Abstract
Autophagy is a cellular survival mechanism that is induced by cancer therapy, among other stresses, and frequently contributes to cancer cell survival during long periods of dormancy and the eventual outgrowth of metastatic disease. Autophagy degrades large cellular structures that, once broken down, contribute to cellular survival through the recycling of their constituent metabolites. However, the extent to which this fuel function of autophagy is key to its role in promoting stemness, dormancy and drug resistance remains to be determined. Other roles for autophagy in determining cell fate more directly through targeted degradation of key transcription factors, such as p53 and FoxO3A, or by enforcing a reversible quiescent growth arrest, are discussed in this review. This review also highlights the need to parse out the roles of different forms of selective autophagy in stemness, CD44 expression and dormancy that, for example, are increasingly being attributed explicitly to mitophagy. The clinical relevance of this work and how an increased understanding of functions of autophagy in stemness, dormancy and drug resistance could be manipulated for increased therapeutic benefit, including eliminating minimal residual disease and preventing metastasis, are discussed.
Keywords: autophagy, mitochondria, stem cells, drug resistance, CD44, FoxO3A, dormancy, quiescence
Introduction
Macro-autophagy (generally referred to as autophagy) is a highly conserved catabolic process in which double-membraned vesicles called autophagosomes form around cellular cargo, including organelles, protein aggregates and intracellular pathogens, leading to their degradation following fusion of the autophagosome with the lysosome [1–3]. Although much of the regulation of autophagy occurs at the post-translational level, ensuring a rapid response to nutrient stress, autophagy-related genes (ATG) genes are also transcriptionally regulated in response to amino acid deprivation and ER stress via the ATF4 and MIT/TFE transcription factors [4,5]. ATG-encoded gene products promote autophagosome formation in three major steps involving the serine kinase activity of the pre-initiation complex, the lipid kinase activity of the initiation complex and the ligase activity of the ATG5/ATG12/ATG16 complex that pulls in processed LC3/ATG8 to nascent phagophores [1].
The pre-initiation complex (containing ATG13, FIP200, ATG101 and the ULK1/ULK2 serine/threonine kinases) is negatively regulated by mammalian target of rapamycin (mTOR) and positively regulated by AMPK, rendering autophagy highly sensitive to both amino acid deprivation and cellular energy deficits [6,7]. ULK1/ATG1 (or ULK2) phosphorylates Beclin1/ATG6 to activate the lipid kinase activity of VPS34 (a class III PI3K), the catalytic component of the initiation complex (also containing ATG14L, VPS15 and other regulatory factors, in addition to Beclin1), increasing phosphoinositol-3-phosphate (PIP3) production. PIP3 promotes recruitment of additional components of the autophagy machinery to the growing phagophore [1], including the ATG5-ATG12/ATG16L-containing conjugation complex that transfers processed LC3-II from ATG3 to phosphatidylethanolamine to permit its integration into the lipid membranes of burgeoning phagophores [1,7].
Processed LC3 at expanding phagophores plays a central role in selecting cargo for degradation through direct interaction with the cargo itself, or indirectly through cargo adaptor molecules that contain specific motifs called LC3-interacting region motifs [8,9]. Selective autophagy includes (but is not limited to) mitophagy [10,11], ribophagy [12,13] and xenophagy [14] in which mitochondria, ribosomes and pathogens, respectively, are selectively targeted for autophagic degradation [3]. Maturation of the autophagosome also requires LC3-related proteins, and this leads to fusion with the lysosome, acid pH-dependent degradation of autophagosomal cargo and recycling of cargo constituents, including nucleotides, fatty acids and amino acids, to the cytosol, where they are now available for various biosynthetic processes that fuel tumor cell growth [1,2].
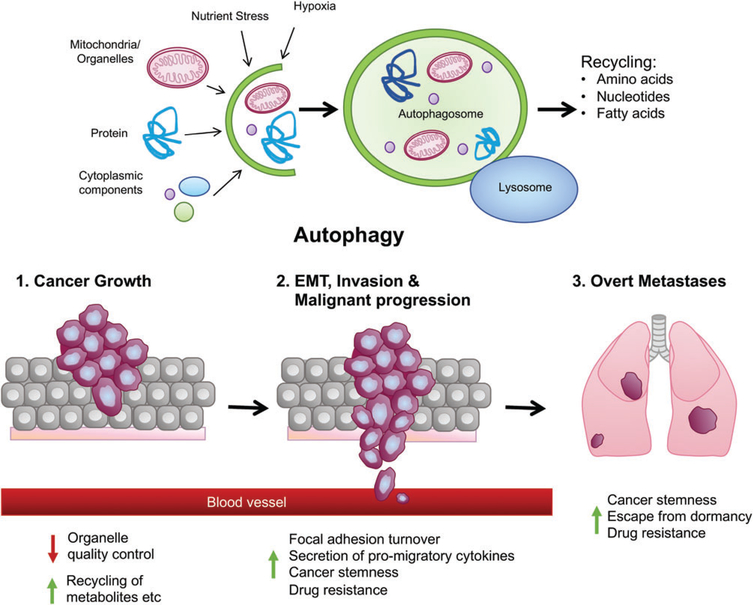
The role of autophagy in cancer is multifaceted [15], with known functions for autophagy in promoting tumor cell survival by supplying recycled metabolites for growth, modulating mitochondrial function via mitophagy [11,16,17] or interesting new functions in tumor cell migration and invasion via control of focal adhesion turnover and secretion of pro-migratory cytokines [18–21]. Autophagy also plays a central role in the tumor microenvironment [22,23] where, for example, autophagy is induced in cancer-associated fibroblasts by their association with tumor cells, resulting in increased fibroblast production of amino acids provided in a paracrine manner to tumor cells to sustain their growth [24–26]. Autophagy was also recently shown to be required in the liver to prevent the production of arginase-1, which degrades circulating arginine, such that loss of autophagy results in reduced circulating arginine, which limits growth of tumors implanted in autophagy-deficient mice [27]. Intriguingly, components of both the innate and adaptive immune systems also rely on autophagy to either sustain tumor growth or inhibit it depending on context [26,28–30] (Figure 1). Various recent reviews have focused on the role of autophagy in the tumor microenvironment, including anti-tumor immunity [23,26,29,30], in addition to comprehensive reviews on other aspects of the function of autophagy in cancer, such as tumor metabolism [16,17], cancer therapy [28,31] and cancer metastasis [21]. The goal of this review is to examine more recently reported roles for autophagy in cancer stem cells (CSCs), tumor cell dormancy and related mechanisms of cancer drug resistance.
Figure 1.
Multifaceted roles of autophagy in cancer. Autophagy is a catabolic process by which cells degrade large cellular cargoes, such as organelles, ribosomes and intracellular pathogens that are captured inside double-membraned autophagosomes before fusing with the lysosome and resultant constituent metabolites (amino acids, nucleotides, fatty acids) released to the cytsosol for reuse in biosynthetic processes and cell growth. This fuel function of autophagy is important in terms of promoting tumor cell survival at many stages in tumorigenesis. Autophagy also performs an organelle quality control function as part of cellular homeostasis that is important in both normal and tumor cells. As cells progress to becoming invasive, autophagy plays a role in promoting cell migration through focal adhesion disassembly and secretion of pro-migratory cytokines, such as IL-6. Autophagy also plays a role in the tumor microenvironment in modulating recruitment and response of T cells to the tumor and providing tumor cells with nutrients via amino acid transfer from cancer-associated fibroblasts to the tumor. Finally, emerging data have identified a role for autophagy in maintaining CSCs and tumor dormancy, both of which may play into drug resistance of cancers, minimal residual disease and metastatic latency.
Autophagy in CSCs
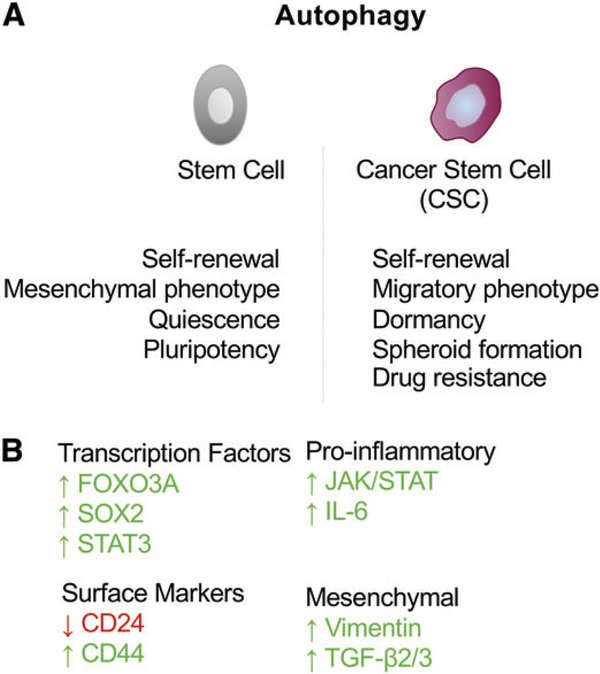
Tumor heterogeneity and progression to therapy-resistant disease in numerous different human cancers has been attributed to the properties of so-called CSCs, which have the capacity to self-renew, to regenerate all aspects of tumor heterogeneity and to evade cell killing due in part to their quiescent state, in addition to increased expression of drug transporters and other resistance genes [32–35]. Autophagy has emerged over the past several years as a requirement for the maintenance of stemness in both normal tissue stem cells [36–41] and CSCs [42–48]. The mechanisms by which autophagy contributes to stemness and why stem cells are more dependent on autophagy than non-stem cells are ongoing research interests of many laboratories (Figure 2A). In normal tissue stem cells, autophagy has been shown to promote neurogenesis through the management of oxidative stress responses and supply of metabolites to neural stem cells [39,49] and to be required for hematopoietic stem cell (HSC) maintenance through a FOXO3A-induced autophagy survival program [40]. Autophagy also promotes the survival of mesenchymal stem cells and human embryonic stem cells [37,38] and is required for the quiescent state of muscle stem cells [41]. Autophagy induces pluripotency with pluripotency factor SOX2 repressing mTOR expression, resulting in the increased autophagy necessary to reprogram somatic cells into induced pluripotent stem cells [50,51]. The requirement for autophagy during somatic cell reprogramming to induced pluripotent stem cells is complex, however, with autophagy-dependent degradation of p62 conversely shown to limit reprogramming [52].
Figure 2.
Autophagy in CSCs. (A) Autophagy plays an important role in maintaining both normal tissue stem cells and CSCs. The survival and quiescence of normal tissue stem cells appears dependent on autophagy and autophagy has also been reported to promote pluripotency. In CSCs, autophagy promotes expression of stem cell markers such as CD44 as well as expression of mesenchymal markers such as vimentin. Autophagy also promotes spheroid formation, in vivo tumorigenesis and drug resistance consistent with a critical role in maintaining CSCs. Inhibition of autophagy limits tumor dormancy and promotes outgrowth of metastases. (B) Key transcription factors have been linked to the induction of autophagy and the stem cell state, including FOXO3A, which induces expression of autophagy genes in stem cells and is itself turned over by autophagy. Also, SOX2 and STAT3 have been shown to modulate autophagy genes and to determine the stemness of CSCs.
Like tissue stem cells, CSCs also show autophagy dependence, with CSCs from primary human ductal carcinoma in situ of the breast reliant on autophagy for mammosphere formation, invasive properties and survival both in vitro and in vivo [53]. Beclin1 expression and autophagic flux are elevated in mammospheres and ALDH+ CSCs derived from mammospheres, compared with tumor cells in the bulk population or grown in 2D culture conditions [54]. Beclin1 and autophagy were also essential for CSC maintenance and tumorigenesis in vivo [54]. Similarly, CD44+CD24−/low breast CSCs were dependent on autophagic flux for survival and stem-like properties, including reduced expression of CD24, increased CD44 expression, vimentin expression and a mesenchymal phenotype induced by TGF-β [42]. Two different shRNA screens identified a key role for autophagy in maintaining breast CSCs with Beclin-1/ATG6 emerging from a shRNA screen for genes that modulate breast CSC plasticity [55] and ATG4A emerging from a screen for genes required for mammosphere formation [56]. Indeed, mammospheres showed increased expression of several autophagy and lysosomal genes and ATG4A was shown to promote CSC numbers and in vivo tumorigenicity [56]. Other genes that came out of this screen for stemness included components of JAK–STAT signaling pathways [56], which is significant given that STAT3 phosphorylation/activation has also been identified as a molecular readout of autophagy dependency in triple-negative breast cancer [57]; and CD44+CD24−/low CSC secretion of IL-6 (which signals through gp130 to JAK–STAT) is autophagy-dependent and required for CSC maintenance [44].
In mouse models of mammary tumorigenesis, autophagy was required to maintain two distinct pools of CSCs, both the highly invasive, mesenchymal CD29hiCD61+ CSCs from MMTV-PyMT and MMTV-Wnt1 transgenic mice and the more luminal ALDH+ CSCs from MMTV-PyMT mice [45,58]. This work showed that autophagy inhibition, through targeted deletion of the FIP200 component of the pre-initiation complex, disrupted both TGF-β/SMAD signaling required for CD29hiCD61+ CSCs and the activation of STAT3 required for ALDH+ CSCs [45]. The authors suggested that autophagy regulates turnover of CREB-related transcription factors known to modulate expression of TGF-β2 and TGF-β3 but did not explain how autophagy was required for STAT3-induced stemness, although it should be noted that IL-6, which is dependent on autophagy for its secretion [18,44], acts via gp130 and JAK2 to activate STAT3 [59]. Interestingly, STAT3 has also been reported to regulate expression of several autophagy genes, including Beclin1 and BNIP3 [18].
Stresses prevalent in the unique tumor microenvironment in which CSCs frequently reside, such as hypoxia and TGF-β, promote epithelial to mesenchymal transition (EMT), leading to increased self-renewal and upregulation of CD44 [60–65]. Induction of EMT promotes a CSC phenotype through transcription factors, including Slug and Twist, that activate self-renewal gene expression programs and tumor-propagating properties [60–65]. Significantly, stresses such as hypoxia and TGF-β also induce autophagy, alongside EMT and stemness [66,67], and transcription factors known to promote EMT, such as MITF in melanomagenesis, activate autophagy gene expression [5,68]. Other transcription factors, including the core stemness factors SOX2 and NANOG, have also been linked to autophagy induction [47]. For example, NANOG was recently shown to bind to the BNIP3L promoter, to induce autophagy under hypoxia and promote tumor cell resistance to immune-mediated killing by cytotoxic T cells [69].
Although the reporting on the role of autophagy in breast cancer CSCs is the most extensive, autophagy has also been implicated in maintaining CSCs in other cancer types, including pancreatic cancer [43,70], bladder cancer [46], colorectal cancer [71], chronic myeloid leukemia [72] and glioblastoma [73]. It remains to be determined to what extent the underlying pathways inducing autophagy in CSCs (Figure 2B), and explaining how autophagy promotes stemness, are conserved from one cancer type to another.
Mitophagy promotes stemness
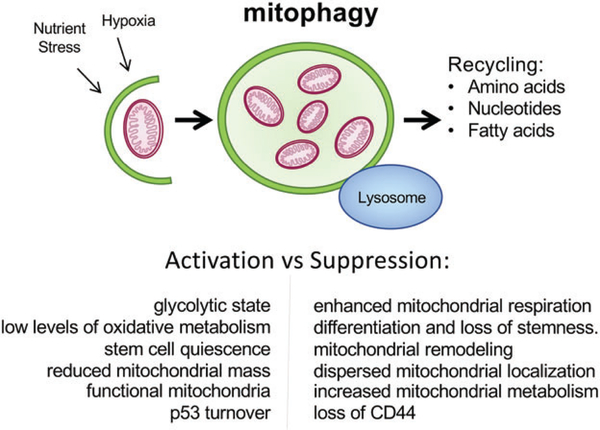
Autophagy is a broadly acting process operating to degrade numerous different cellular components and until recently the different functions of autophagy in stem cells had not been parsed out [21]. However, the selective degradation of mitochondria, or mitophagy as it is most commonly referred to, has now been directly implicated in stem cell self-renewal [74–77]. Mitophagy was shown to be required for the self-renewal of HSCs by turning over respiring mitochondria to maintain HSCs in a glycolytic state with low levels of oxidative metabolism [78–80]. The balance between glycolysis and oxidative metabolism has been reported in numerous systems to determine rates of stem cell quiescence versus differentiation [75,81–83]. Reducing mitochondrial mass through mitophagy limits oxidative metabolism, making stem cells dependent on glycolysis, which is less efficient in generating ATP than oxidative phosphorylation, thereby contributing to the slow cycling, self-renewing state that phenotypically defines stem cells [82]. Mitophagy also promotes preferential segregation of younger, more functional mitochondria to daughter CSCs and older mitochondria to daughter non-stem cells [74]. Conversely, suppression of mitophagy (or indeed increased mitochondrial biogenesis) enhances mitochondrial respiration to promote differentiation and loss of stemness. This is associated with mitochondrial remodeling, dispersed cytoplasmic localization of mitochondria and increased expression of enzymes involved in respiration and mitochondrial metabolism (Figure 3) [81,83–86].
Figure 3.
Mitophagy promotes stemness. Mitophagy is a selective form of autophagy in which mitochondria are specifically targeted for degradation at the autophagosome. Recently, mitophagy has been specifically implicated in maintaining the stem cell state by promoting turnover of mitochondria and limiting the capacity of the stem cell for oxidative phosphorylation and making stem cells more dependent on glycolysis for energy demands. This has also been proposed to contribute to the quiescent state of stem cells. Inhibition of mitophagy suppressed CD44 expression and also promoted translocation of p53 to the nucleus, where it has been reported to antagonize expression of stem cell genes.
Increased mitophagy was detected in esophageal squamous cell carcinoma cells undergoing EMT and inhibition of Parkin-dependent mitophagy in esophageal squamous cell carcinoma cells caused loss of expression of the stem cell marker CD44, leading to cell death [48]. Consistent with these findings, reduced mitochondrial mass distinguishes CSCs from non-CSCs in lung cancer and head and neck cancer [87,88]. In liver cancer, mitophagy has been reported to be required for the maintenance of hepatic CSCs [77]. This was achieved by eliminating p53 localized to mitochondria that was degraded in a mitophagy-dependent manner. When mitophagy was inhibited, PINK1 phosphorylated p53, resulting in its translocation to the nucleus, where it antagonized OCT4 and SOX2 induction of NANOG, a critical transcription factor required for stemness [77]. This intriguing finding suggests that mitophagy may regulate p53 localization and activity more broadly and, given that the stem cell marker CD44 is p53-regulated [89], it will be interesting to determine whether mitophagy-dependent regulation of CD44 levels [48] is p53-dependent. This work also prompts consideration of whether other key transcription factors that localize to the mitochondria, for example STAT3 and FOXO3A, also have their subcellular localization and activity modulated by mitophagy.
Autophagy and tumor dormancy
Disseminated tumor cells (DTCs) at secondary sites can remain dormant for decades, as is apparent from their outgrowth as overt metastatic lesions in breast cancer and prostate cancer patients who were treated effectively years before for their primary disease [90–93]. The insidious nature of dormant cancer cells has lent urgency to efforts to understand the mechanistic basis of dormancy. Autophagy is activated by nutrient deprivation and other stressful conditions that DTCs are probably exposed to when seeding new metastatic sites, leading investigators to test whether autophagy sustains tumor cell viability during dormancy [93]. Indeed, autophagy has been shown to promote the survival of dormant disseminated breast cancer cells and to be required for metastasis following dormancy in preclinical models of breast cancer [94]. Autophagy inhibition effectively reduced the metastatic burden in the lungs of transplanted mice and it was proposed that autophagy is required for the switch from dormancy to tumor cell growth, as autophagy inhibition specifically depleted dormant cells from tumors, leaving the proliferative tumor cells intact [94]. Also, inhibition of autophagy prevented dormant ovarian tumors expressing the ARHI (aplasia Ras homolog member I) tumor suppressor from growing out [95]. ARHI expression is lost in a significant proportion of ovarian cancers [96] and re-expression of ARHI in ARHI-deficient SKOv3 ovarian cancer cells induced autophagy and blocked tumor growth in transplanted mice [95]. Knocking down ARHI in these tumors allowed them to then grow out in an autophagy-dependent manner, indicating that the dormancy enforced by ARHI expression was autophagy-dependent [95]. Similarly, autophagy inhibition in the Eμ-Myc mouse model of B-cell lymphoma following treatment with alkylating agents blocked tumor recurrence consistent with a role for autophagy in both tumor dormancy and drug resistance [97]. These various studies provide powerful justification for the use of autophagy inhibitors in combination with conventional therapies to eliminate DTCs, minimal residual disease and prevent metastasis.
The relatively quiescent and motile state of CSCs that, like dormant tumor cells are dependent on autophagy for survival, has linked CSCs to dormancy and indeed CSC markers are upregulated on DTCs in the bone marrow of breast cancer patients [98], leading to the suggestion that dormant tumor cells are in fact CSCs [93,95]. In a switchable mouse model of pancreatic ductal adenocarcinoma (PDAC), dormant tumor cells that survived K-Ras inactivation to promote tumor regrowth upregulated autophagy and showed features of CSCs, including the ability to form tumor spheroids, high CD44 expression and increased tumor initiation properties in vivo [70]. Interestingly, these dormant PDAC CSCs had an increased dependence on autophagy and mitochondrial function, including β-oxidation of fatty acids, than non-CSCs for spheroid formation and survival, with dormant PDAC CSCs being more sensitive to inhibition of either autophagy or oxygen consumption than non-CSCs [70].
Autophagy may promote the dormancy of DTCs by supplying key metabolites or, as discussed above for CSCs, autophagy may play a more instructive role in dormancy by turning over key transcription factors that modulate the dormant stem-like state [93,99,100]. Alternatively, autophagy may promote tumor cell dormancy by ensuring a reversible quiescent state and preventing irreversible senescence, as was previously reported in muscle stem cells and HSCs [40,41,79]. The LKB1-AMPK axis is a major modulator of HSC homeostasis, acting to promote stem cell quiescence, mitochondrial function, lipid metabolism and survival [101–103] and, as previously mentioned, LKB1-AMPK signaling is also a potent activator of autophagy [6]. Quiescence and cell survival are coordinated downstream of LKB1-AMPK activation via a p27Kip1-dependent growth arrest in G1 of the cell cycle and the aforementioned AMPK-induced activation of the pre-initiation complex and AMPK-dependent phosphorylation of ULK1 [6,104]. Deletion of p27Kip1 results in rapid apoptotic cell death under metabolic stress and LKB1-AMPK signaling [104], suggesting a mechanism by which autophagy induction is linked to growth arrest to promote survival. Interestingly, loss of LKB1 was also associated with survival of aneuploid HSCs [102], with autophagy inhibition shown in separate studies to preferentially kill aneuploid cells due to accumulation of autophagic cargo in the lysosome [105,106]. Given the role of CSCs in therapy resistance and disease recurrence [35,107], and that dormant DTCs contribute to the metastatic outgrowth of cancers over time [91], understanding how autophagy can be effectively inhibited to suppress both of these phenotypes is a major challenge for translational cancer research.
Autophagy mediates cancer drug resistance
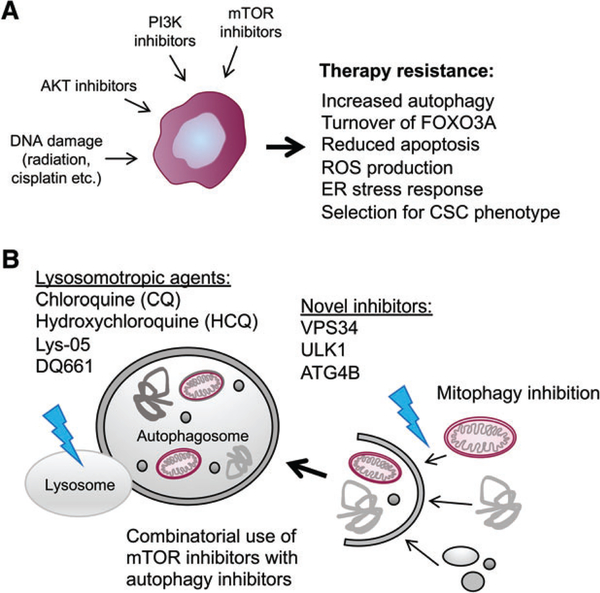
A wide range of cancer therapies has been shown to induce autophagy and in most cases, although not all, autophagy has been shown to promote tumor cell survival and contribute to therapy resistance (Figure 4A) [108–110]. For example, in estrogen receptor-positive breast cancer, inhibition of autophagy sensitized resistant tumors to tamoxifen-induced killing [111,112]. Similarly, in prostate cancer, autophagy inhibition overcame resistance to enzalutamide [113]. Autophagy is also induced in response to treatment of gastrointestinal stromal tumor (GIST) cells with Imatinib™ and inhibition of autophagy, including with the lysosomotropic agent chloroquine (CQ), caused tumor cell apoptosis [114]. Numerous studies in other cancer types and in response to other cancer therapies confirm that autophagy is both induced by treatments used and confers resistance to the treatment [28,110].
Figure 4.
Autophagy promotes cancer drug resistance. (A) Autophagy is induced in tumors by many different cancer therapeutic approaches, including irradiation, inhibition of PI3K, AKT or mTOR, as well as other conventional and targeted therapies. As a result of autophagy induction, tumor cells are more resistant to apoptosis, with an interesting mechanism revealed recently showing targeted turnover of FOXO3A by autophagy to prevent FOXO3A-dependent induction of Puma, a BH3-only pro-apoptotic protein [119]. Autophagy may also promote drug resistance by promoting selection for a CSC phenotype, as has been suggested by work in breast cancer [117] and glioblastoma [118]. (B) CQ and its derivatives are the mainstay of efforts to inhibit autophagy in a clinical setting and this has seen some efficacy in combination with conventional therapies for some cancers and drug combinations. Clinical trials with the combination of CQ and mTOR inhibitors are ongoing and there is particular interest in testing DQ661, which has the dual activity of inhibiting autophagy and mTOR at the lysosome. New generation autophagy inhibitors include small molecules targeted at catalytic components of autophagosome biogenesis, including ULK1, VPS34 and ATG4B.
The therapeutic induction of autophagy is frequently attributed to reduced mTOR activity leading to autophagy derepression, and this is most obvious with therapies targeted at inhibiting PI3K, AKT or indeed mTOR itself [109]. However, the induction of autophagy by other conventional and non-conventional treatments is varied and not completely understood. DNA damage-induced p53 activity may explain how autophagy is induced by conventional genotoxic agents, such as radiation or cisplatin, as a result of p53-mediated induction of autophagy regulators, such as DRAM1 [115]. However, the role of p53 in these responses is complicated by the fact that p53 is also pro-apoptotic and, depending on context, can also inhibit autophagy [116]. Other aspects of cancer therapy, including increased production of reactive oxygen species due to mitochondrial damage and an ER stress response due to protein aggregation may explain activation of autophagy via induction of the activity of the FOXO and ATF4 transcription factors, respectively, that are known to induce autophagy genes, such as ATG5, LC3 and others [4,40,117].
Such direct molecular mechanisms may explain how autophagy is upregulated in response to different therapies, but an alternative or parallel explanation is that therapy is selecting for those cancer cells that already have high levels of autophagy, namely therapy refractory CSCs. For example, different tumor types showed increased autophagy in response to irradiation treatment, and inhibition of autophagy reduced clonogenic survival of breast, lung and cervical cancer cell lines following irradiation [118]. Significantly, autophagy inhibition specifically reduced clonogenic survival of radioresistant tumor cells but not radiosensitive subclones, consistent with the radioresistant cells being more autophagy dependent [118]. Autophagic flux was also selectively higher in cisplatin-resistant bladder cancer cells and autophagy inhibition specifically depleted drug-resistant bladder CSCs [46]. In primary human glioblastoma (GBM), MST4 kinase (encoded by STK26) was upregulated due to promoter hypomethylation in glioblastoma stem cells (GSCs) in response to irradiation [119]. Elevated MST4 activity induced phosphorylation and activation of the ATG4B protease, leading to increased autophagic flux in GSCs, increased self-renewal properties and sphere formation, in addition to increased tumorigenicity in vivo [119]. Direct targeting of ATG4B or autophagy inhibition with CQ promoted the therapeutic effects of radiation in a GBM transplant model. This was associated with loss of GSC self-renewal capacity. Consistent with these findings, levels of MST4, phospho-ATG4B and LC3B staining correlated negatively with patient outcome for GBM. These findings suggest that there are indeed specific molecular mechanisms promoting autophagy induction in tumor cells, but also that at least some of these mechanisms are specific to CSCs and could contribute to explaining how CSCs are key mediators of drug resistance.
Recent work has identified a negative feedback loop wherein FoxO3A, a key transcriptional inducer of ATG genes in response to nutrient stress and reactive oxygen species, particularly in stem cells [40], is itself turned over by autophagy [120]. When autophagy was inhibited using CQ, FoxO3a accumulated in tumor cells, leading to increased expression of its pro-apoptotic target gene Puma, resulting in programmed cell death [120]. Indeed, this study showed an essential role for FoxO3A in binding to the gene regulatory region of Puma to promote synergistic tumor cell killing by genotoxic agents, including doxorubicin and etoposide, in combination with CQ [120]. Interestingly, CQ and autophagy inhibition also synergized with Nutlin-1 to activate p53 to super-induce Puma and cause cell death [120]. Together, this ever-growing body of research showing autophagy playing a cytoprotective effect provides a strong rationale to combine cancer therapeutic approaches with agents that inhibit autophagy [110,121].
Targeting autophagy for improved cancer treatment
As already alluded to, treatment with CQ or hydroxychloroquine (HCQ) is one of the most commonly used approaches to inhibit autophagy in the clinic to promote tumor cell killing by conventional chemotherapeutics (Figure 4B) [110,121]. CQ is a FDA-approved drug initially derived from the bark of the cinchona tree and used to treat malaria, arthritis and lupus, making it relatively cheap and accessible. Although CQ and HCQ inhibit autophagy, their mode of action is at the lysosome where they are trapped by protonation, leading to alkalinization of the lysosomes and inhibition of lysosomal acid protease activity [121]. Interestingly, metastatic tumor cells appear to be preferentially sensitive to CQ, and this was attributed to greater dependence on lysosomal function than non-metastatic tumor cells, but whether these metastatic cancer cells also exhibited increased stem-like properties was not examined [122]. Multiple clinical trials have now reported on the efficacy of combining CQ with conventional chemotherapies [110], such as in GBM treatment, where CQ in combination with temozolomide more than doubled patient survival times compared with temozolomide alone [121,123,124]. Similarly, CQ in combination with doxorubicin for the treatment of non-Hodgkin lymphoma in dogs showed improved overall drug response and progression-free survival compared with animals receiving doxorubicin alone [125]. CQ has now been tested in a range of different human cancers, for example in combination with gemcitabine for the treatment of PDAC [126], in combination with radiation for the treatment of metastatic breast cancer [127] and in combination with rapamycin analogs for different types of solid tumor and melanoma [128,129], in addition to ongoing trials comparing HCQ to CQ in adjuvant therapies [110].
A central challenge in assessing the efficacy of autophagy inhibitors in cancer therapy is to develop better, more reliable markers of autophagic flux in vivo to determine whether drug combinations are indeed effectively inhibiting autophagy as part of the treatment modality and outcome response [110]. This is particularly important as CQ has autophagy-independent effects, including inhibition of the ATM kinase [130] and effects on vasculature [121]. For example, recent studies showed that the growth suppressive effect of CQ was independent of autophagic flux in metastatic cancer cells and attributed to lysosomal dysfunction more specifically [122]. Added to this, CQ is currently used at micromolar doses, and such doses have been linked to toxicities, including fatal blood loss [110,121,131]. Furthermore, as autophagy is required systemically for tissue homeostasis in response to stress [132], one of the major concerns with autophagy inhibition as a therapeutic approach is whether the adverse consequences of systemic autophagy inhibition can be tolerated. This has spurred investigators to develop more potent analogs of CQ that are active at lower doses and have fewer side-effects, including Lys05, a dimeric version of CQ [131]. Interestingly, as autophagy and mTOR are both critically regulated at the lysosome [133,134] and the potent pro-growth activity of mTOR can dampen autophagy induction, Amaravadi and colleagues [135] recently took the innovative approach of screening for novel drugs that target the lysosome to both block autophagy and simultaneously inhibit mTOR activity. The lead compound they developed is a dimeric quinacrine (DQ661), which is derived from Lys05, has improved lysosomal targeting capability and impairs the activity of palmitoyl-protein thioesterase (PPT1), which is required for mTOR interaction with Rheb at the lysosome [135]; inhibition of PPT1 also prevents mTORC1 from associating with the lysosomal membrane, causing mTOR inhibition [136]. DQ661 appears to function much more effectively than existing mTOR inhibitors to suppress tumor growth and concomitantly inhibits autophagy by inhibiting the lysosome in a mouse melanoma model, a human colon cancer model and an orthotopic PDAC model [135]. Interestingly, PPT1 appears to be the common molecular target at the lysosome of CQ, HCQ and Lys05, in addition to DQ661, and deletion of PPT1 inactivated the ability of CQ or CQ derivatives to block autophagy [136]. Increased PPT1 expression in human cancers was linked to poor prognosis and, conversely, knockout of PPT1 in tumor cells inhibited tumor growth, tumor spheroid formation and tumorigenesis in vivo, suggesting that PPT1 may be a good therapeutic target [136].
As scientific understanding of autophagy has increased, the development of targeted small molecule inhibitors to key regulatory nodes in autophagy pathways has emerged as an alternative therapeutic approach, beyond CQ and related antimalarials (Figure 4B). For example, development of targeted inhibitors of ULK1 [137,138], VPS34 [139] and other enzymes required for autophagy [140] is a work in progress and Vescor Therapeutics LLC has been set up by leaders in the field to specifically develop new drugs along these lines (https://vescortx.com/pipeline/). Beyond the development of autophagy inhibitors, the development of specific mitophagy inhibitors also seems that it could prove productive, given the concerns about global autophagy inhibition for tissue homeostasis and that mitophagy appears to underlie many of the relevant functions previously attributed to general autophagy. Use of specific inhibitors will also be powerful moving forward to explore how autophagy determines stemness, whether dormant tumor cells are autophagy-dependent CSCs and which autophagy functions are key to understanding how autophagy promotes drug resistance and cancer recurrence.
Acknowledgements
This work was supported by T32 CA009594 (AGS) and RO1 CA216242 (KFM).
Footnotes
No conflicts of interest were declared.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011; 147 728–741. [DOI] [PubMed] [Google Scholar]
- 2.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J 2017; 36 1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rouschop KM, van den Beucken T, Dubois L, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010; 120 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of the autophagy-lysosome system in pancreatic cancer. Nature 2015; 524 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci 2015; 128 193–205. [DOI] [PubMed] [Google Scholar]
- 8.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80 125–156. [DOI] [PubMed] [Google Scholar]
- 9.Rogov V, Dotsch V, Johansen T, et al. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53 167–178. [DOI] [PubMed] [Google Scholar]
- 10.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 2014; 157 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake LE, Springer MZ, Poole LP, et al. Expanding perspectives on the significance of mitophagy in cancer. Semin Cancer Biol 2017; 47 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An H, Harper JW. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 2018; 20 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyant GA, Abu-Remaileh M, Frenkel EM, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 2018; 360 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X, Levine B. Autophagy and viruses: adversaries or allies? J Innate Immun 2013; 5 480–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016; 30 1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell 2016; 61 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab 2017; 25 1037–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lock R, Kenific CM, Leidal AM, et al. Autophagy dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer Discov 2014; 4 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharifi MN, Mowers EE, Drake LE, et al. Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of Paxillin with LC3. Cell Rep 2016; 15 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenific CM, Stehbens SJ, Goldsmith J, et al. NBR1 enables autophagy-dependent focal adhesion turnover. J Cell Biol 2016; 212 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene 2017; 36 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maes H, Rubio N, Garg AD, et al. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med 2013; 19 428–446. [DOI] [PubMed] [Google Scholar]
- 23.Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J 2018; 285 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katheder NS, Khezri R, O’Farrell F, et al. Microenvironmental autophagy promotes tumour growth. Nature 2017; 541 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sousa CM, Biancur DE, Wang X, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 2016; 536 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang A, Herter-Sprie G, Zhang H, et al. Autophagy sustains pancreatic cancer growth through both cell autonomous and non-autonomous mechanisms. Cancer Discov 2018; 8 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poillet-Perez L, Xie X, Zhan L, et al. Autophagy maintains tumour growth through circulating arginine. Nature 2018; 563 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galluzzi L, Bravo-San Pedro JM, Levine B, et al. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov 2017; 16 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Galluzzi L, Zitvogel L, et al. Autophagy and cellular immune responses. Immunity 2013; 39 211–227. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell 2016; 166 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulcahy Levy JM, Zahedi S, Griesinger AM, et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. Elife 2017; 6 e19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013; 501 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014; 14 275–291. [DOI] [PubMed] [Google Scholar]
- 34.Ramos EK, Hoffmann AD, Gerson SL, et al. New opportunities and challenges to defeat cancer stem cells. Trends Cancer 2017; 3 780–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017; 14 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Lee JY, Wei H, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 2010; 116 4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliver L, Hue E, Priault M, et al. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev 2012; 21 2779–2788. [DOI] [PubMed] [Google Scholar]
- 38.Tra T, Gong L, Kao LP, et al. Autophagy in human embryonic stem cells. PLoS One 2011; 6 e27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez P, Arroba AI, Cecconi F, et al. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 2012; 8 187–199. [DOI] [PubMed] [Google Scholar]
- 40.Warr MR, Binnewies M, Flach J, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 2013; 494 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Prat L, Martinez-Vicente M, Perdiguero E, et al. Autophagy maintains stemness by preventing senescence. Nature 2016; 529 37–42. [DOI] [PubMed] [Google Scholar]
- 42.Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, et al. Autophagy positively regulates the CD44(+) CD24(−/low) breast cancer stem-like phenotype. Cell Cycle 2011; 10 3871–3885. [DOI] [PubMed] [Google Scholar]
- 43.Rausch V, Liu L, A A, et al. Autophagy mediates survival of pancreatic tumour-initiating cells in a hypoxic microenvironment. J Pathol 2012; 227 325–335. [DOI] [PubMed] [Google Scholar]
- 44.Maycotte P, Jones KL, Goodall ML, et al. Autophagy supports breast cancer stem cell maintenance by regulating IL6 secretion. Mol Cancer Res 2015; 13 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo SK, Wen J, Chen S, et al. Autophagy differentially regulates distinct breast cancer stem-like cells in murine models via EGFR/Stat3 and Tgfbeta/Smad signaling. Cancer Res 2016; 76 3397–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ojha R, Singh SK, Bhattacharyya S. JAK-mediated autophagy regulates stemness and cell survival in cisplatin resistant bladder cancer cells. Biochim Biophys Acta 2016; 1860 2484–2497. [DOI] [PubMed] [Google Scholar]
- 47.Sharif T, Martell E, Dai C,et al. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy 2017; 13 264–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan KA, Chandramouleeswaran PM, Tanaka K, et al. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene 2017; 36 4843–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Liang CC, Bian ZC, et al. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 2013; 16 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Xia P, Ye B, et al. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell 2013; 13 617–625. [DOI] [PubMed] [Google Scholar]
- 51.Ma T, Li J, Xu Y, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol 2015; 17 1379–1387. [DOI] [PubMed] [Google Scholar]
- 52.Wu Y, Li Y, Zhang H, et al. Autophagy and mTORC1 regulate the stochastic phase of somatic cell reprogramming. Nat Cell Biol 2015; 17 715–725. [DOI] [PubMed] [Google Scholar]
- 53.Espina V, Mariani BD, Gallagher RI, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One 2010; 5 e10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong C, Bauvy C, Tonelli G, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene 2013; 32 2261–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta PB, Fillmore CM, Jiang G, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 2011; 146 633–644. [DOI] [PubMed] [Google Scholar]
- 56.Wolf J, Dewi DL, Fredebohm J, et al. A mammosphere formation RNAi screen reveals thatATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res 2013; 15 R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maycotte P, Gearheart CM, Barnard R, et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res 2014; 74 2579–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeo SK, Guan JL. Hierarchical heterogeneity in mammary tumors and its regulation by autophagy. Autophagy 2016; 12 1960–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.You L, Wang Z, Li H, et al. The role of STAT3 in autophagy. Autophagy 2015; 11 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo W, Keckesova Z, Donaher JL, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012; 148 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011; 331 1559–1564. [DOI] [PubMed] [Google Scholar]
- 62.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A 2011; 108 7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mani SA, Guo W, Liao MJ, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.May CD, Sphyris N, Evans KW, et al. Epithelial–mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res 2011; 13 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye X, Tam WL, Shibue T, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015; 525 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiyono K, Suzuki HI, Matsuyama H, et al. Autophagy is activated by TGF-b and potentiates TGF-b-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res 2009; 69 8844–8852. [DOI] [PubMed] [Google Scholar]
- 67.Li J, Yang B, Zhou Q, et al. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial–mesenchymal transition. Carcinogenesis 2013; 34 1343–1351. [DOI] [PubMed] [Google Scholar]
- 68.Caramel J, Papadogeorgakis E, Hill L, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013; 24 466–480. [DOI] [PubMed] [Google Scholar]
- 69.Hasmim M, Janji B, Khaled M, et al. Cutting edge: NANOG activates autophagy under hypoxic stress by binding to BNIP3L promoter. J Immunol 2017; 198 1423–1428. [DOI] [PubMed] [Google Scholar]
- 70.Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014; 514 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kantara C, O’Connell M, Sarkar S, et al. Curcumin promotes autophagic survival of a subset of colon cancer stem cells, which are ablated by DCLK1-siRNA. Cancer Res 2014; 74 2487–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bellodi C, Lidonnici MR, Hamilton A, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest 2009; 119 1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Galavotti S, Bartesaghi S, Faccenda D, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene 2013; 32 699–712. [DOI] [PubMed] [Google Scholar]
- 74.Katajisto P, Dohla J, Chaffer CL, et al. Stem cellsAsymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science 2015; 348 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adams WC, Chen YH, Kratchmarov R, et al. Anabolism-associated mitochondrial stasis driving lymphocyte differentiation over self-renewal. Cell Rep 2016; 17 3142–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sin J, Andres AM, Taylor DJ, et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016; 12 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu K, Lee J, Kim JY, et al. Mitophagy controls the activities of tumor suppressor p53 to regulate hepatic cancer stem cells. Mol Cell 2017; 68 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ito K, Turcotte R, Cui J, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 2016; 354 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho TT, Warr MR, Adelman ER, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature 2017; 543 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vannini N, Girotra M, Naveiras O, et al. Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun 2016; 7 13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu W, Gaeta X, Sahakyan A, et al. Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell 2016; 19 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 2014; 15 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu X, Duan S, Yi F, et al. Mitochondrial regulation in pluripotent stem cells. Cell Metab 2013; 18 325–332. [DOI] [PubMed] [Google Scholar]
- 84.Chung S, Dzeja PP, Faustino RS, et al. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 2007; 4(suppl 1): S60–S67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Varum S, Rodrigues AS, Moura MB, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 2011; 6 e20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 2011; 14 264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ye XQ, Li Q, Wang GH, et al. Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer 2011; 129 820–831. [DOI] [PubMed] [Google Scholar]
- 88.Shen YA, Wang CY, Hsieh YT, et al. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 2015; 14 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Godar S, Ince TA, Bell GW, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008; 134 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karrison TG, Ferguson DJ, Meier P. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 1999; 91 80–85. [DOI] [PubMed] [Google Scholar]
- 91.McGowan PM, Kirstein JM, Chambers AF. Micrometastatic disease and metastatic outgrowth: clinical issues and experimental approaches. Future Oncol 2009; 5 1083–1098. [DOI] [PubMed] [Google Scholar]
- 92.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell 2013; 155 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014; 14 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vera-Ramirez L, Vodnala SK, Nini R, et al. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018; 9 1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu Z, Luo RZ, Lu Y, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cells. J Clin Invest 2008; 118 3917–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosen DG, Wang L, Jain AN, et al. Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res 2004; 10 6559–6566. [DOI] [PubMed] [Google Scholar]
- 97.Amaravadi RK, Yu DS, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 2007; 117 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Balic M, Lin H, Young L, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res 2006; 12 5615–5621. [DOI] [PubMed] [Google Scholar]
- 99.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 2005; 120 237–249. [DOI] [PubMed] [Google Scholar]
- 100.Galluzzi L, Pietrocola F, Levine B, et al. Metabolic control of autophagy. Cell 2014; 159 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gurumurthy S, Xie SZ, Alagesan B, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature 2010; 468 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature 2010; 468 653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gan B, Hu J, Jiang S, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature 2010; 468 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liang J, Saho SH, Xu ZX, et al. The energy sensing LKB1-AMPK pathway regulates p27kip1 phosphorylation mediatiing the decision to enter autophagy or apoptosis. Nat Cell Biol 2007; 9 218–224. [DOI] [PubMed] [Google Scholar]
- 105.Tang YC, Williams BR, Siegel JJ, et al. Identification of aneuploidy-selective antiproliferation compounds. Cell 2011; 144 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Santaguida S, Vasile E, White E, et al. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev 2015; 29 2010–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011; 17 1086–1093. [DOI] [PubMed] [Google Scholar]
- 108.Kondo Y, Kanzawa T, Sawaya R, et al. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer 2005; 5 726–734. [DOI] [PubMed] [Google Scholar]
- 109.Amaravadi RK, Lippincott-Schwartz J, Yin XM, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 2011; 17 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017; 17 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qadir MA, Kwok B, Dragowska WH, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat 2008; 112 389–403. [DOI] [PubMed] [Google Scholar]
- 112.Samaddar JS, Gaddy VT, Duplantier J, et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol Cancer Ther 2008; 7 2977–2987. [DOI] [PubMed] [Google Scholar]
- 113.Nguyen HG, Yang JC, Kung HJ, et al. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene 2014; 33 4521–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta A, Roy S, Lazar AJF, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST). Proc Natl Acad Sci U S A 2010; 107 14333–14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crighton D, Wilkinson S, O’Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126 121–134. [DOI] [PubMed] [Google Scholar]
- 116.Simon HU, Friis R, Tait SW, et al. Retrograde signaling from autophagy modulates stress responses. Sci Signal 2017; 10 eaag2791. [DOI] [PubMed] [Google Scholar]
- 117.Ranganathan AC, Zhang L, Adam AP, et al. Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res 2006; 66 1702–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Apel A, Herr I, Schwarz H, et al. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res 2008; 68 1485–1494. [DOI] [PubMed] [Google Scholar]
- 119.Huang T, Kim CK, Alvarez AA, et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell 2017; 32 840–855.e848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fitzwalter BE, Towers CG, Sullivan KD, et al. Autophagy inhibition mediates apoptosis sensitization in cancer therapy by relieving FOXO3a turnover. Dev Cell 2018; 44 555–565.e553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pascolo S Time to use a dose of chloroquine as an adjuvant to anti-cancer chemotherapies. Eur J Pharmacol 2016; 771 139–144. [DOI] [PubMed] [Google Scholar]
- 122.Morgan MJ, Fitzwalter BE, Owens CR, et al. Metastatic cells are preferentially vulnerable to lysosomal inhibition. Proc Natl Acad Sci U S A 2018; 115 E8479–E8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg Focus 2003; 14 e3. [DOI] [PubMed] [Google Scholar]
- 124.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med 2006; 144 337–343. [DOI] [PubMed] [Google Scholar]
- 125.Barnard RA, Wittenburg LA, Amaravadi RK, et al. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy 2014; 10 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boone BA, Bahary N, Zureikat AH, et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann Surg Oncol 2015; 22 4402–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rojas-Puentes LL, Gonzalez-Pinedo M, Crismatt A, et al. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiat Oncol 2013; 8 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rangwala R, Leone R, Chang YC, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 2014; 10 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rangwala R, Chang YC, Hu J, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014; 10 1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Loehberg CR, Thompson T, Kastan MB, et al. Ataxia telangiectasia-mutated and p53 are potential mediators of chloroquine-induced resistance to mammary carcinogenesis. Cancer Res 2007; 67 12026–12033. [DOI] [PubMed] [Google Scholar]
- 131.McAfee Q, Zhang Z, Samanta A, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A 2012; 109 8253–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karsli-Uzunbas G, Guo JY, Price S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 2014; 4 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wyant GA, Abu-Remaileh M, Wolfson RL, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 2017; 171 642–654.e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rebecca VW, Nicastri MC, McLaughlin N, et al. A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov 2017; 7 1266–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rebecca VW, Nicastri MC, Fennelly C, et al. PPT1 promotes tumor growth and is the molecular target of chloroquine derivatives in cancer. Cancer Discov 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Egan DF, Chun MG, Vamos M, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell 2015; 59 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Petherick KJ, Conway OJ, Mpamhanga C, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem 2015; 290 11376–11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 2014; 10 1013–1019. [DOI] [PubMed] [Google Scholar]
- 140.Akin D, Wang SK, Habibzadegah-Tari P, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 2014; 10: 2021–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]