not to its non-phosphorylated counterpart, as shown in Fig. 3B. Additionally, an intracellular staining assay was introduced to evaluate cytokine expression within a heterogeneous population of cells. After fixing the cell membrane and Golgi apparatus, the accumulated IFN-γ was detected in the CD8+ T cell population (3.7%) of splenocytes from KLIpS-immunized AAD transgenic mice, which made up a relatively smaller population in AAD transgenic mice immunized with KLIS (0.5%) or VYC (0.7%), as shown in Fig. 3C and 3D. The cytokine secretion assay confirmed that KLIpS is immunogenic and capable of inducing effector CD8+ T cells through the HLA-AAD molecule in transgenic mice.
Fig. 3.
Immunization with KLIpS induces antigen-specific IFN-γ-secreting CD8+ T cell population. (A) AAD transgenic and wild-type (WT) mice (n = 6) were immunized twice via footpad with 50 μg KLIpS formulated with PADRE in IFA adjuvant. Seven days after final immunization, 5 × 105 lymph nodes were harvested and cocultured with peptide for 48 hrs in ELISPOT plate. The antigen-specific IFN-γ-secreting spots were developed and analyzed using ELISPOT reader. (Student’s t-test, AAD vs WT, **, p < 0.01). The error bar represented the standard deviation (SD) of duplicate experiments. (B) Six AAD transgenic mice were vaccinated with KLIpS, as described in the legend of Fig. 2A. Cross antigen-restimulation was performed to assay the specificity of peptide-induced IFN-γ-secreting cells from peptide-immunized AAD transgenic mice. (Student’s t-testfor KLIpS restimulation: KLIS vs KLIpS immunization; for KLIpS immunization: KLIS vs KLIpS restimulation, **, p < 0.01). The error bar represented the standard deviation (SD) of triplicate experiments. (C) Six AAD transgenic mice were vaccinated with KLIpS, as described in the legend of Fig. 2A. Detection of the effector T cell population in 1 × 105 splenocytes of peptide-immunized AAD-transgenic mouse using flow cytometry. The upper-right corners revealed a significant population of stimulated IFN-γ-secreting CD8+ T cells among the lymphocytes of KLIpS-immunized AAD transgenic mice. (D) The bar graph shows the percentage of IFN-γ-secreting cells in CD8+ T cells (Student’s t-test, KLIS vs KLIpS, **, p < 0.01). The error bar represented the standard deviation (SD) of triplicate experiments.
Supplementary Fig. S5.
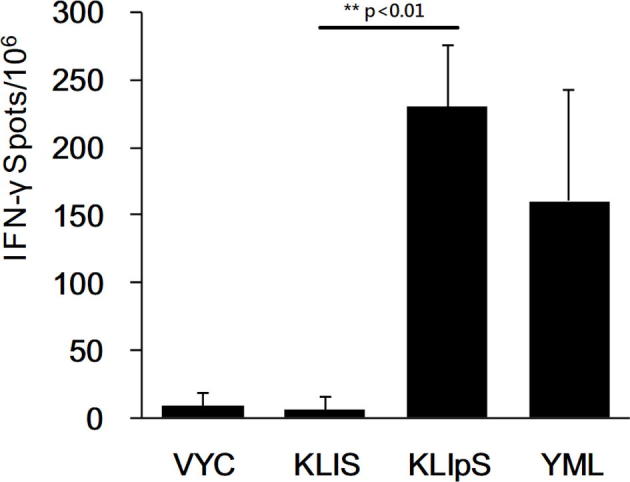
Monitoring T cell induction in response to peptide immunization in transgenic mice by the ELISPOT assay. AAD transgenic mice (n=5) were immunized 50 μg peptide formulated with PADRE in IFA adjuvant. Seven days after final immunization, 5 x 105 lymph nodes were harvested and cocultured with peptide for 48 hrs in ELISPOT plate, and then inguinal lymph nodes were collected for detection of the IFN-γ-secreting cell population. Under antigen restimulation, adequate IFN-γ-secreting cells were detected in the lymph nodes of AAD transgenic mice that were immunized with KLIpS and YML (positive control), but they were rarely observed in mice immunized with KLIS and VYC (negative control). (Student t-test, KLIS vs KLIpS, **, p<0.01)
3.5. Cytolytic activity of KLIpS-specific CTLs
In addition to the cytokine response of peptide restimulation, we further evaluated the cytolytic activity of KLIpS-specific CTLs on different cell targets in vivo and in vitro. In the in vivo killing assay, the naïve splenocytes of transgenic mice was pulsed with phosphopeptide to mimic the cancer cell in native-like condition. A mixture of equal amounts of epitope-pulsed (CFSEhigh) and untreated (CFSElow) splenocytes was injected into the KLIpS-immunized AAD transgenic mice through the tail vein. Then the splenocytes of recipient were then harvested to determine the percentage of CFSEhigh and CFSElow among the total CFSE+ cells. As shown in Fig. 4A, nearly half of the CFSEhigh were diminished in mice immunized with either KLIpS (48.11 ± 18.36%) or the CTL peptide (61.7 ± 41.34%, YML is the positive control). As such, the results evidenced the stimulated antigen-specific T cell can recognize and induce cytolysis of target cells with the HLA-AAD-KLIpS complex in vivo.
Fig. 4.
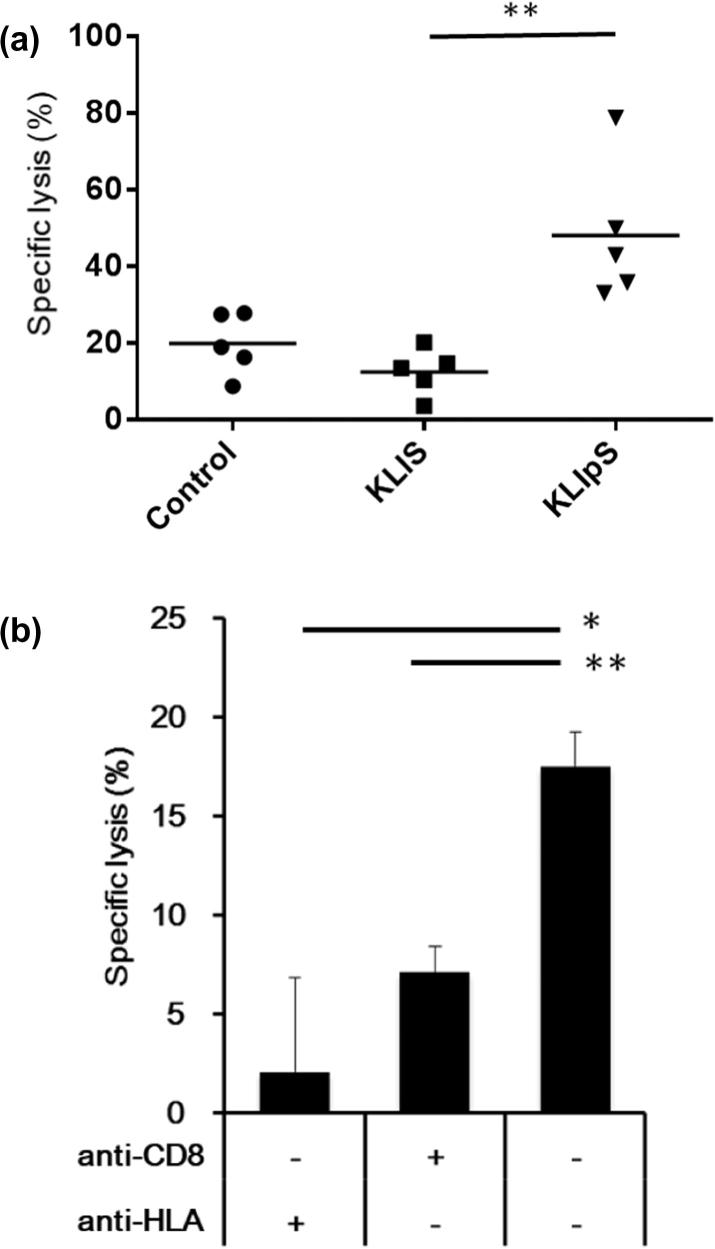
KLIpS immunization could induce specific cytolytic activity. (A) AAD transgenic mice (n = 5) were immunized twice with KLIpS (triangle), KLIS (square) or PBS (control, circle) formulated with PADRE in IFA. Severn days after immunization, the immunized mice were injected with CFSE-labeled peptide-pulsed spleen cells to exam the in vivo cytolytic activity of CTLs. The specific lysis percentage represented the reduced amount of peptide-pulsed spleen cells after 18 hrs post-adoptive transfer. (Student’s t-test, KLIS vs KLIpS, **, p < 0.01). (B) 5 × 105 CD8+ T cells were purified from spleens of KLIpS-immunized transgenic mice and co-cultured with 5 × 103 51Cr-labeled H2981 for 18 h. The 51Cr-release assay result shows that KLIpS-stimulated mouse CD8+ T cells could cytolysis human lung H2981 cancer cell. 51Cr-labeled H2981 cells were incubated with 20 μg/ml anti-HLA Ab (W6/32), or purified CD8+ T cells were incubated with 10 μg/ml anti-CD8 Ab (53–6.7) for 15 min before co-culture. Addition of specific antibody could significantly inhibit the cytolysis activity of mouse CD8+ T cell. (w/o Ab vs anti-HLA Ab, *, p < 0.05; w/o Ab vs anti-CD8 Ab, **, p < 0.01). Specific lysis percentage was calculated using formula: 100 × [(experimental release − spontaneous release)/(maximal release - spontaneous release)].
Apart from phosphopeptide-pulsed splenocytes, the CTL recognition on human cancer cell line was examined in the study. H2981 is an HLA-A2 positive human lung cancer cell line that expresses high abundant TRAP1 proteins as those found in TC1/AAD cell, which suggested that KLIpS could be expressed in H2981 cells as TC1/AAD cell line. We, therefore, extracted the CD8+ T cells from KLIpS immunized mice to evaluate the cytolytic activity on H2981 using an in vitro chromium (51Cr) release assay. In Fig. 4B, the results demonstrated that up to 20% of 51Cr-labeled H2981(target, T) was lysed when incubating with mouse effector (effector, E) at the ratio of 100:1(E:T), while this cytolytic activity was markedly inhibited when either TCR or HLA was blocked with anti-CD8 or anti-HLA antibody, respectively. As such, the results revealed that the recognition of effector CD8+ T cell was mediated through the interaction of HLA and T cell receptor (TCR) molecules, but also demonstrated the capability of KLIpS-stimulated T cell to cytolysis the target cell with HLA-A2 restricted KLIpS peptide.
3.6. Anti-tumor effect of KLIpS-specific CTLs
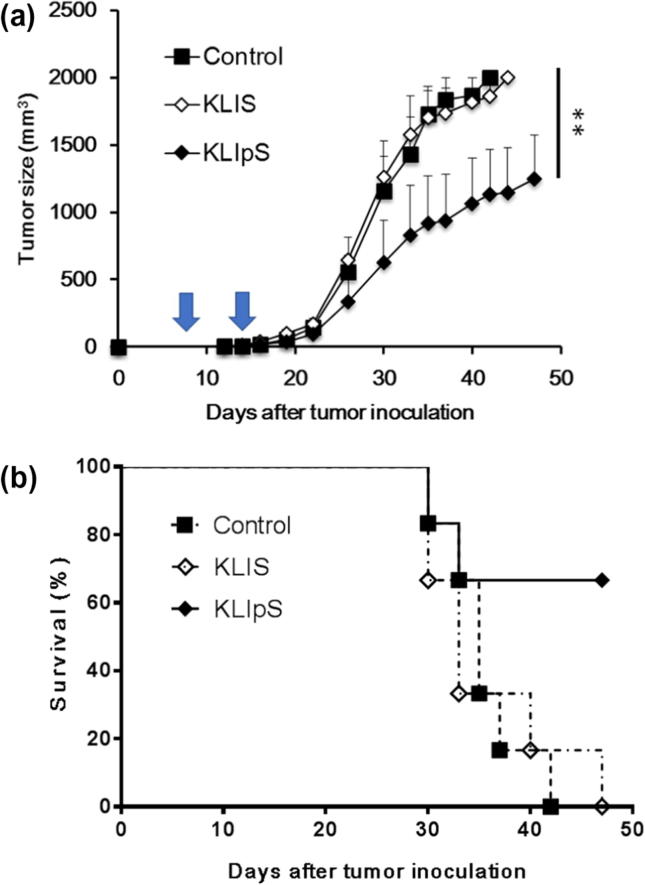
Based on the killing effect on cancer cells, a tumor challenge model was developed to examine the cytolytic effect of KLIpS-specific CTLs in tumor tissue. Fig. 5 profiles the tumor size in mice immunized with peptide antigen and PBS (control), respectively. In the tumor challenge assay, tumor-bearing mice (n = 5) immunized with KLIS or PBS buffer were all sacrificed by 7 weeks. Immunization of KLIpS peptide caused a significant reduction of tumor growth and prolonged the survival period of tumor-bearing mice for more than 7 weeks, demonstrating the potency of KLIpS vaccination as a therapeutic strategy for cancer treatment.
Fig. 5.
Vaccination of KLIpS presented the anti-cancer effect to suppress the tumor growth. (A) AAD transgenic mice (n = 6) were inoculated with 2 × 105 TC1/AAD cancer cell for tumor development. Seven days after cancer cell injection, the tumor-bearing mice were immunized twice via footpad injection with either 50 μg KLIpS, KLIpS or PBS (control) formulated with PADRE in IFA on Day 7 and 14 indicated with arrow in figure. Tumor size was measured by palpation with calipers until the tumor volume exceeded 2000 mm3. (ANOVA test, KLIS vs KLIpS, **, p < 0.01). (B) Mice survival time was monitored and analyzed.
4. Discussion
Lung cancer is one of the most prevalent cancers worldwide and also a leading cause of malignant-related death because of its highly metastatic potential. Systemic chemotherapy is commonly used for lung cancer treatment, which could improve the survival rate but might provoke serious side effect. Currently, immunotherapy is considered as a candidate strategy since increasing evidences showed that host immune system can confer significant survival advantages. To meet the success of immunotherapeutic treatment, ideal targets are essential and critical. In the study, we demonstrated that TRAP1 was highly expressed in variety of lung cancer cell lines and patient tissues, but nearly silenced in normal cell. With the aid of MS analysis, one novel TRAP1-derived HLA-A2-restricted phosphopeptide, KLIpS, was identified from the AAD gene-transfected mouse lung epithelial cancer cell. The phosphorylation site of KLIpS fits the consensus motif of kinases, R/KxxS/T. To our knowledge, it was the first HLA-A2 restricted phosphopeptide, KLIpSVETDI, identified from the tumor-associated antigen of TRAP1. The correlation between the new phosphorylation site and TRAP1 functionality was not clear, but increasing phosphorylation of the protein suggested an altered kinase activity in mitochondria [44].
Other than the phosphate dynamics of the protein, the phosphate moiety on the fourth N-terminal residue of KLIpS contributes unique features for peptide-specific T cell induction. First, the phosphate enhanced the binding affinity of KLIpS with the HLA-A2 molecule. A structural investigation on HLA-peptide binding has indicated that the phosphate conjugated on the forth N-terminal residue could form an electrostatic interaction with polar residues of the HLA-A2 molecule [43]. Our study is consistent with previous observations that assay did present that KLIpS could form a stabilized complex with HLA-A2 molecule in the TAP-deficient as compared to the non-phosphorylated counterpart [43]. Apart from complex assembly, we also demonstrated that immunization of this phosphopeptide in HLA-AAD transgenic mice could induce IFN-γ-secreting CD8+ T cell in splenocytes during antigen stimulation. The cytokine IFN-γ increases MHC expression on target cells and mobilizes the cytotoxic lymphocytes to tumor-associated antigens [45], [46]. In the cytolysis assay, KLIpS showed the immunogenicity to induce the antigen-specific T cell that could lyse the KLIpS-pulsed splenocytes in vivo and KLIpS-presented human cancer cell line, H2981, in vitro. It is noteworthy that the stimulated CTLs showed a high specificity against KLIpS but were not cross-recognized with the non-phosphorylated counterpart of KLIS, suggesting this phosphate group play a role in antigen presentation. Such an intense cytolytic effect was also observed in tumor tissue. In the tumor challenge assay, KLIpS-immunization induced persistent CTL activity for tumor suppression, in which three out of five tumor-bearing mice survived more than 7 weeks with a reduction of the tumor size. Overall, these results revealed that vaccination of KLIpS could induced an intense and antigen-specific CTLs to lysed the cancer cells with this antigenic epitope or inhibit the tumor growth in the animal model, which highlighted this phosphopeptide is an ideal target for immunotherapy. An interesting finding was observed in the in vitro killing assy (Fig. 4B) and tumor challenge model, both results showed that the KLIpS-specific CD8+ T cells were not able to eradicate the tumor cells. One of the possible factors is the suppression of tumor microenvironment [47]. In order to overcoming the suppression, integrating the treatment of immune checkpoint antibody or protease inhibitor with the peptide vaccination can be an effective strategy to enhance the antitumor immunity. Another possible factor to affect the killing effect of CTL response could be the low density of KLIpS distributes on cancer cell. As a result, quantifying the abundance of KLIpS on cancer cell lines could provide useful information to assess the cytolysis potency of CTL on cell targets. Based on the peptide density information and procedure optimization, we expect to develop a novel peptide-based vaccination approach for improving the outcome of CTL responses on the treatment of cancer therapy.
Author contributions
Concept and design: SJ Liu, WC Sung.
Development of methodology: Y-P Sher, S-J Liu, W-C Sung.
Acquisition of data (provided animals, acquired and managed patients, provided facilities): I-H Chen, S-J Liu.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M-H Lin, K-Y Shen, BS Liu, Y-P Sher, G-C Tseng.
Writing, review, and/or revision of manuscript: M-H Lin, K-Y Shen, Y-P Sher, S-J Liu, W-C Sung.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M-H Lin, K-Y Shen, Y-P Sher, S-J Liu, W-C Sung.
Study supervision: S-J Liu, W-C Sung.
Acknowledgments
Acknowledgements
The study was supported by the Minister of Science and Technology, Taiwan (Grant no. MOST 107-2314-B-400-027) and National Research Program for Biopharmaceuticals grants from the Ministry of Science and Technology, Taiwan (MOST 105-2325-B-400-004).
Conflict of interest
No potential conflicts of interest were disclosed.
References
- 1.Lavery L.A., Partridge J.R., Ramelot T.A., Elnatan D., Kennedy M.A., Agard D.A. Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol Cell. 2014;53:330–343. doi: 10.1016/j.molcel.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montesano Gesualdi N., Chirico G., Pirozzi G., Costantino E., Landriscina M., Esposito F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress. 2007;10:342–350. doi: 10.1080/10253890701314863. [DOI] [PubMed] [Google Scholar]
- 3.Gao J.Y., Song B.R., Peng J.J., Lu Y.M. Correlation between mitochondrial TRAP-1 expression and lymph node metastasis in colorectal cancer. World J Gastroenterol. 2012;18:5965–5971. doi: 10.3748/wjg.v18.i41.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agorreta J., Hu J., Liu D., Delia D., Turley H., Ferguson D.J. TRAP1 regulates proliferation, mitochondrial function, and has prognostic significance in NSCLC. Mol Cancer Res. 2014;12:660–669. doi: 10.1158/1541-7786.MCR-13-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B., Wang J., Huang Z., Wei P., Liu Y., Hao J. Aberrantly upregulated TRAP1 is required for tumorigenesis of breast cancer. Oncotarget. 2015;6:44495–44508. doi: 10.18632/oncotarget.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang F., Huang Y.S., Shi X.H., Zhang Q. Mitochondrial chaperone tumour necrosis factor receptor-associated protein 1 protects cardiomyocytes from hypoxic injury by regulating mitochondrial permeability transition pore opening. Febs J. 2010;277:1929–1938. doi: 10.1111/j.1742-4658.2010.07615.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida S.T.S., Muhlebach G., Sourbier C., Lee M.J., Lee S., Vartholomaiou E. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondria and aerobic glycolysis. Proc Natl Acad Sci U S A. 2013;110:e1604. doi: 10.1073/pnas.1220659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg F., Hamanaka R., Wheaton W.W., Weinberg S., Joseph J., Lopez M. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sciacovelli M., Guzzo G., Morello V., Frezza C., Zheng L., Nannini N. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. CellMetab. 2013;17:988–999. doi: 10.1016/j.cmet.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegelin M.D. Inhibition of the mitochondrial Hsp90 chaperone network: a novel, efficient treatment strategy for cancer? Cancer Lett. 2013;333:133–146. doi: 10.1016/j.canlet.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 11.Neckers L., Ivy S.P. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Lee C., Park H.K., Jeong H., Lim J., Lee A.J., Cheon K.Y. Development of a mitochondria-targeted Hsp90 inhibitor based on the crystal structures of human TRAP1. J Am Chem Soc. 2015;137:4358–4367. doi: 10.1021/ja511893n. [DOI] [PubMed] [Google Scholar]
- 13.Drysdale M.J., Brough P.A., Massey A., Jensen M.R., Schoepfer J. Targeting Hsp90 for the treatment of cancer. Curr Opin Drug Discov Devel. 2006;9:483–495. [PubMed] [Google Scholar]
- 14.Rock K.L., Goldberg A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 15.Engelhard V.H. Structure of peptides associated with MHC class I molecules. Curr Opin Immunol. 1994;6:13–23. doi: 10.1016/0952-7915(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 16.Shastri N., Schwab S., Serwold T. Producing nature's gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 17.Skipper J.C., Hendrickson R.C., Gulden P.H., Brichard V., Van Pel A., Chen Y. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sykulev Y., Joo M., Vturina I., Tsomides T.J., Eisen H.N. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 19.Condelli V., Piscazzi A., Sisinni L., Matassa D.S., Maddalena F., Lettini G. TRAP1 is involved in BRAF regulation and downstream attenuation of ERK phosphorylation and cell-cycle progression: a novel target for BRAF-mutated colorectal tumors. Cancer Res. 2014;74:6693–6704. doi: 10.1158/0008-5472.CAN-14-1331. [DOI] [PubMed] [Google Scholar]
- 20.Masgras I., Ciscato F., Brunati A.M., Tibaldi E., Indraccolo S., Curtarello M. Absence of neurofibromin induces an oncogenic metabolic switch via mitochondrial ERK-mediated phosphorylation of the chaperone TRAP1. Cell Rep. 2017;18:659–672. doi: 10.1016/j.celrep.2016.12.056. [DOI] [PubMed] [Google Scholar]
- 21.Andersen M.H., Bonfill J.E., Neisig A., Arsequell G., Sondergaard I., Valencia G. Phosphorylated peptides can be transported by TAP molecules, presented by class I MHC molecules, and recognized by phosphopeptide-specific CTL. J Immunol. 1999;163:3812–3818. [PubMed] [Google Scholar]
- 22.Zaring A.L., Ficarro S.B., White F.M., Shabanowitz J., Hunt D.F., Engelhard V.H. Phosphorylated peptide are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J Exp Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammed F., Cobbold M., Zarling A.L., Salim M., Barrett-Wilt G.A., Shabanowitz J. Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol. 2008;9:1236–1243. doi: 10.1038/ni.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Admon A., Barnea E., Ziv T. Tumor antigens and proteomics from the point of view of the major histocompatibility complex peptides. Mol Cell Proteomics. 2003;2:388–398. doi: 10.1074/mcp.R300004-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Newberg M.H., Smith D.H., Haertel S.B., Vining D.R., Lacy E., Engelhard V.H. Importance of MHC class 1 alpha2 and alpha3 domains in the recognition of self and non-self MHC molecules. J Immunol. 1996;156:2473–2480. [PubMed] [Google Scholar]
- 26.Peng S.T.C., He L., Tsai Y.-C., Lin C.-T., Boyd D., Pardoll D. Characterization of HLA-A2-restricted HPV 16 E7 specific CD8+ T cell immune responses induced by DNA vaccines in HLA-A2 transgenic mice. GeneTher. 2006;13:67–77. doi: 10.1038/sj.gt.3302607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarling A., Polefrone J.M., Evans A.M., Mikesh L.M., Shabanowitz J., Lewis S.T. identification of class I MHC-associated phosphopeptides as targets for cancer immunotherpy. Proc Natl Acad Sci USA. 2006;103:14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmel C., Stevanovic S. The use of HPLC-MS in T-cell epitope identification. Methods. 2003;29:248–259. doi: 10.1016/s1046-2023(02)00347-x. [DOI] [PubMed] [Google Scholar]
- 29.Sher Y.P., Lin S.I., Chen I.H., Liu H.Y., Lin C.Y., Chiang I.P. A HLA-A2-restricted CTL epitope induces anti-tumor effects against human lung cancer in mouse xenograft model. Oncotarget. 2016;7:671–683. doi: 10.18632/oncotarget.6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin C.Y., Chen H.J., Huang C.C., Lai L.C., Lu T.P., Tseng G.C. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014;74:5229–5243. doi: 10.1158/0008-5472.CAN-13-2995. [DOI] [PubMed] [Google Scholar]
- 31.Meyer V.S., Drews O., Gunder M., Hennenlotter J., Rammensee H.G., Stevanovic S. Identification of natural MHC class II presented phosphopeptides and tumor-derived MHC class I phospholigands. J Proteome Res. 2009;8:3666–3674. doi: 10.1021/pr800937k. [DOI] [PubMed] [Google Scholar]
- 32.Zarling A.L., Ficarro S.B., White F.M., Shabanowitz J., Hunt D.F., Engelhard V.H. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo. J Exp Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen M., Lundegaard C., Worning P., Lauemoller S.L., Lamberth K., Buus S. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O., Nielsen M. NetMHC- 3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweitzer S., Schneiders A.M., Langhans B., Kraas W., Jung G., Vidalin O. Flow cytometric analysis of peptide binding to major histocampatibility complex class I for hepatitis C virus core T-cell epitopes. Cytometry. 2000;41:271–278. doi: 10.1002/1097-0320(20001201)41:4<271::aid-cyto5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 37.Tu S.H., Huang H.I., Lin S.I., Liu H.Y., Sher Y.P., Chiang S.K. A novel HLA-A2-restricted CTL epitope of tumor-associated antigen L6 can inhibit tumor growth in vivo. J Immunother. 2012;35:235–244. doi: 10.1097/CJI.0b013e318248f2ae. [DOI] [PubMed] [Google Scholar]
- 38.Alexander J., Sidney J., Southwood S., Ruppert J., Oseroff C., Maewal A. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 39.Wu C.Y., Monie A., Pang X., Hung C.F., Wu T.C. Improving therapeutic HPV peptide-based vaccine potency by enhancing CD4+ T help and dendritic cell activation. J Biomed Sci. 2010;17:88. doi: 10.1186/1423-0127-17-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agorreta J., Hu J., Liu D., Delia D., Turley H., Ferguson D.J.P. TRAP1 regulates proliferation, mitochondria function, and has prognostic significance in NSCLC. Mol Caner Res. 2014;12:660–669. doi: 10.1158/1541-7786.MCR-13-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Los G., Verdegaal E., Noteborn H.P., Ruevekamp M., de Graeff A., Meesters E.W. Cellular pharmacokinetics of carboplatin and cisplatin in relation to their cytotoxic action. Biochem Pharmacol. 1991;42:357–363. doi: 10.1016/0006-2952(91)90723-i. [DOI] [PubMed] [Google Scholar]
- 42.Slingluff C.L., Jr., Colella T.A., Thompson L., Graham D.D., Skipper J.C., Caldwell J. Melanomas with concordant loss of multiple melanocytic differentiation proteins: immune escape that may be overcome by targeting unique or undefined antigens. CancerImmunol Immunother. 2000;48:661–672. doi: 10.1007/s002620050015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen J., Wurzbacher S.J., Williamson N.A., Ramarathnam S.H., Reid H.H., Mair A.K.-N. Phosphorylated self peptides alter human leukocyte antigen class I-restricted antigen presentation and generate tumor-specific epitopes. Proc Natl Acad Sci USA. 2009;106:2776–2781. doi: 10.1073/pnas.0812901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantley L.C., Auger K.R., Carpenter C., Duckworth B., Graziani A., Kapeller R. Oncogenes and signal transduction. Cell. 1991;64:281–302. doi: 10.1016/0092-8674(91)90639-g. [DOI] [PubMed] [Google Scholar]
- 45.Bohm W., Thoma S., Leithauser F., Moller P., Schirmbeck R., Reimann J. T cell-mediated, IFN-gamma-facilitated rejection of murine B16 melanomas. J Immunol. 1998;161:897–908. [PubMed] [Google Scholar]
- 46.Krasagakis K., Garbe C., Zouboulis C.C., Orfanos C.E. Growth control of melanoma cells and melanocytes by cytokines. Recent Res Cancer Res. 1995;139:169–182. doi: 10.1007/978-3-642-78771-3_12. [DOI] [PubMed] [Google Scholar]
- 47.Mougiakakos D., Choudhury A., Lladser A., Kiessling R., Johansson C.C. Regulatory T cells in cancer. Adv Cancer Res. 2010;107:57–117. doi: 10.1016/S0065-230X(10)07003-X. [DOI] [PubMed] [Google Scholar]