Abstract
Tripartite motif‐containing 14 (TRIM14) is a mitochondrial adaptor that promotes innate immune signaling and plays important roles in antiviral defense. Expression of TRIM14 is induced by interferon (IFN)‐I. However, the mechanism by which IFN‐I induces TRIM14 production is not yet determined. In this study, we have examined the function of TRIM14 promoter and found that a GC box and an IFN‐stimulated response element (ISRE) are necessary for the basal level transcription of TRIM14. We further observed that IFN‐I activates the TRIM14 promoter through the ISRE. In particular, interferon regulatory factor (IRF)‐1 and IRF‐2 bind to the TRIM14 promoter and activate transcription of TRIM14. Moreover, knockdown of IRF‐1 reduces the stimulation of TRIM14 transcription by IFN‐α, suggesting that IRF‐1 is involved in the activation of TRIM14 by IFN‐I. IRF‐2 has little effect on IFN‐α‐induced TRIM14 transcription but is essential for the basal transcription of TRIM14.
Keywords: IRF‐1, IRF‐2, ISRE, promoter, TRIM14
Abbreviations
- IFN
interferon
- IRF
interferon regulatory factor
- ISG
interferon‐stimulated gene
- ISRE
IFN‐stimulated response element
- SD
standard deviation
- STAT
signal transducer and activator of transcription
- TRIM14
tripartite motif‐containing 14
- WT
wild‐type
Tripartite motif (TRIM) proteins contain a RING finger, one or two B‐box motifs and a coiled‐coil motif. Ample studies have demonstrated that TRIM family proteins play important roles in antiviral innate immune response. For example, TRIM22 activates nuclear factor‐κB signaling 1, and TRIM56 promotes double‐stranded DNA‐stimulated interferon induction by ubiquitination of stimulator of interferon gene (STING) 2, 3.
Tripartite motif‐containing 14 (TRIM14) is a member of the tripartite‐motif protein family; it is expressed in a variety of tissues 4. As a regulatory factor, it plays an important role in the innate immune response. In the retinoic acid‐inducible gene I (a sensor of virus double‐stranded RNA) signaling pathway, TRIM14 acts as a link protein in activating the expression of interferon (IFN)‐I. TRIM14 also enhances the IFN‐I signaling pathway by stabilizing cyclic GMP–AMP synthase (a DNA virus sensor). IFN‐I in turn upregulates the expression of TRIM14, thus enhancing the innate immune response mediated by the virus 5, 6. In addition, TRIM14 is necessary for retinoic acid‐inducible gene I‐mediated innate antiviral immunity by forming a WHIP–TRIM14–PPP6C mitochondrial complex 7.
Interferons regulate downstream genes through the Janus kinase–signal transducer and activator of transcription (STAT) pathway 8. IFN‐I binds to the interferon α and β receptor and activates the Janus kinase–STAT pathway to enhance the transcription of interferon‐stimulated genes (ISGs) 9, 10, 11. IFN regulatory factors (IRFs) are also involved in IFN‐mediated signaling pathways. The IRF family consists of nine members (IRF‐1–IRF‐9), which were characterized as transcriptional regulators of IFN and IFN‐inducible genes 12. IRF‐1 was the first IRF identified and is characterized by its ability to bind to the IFN‐β promoter and activate IFN‐β expression 13. IRF‐1 can regulate some IFN‐regulated genes by directly binding to the IFN‐stimulated response element (ISRE) of their promoters, including RANTES/CC15 14 and low molecular weight protein 7 15. IRF‐2 has a similar structure to IRF‐1; it is thus considered as a transcriptional inhibitor of IRF‐1‐mediated transcriptional activation. However, IRF‐2 also has a transcriptional activation function; for example, it can activate the expression of histone H4 16.
Many TRIM genes are regulated by IFN‐I or IFN‐II 17, 18, and IFN‐I can induce the expression of TRIM14. However, it has not been determined which elements and transcription factors activate TRIM14 expression following IFN‐I induction. In this study, we have determined that the ISRE in the TRIM14 promoter is necessary for the regulation of TRIM14 by IFN‐I. Furthermore, we found that IRF‐1 and IRF‐2 bind to the ISRE and mediate transcriptional activation of TRIM14 at different stages. IRF‐1 is involved in the activation of TRIM14 by IFN‐I, whereas IRF‐2 is essential for the basal transcription of TRIM14.
Results
Promoter activity analysis of TRIM14
In order to understand the mechanism by which TRIM14 expression is induced by IFNs, we analyzed its promoter activity. We determined the transcriptional initiation site of TRIM14 through 5′‐rapid amplification of cDNA ends (5′‐RACE) PCR. The products were cloned into pMD18‐T and sequenced. Sequencing results show that seven clones contain two transcriptional start sites and five of these clones started with cytosine, and we thus defined this cytosine as the transcription start site and mark it as +1 (Fig. 1A). Then, we cloned the 2020 bp DNA upstream of the transcription initiation site and performed sequence analysis. The results show that it contains three potential cis‐acting components, including two GC boxes (GC box 1 and GC box 2) and one ISRE (Fig. 1A).
Figure 1.
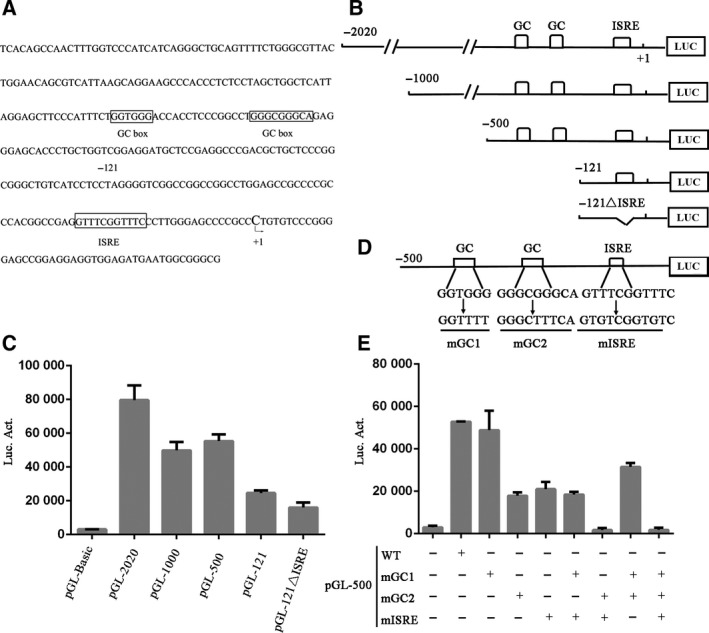
Identification and analysis of TRIM14 promoter. (A) Arrow indicates the starting point of transcription, and the C is designed as +1. The potential cis‐elements are boxed. (B) The 5′‐truncated plasmids of TRIM14 promoter. (C) The promoter constructs were transfected into HeLa cells. After transfection for 48 h, a luciferase assay was performed and β‐gal activity was used as a normalization control for the luciferase activity. (D) TRIM14 promoter core region mutation in pGL‐500. (E) The wild‐type (WT) or mutant PGL‐500 was transfected into HeLa cells; luciferase was detected 48 h after transfection. Results are presented as mean ± standard deviation (SD), and data used for the analysis were from three independent experiments.
To verify whether the predicted cis‐acting elements are involved in regulating TRIM14 promoter activity, we constructed a series of 5′ truncated plasmids (Fig. 1B), and then transfected these reporter plasmids into HeLa cells and measured the luciferase activity 48 h post‐transfection. The basal transcriptional activity of truncated TRIM14 promoter from −500 to −121 decreased significantly and the basal transcriptional activity of the TRIM14 promoter was further reduced by further truncations (Fig. 1C). The −500 to +1 bp region is consistent with our predicted positions of the cis‐acting elements. In order to further confirm whether the two GC boxes and the ISRE are involved in regulating the basic transcriptional activity of the TRIM14 promoter, we constructed a series of mutations on pGL‐500 (Fig. 1D). As shown in Fig. 1E, the basal transcriptional activity decreased by 75% (GC2 mutation) and 70% (ISRE mutation). However, GC1 mutation does not affect the basal transcriptional activity of the TRIM14 promoter (Fig. 1E, column 3, 7, 9). Therefore, we conclude that GC2 and ISRE are essential elements in the basal transcription of TRIM14.
The ISRE is essential for IFNs to activate TRIM14 promoter
To verify whether the TRIM14 promoter is regulated by IFNs, HeLa cells were transfected with pGL‐2020 or pGL‐121. Thirty‐two hours post‐transfection, cells were treated with IFN‐α or IFN‐γ for 16 h. As shown in Fig. 2A, both IFN‐α and IFN‐γ can upregulate gene expression from the TRIM14 promoter, and IFN‐α is more potent than IFN‐γ. pGL‐2020 and pGL‐121 are upregulated to almost the same extent by IFNs, indicating that only −121 to +1 bp is the region that responds to IFN activation. The region −121 to +1 contains an ISRE. To further confirm whether IFNs activate the promoter via the ISRE, we constructed two ISRE mutation plasmids (pGL‐2020mISRE, pGL‐121mISRE). We found that the TRIM14 promoter does not respond to IFNs when the ISRE is mutated (Fig. 2B,C). These results demonstrate that the ISRE is essential for IFNs to activate the TRIM14 promoter.
Figure 2.
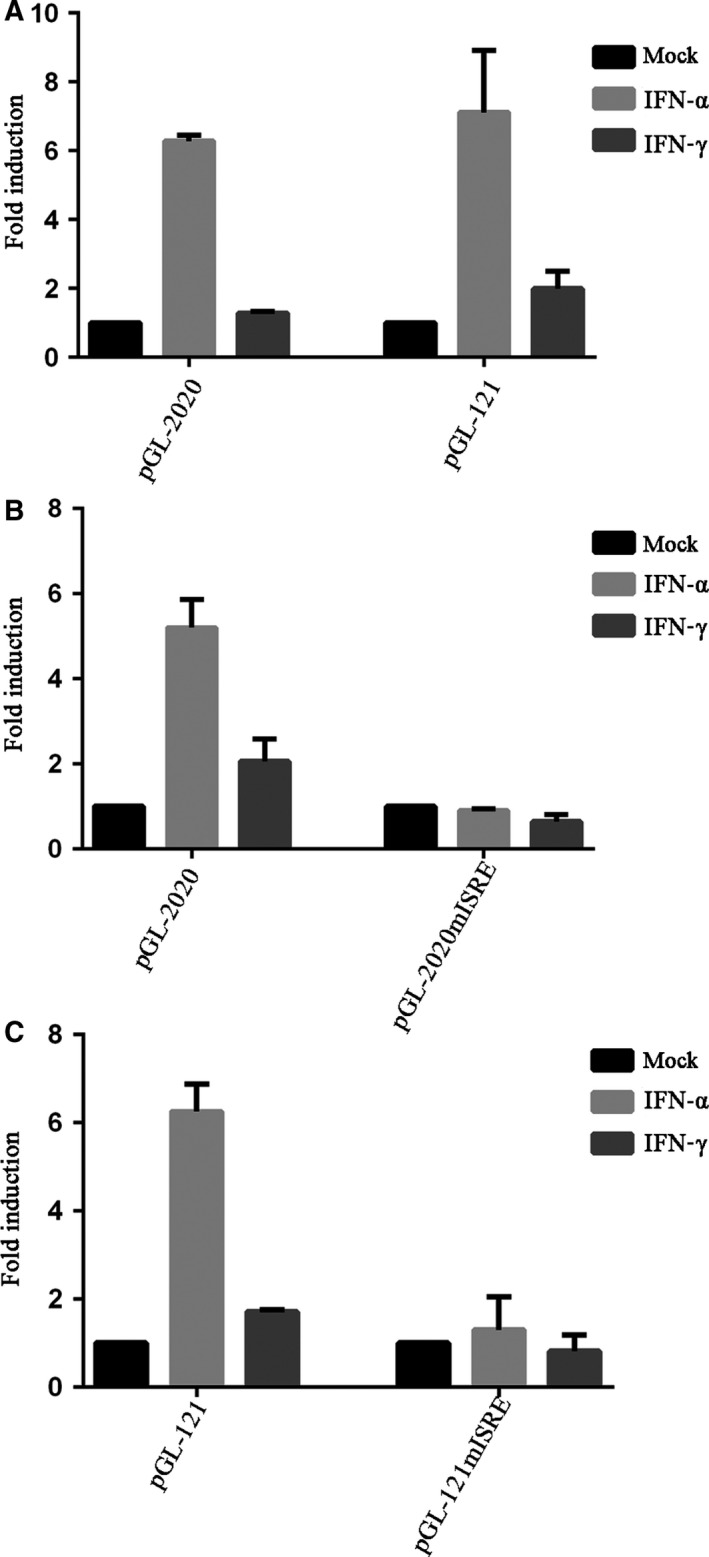
IFNs enhance TRIM14 transcription through ISRE. (A) pGL‐2020 or pGL‐121 was transfected into HeLa cells. After 32 h of transfection, the cells were treated with IFN‐α or IFN‐γ (10 ng·mL−1) for 16 h. Luciferase activity was determined after 48 h transfection. (B, C) pGL‐2020 or pGL‐2020ISRE or pGL‐121 or pGL‐121mISRE was transfected into HeLa cells and HeLa cells were stimulated with IFN‐α or IFN‐γ (10 ng·mL−1) 16 h before luciferase detection. Results are presented as mean ± SD, and data used for the analysis were from three independent experiments.
IRF‐1 and IRF‐2 bind to the ISRE to increase TRIM14 expression
Many members of the IRF family can regulate transcription of IFN‐stimulated genes. To determine whether the IRF family is involved in regulating TRIM14, we co‐transfected Myc–IRF‐1, Myc–IRF‐2, Myc–IRF‐3, Myc–IRF‐5 and Myc–IRF‐7 encoding plasmids with pGL‐121 into HeLa cells and examined pGL‐121 promoter activity. As shown in Fig. 3A, overexpression of IRF‐1 and IRF‐2 can significantly activate the TRIM14 promoter, and the activation by IRF‐1 is greater than that by IRF‐2. Because Myc‐IRF‐2 plasmid did not express (Fig. 3A), we co‐transfected pGL‐121 with pCDNA3.1‐IRF‐1 or pCDNA3.1‐IRF‐2 (expressed well) into HeLa cells and pGL‐121 promoter activity was examined. The results showed that IRF‐1 and IRF‐2 activated the TRIM14 promoter and increased TRIM14 protein expression (Fig. 3B).
Figure 3.
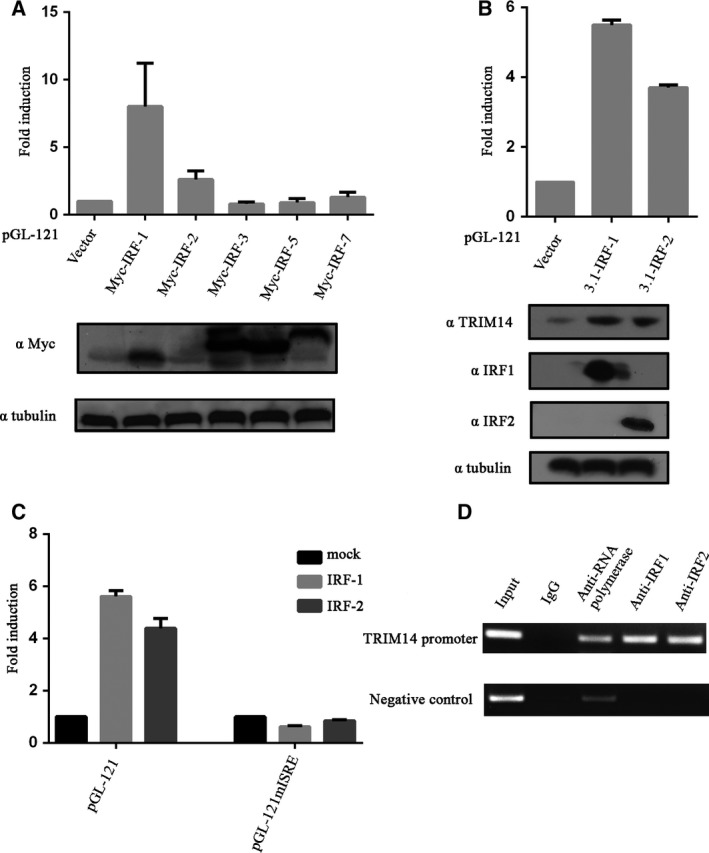
IRF‐1 and IRF‐2 bind to the ISRE to increase TRIM14 expression. (A, B) pGL‐121 with IRFs transfected into HeLa cells; luciferase activity was measured after transfection for 48 h and western blot analysis was performed. (C) PGL‐121 or PGL‐121mISRE was co‐transfected with pcDNA3.1 or pcDNA3.1‐IRF‐1 or pcDNA3.1‐IRF‐2 into HeLa cells. Luciferase activity was measured 48 h after transfection. (D) HeLa cells were treated with IFN‐α (10 ng·mL−1) for 12 h and detected with anti‐IRF‐1, anti‐IRF‐2 or control IgG and anti‐RNA polymerase. The designed TRIM14 ISRE primer was used to amplify precipitated DNA by PCR. Results are presented as mean ± SD, and data used for the analysis were from three independent experiments.
ISRE is a known binding site for many IRFs. To demonstrate whether IRF‐1 and IRF‐2 regulate TRIM14 transcription by binding to the ISRE in the TRIM14 promoter, we co‐transfected IRF‐1 or IRF‐2 with pGL‐121 or pGL‐121mISRE into HeLa cells. The results showed that IRF‐1 and IRF‐2 did not activate the pGL‐121mISRE (Fig. 3C), indicating that IRF‐1 and IRF‐2 activate TRIM14 transcription via the ISRE. To further demonstrate whether IRF‐1 and IRF‐2 both bind to the ISRE and regulate the TRIM14 promoter, we performed a ChIP assay to determine whether endogenous IRF‐1 and IRF‐2 can bind to ISRE with IFN‐α stimulation. The PCR primers cover the region from −150 to +1, which only contains the ISRE. The results showed that endogenous IRF‐1 and IRF‐2 bound to the ISRE (Fig. 3D), suggesting that IRF‐1 and IRF‐2 regulate TRIM14 expression by binding to the ISRE.
IRF‐1 and IRF‐2 differentially upregulate TRIM14 expression with IFN‐α treatment
To determine whether IRF‐1 and IRF‐2 are necessary for IFN‐α‐induced TRIM14 expression, we knocked down IRF‐1 and IRF‐2 by shIRF‐1 and shIRF‐2 in HeLa cells. We co‐transfected shIRF‐1 or shIRF‐2 and pGL‐121 into HeLa cells. pGL‐121 transcriptional activity and levels of endogenous IRF‐1 and IRF‐2 were measured. As shown in Fig. 4A, when the endogenous IRF‐1 in HeLa cells was knocked down, basal transcriptional activity of TRIM14 decreased moderately, but transcriptional activity decreased by 47% after IFN‐α stimulation. In contrast, knockdown of IRF‐2 resulted in the suppression of basal TRIM14 expression, but did not affect the IFN‐inducible expression (Fig. 4B). Overall, these data indicate that IRF‐1, but not IRF‐2, is involved in the activation of TRIM14 by IFN‐α.
Figure 4.
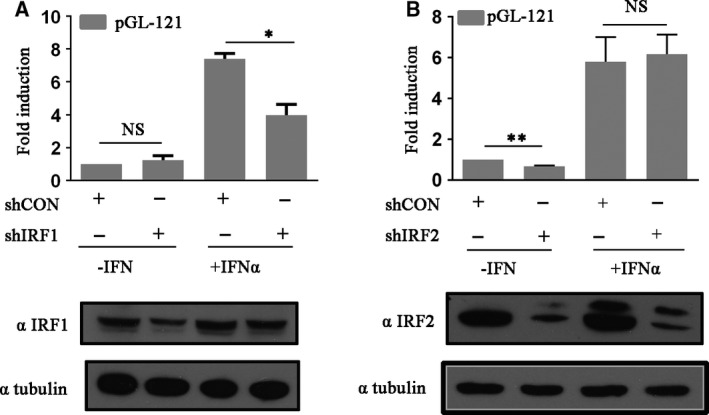
IRF‐1 and IRF‐2 differentially upregulate TRIM14 expression on IFN‐α treatment. (A, B) shcontrol, shIRF‐1 or shIRF‐2 co‐transfected HeLa cells with pGL‐121. Cells were treated with or without IFN‐α (10 ng·mL−1) for 16 h; luciferase activity was measured and western blot analysis was performed. Results are presented as mean ± SD, and data used for the analysis were from three independent experiments. Values are the mean and standard error of three independent experiments, *P < 0.05, **P < 0.01.
Discussion
Interferons play an important role in the antiviral innate immune response. IFNs can activate the expression of ISGs, which inhibit viruses at the different replication stages. TRIM14 is an ISG and antagonizes a variety of viruses, including mouse leukemia virus, sindbis virus, hepatitis C virus and others 19, 20, 21. TRIM14 can be induced by IFN‐I and enhances the host's immune response to virus infection 5, 6. In this study, we determined the specific mechanism of IFN‐I activation of TRIM14 expression.
First, we cloned and analyzed the features of the TRIM14 promoter. The TRIM14 promoter has a GC box and an ISRE, but no TATA box, and transcription initiates at two start sites. Base composition analysis showed that the GC content near the initiator is 60.2% (−500 to +1). There are two typical promoter types in the mammalian genome, which are classified mainly by GC content and CpG dinucleotide frequency 22. The high CG promoters (high GC content and high CpG dinucleotide frequency) typically contain multiple transcription start sites and do not contain a TATA box, and genes with high CG promoters are widely expressed throughout the biological cycle. The low CG promoters (low GC content and low CpG dinucleotide frequency) contain a single transcriptional start site and TATA‐box enrichment; low CG promoters are associated with tissue‐specific expression of genes 23. Based on the features of the TRIM14 promoter, it belongs to the high CG promoters. This is also consistent with its expression profiles in many tissues 4.
ISREs control the expression of many ISGs. Similar to some IFN‐induced genes, such as BAFF, TRIM21 and TLR3 24, 25, 26, the promoter of TRIM14 contains an ISRE located from −27 to −17. We determined that IFN‐I activates TRIM14 transcription through the ISRE. ISREs are recognized by different IRFs, and IRF‐1 can enhance transcription of many ISGs through an ISRE 27, such as ISG20 and interleukin‐7 28, 29. IRF‐2 is also known to recognize ISREs 30, and IRF‐2 is considered as a transcription inhibitor and antagonist of IRF‐1 31, 32. In this study, we observed that IRF‐1 and IRF‐2 bound to the ISRE and both activated the transcription of TRIM14, although the activation by IRF‐1 is stronger than that of IRF‐2. Our data also show that IRF‐1 and IRF‐2 differentially upregulate TRIM14 expression upon IFN‐I treatment. IRF‐2 maintains a basal level expression and IRF‐1 is involved in the IFN‐I‐inducible expression of TRIM14. In addition to regulating the transcription of TRIM21, TLR3, IFITM3 and other ISGs by IRF‐1 and IRF‐2 33, our data support further that IRFs play a crucial role in coordinating the transcriptional activation in the cellular antiviral response.
Previous studies have shown that STAT‐1 is essential for IFN‐I induction of TRIM14 34. They found that TRIM14 was not induced by IFN‐α in STAT‐1 knockout cells, while our work showed that knocking down IRF‐1 also reduce the IFN‐α activation of TRIM14. Considering that STAT‐1 can upregulate IRF‐1 and enhance IRF‐1 translocation into the nucleus and binding to ISRE 35, we propose that IRF‐1 enhances IFN activation of TRIM14 through STAT‐1, creating a positive feedback effect. IRF‐1 is an auxiliary amplifier of IFN‐I activation of TRIM14. This model is illustrated in Fig 5.
Figure 5.
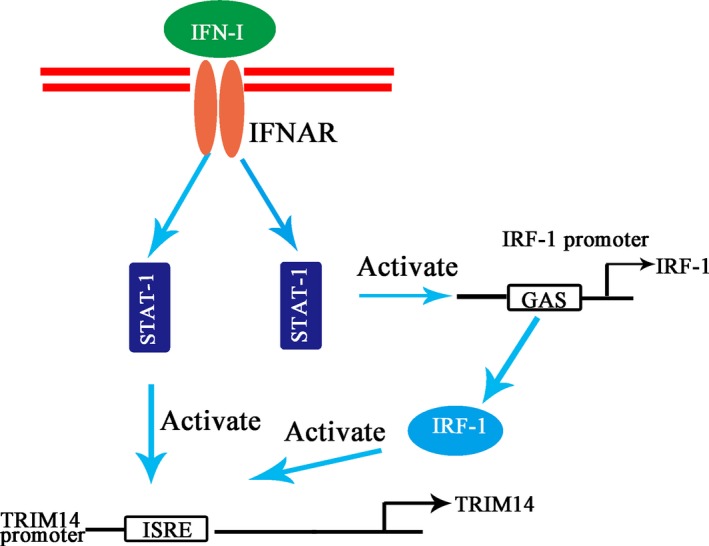
Model of regulation of TRIM14 transcription by IFN‐I. GAS, gamma activation sequence; IFNAR, Interferon‐alpha/beta receptor.
In conclusion, our study demonstrates that IFN‐I upregulates TRIM14 expression by recruiting IRF‐1 to the ISRE of the TRIM14 promoter. Our findings on the specific regulation mechanism of TRIM14 expression not only advance our understanding of the physiological function of TRIM14, but also elucidate how TRIM14 is regulated in the innate immune response.
Materials and methods
Constructs and antibodies
Plasmids pGL‐2020, pGL‐1000, pGL‐500, pGL‐121 and pGL‐121∆ISRE were constructed by cloning the PCR‐amplified fragment of the TRIM14 promoter into pGL3‐basic (Promega, Madison, WI, USA), and the primers are listed in Table 1. pGL‐500 mutants were constructed using a site‐directed mutagenesis kit (Toyobo, Osaka, Japan), and the primers used are listed in Table 1. Plasmids IRF‐1, IRF‐2, IRF‐3, IRF‐5 and IRF‐7 were constructed by inserting the coding sequence into the pCDNA3.1 (+) (Thermo Fisher Scientific, Waltham, MA, USA) or pCMV‐Tag3B (Agilent, Santa Clara, CA, USA) vector. The shRNA constructs for IRF‐1 and IRF‐2 were constructed using the pSIREN‐RetroQ vector (Clontech, Mountain View, CA, USA). The target sequence for IRF‐1 was: 5′‐GGGGTACCTACTCAATGAACCT‐3′; and for IRF‐2: 5′‐GGGGTACCTACTCAATG‐AACCT. Sequences of all of the constructs were confirmed by sequencing. Antibodies against IRF‐1, IRF‐2, Myc and Flag were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), α‐tubulin from Sigma‐Aldrich (St. Louis, MO, USA), and TRIM14 antibody from Abcam (Cambridge, UK).
Table 1.
Primers for amplifying TRIM14 promoter fragments and site‐directed mutagenesis.
| Primer | Sequence (5′–3′) |
|---|---|
| P2020 | CGACGCGTCCACCTTAACGCTACAATAATTGTGCTTCC |
| p1000 | GACGCGTCTGCAACCTTGCACAAAGAACC |
| P500 | GACGCGTGTTTTGCCTTTAAAAGCC |
| P121 | GACGCGTGAGGCCCGACGCTGCTCCCG |
| Plow | CAGATCTCGCCATTCATCTCCACCTCCTCC |
| P121∆ISRE‐F | GACGCGTGAGGCCCGACGCTGCTCCCG |
| P121∆ISRE‐R | CAGATCTCGCCATTCATCTCCACCTCCTCCGGCTCCCCGGGACACAGGGCGGGGCTCCCAAGGCTCGGCCGTGGGCGGGGCGGCTCC |
| mGC1‐F | CTTCCCATTTCTGGTTTTACCACCTCCCGGCC |
| mGC1‐R | GGCCGGGAGGTGGTAAAACCAGAAATGGGAAG |
| mGC2‐F | CCTCCCGGCCTGGGCTTTCAGAGGGAGCACCCTG |
| mGC2‐R | CAGGGTGCTCCCTCTGAAAGCCCAGGCCGGGAGG |
| mISRE‐F | ACGGCCGAGGTGTCGGTGTCCCTTGGGAGCCCCGC |
| mISRE‐R | TCCCAAGGGACACCGACACCTCGGCCGTGGGCG |
Cell culture and transfection
HeLa cells (maintained in our lab) were grown in Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) medium supplemented with 10% FBS (BI) in 5% CO2 at 37°. Transfection was performed using polyetherimide reagent (Sigma‐Aldrich).
5′‐Rapid amplification of cDNA ends
5′‐Rapid amplification of cDNA ends was used to characterize the 5′‐end of TRIM14 transcript using Firstchoice RLM‐RACE Kit (Ambion, Austin, TX, USA). Total RNA was extracted from HeLa cells (Thermo Fisher Scientific). DNA fragments were amplified by nested PCR. The sequence of the first round of PCR specific primer was 5′‐TGCCAGCTGCTTTAAACATTC‐3′, and primer used in the second round was 5′‐GCGGCAGCGGCGACAGAAG‐3′. PCR products were cloned into the pMD18‐T vector for further analysis.
Chromatin immunoprecipitation
ChIP analysis was conducted based on the manufacturer's instructions using an EZ‐chip kit (Millipore, Burlington, MA, USA). PCR primers used for amplifying the TRIM14 promoter are as follows: forward, 5′‐GAGGCCCGACGCTGCTCCCG‐3′ and reverse: 5′‐CGCCATTCATCTCCACCCCTCC.
Luciferase reporter assay
Luciferase plasmids and β‐galactosidase expression plasmids were transfected into cells. Luciferase activity was determined with a luciferase report system (Promega) and normalized to β‐galactosidase activity.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
JC, JT and WQ participated in the design of the study; JC, XX, YL, XH and YX helped in data collection and the interpretation of data. JC drafted the article. JT and WQ revised the article and gave final approval of the version to be submitted.
Acknowledgement
This work was supported by National Natural Science Foundation of China (31870161 and 81571988).
References
- 1. Yu S, Gao B, Duan Z, Xu W and Xiong S (2011) Identification of tripartite motif‐containing 22 (TRIM22) as a novel NF‐kappaB activator. Biochem Biophys Res Commun 410, 247–251. [DOI] [PubMed] [Google Scholar]
- 2. Uchil PD, Hinz A, Siegel S, Coenen‐Stass A, Pertel T, Luban J and Mothes W (2013) TRIM protein‐mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol 87, 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T and Akira S (2010) The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double‐stranded DNA. Immunity 33, 765–776. [DOI] [PubMed] [Google Scholar]
- 4. Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K et al (2014) Analysis of the human tissue‐specific expression by genome‐wide integration of transcriptomics and antibody‐based proteomics. Mol Cell Proteomics 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou Z, Jia X, Xue QH, Dou ZX, Ma YJ, Zhao ZD, Jiang ZF, He B, Jin Q and Wang JW (2014) TRIM14 is a mitochondrial adaptor that facilitates retinoic acid‐inducible gene‐I‐like receptor‐mediated innate immune response. Proc Natl Acad Sci USA 111, E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen MX, Meng QC, Qin YF, Liang PP, Tan P, He L, Zhou YB, Chen YJ, Huang JJ, Wang RF et al (2016) TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell 64, 105–119. [DOI] [PubMed] [Google Scholar]
- 7. Tan P, He L, Cui J, Qian C, Cao X, Lin M, Zhu Q, Li Y, Xing C, Yu X et al (2017) Assembly of the WHIP‐TRIM14‐PPP6C mitochondrial complex promotes RIG‐I‐mediated antiviral signaling. Mol Cell 68 , e5. [DOI] [PubMed] [Google Scholar]
- 8. Sadler AJ and Williams BRG (2008) Interferon‐inducible antiviral effectors. Nat Rev Immunol 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Honda K, Takaoka A and Taniguchi T (2006) Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360. [DOI] [PubMed] [Google Scholar]
- 10. Hervas‐Stubbs S, Perez‐Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A and Melero I (2011) Direct effects of type I interferons on cells of the immune system. Clin Cancer Res 17, 2619–2627. [DOI] [PubMed] [Google Scholar]
- 11. Darnell JE (1998) Studies of IFN‐induced transcriptional activation uncover the Jak‐Stat pathway. J Interf Cytok Res 18, 549–554. [DOI] [PubMed] [Google Scholar]
- 12. Tamura T, Yanai H, Savitsky D and Taniguchi T (2008) The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26, 535–584. [DOI] [PubMed] [Google Scholar]
- 13. Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T and Taniguchi T (1989) Structurally similar but functionally distinct factors, IRF‐1 and IRF‐2, bind to the same regulatory elements of IFN and IFN‐inducible genes. Cell 58, 729–739. [DOI] [PubMed] [Google Scholar]
- 14. Liu JG, Guan XQ and Ma XJ (2005) Interferon regulatory factor 1 is an essential and direct transcriptional activator for interferon gamma‐induced RANTES/CC15 expression in macrophages. J Biol Chem 280, 24347–24355. [DOI] [PubMed] [Google Scholar]
- 15. Namiki S, Nakamura T, Oshima S, Yamazaki M, Sekine Y, Tsuchiya K, Okamoto R, Kanai T and Watanabe M (2005) IRF‐1 mediates upregulation of LMP7 by IFN‐gamma and concerted expression of immunosubunits of the proteasome. FEBS Lett 579, 2781–2787. [DOI] [PubMed] [Google Scholar]
- 16. Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL and Stein GS (1995) Activation of a cell‐cycle‐regulated histone gene by the oncogenic transcription factor IRF‐2. Nature 377, 362–365. [DOI] [PubMed] [Google Scholar]
- 17. Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin‐Goguet F, Mothes W, Hazan U, Transy C, Pancino G et al (2009) Human TRIM gene expression in response to interferons. PLoS ONE 4, e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rajsbaum R, Stoye JP and O'Garra A (2008) Type I interferon‐dependent and ‐independent expression of tripartite motif proteins in immune cells. Eur J Immunol 38, 619–630. [DOI] [PubMed] [Google Scholar]
- 19. Uchil PD, Quinlan BD, Chan WT, Luna JM and Mothes W (2008) TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 4, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nenasheva VV, Kovaleva GV, Uryvaev LV, Ionova KS, Dedova AV, Vorkunova GK, Chernyshenko SV, Khaidarova NV and Tarantul VZ (2015) Enhanced expression of trim14 gene suppressed Sindbis virus reproduction and modulated the transcription of a large number of genes of innate immunity. Immunol Res 62, 255–262. [DOI] [PubMed] [Google Scholar]
- 21. Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Windisch MP, Trippler M, Lohmann V et al (2012) Identification of type I and type II interferon‐induced effectors controlling hepatitis C virus replication. Hepatology 56, 2082–2093. [DOI] [PubMed] [Google Scholar]
- 22. Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC et al (2006) Genome‐wide analysis of mammalian promoter architecture and evolution. Nat Genet 38, 626–635. [DOI] [PubMed] [Google Scholar]
- 23. Lenhard B, Sandelin A and Carninci P (2012) Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 13, 233–245. [DOI] [PubMed] [Google Scholar]
- 24. Sjostrand M, Johansson A, Aqrawi L, Olsson T, Wahren‐Herlenius M and Espinosa A (2016) The expression of BAFF is controlled by IRF transcription factors. J Immunol 196, 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sjostrand M, Ambrosi A, Brauner S, Sullivan J, Malin S, Kuchroo VK, Espinosa A and Wahren‐Herlenius M (2013) Expression of the immune regulator tripartite‐motif 21 is controlled by IFN regulatory factors. J Immunol 191, 3753–3763. [DOI] [PubMed] [Google Scholar]
- 26. Sun L, Jiang ZF, Acosta‐Rodriguez VA, Berger M, Du X, Choi JH, Wang JH, Wang KW, Kilaru GK, Mohawk JA et al (2017) HCFC2 is needed for IRF1‐and IRF2‐dependent Tlr3 transcription and for survival during viral infections. J Exp Med 214, 3263–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saha B, Prasanna SJ, Chandrasekar B and Nandi D (2010) Gene modulation and immunoregulatory roles of interferon gamma. Cytokine 50, 1–14. [DOI] [PubMed] [Google Scholar]
- 28. Gongora C, Degols G, Espert L, Hua TD and Mechti N (2000) A unique ISRE, in the TATA‐less human Isg20 promoter, confers IRF‐1‐mediated responsiveness to both interferon type I and type II. Nucleic Acids Res 28, 2333–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oshima S, Nakamura T, Namiki S, Okada E, Tsuchiya K, Okamoto R, Yamazaki M, Yokota T, Aida M, Yamaguchi Y et al (2004) Interferon regulatory factor 1 (IRF‐1) and IRF‐2 distinctively up‐regulate gene expression and production of interleukin‐7 in human intestinal epithelial cells. Mol Cell Biol 24, 6298–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh SI, Kimura T, Green SJ et al (1994) Requirement for transcription factor IRF‐1 in NO synthase induction in macrophages. Science 263, 1612–1615. [DOI] [PubMed] [Google Scholar]
- 31. Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T and Taniguchi T (1990) Absence of the type I IFN system in EC cells: transcriptional activator (IRF‐1) and repressor (IRF‐2) genes are developmentally regulated. Cell 63, 303–312. [DOI] [PubMed] [Google Scholar]
- 32. Fujita T, Kimura Y, Miyamoto M, Barsoumian EL and Taniguchi T (1989) Induction of endogenous IFN‐alpha and IFN‐beta genes by a regulatory transcription factor, IRF‐1. Nature 337, 270–272. [DOI] [PubMed] [Google Scholar]
- 33. Ren G, Cui KR, Zhang ZY and Zhao KJ (2015) Division of labor between IRF1 and IRF2 in regulating different stages of transcriptional activation in cellular antiviral activities. Cell Biosci 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan GY, Xu FC, Song HX, Yuan Y, Xiao QF, Ma F, Qin FXF and Cheng GH (2018) Identification of TRIM14 as a Type I IFN‐stimulated gene controlling hepatitis B virus replication by targeting HBx. Front Immunol 9, 1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reich NC (2002) Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J Interferon Cytokine Res 22, 103–109. [DOI] [PubMed] [Google Scholar]


