Abstract
The pancreatic acinar-enriched miR-216a, miR-216b and miR-217 are encoded within the miR217HG. These miRNAs have been purported to play a tumor suppressive role as their expression is reduced in both human and mouse pancreatic ductal adenocarcinoma (PDAC). To examine this possibility, we generated individual, germline knockout (KO) mice of miR-216a, miR-216b or miR-217. Unlike our previous study showing germline deletion of the miR217HG was embryonic lethal, CRISPR-Cas9 deleted portions of the 5’ seed region of the miRNAs produced live births. To investigate possible phenotypes during pancreatic acinar ductal metaplasia (ADM), pancreatic acini from wild type and KO mice were plated on collagen and allowed to transdifferentiate over 4 days. Acini from each of the three miRNA KO mice produced greater numbers of ducts compared to controls. Evaluation of the gene expression during in vitro ADM demonstrated an increase in Krt19 and a reduction in acinar genes (Carboxypeptidase A1, Amylase2a) on day 4 of the transdifferentiation. Recovery was delayed for the miR-216a and miR-216b KOs following caerulein-induced acute pancreatitis. Also predominate in the caerulein treated miR-216a and miR-216b KO mice was the presence of pancreatic duct glands (PDGs). To further establish a phenotype, miRNA KO mice were crossed with EL-KRASG12D (EK) mice and followed up to 13 months of age. While all mice developed severe dysplasia and cystic papillary neoplasms, there existed no apparent phenotypic difference in the miRNA KO/EK mice compared to EK mice. Our data does not support a tumor suppressor role for miR-216a, miR-216b or miR-217 in PDAC and emphasizes the need for phenotypic evaluation of miRNAs in complex in vivo models beyond that performed using cell culture.
Subject terms: Health sciences, Pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the third most lethal cancer in the USA and is predicted to become the second most deadly cancer in the country by 20301,2. It is widely understood that the cells of origin for PDAC are the exocrine acini3–5. Pancreatic acini display remarkable plasticity6 and exhibit a natural ability to dedifferentiate and then transdifferentiate into ductal-like cells following pancreatic injury by a reversible process known as acinar ductal metaplasia (ADM). The hallmarks of ADM include loss of acinar gene expression (e.g. amylase) and gain of ductal epithelial morphology and gene expression including cytokeratin 19 (KRT19)7–9. Pancreatic intraepithelial neoplasm (PanINs), one of the main precursor lesions to PDAC, arise from ADM4. ADM is the earliest known precursor lesion for PDAC making acinar to ductal transdifferentiation a key phase in the initiation of pancreatic cancer.
microRNAs (miRNAs) display altered patterns of expression in clinical specimens of pancreatic ductal adenocarcinoma (PDAC), with some miRNAs increased and others decreased in the tumors10–12. One particular miRNA cluster that has generated interest in PDAC are those miRNAs encoded within the miR217 host gene (miR217HG i.e. miR-216a, miR-216b and miR-217). miR217HG is a lncRNA that is located within chromosome 2p16.1 in humans and 11qA3.3 in mice. The processed, mature miR-216a, miR-216b and miR-217 are pancreas enriched13,14 with predominate, abundant expression in pancreatic acini15. miR-216a, miR-216b and miR-217 are also expressed in brain16 and to a lesser degree in kidneys, lung and small intestine17. miR-216a, miR-216b and miR-217 expression is reduced in mice and humans with pancreatitis18–20, during PanIN progression21,22 and in PDAC in both mice and humans12,19,23,24.
We and others posited that the miR-216/−217 cluster may be tumor suppressive in pancreas since other tissue specific/enriched miRNAs such as miR-122 in liver25,26 and miR-126 in muscle27 are tumor suppressive. Further evidence supporting a potential tumor suppressive role for these miRNAs in PDAC comes from an evaluation of their reported target genes that include KRAS28,29, ANLN30, E2F331, JAK232 and TPT133. The oncogenic lncRNA MALAT1 enhanced KRAS activity and PDAC development by sponging miR-217 and altering the miR-217 nucleus/cytoplasm ratio34. Validation of each of these miRNA/target gene interactions and their effects were obtained using cell culture systems and have not been replicated in transgenic mouse models to our knowledge.
In order to establish a functional role for the three mature miRNAs, we deleted a 27.9 kbp region of miR217HG in mice, however the deletion was embryonic lethal at around E9.523. While the specific cause(s) of the lethality are unknown, possibilities include simultaneous deletion of all three miRNAs or deletion of a functional miR217HG lncRNA. To evaluate these possibilities in greater detail, we generated individual germ line KO mice for miR-216a, miR-216b and miR-217 using CRISPR/Cas9 gene editing. Portions of each mature miRNA including the 5′ seed regions were successfully deleted, and viable mice ensued. Phenotypic evaluation of the KOs, including effects on in vitro transdifferentiation, caerulein-induced acute pancreatitis and crossing KO mice with those harboring a mutant KRAS, produced subtle changes. The miRNA KO did not enhance metaplasia beyond advanced cystic papillary neoplasms (CPN), which was not different from control. We conclude that miR-216a, miR-216b and miR-217 are not tumor suppressive in pancreas and our work emphasizes the need to evaluate miRNA function beyond in vitro systems.
Materials and Methods
All experimental methods were performed in accordance with the relevant guidelines and regulations.
Generation of CRISPR/Cas9 miRNA knockout mice
All experiments conducted in this study were approved by the Institutional Animal Care and Use Committee (IACUC) at both the Ohio State University and the University of Florida. Single guide RNA sequences targeting miR-216a, miR-216b and miR-217 genes were designed using the CRISPR/Cas9 design tool (crispr.mit.edu, SFig. 1). Using the primer pairs for cloning (Supplemental Table 1), we annealed, phosphorylated and cloned the miRNA targeting sgRNA sequences within the px330 bicistronic expression plasmid (Addgene). PCR amplification was performed to add the T7 promoter sequence to the sgRNA template using primers 217 F and R, 216a-1 F and R, 216a-2 F and R, 216b-1 F and R and 216b-2 F and R (Supplemental Table 1). The T7-sgRNA PCR product was purified using Qiagen Gel purifying kit and served as a template for in vitro transcription using ampliscribe T-flash transcription kit (Epicentre, Madison, WI). The reaction was purified using Centri-Spin columns (Princeton separations, NJ) and sgRNAs were eluted in RNAse-free water and stored at −80 °C. Cas9 mRNA (Trilink Biotechnologies, San Diego, CA) was codon optimized, capped and polyadenylated mimicking the fully processed mature mRNA. Mutant mice were generated at the Genetically Engineered Mouse Modeling Core of the Ohio State University according to standard procedure35,36. Briefly, a mix of Cas9 mRNA (100 ng/uL, CleanCap™ Cas9 mRNA TriLink BioTechnologies) and miR-specific sgRNA (50 ng/uL) was injected into the cytoplasm of fertilized C57Bl/6N zygotes. Following micromanipulation, zygotes were transferred to pseudo-pregnant recipient mothers and allowed to develop to term. To mitigate off-targets effects, mutant mice were crossed into C57Bl/6 for 5 generation before use for experimental purposes. The mfold web server determined if the deletions produced any changes in the folding of the pre-miRNA sequences. Based on the mfold results and TOPO sequencing, the mutant pups that exhibited the largest length of deletions were selected for breeding to obtain a pure biallelic miRNA KO.
High resolution melt analysis and Sanger sequencing
Genomic DNA isolated from the mouse’s tails that were born following the injection of the sgRNA and Cas9 mRNA was PCR amplified using primers (Supplemental Table 1) for 98 °C for 2 mins; 35 × (98 °C for 5 secs, 60 °C for 5 secs); 65–95 °C (in 0.5 °C increments) 5 sec/step. The PCR reaction was performed using Evagreen dye on a Biorad CFX96 PCR system. Several mutants were identified from the pool of pups that were obtained after CRISPR/Cas9 injections. PCR amplification was carried out on the positive mutants that were identified through high resolution melt analysis and were cloned into a TOPO vector using Original TA cloning kit (Invitrogen, USA). Sanger sequencing was performed to verify and determine the mutations that occurred in the miRNA transcript.
qRT-PCR
RNA was isolated from the mouse pancreata using Trizol reagent37. All RNA samples had an RNA integrity number (RIN) of 7.5 or greater. cDNA was synthesized to the mature miR-216a, miR-216b and miR-217 as described38, 18S rRNA was used as the internal control and data were analyzed using the comparative CT method. Situations did not exist where a significant change was observed in the 18S rRNA expression between test and control groups (fold change <1.5, P > 0.05). miRNA expression was considered not expressed if the mean CT ≥ 36.
Acinar-ductal transdifferentiation
Pancreatic acinar cell culture was performed as previously described by Shi et al., with several modifications39. Rat tail collagen (Invitrogen) was neutralized and 250 µl was added per well of a 24 well plate to provide a firm attachment base. Freshly isolated mouse pancreata were washed in cold HBSS and dissected into small pieces. Acini were digested with 5 ml of 0.2 mg/ml Collagenase P (Roche) and incubated at 37 °C for 30 mins. HBSS/5% FBS was added to inactivate the collagenase, isolated acini were washed three times with HBSS and filtered through a 100 µm mesh. Cell suspensions were layered onto HBSS/30% FBS and centrifuged at 180 x g for 3 mins. Pellets were resuspended in Waymouth’s media supplemented as described39. Acini were mixed 1:1 with neutralized collagen and 0.5 ml of this mixture was plated onto the collagen layer in the 24 well plate. Following 1 hr at 37 °C, 1 ml of culture media was added to each well. An eight mm2 adhesive grid (Sigma) was affixed to the bottom of each well and microscopic acini and ducts were manually counted. To isolate RNA from the cultures, collagen was digested in collagenase P solution at 37 °C for 30 mins with shaking. Acini were pelleted and total RNA was isolated using the RNAesy Kit (Qiagen). Random cDNA was prepared using Superscript II polymerase (Invitrogen) and qPCR was performed using SYBR green reagent detection. Primers used in the study are provided in Supplemental Table 1.
Acute pancreatitis
Acute pancreatitis was induced in the mice using the cholecystokinin analogue caerulein, purchased as the ammonium salt (Bachem, Torrance, CA). miRNA KO and control C57/B6 mice were injected hourly with 8 injections of caerulein (100 µg/kg) on two consecutive days, for a total of 16 injections. Mice were euthanized at day 2, day 4 and day 7 following the final caerulein injection. Pancreas tissues were removed from the animals, fixed in formalin, embedded into paraffin, sectioned and stained with Hematoxylin and Eosin. For all of the caerulein treated and saline controls, serial sectioning was performed of the entire pancreas tissue and H&E staining was carried out on every fifth slide to confirm the presence or absence of PDGs. For semi-quantification, slides were analyzed by two different investigators in a blinded fashion. A total of 6 mice were used for each treatment arm (caerulein or saline). The entire slide section was evaluated. The percentage of ADM was estimated by the presence of ADM compared to the overall tissue. The degree of inflammation, fibrosis and atrophy was scored as +, ++, +++.
Tissue histology analysis of miRNA-KO Kras crossed mice
All the mice during the study were bred and maintained at the University of Florida. Genetically engineered mice of the following genotypes were used: EL-KRASG12D (or EK), miR-216a/EK, miR-216b/EK and miR-217/EK. Where EL stands for Elastase promoter; Kras stands for activating Kras mutation (G12D), EK for EL-KRASG12D; miR-216a, miR-216b and miR-217 have deletions on those microRNAs, respectively. Mice were euthanized at 3, 6 and 13 months of age. A minimum of 5 mice were used for each comparison. The pancreas was removed, fixed in 10% formalin, processed, sectioned at 4 µm, and stained with Hematoxylin and Eosin for histological evaluation. Periodic acid Schiff (PAS) staining was done to confirm the presence of mucins within the PDGs. To evaluate the differences in the pancreatic neoplastic phenotypes, tissue slides were scanned using Aperio Microscope and then blinded for evaluation by two different investigators using the Aperio Image Scope software (Leica, Wetzlar, Germany) which provides tools to measure and quantify selected areas. The whole area of the tissue was measured and used to normalize the frequency of lesions (total number of lesions, CPNs and cysts, per tissue area in mm2). The percentage of parenchyma replaced by fat (lipoatrophy) was also obtained by dividing total lipoatrophy areas (mm2) by the total tissue area (mm2) To estimate the percentage of normal tissue, ADM and fibrosis present in each sample, a 4x magnification of the tissue was used and five different fields of views were assessed, quantified and averaged. The area of the five largest lesions (mm2) was measured and averaged. Each group was compared with the age-matched EL-Kras control group. Possible outliers were identified using GraphPad and differences between the groups were assessed by One-Way ANOVA and only values p < 0.05 were considered significant. For comparing all groups and ages, Two-Way ANOVA was used for statistical purposes and only values p < 0.05 were considered significant.
Results
Development of miR-216/−217 knockout mice
The miRNAs encoded within the miR-217HG have been reported as enriched in pancreas13–15. The NIH Genotype-Tissue Expression (GTEx) project data set on 51 normal human tissues was mined to confirm these findings (SFig. 2). To evaluate a possible role for these miRNAs in maintaining acinar cell identity or tumor suppression, we generated individual germ line KOs of miR-216a, miR-216b and miR-217. While conditional gene deletion is the preferred method to study a tissue specific phenotype, we presumed that since the miR217HG is predominately expressed in pancreas (SFig. 2), germ line deletion would serve as a de facto conditional deletion in pancreas. CRISPR/Cas9 gene editing was used to delete a small portion of each mature miRNA that included the 5’ seed region. High resolution melt analysis showed that 2 out of 23 pups were positive for miR-217 mutations, 6 out of 48 mice pups were positive for miR-216a mutations and only one pup out of 15 pups was identified as a mutant for miR-216b targeting (SFig. 3A). TOPO cloning and Sanger sequencing verified the deletions that occurred in the miRNA transcripts (SFig. 3B).
miRNA and mRNA expression in KO mice
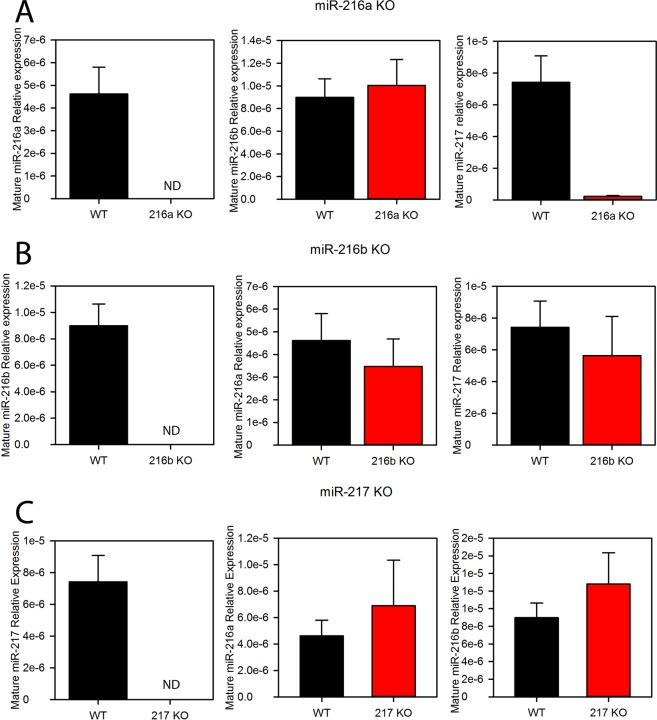
Successful KO of each miRNA in mouse pancreata were confirmed by qPCR (Fig. 1). In mouse, miR-216a and miR-217 lie on the same pre-miRNA while miR-216b is on a separate transcript (SFig. 1). We wondered if deletion of one miRNA would alter the expression of another miRNA in the miR217HG cluster. In the miR-216a KO, miR-217 was also reduced by >95%. These phenomena existed in both young (1 month, not shown) and old (13 month, Fig. 1) KO mice. However, in both the miR-217 and miR-216b KOs, only the affected miRNA was reduced (Fig. 1). Since miR-216a lies upstream of miR-217, deletion of the upstream miRNA possibly affected processing of the downstream miRNA. We next performed an unbiased, gene expression profiling array that contained both coding and noncoding RNAs on RNA extracted from the pancreas of 6 week old miR-217 KO and wild type control mice. Unexpectedly, there was little significant change in gene expression from the cDNA array analysis of the miR-217 KO and wild type mice (SFig. 4). miR-217 was one of the few genes that were differentially expressed in the KO animals (fold change <1.5, P < 0.05). Three members of the Reg3 family (Reg3a, b and g) were increased in the miR-217 KO mice (SFig. 4).
Figure 1.
miRNA expression in the pancreata of individual knockout mice. TaqMan qRT-PCR was performed on miRNAs isolated from the pancreata of 13 month old A. miR-216a, B. miR-216b and C. miR-217 knockout mice. The expression of each individual miRNA, relative to 18S rRNA, is depicted on the y-axis of each graph. ND, not detectable.
KO of miR-216a, miR-216b and miR-217 alters in vitro ADM
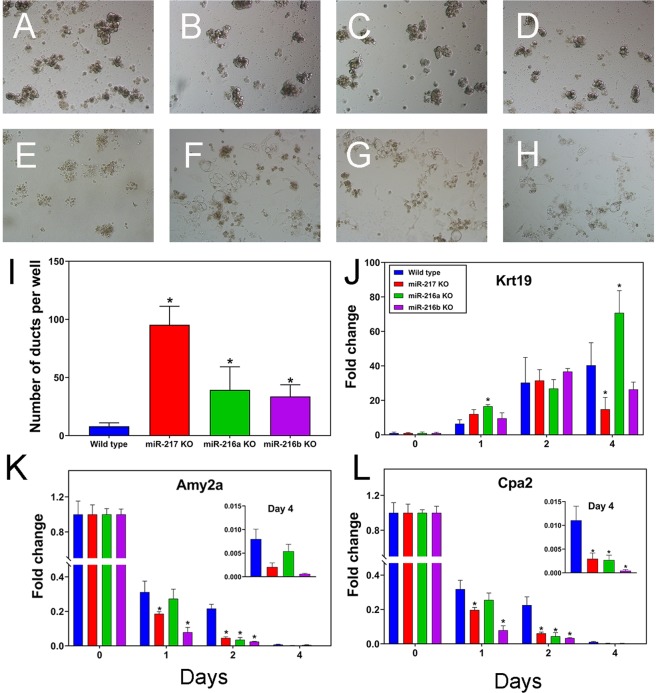
In transgenic mouse models of PDAC, neoplasia formation is proceeded by the transdifferentiation of acinar cells into cells that display ductal epithelial gene expression and morphology (i.e. acinar ductal metaplasia or ADM). Changes in gene expression include reduction in acinar (Amy2a, Cpa2) and increased expression of ductal epithelial genes such as Krt19. ADM was studied experimentally by culturing the pancreatic acini on collagen from each of the three miRNA KO mice as well as control mice. When cultured on collagen in the absence of additional growth factors such as TGFα, ADM should be greatly reduced in wild type mice. Since miR-216a, miR-216b and miR-217 are highly enriched in pancreatic acini, we hypothesized that ADM would be enhanced in the KOs compared to wild type mice. As anticipated, the number of ducts increased for the three KO mice compared to wild type control (Fig. 2A–I). To further explore the effects of the miRNAs on ADM we quantified the expression in both acinar and ductal genes by qPCR. In the wild type mice, acinar genes decreased while Krt19 increased over the 4 day ADM experiment (Fig. 2J–L). While the trends were not consistent, there was a tendency for the acinar (Amy2a, Cpa2, Cpb1 and Ela1) gene expression to be reduced while Krt19 and Krt7 expression increased in the miRNA KOs over the 4 day ADM period (Fig. 2J–L, SFig. 5).
Figure 2.
miRNA knockout accelerates in vitro pancreatic acinar ductal metaplasia. Mouse pancreatic acinar cells were plated onto collagen and the transdifferentiation occurred over 4 days. Bright field images were captured at Day 1 (A–D) and Day 4 (E–H) of culture for the wild type (A,E), miR-217 (B,F), miR-216a (C,G) and miR-216b (D,H) KOs. (I) Number of ducts after 4 days of culture in the wild type and individual knockout mice. RNA was isolated from the cultures at days 1 to 4 of the transdifferentiation and the expression of J. cytokeratin 19, K. amylase 2A and L. carboxypeptidase A2 was determined by qRT-PCR. Data are presented relative to 18S rRNA (mean ± SEM) and were normalized to the day 0 controls. *P < 0.05 (Student’s t-test).
Acute pancreatitis is enhanced in miRNA KO mice
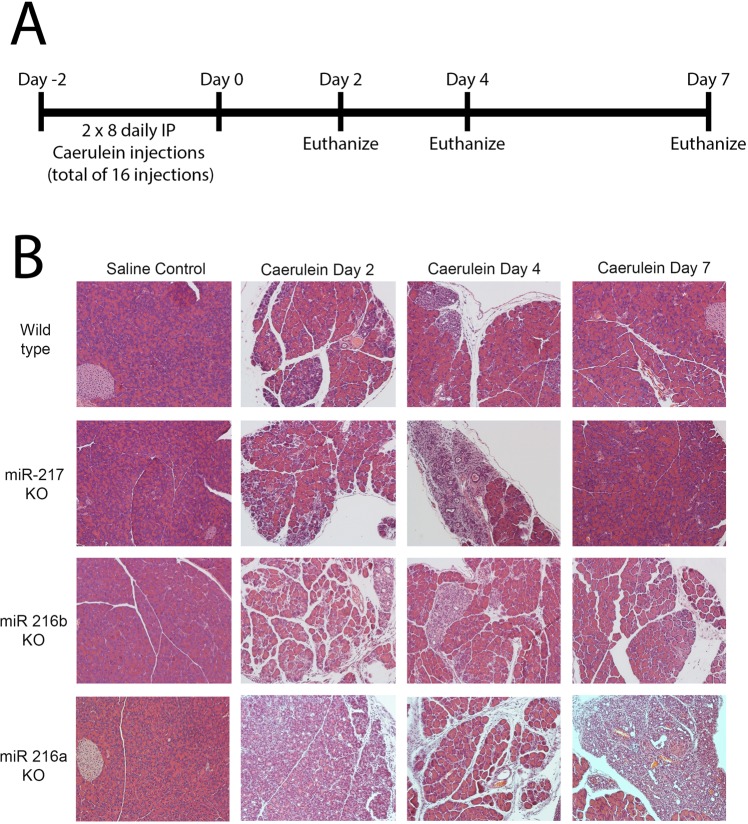
Acute pancreatitis was induced following 16 intraperitoneal injections of caerulein in the wild type control and 3 miRNA KO mice (Fig. 3A). Pancreata were harvested at three different time-points and H&E staining was performed to evaluate histological changes (Fig. 3B). Caerulein-induced acute pancreatitis is a reversible phenomenon and complete acinar regeneration occurs by day 7. Caerulein induced extensive acinar injury, atrophy and increased inflammation in all the three KOs compared to WT mice (Fig. 3B, Supplemental Table 2). miRNA KO mice also displayed an increase in acinar cells undergoing ADM (Supplemental Table 2). The three KO mice exhibited increased ductal formation which was highly evident at day 2 in miR-216a and 216b KO and day 4 in miR-217 KO (Fig. 3B). By day 7, pancreata of the miR-217 and miR-216b KOs completely regenerated, however the pancreas of miR-216a KO did not (Fig. 3B).
Figure 3.
Recovery from acute pancreatitis. A. Wild type or miR-217, miR-216b or miR-216a knockout mice (2 month old) were dosed daily with eight injections of caerulein for two days. B. Mice were sacrificed at 2, 4 and 7 days following the last caerulein dose and the pancreata were processed for histology. 20X magnification.
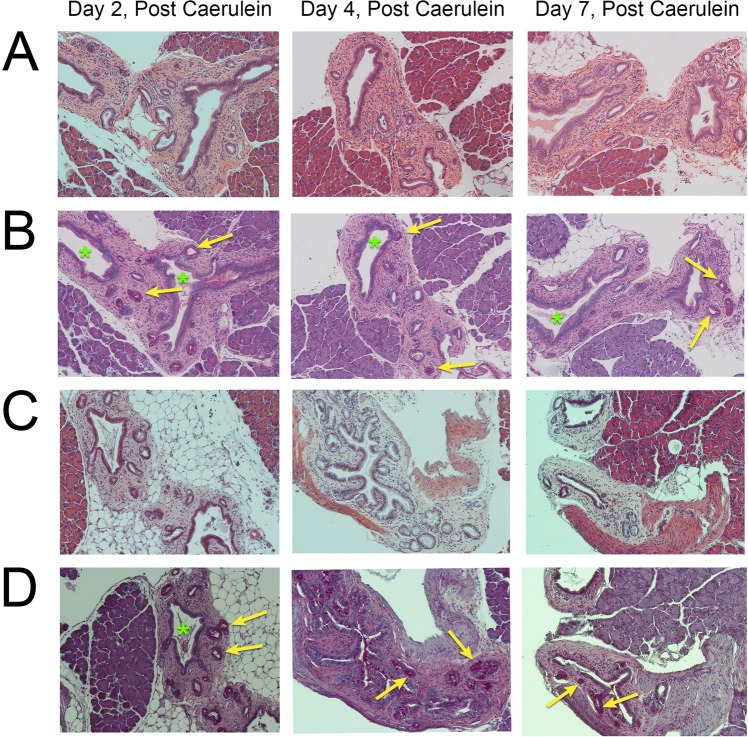
Pancreatic ductal glands (PDGs) are a distinct compartment that have been associated with chronic pancreatic injury and may be linked to PDAC development through the formation of intraductal papillary mucinous neoplasms (IPMNs)40. While PDGs were present in one of the caerulein treated wild type mice at day 4 (not shown), caerulein increased the PDGs formed in several of the miR-216a and miR-216b KOs (Fig. 4). PDGs were confirmed in the mouse pancreata for the presence of mucins by positive PAS staining (Fig. 4B,D). While the miR-216b KO exhibited PDGs at all three time points, we did not observe PDG structures in vehicle controls for miR-216a KO. The PDG structures observed on day 4 in miR-216a KO mice appeared to be undergoing hyperplasia.
Figure 4.
Presence of pancreatic ductal glands during acute pancreatitis. The presence of pancreatic ductal glands (PDGs, yellow arrows) was observed in the miR-216b and miR-216a knockout mice 2, 4 and 7 days, following caerulein-induced acute pancreatitis. Pancreata were stained with A,C hematoxylin and eosin or B,D periodic acid Schiff (PAS). Green asterisks represent enlarged ducts on the PAS stained sections. 20X magnification.
Lack of consistent phenotype when miRNA KO mice are crossed with EK mice
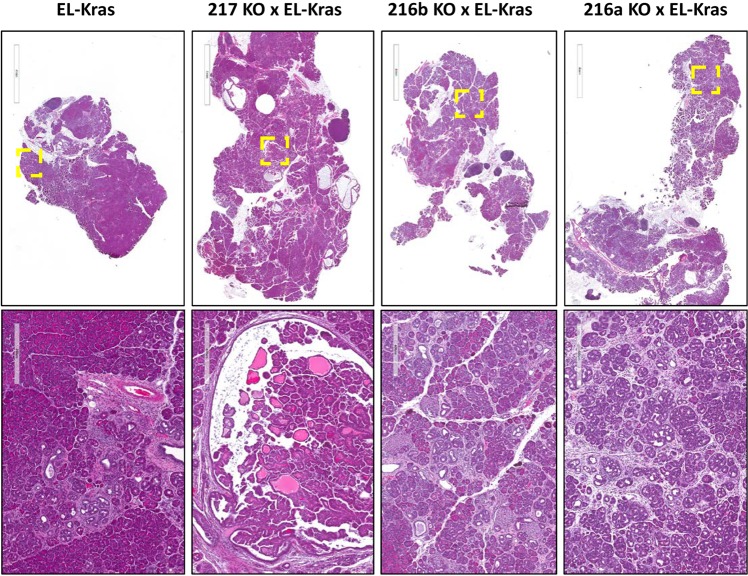
We next questioned if a phenotype may be enhanced when the miRNA KO mice were bred to mice harboring an activating Kras mutation. EK mice were used as these mice have been shown to develop CPNs by 2–3 months of age that progressively become larger though only infrequently more severe41. miR-216a/EK, miR-216b/EK and miR-217/EK mice as well as EK controls were sacrificed at 3, 6, and 13 months of age. Upon histological evaluation, widespread dysplasia was apparent in the EK miRNA KO crosses by 3 months of age (Fig. 5). Dysplasia was also observed in the 6 and 13 month old mice (SFigs 7 and 8). A variety of histopathological lesions were quantified (e.g. total lesions, total lesion area, % normal tissue, % fibrosis, % ADM, % lipoatrophy and lesion size) in a blinded fashion on all three age groups of mice, however none of these differed between the KC and miR-216a/EK, miR-216b/EK or miR-217/EK mice with the exception of one lipoatrophy comparison (SFig. 9, Supplemental Table 3).
Figure 5.
Histopathology of miRNA knockout and KrasG12D mouse crosses. The miR-216a, miR-216b and miR-217 KO mice were crossed with the EL-KRASG12D mice. The histopathology from the pancreata of 3 month old mice are shown at 4× (upper panels) and 20× (lower panels) magnification.
Discussion
Many studies have identified differential miRNA expression in the tumors of patients with PDAC [reviewed in42]. Studies using cancer cell lines describe relationships between differential miRNA expression and alterations in cell proliferation, colony formation, induction of apoptosis and migration/invasion, suggesting that miRNAs may play a tumor suppressive or promotion role in PDAC28,34,43–45. More definitive evidence for such a role is generally gathered following gene knockin or knockout experiments in mice. We are aware of only one miRNA KO mouse that was developed in the context of pancreatic cancer, miR-324, which produced no phenotype towards the development of pancreatic acinar carcinoma46.
For simplicity, we generated germline miRNA KO mice rather than conditional KOs as the miR217HG cluster is predominately expressed in pancreas (SFig. 2) and13,14. One disadvantage of our approach is that compared to conditional KOs engineered to be inducible at the adult stage, the deleted miRNA gene may undergo compensation at the germline level. Compensation occurred between miR-34b/c and miR-449; miRNAs that are present on different chromosomes but share the identical 5′ seed sequence47. Evidence to support gene compensation in our study was mixed. Germline deletion of the highly pancreas-enriched miR-217 produced very little differential gene expression in the pancreas (SFig. 4), so if compensation was present, it was not evident from the gene expression profiling data. qPCR analysis of the other two miRNAs in the cluster that were not deleted only showed differential expression for miR-217 in the miR-216a KO, however its expression was reduced and not increased (Fig. 1). Of interest is that three members of the Reg3 family (Reg3a, b and g) were increased in the pancreata of the miR-217 KO mice (SFig. 4). Expression of the REG3 family are enriched in the pancreas and terminal ileum. Reg3g promoted PDAC in mice48,49 and REG3A/G were highly expressed in the ADM lesions of PDAC patients50.
Our data suggest some acceleration of ADM in the miRNA KO mice in both cell cultures and in caerulein treated animals. While we are unsure of a specific mechanism to describe this phenomenon, increased expression of Reg3a, b and g is a possibility, at least for the miR-217 KO mice (SFig. 4). Addition of Reg3a protein to acinar cultures promoted duct formation through activation of the MAPK pathway50. We do not believe that miR-217 directly targets the 3′ UTR of the Reg proteins since these interactions are not predicted using miRNA target algorithms such as TargetScan, though indirect regulation is possible.
While some of the older mice in our study developed neoplasia, we failed to demonstrate a pronounced and consistent phenotype in mice even among the 13 month-old miRNA KO/EK crosses. So, our findings paralleled those of Morris, et al., who deleted the miRNA processing enzyme Dicer in the pancreas using a Pdx1 driven Cre strain51. They report that Dicer knockout resulted in Kras associated ADM however Dicer loss did not accelerate Kras driven PDAC. It is possible that while KO of the miRNAs promote ADM and some PanIN lesions, it does not promote PDAC. Another explanation for a lack of phenotype in our miRNA KO/EK crosses is that the Kras mini gene used in developing the EK mice uses the metallothionein 3′UTR rather than the Kras 3′UTR41. As both miR-21729 and miR-216b28 bound to the 3′UTR of Kras and regulated Kras levels in vitro, any potential regulation of mutant Kras by these miRNAs would be lost in our miRNA KO/EK mice. In conclusion, our findings highlight the complexity associated with assigning tumor-promoting phenotypes with miRNAs and emphasize the need for studying these molecular events using in vivo models of cancer.
Supplementary information
Acknowledgements
We thank Dr. Xin-an Pu of the Genetically Engineered Mouse Modeling Core of the Ohio State University Comprehensive Cancer Center for mouse zygote CRISPR injections. Dr. Coppola and the Genetic Engineering Mouse Modeling Core are supported by NCI grant P30 CA016058. Julie Bray is supported by grant F31 CA220937.
Author Contributions
D.S.S., designed and performed experiments and wrote the manuscript; J.J., L.W. and J.A.B. performed experiments; A.C.A.P., P.J.G. and V.C. designed experiments, K.F., X.L., J.L., C.T. and G.M. data analysis and interpretation of histopathology; T.D.S., designed experiments and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47566-x.
References
- 1.Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Grippo PJ, Sandgren EP. Acinar-to-ductal metaplasia accompanies c-myc-induced exocrine pancreatic cancer progression in transgenic rodents. Int J Cancer. 2012;131:1243–1248. doi: 10.1002/ijc.27322. [DOI] [PubMed] [Google Scholar]
- 4.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Murtaugh LC, Leach SD. A case of mistaken identity? Nonductal origins of pancreatic “ductal” cancers. Cancer Cell. 2007;11:211–213. doi: 10.1016/j.ccr.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 6.Stanger BZ, Hebrok M. Control of cell identity in pancreas development and regeneration. Gastroenterology. 2013;144:1170–1179. doi: 10.1053/j.gastro.2013.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halbrook CJ, et al. Mitogen-activated Protein Kinase Kinase Activity Maintains Acinar-to-Ductal Metaplasia and Is Required for Organ Regeneration in Pancreatitis. Cell Mol Gastroenterol Hepatol. 2017;3:99–118. doi: 10.1016/j.jcmgh.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou GY, et al. Protein kinase D1 drives pancreatic acinar cell reprogramming and progression to intraepithelial neoplasia. Nat Commun. 2015;6:6200. doi: 10.1038/ncomms7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, Y. et al. Epithelial-Myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife6 (2017). [DOI] [PMC free article] [PubMed]
- 10.Bloomston M, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 11.Lee EJ, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szafranska AE, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 13.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 14.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endo K, Weng H, Kito N, Fukushima Y, Iwai N. MiR-216a and miR-216b as markers for acute phased pancreatic injury. Biomedical research. 2013;34:179–188. doi: 10.2220/biomedres.34.179. [DOI] [PubMed] [Google Scholar]
- 16.Majumdar A, et al. miR-217-casein kinase-2 cross talk regulates ERK activation in ganglioglioma. Journal of molecular medicine. 2017;95:1215–1226. doi: 10.1007/s00109-017-1571-z. [DOI] [PubMed] [Google Scholar]
- 17.Kito N, Endo K, Ikesue M, Weng H, Iwai N. miRNA Profiles of Tubular Cells: Diagnosis of Kidney Injury. Biomed Res Int. 2015;2015:465479. doi: 10.1155/2015/465479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng S, et al. Chronic pancreatitis and pancreatic cancer demonstrate active epithelial-mesenchymal transition profile, regulated by miR-217-SIRT1 pathway. Cancer Lett. 2014;355:184–191. doi: 10.1016/j.canlet.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin D, et al. Evaluation of miR-216a and miR-217 as potential biomarkers of acute pancreatic injury in rats and mice. Biomarkers: biochemical indicators of exposure, response, and susceptibility to chemicals. 2014;19:517–529. doi: 10.3109/1354750X.2014.944217. [DOI] [PubMed] [Google Scholar]
- 20.Vychytilova-Faltejskova P, et al. MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:38. doi: 10.1186/s13000-015-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang X, Yu C, Li J, Yu S, Chen J. hsa-miR-96 and hsa-miR-217 Expression Down-Regulates with Increasing Dysplasia in Pancreatic Intraepithelial Neoplasias and Intraductal Papillary Mucinous Neoplasms. Int J Med Sci. 2017;14:412–418. doi: 10.7150/ijms.18641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Li A, Hong SM, Hruban RH, Goggins M. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res. 2012;18:981–992. doi: 10.1158/1078-0432.CCR-11-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azevedo-Pouly Ana Clara P., Sutaria Dhruvitkumar S., Jiang Jinmai, Elgamal Ola A., Amari Foued, Allard David, Grippo Paul J., Coppola Vincenzo, Schmittgen Thomas D. miR-216 and miR-217 expression is reduced in transgenic mouse models of pancreatic adenocarcinoma, knockout of miR-216/miR-217 host gene is embryonic lethal. Functional & Integrative Genomics. 2016;17(2-3):203–212. doi: 10.1007/s10142-016-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz NA, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012;25:1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 25.Hsu SH, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai WC, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taulli R, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu X, et al. Archives of medical science: AMS. 2018. MiR-216b inhibits pancreatic cancer cell progression and promotes apoptosis by down-regulating KRAS; pp. 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao WG, et al. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 30.Idichi T, et al. Regulation of actin-binding protein ANLN by antitumor miR-217 inhibits cancer cell aggressiveness in pancreatic ductal adenocarcinoma. Oncotarget. 2017;8:53180–53193. doi: 10.18632/oncotarget.18261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Zhang HF, Qin CF. MicroRNA-217 functions as a prognosis predictor and inhibits pancreatic cancer cell proliferation and invasion via targeting E2F3. Eur Rev Med Pharmacol Sci. 2017;21:4050–4057. [PubMed] [Google Scholar]
- 32.Hou BH, et al. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 33.You Y, et al. MicroRNA-216b-5p Functions as a Tumor-suppressive RNA by Targeting TPT1 in Pancreatic Cancer Cells. J Cancer. 2017;8:2854–2865. doi: 10.7150/jca.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P, et al. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:5186. doi: 10.1038/s41598-017-05274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azevedo-Pouly, A. C., Elgamal, O. A. & Schmittgen, T. D. RNA isolation from mouse pancreas: a ribonuclease-rich tissue. Journal of visualized experiments: JoVE, e51779 (2014). [DOI] [PMC free article] [PubMed]
- 38.Schmittgen TD, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi G, et al. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene. 2013;32:1950–1958. doi: 10.1038/onc.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strobel O, et al. Pancreatic duct glands are distinct ductal compartments that react to chronic injury and mediate Shh-induced metaplasia. Gastroenterology. 2010;138:1166–1177. doi: 10.1053/j.gastro.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grippo PJ, Nowlin PS, Demeure MJ, Longnecker DS, Sandgren EP. Preinvasive pancreatic neoplasia of ductal phenotype induced by acinar cell targeting of mutant Kras in transgenic mice. Cancer Res. 2003;63:2016–2019. [PubMed] [Google Scholar]
- 42.Yonemori K, Kurahara H, Maemura K, Natsugoe S. MicroRNA in pancreatic cancer. J Hum Genet. 2017;62:33–40. doi: 10.1038/jhg.2016.59. [DOI] [PubMed] [Google Scholar]
- 43.Park, J. K., Doseff, A. I. & Schmittgen, T. D. MicroRNAs Targeting Caspase-3 and -7 in PANC-1 Cells. Int J Mol Sci19 (2018). [DOI] [PMC free article] [PubMed]
- 44.Park JK, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406:518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38:e190–199. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 46.Dooley J, et al. No Functional Role for microRNA-342 in a Mouse Model of Pancreatic Acinar Carcinoma. Front Oncol. 2017;7:101. doi: 10.3389/fonc.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bao J, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, et al. Acceleration of pancreatic tumorigenesis under immunosuppressive microenvironment induced by Reg3g overexpression. Cell Death Dis. 2017;8:e3033. doi: 10.1038/cddis.2017.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin G, et al. Reg3g Promotes Pancreatic Carcinogenesis in a Murine Model of Chronic Pancreatitis. Dig Dis Sci. 2015;60:3656–3668. doi: 10.1007/s10620-015-3787-5. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, et al. Reg proteins promote acinar-to-ductal metaplasia and act as novel diagnostic and prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget. 2016;7:77838–77853. doi: 10.18632/oncotarget.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris JPT, et al. Dicer regulates differentiation and viability during mouse pancreatic cancer initiation. PLoS One. 2014;9:e95486. doi: 10.1371/journal.pone.0095486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.