Abstract
A plethora of data points towards a role of the gastrointestinal (GI) microbiota of neonatal and young vertebrates in supporting the development and regulation of the host immune system. However, knowledge of the impact that infections by GI helminths exert on the developing microbiota of juvenile hosts is, thus far, limited. This study investigates, for the first time, the associations between acute infections by GI helminths and the faecal microbial and metabolic profiles of a cohort of equine youngstock, prior to and following treatment with parasiticides (ivermectin). We observed that high versus low parasite burdens (measured via parasite egg counts in faecal samples) were associated with specific compositional alterations of the developing microbiome; in particular, the faecal microbiota of animals with heavy worm infection burdens was characterised by lower microbial richness, and alterations to the relative abundances of bacterial taxa with immune-modulatory functions. Amino acids and glucose were increased in faecal samples from the same cohort, which indicated the likely occurrence of intestinal malabsorption. These data support the hypothesis that GI helminth infections in young livestock are associated with significant alterations to the GI microbiota, which may impact on both metabolism and development of acquired immunity. This knowledge will direct future studies aimed to identify the long-term impact of infection-induced alterations of the GI microbiota in young livestock.
Subject terms: Bacteriology, Parasite host response
Introduction
A wealth of data supports the primary role(s) that the vertebrate gastrointestinal (GI) microbiome plays in overall host health1–3. In humans, microbial populations inhabiting the gut are acquired during early life, and a ‘stable’ gut microbiome is established by 31 months of age4,5. A similar process has been reported in other animals, with the timing of maturation and development of the gut microbiome depending on species lifespan and average age at weaning6,7. A balanced gut microbiota is key to vertebrate long-term health and wellbeing; nevertheless, several factors and life events may impact on the establishment of a ‘healthy’ gut flora in early life. Such factors include, but are not limited to, early life nutrition, use of antimicrobials during gestation and/or in young animals, mode of delivery and infectious diseases8–23.
In neonates and young animals, perturbations of the developing GI microbiota are known to pre-dispose to the onset of a number of systemic conditions, such as allergies, autoimmunity, obesity and colonisation by infectious agents, such as parasites24–29. The intimate mechanisms governing the relationships between host, gut microbiota and parasites are complex and, in many cases, not fully understood; however, experimental evidence points towards a role of the microbiota in supporting the development and regulation of the host immature immune system, e.g. ensuring that adequate responses are mounted against pathogenic stimuli1,30. Indeed, recent data generated from an amphibian model of parasite infection demonstrated that susceptibility to colonisation by the helminth Aplectana hamatospicula of adult animals was directly attributable to a reduced diversity of the microbial populations inhabiting the gut of juveniles27. On the other hand, helminth parasites have been shown to interact, directly and/or indirectly, with the gut flora of their vertebrate hosts31–42, and thus could potentially interfere with the establishment of a ‘healthy’ microbiome in young humans and animals from endemic areas. For instance, experimental infections of rodents with the whipworm Trichuris muris (a model for human infections caused by T. trichiura) and the human blood fluke Schistosoma mansoni have been associated with drastic reductions in GI microbial diversity36–43, with likely negative implications for microbiome maturation in juvenile hosts, and thus for long term immune and metabolic homeostasis. Beside humans, such effects are likely to have particularly severe repercussions in managed livestock herbivore species, e.g. ruminants and equines, that often harbour high burdens of GI helminths due to slow development of acquired immunity and high transmission rates within herds44,45. Thus far, very few studies have examined the impact of helminth infections on the GI microbiota of herbivorous livestock, with inconsistent findings40,46–48. In particular, infection of calves (3–4 months old (mo)) and goats (3 mo) with the abomasal parasites Ostertagia ostertagi47 and Haemonchus contortus46, respectively, showed no effect of the infection on microbial diversity at the site of parasite establishment, whilst a study of H. contortus infection in lambs (3 mo), reported a transient increase in gut bacterial alpha diversity associated with parasite colonisation48. Clearly, further studies are necessary to elucidate the impact that infections with large burdens of parasitic helminths exert on the developing microbiome of juvenile vertebrates and, in turn, on host long-term health and wellbeing.
Managed equine youngstock provide an ideal system for such investigations; indeed, domestic horses are characterised by long lifespans when compared to other livestock species, thus enabling comparisons between the microbiota of young versus adult stock within herds and long-term monitoring of health parameters. In addition, equines are often subjected to strict diets, which provides an opportunity to overcome confounding factors linked to diet variability. Finally, the equine GI microbiome has been extensively characterised in both health and disease7,49–69, thus providing a benchmark for comparative analyses with microbiome composition in parasite-infected horses.
Therefore, in order to identify changes to GI microbial composition, diversity and function associated with GI helminth infection in young herbivorous livestock, our study characterised, for the first time, the faecal microbiome and metabolome of a cohort of thoroughbred (TB) youngstock acutely infected with an economically important group of GI helminths (i.e. the Cyathostominae, strongyle parasites characterised by a direct, non-migratory life cycle with oro-faecal transmission70,71) pre- and post-treatment with anthelmintic compounds.
Results
Comparison of faecal microbiota composition in equine youngstock with high versus low parasite burdens, prior to and following anthelmintic treatment
A cohort of 53 TB equine youngstock were examined for cyathostomin infection and allocated to high- and low-infection burden groups according to the following criteria: i) faecal parasite egg count (FEC – a proxy of parasite infection burden) of ≥100 eggs per gram (e.p.g.) (=C-high) or ≤10 e.p.g. (C-low) in duplicate faecal samples on day 0 (D0) of the study; (ii) negative for co-infections with other GI helminths; (iii) no antibiotic treatment for at least 2 months prior to sampling. Out of the 53 animals screened, 23 matched these criteria, of which 9 were enrolled into the C-high group and 14 into the C-low (Supplementary Table S1). Samples were collected for analyses of faecal microbiota and metabolites from all animals in the C-high and C-low groups immediately prior to treatment with ivermectin (0.2 mg/kg) at D0, as well as at two (D2) and 14 (D14) days post-treatment. FEC analysis performed on samples collected at D2 and D14 showed FEC reduction rates (FECR) of 100% by D14 in all treated animals (Supplementary Table S1). Following total DNA extraction from faecal samples, bacterial 16S rRNA high-throughput sequencing was performed on each sample. A total of 5,201,731 raw paired-end reads were generated from 74 DNA faecal extracts of C-high and C-low yearlings collected on D0, D2 and D14, respectively, and subjected to further processing. Following primer trimming, joining of paired-end reads, filtering of low-quality sequences and removal of ‘contaminant’ and singleton Operational Taxonomic Units (OTUs), a total of 3,048,051 (mean 43,543; range 21,992–58,947) high-quality sequences were retained for further bioinformatics analyses. The rarefaction curves generated following in silico subtraction of low-quality and contaminant sequences indicated that the majority of faecal bacterial communities were represented in the remaining sequence data, thus allowing us to undertake further analyses (Supplementary Fig. S1). These sequences were assigned to 9,972 OTUs and 15 bacterial phyla, respectively. The phyla Bacteroidetes (42.303%) and Firmicutes (42.108%) were predominant in all samples, followed by the phyla Verrucomicrobia (1.319%), Spirochaetes (0.810%), Actinobacteria (0.402%), Proteobacteria (0.3908%), Tenericutes (0.073%), Fibrobacteres (0.0552%), TM7 (0.012%), Lentisphaerae (0.0065%), Synergistetes (0.001%), Fusobacteria (0.001%), Deferribacteres (0.001%), Cyanobacteria (0.001%) and Chlamydiae (0.0001%) (Supplementary Fig. S2), while 11.1% of OTUs could not be assigned to any bacterial group. Predominant sub-taxa were class Bacteroidia, order Bacteroidales within the phylum Bacteroidetes; and class Clostridia, order Clostridiales, families Lachnospiraceae and Ruminococcacae within the Firmicutes. Given the male-gender bias in the C-high group (Supplementary Table S1), a supervised multivariate Canonical Correlation Analysis (CCA) was performed on the microbial communities detected in samples collected from the whole cohort (C-high and C-low) at D0 with ‘gender’ as an explanatory variable; based on the results of this analysis, gender did not impact significantly on the overall gut microbial composition or alpha diversity of equines enrolled in this study (P = 0.826; F = 0.48) (Supplementary Fig. S3).
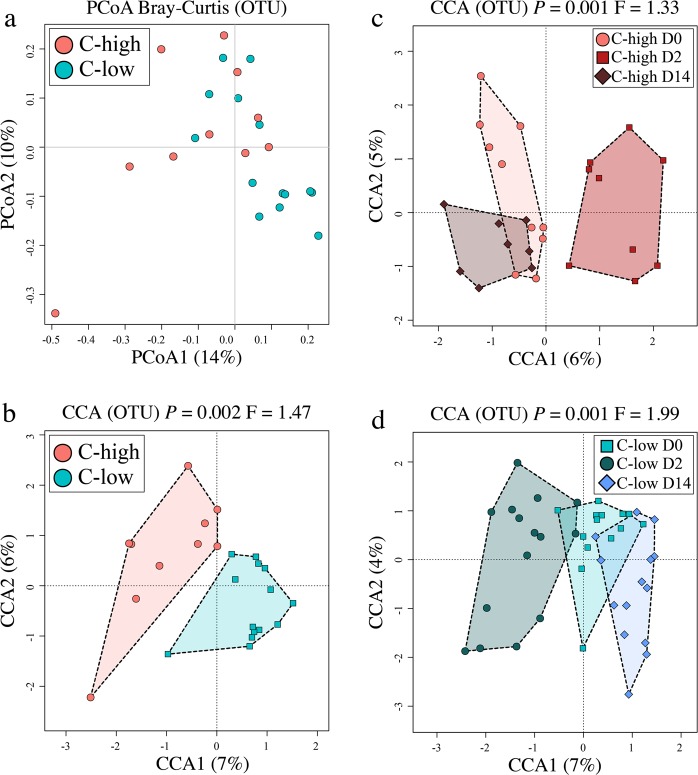
Global analyses of faecal microbial composition of C-high versus C-low at D0 (pre-treatment), using Principal Coordinates analyses (PCoA) with Bray-Curtis distance estimates, did not show marked clustering according to FEC (Fig. 1a); however, a significant difference between these two groups was detected using supervised CCA (P = 0.002; F = 1.47) (Fig. 1b), whilst the effect of gender was insignificant (P = 0.105; F = 1.13), and on a different axis, in the same model (Supplementary Fig. S4). Additionally, the microbial profiles of both C-high and C-low clustered separately according to sample collection time points (D0, D2 and D14) (C-high P = 0.001, F = 1.33; C-low P = 0.001, F = 1.99) (Fig. 1c,d).
Figure 1.
The faecal microbial profiles of youngstock with high (C-high) and low (C-low) parasite infection burdens prior to and following anthelmintic treatment. Multivariate analyses of the faecal microbial composition (based on Operational Taxonomic Unit [OTU] presence and abundance) including (a) Principal Coordinates Analyses (PCoA) of faecal microbial profiles from animals in C-high (≥100 eggs per gram (e.p.g.)) versus C-low (≤10 e.p.g.), (b) Canonical Correlation Analyses (CCA) of faecal microbial profiles from animals in C-high versus C-low, (c) C-high at day 0 (D0) versus day 2 (D2) and day 14 (D14), and (d) C-low at D0 versus D2 and D14.
Microbial richness is reduced in animals with large parasite burdens, and increased following anthelmintic treatment
Microbial richness was lower in C-high at D0 (P = 0.05) than in C-low at the same time point (Fig. 2a). Furthermore, this parameter increased post-treatment (at D2 and D14) to levels comparable to those detected in samples from C-low at D0, although this increase was not statistically significant using ANOVA (P = 0.1) (Fig. 2b). No differences in faecal microbial richness were observed between samples from C-low collected at D0, D2 and D14 (P = 0.87) (Fig. 2c). No statistically significant differences in microbial evenness and Shannon Index were detected between C-high and C-low at D0 (Fig. 2a); however, a significant increase in microbial evenness and Shannon index was observed in both C-high and C-low at D2 and D14 when compared with samples collected at D0 (evenness C-high P = 0.027, C-low P = 0.004; Shannon Index C-high P = 0.028, C-low P = 0.026) (Fig. 2b,c). No significant differences in faecal microbial beta diversity (measured via PERMDISP) were observed between C-high and C-low, according to parasite infection burden and/or sample collection time point post-anthelmintic administration (Supplementary Fig. S5). Given the gender bias towards males in the C-high group, analysis of microbial alpha diversity was repeated for C-high and C-low at D0 with ‘gender’ as an explanatory variable; no significant difference was observed between male and female subjects (Shannon Index P = 0.66; evenness P = 0.7; richness P = 0.69) (Supplementary Fig. S3).
Figure 2.
Faecal microbial richness is reduced in animals with high parasite burdens, and increased post-anthelmintic treatment. Microbial alpha diversity (based on Operational Taxonomic Unit [OTU] presence and abundance), measured by richness, evenness and Shannon index, in faecal samples from (a) C-high (≥100 eggs per gram (e.p.g.)) versus C-low (≤10 e.p.g.), (b) C-high at day 0 (D0) versus day 2 (D2) and day 14, and (c) C-low at D0 versus D2 and D14.
The relationships between infection burdens, anthelmintic treatment and altered abundances of specific microbial taxa
Differences in the relative abundance of specific microbial taxa between groups were evaluated using Linear discriminant analysis Effect Size (LEfSe) and correlations between individual bacterial families, parasite infection burden and sample collection time points were represented by Pearson’s correlation network analysis (Tables 1–3; Fig. 3). At D0, Bacteroidetes were reduced in C-high compared to C-low (LDA effect size 4.66), which was largely attributable to a reduced abundance of bacteria belonging to the families Prevotellaceae and Paraprevotellaceae, and genus Alistipes (Family Rikenellaceae) (Table 1; Fig. 3a). However, in C-high, a relative expansion of bacteria belonging to the families Mogibacteriaceae and Leuconostocaceae (genus Weissella), and genus Paraeggerthella belonging to the class Clostridia (phylum Firmicutes) was observed when compared with C-low (Table 1; Fig. 3a). In the latter group, several taxa belonging to the class Clostridia, including Anaerotruncus, Pseudobutyrivibrio and Unclassified Acidaminobacteraceae, were increased compared with C-high (Table 1; Fig. 3a). When assessing the impact of anthelmintic treatment on the faecal microbial composition of yearlings enrolled in our study, LEfSe analysis revealed a significant decrease of bacteria belonging to the phylum Firmicutes in C-high at D14 when compared to D0, which was largely attributable to a reduction in Mogibacteriaceae, Dehalobacteriaceae, Lactobacillaceae, Streptococcaceae and Enterococcaceae (Table 2; Fig. 3b); in contrast, Prevotellaceae were increased in faecal samples collected from C-high at D14 compared with D0 (Table 2; Fig. 3b). A transient expansion of bacteria belonging to the families Lachnospiraceae, Clostridiaceae and Succinovibrionaceae was also observed in C-high at D2 (Table 2; Fig. 3b). In C-low, Betaproteobacteria (order) and Verrucomicrobia (phylum) were reduced and increased, respectively, at D14 when compared to D0 and D2 (Table 3; Fig. 3c). Similar to samples collected from C-high, bacteria belonging to the families Lachnospiracaeae, Clostridiaceae and Succinivibrionaceae were increased at D2 compared with D0 and D14, respectively (Table 3; Fig. 3c). Of the several bacterial taxa whose abundance was significantly different in samples from C-high and C-low collected pre- and post-anthelmintic treatment, four (i.e. Mogibacteriaceae, Prevotellaceae, Paraprevotellaceae and Rikenellaceae; analysed individually using ANOVA at family level) were significantly affected at D14 following the administration of ivermectin in C-high, whilst remaining unchanged in C-low at the same time point (Fig. 4).
Table 1.
Increased abundance of Mogibacteriaceae, Leuconostocaceae and Eubacteriaceae, and reduced abundance of Prevotellaceae, Paraprevotellaceae and Rikenellaceae in faecal samples from youngstock with high parasite infection burdens.
| Phylum | Class | Order | Family | Genus | Species | C-high | C-low |
|---|---|---|---|---|---|---|---|
| Bacteroidetes |  |
||||||
| Bacteroidia | |||||||
| Bacteroidales | |||||||
| Prevotellaceae |  |
||||||
| Paraprevotellaceae | |||||||
| Rikenellaceae | Alistipes | ||||||
| Bacteroidaceae |  |
||||||
| BF311 | |||||||
| Firmicutes | Bacilli | Lactobacillales | Leuconostocaceae |  |
|||
| Weissella | |||||||
| Clostridia | Clostridiales | Mogibacteriaceae | |||||
| Mogibacterium | |||||||
| Mogibacterium (unclassified) | |||||||
| Eubacteriaceae | Paraeggerthella | Paraeggerthella hongkongensis | |||||
| Lachnospiracaeae | Pseudobutyrivibrio |  |
|||||
| Acidaminobacteraceae | |||||||
| Acidaminobacteraceae (unclassified) | |||||||
| Acidaminobacteraceae (unclassified) | |||||||
| Clostridiaceae | Anaerotruncus | Anaerotruncus unclassified | |||||
| Proteobacteria | Gammaproteobacteria | Pasteurellales |  |
||||
| Pasteurellaceae | |||||||
| Epsilonproteobacteria | Campylobacterales | Camplylobacteraceae | Campylobacter | Campylobacter jejuni | |||
| Alphaproteobacteria | RF32 |  |
|||||
| RF32 (unclassified) | |||||||
| RF32 (unclassified) | |||||||
| Betaproteobacteria | Burkholderiales |
Differences in relative abundance of selected microbial taxa in faecal samples from C-high (faecal egg count (FEC) > 100 eggs per gram (e.p.g.)) and C-low (FEC < 10 e.p.g.). Light grey: effect size of 3.5–4.5; dark grey: effect size >4.5.
Table 3.
Increased abundance of Clostridiales, Bacteroidales and Verrucomicrobia, and reduced α- and β-Proteobacteria in faecal samples from youngstock with low parasite infection burdens prior to and following anthelmintic treatment.
| Phylum | Class | Order | Family | Genus | Species | D0 | D2 | D14 |
|---|---|---|---|---|---|---|---|---|
| Bacteroidetes | Bacteroidia |  |
||||||
| Bacteroidales | ||||||||
| Verrucomicrobia |  |
|||||||
| Verruco5 |  |
|||||||
| WCHB141 | ||||||||
| RFP12 | ||||||||
| RFP12 unclassified | RFP12 unclassified | |||||||
| Firmicutes | Bacilli |  |
||||||
| Lactobacillales | ||||||||
| Lactobacillaceae | Lactobacillus | Lactobacillus ruminis | ||||||
| Enterococcaceae | Enterococcus | Enterococcus casseliflavus |  |
|||||
| Enterococcus |  |
|||||||
| Clostridia | Clostridiales | Lachnospiraceae |  |
|||||
| Roseburia |  |
|||||||
| Roseburia inulinivorans | ||||||||
| Pseudobutyrivibrio | ||||||||
| Pseudobutyrivibrio unclassified | ||||||||
| Clostridiaceae | Clostridium |  |
||||||
| Clostridium butyricum | ||||||||
| Acidaminobacteraceae | Mitsuokella |  |
||||||
| Mitsuokella multacida | ||||||||
| Eubacteriaceae | Eubacterium | |||||||
| Eubacterium unclassified | ||||||||
| Eubacteriaceae |  |
|||||||
| Ruminococcaceae | ||||||||
| Oscillospira | ||||||||
| Oscillospira guillermondi | ||||||||
| Ruminococcus | Ruminococcus bromii |  |
||||||
| Ruminococcaceae unclassified |  |
|||||||
| Ruminococcus | ||||||||
| Ruminococcus callidus |  |
|||||||
| Proteobacteria | Gammaproteobacteria | |||||||
| Aeromonadales | ||||||||
| Succinivibrionacae | ||||||||
| Betaproteobacteria |  |
|||||||
| Burkholderiales | ||||||||
| Alphaproteobacteria | RF32 | |||||||
| RF32 unclassified | ||||||||
| RF32 unclassified |
Differences in relative abundance of selected microbial taxa in faecal samples from C-low (FEC < 10 egg per gram (e.p.g.)) prior to treatment (D0), and 2 (D2) and 14 (D14) days post-treatment. Light grey: effect size of 3.5–4.5; dark grey: effect size >4.5.
Figure 3.
Network analyses reveal associations between faecal microbial composition, parasite infection burden, and time pre- and post-anthelmintic treatment. Pearson’s correlation network analyses showing bacterial taxa (at family level) that were positively associated to faecal samples from (a) C-high (≥100 eggs per gram (e.p.g.)) (in red) versus C-low (≤10 e.p.g.) (in blue), (b) C-high at Day 0 (D0) (in red), day 2 (D2) (in yellow) and day 14 (D14) (in green) post-treatment, and (c) C-low at D0 (in blue), D2 (in yellow) and D14 (in green). For taxa associated with multiple sample groups, the respective circle colors are mixed according to the strength of the association.
Table 2.
Increased abundance of Clostridiales and Prevotellaceae, and reduced Lactobacillaceae and Mogibacteriaceae in faecal samples from youngstock with high infection burdens following anthelmintic treatment.
| Phylum | Class | Order | Family | Genus | Species | D0 | D2 | D14 |
|---|---|---|---|---|---|---|---|---|
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae |  |
||||
| Prevotellaceae unclassified |  |
|||||||
| Prevotellaceae unclassified |  |
|||||||
| Firmicutes | ||||||||
| Bacilli | ||||||||
| Lactobacillales | ||||||||
| Lactobacillaceae | ||||||||
| Lactobacillus | ||||||||
| Enterococcaceae | Enterococcus |  |
||||||
| Enterococcus casseliflavus | ||||||||
| Streptococcaceae | ||||||||
| Streptococcus | ||||||||
| Erysipelotrichales | Erysipelotrchaceae | RFN20 |  |
|||||
| RFN20 unclassified | ||||||||
| Clostridia | Clostridiales |  |
||||||
| Lachnospiraceae | ||||||||
| Roseburia |  |
|||||||
| Roseburia inulinivorans | ||||||||
| Pseudobutyrivibrio | ||||||||
| Pseudobutyrivibrio unclassified | ||||||||
| Clostridiaceae | Clostridium |  |
||||||
| Mogibacteriaceae |  |
|||||||
| Mogibacterium | ||||||||
| Mogibacterium unclassified | ||||||||
| Dehalobacteriaceae | Dehalobacteriaceae unclassified | |||||||
| Dehalobacteriaceae unclassified | ||||||||
| Ruminococcaceae | ||||||||
| Oscillospira | ||||||||
| Oscillospira guillermondi | ||||||||
| Ruminococcus | Ruminococcus bromi |  |
||||||
| Ruminococcaceae unclassified | ||||||||
| Proteobacteria | Gammaproteobacteria |  |
||||||
| Aeromonadales |  |
|||||||
| Succinivibrionacae |
Differences in relative abundance of selected microbial taxa in faecal samples from C-high (FEC > 100 egg per gram (e.p.g.)) prior to treatment (D0), and 2 (D2) and 14 (D14) days post-treatment. Light grey: effect size of 3.5–4.5; dark grey: effect size >4.5.
Figure 4.
The abundances of faecal Paraprevotellaceae, Rikinellaceae, Prevotellaceae and Mogibacteriaceae prior to and following anthelmintic treatment. The relative abundances of Paraprevotellaceae, Rikinellaceae, Prevotellaceae and Mogibacteriaceae in (a) C-high and C-low prior to anthelmintic treatment and (b) C-high (≥100 eggs per gram (e.p.g.)) and (c) C-low (≤10 e.p.g.) prior to and following anthelmintic treatment (ANOVA).
Faecal metabolite changes associated with high- and low infection burdens and anthelmintic treatment
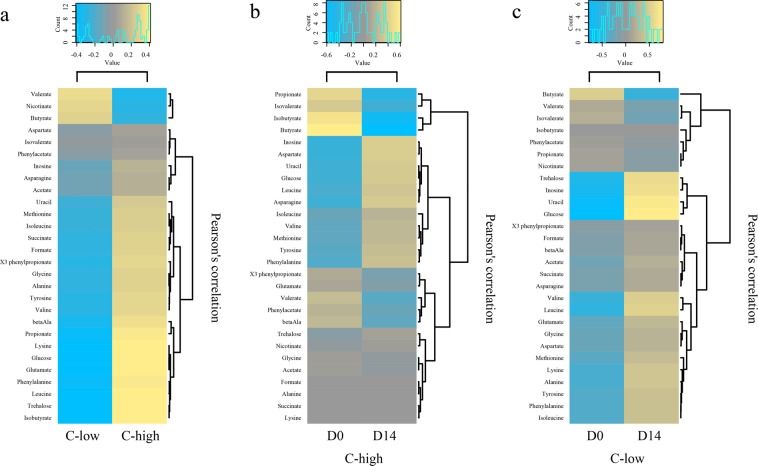
A total of 28 faecal metabolites in all samples from C-high and C-low collected at D0 and D14 were identified and quantified by Proton Nuclear Magnetic Resonance (1H-NMR). These were subjected to PCoA (Supplementary Fig. S6) and CCA; the latter allowed us to identify differences between the faecal metabolomes of C-high and C-low at D0 (P = 0.059; F = 1.97) (Fig. 5a). Pearson’s correlation heatmap analyses demonstrated increased abundances of isobutyrate, trehalose, leucine, phenylalanine, glutamate, glucose, lysine and propionate, and decreased nicotinate, valerate and butyrate, in C-high versus C-low (Fig. 6a); however, these differences were not statistically significant according to ANOVA following False Discovery Rate (FDR) correction for multiple testing (P > 0.05 for all metabolites) (Supplementary Table S2). CCA also detected a difference between the faecal metabolomes of C-high at D0 and D14 (P = 0.001; F = 1.2), that however, for individual metabolites, was not statistically significant using ANOVA following FDR correction (P > 0.05 for all metabolites) (Fig. 5b; Supplementary Table S3); nevertheless, a relative reduction in the short chain fatty acids (SCFAs) butyrate, isobutyrate and propionate was observed in samples from this group at D14 versus D0 via Pearson’s correlation heatmaps (Fig. 6b). Conversely, in addition to a greater effect size according to CCA (P = 0.001; F = 5.88), clear differences were detected between the faecal metabolites identified in samples from C-low at D0 and D14 (Fig. 5C; Supplementary Table S4). In particular, glucose, uracil, inosine, trehalose, leucine, butyrate and valine were more abundant in samples collected at D14 when compared to D0 (ANOVA; P < 0.001, P < 0.001, P = 0.001, P = 0.003, P = 0.022, P = 0.041 and P = 0.044, respectively) (Fig. 6c; Supplementary Table S4). Partial Least Squares (PLS) analysis of metabolite and bacterial OTU data obtained from faecal samples at D0, with ‘metabolites’ as the dependent matrix and ‘bacterial OTUs’ as the independent matrix, did not identify any significant association between these two datasets (P = 0.96)
Figure 5.
The faecal metabolic profiles of youngstock with high and low parasite infection burdens prior to and following anthelmintic treatment. Canonical Correlation Analyses (CCA) plots depicting differences between global faecal metabolic profiles of samples from (a) C-high (≥100 eggs per gram (e.p.g.)) (in red) versus C-low (≤10 e.p.g.) (in blue); (b) C-high at day 0 (D0) (in red) and day 14 (D14) (in brown); and (c) C-low at D0 (in green) and D14 (in blue).
Figure 6.
Faecal metabolic profiles of youngstock with high and low parasite infection burdens prior to and following anthelmintic treatment. Pearson’s correlation heatmaps depicting differences in the relative abundances of faecal metabolites between (a) C-high (≥100 eggs per gram (e.p.g.)) and C-low (≤10 e.p.g.) at day 0 (D0); (b) C-high at D0 and day 14 (D14); and (c): C-low at D0 and D14.
Discussion
In this study, acute helminth infections in a cohort of equine youngstock were associated with significant alterations in gut microbial profiles and diversity, which were reversed following parasiticide treatment. The faecal microbial profiles of the yearlings in our study were in line with those described previously for managed equines54,67,69,72,73, with the predominance of OTUs assigned to Bacteroidetes (42.3%) and Firmicutes (42.1%); nevertheless, a greater interindividual variation in taxonomic composition was observed in this group of animals when compared to a cohort of adult broodmares hosted on the same farm40. This discrepancy may be suggestive of a ‘developing microbiome’ in these young animals. Indeed, a recent study in human infants demonstrated the occurrence of three stages of microbial development in the GI tract during early life, which include an early phase characterised by rapid changes in the core GI microbiota over time, an intermediate transitional phase characterised by fewer changes over time, and a final and stable ‘adult’ microbial profile4. In support of our findings, a previous study conducted in equine youngstock demonstrated that, although gut microbiome re-modelling occurs mostly during the first 60 days of life, the gut microbial profiles of horses of 9 months of age is still significantly different from those of adult animals7. Of note, environmental and pathogenic stimuli which impact on the order of colonisation of the GI tract during the developmental phase may have significant repercussions on adult microbiome composition and homeostasis1,28,74. Hence, given the known roles of the gut microbiome in immune regulation and metabolism, it is likely that the alterations in faecal microbiome composition of equine youngstock observed in association with GI helminth infections may result in long-term implications for animal health and wellbeing.
Indeed, the abundance of several bacterial taxa with key roles in metabolism and immune-regulation differed significantly between C-high and C-low. For instance, bacterial families belonging to the classes Clostridia and Bacteroidetes, were increased and decreased, respectively, in C-high when compared to C-low prior to anthelmintic treatment. Of note, several differences in faecal microbial profiles observed between groups before treatment were reversed following anthelmintic administration; for instance, bacteria belonging to the families Mogibacteriaceae (phylum Firmicutes; class Clostridia), Prevotellaceae (phylum Bacteroidetes; class Bacteroidetes) were similar in abundance in faecal samples from C-low and C-high by D14. A reduction in Bacteroidetes (genera Prevotella and Parabacteroides) was also observed in mice experimentally infected with the murine whipworm, T. muris36. Members of the family Prevotellaceae are commensals of mammalian mucosal surfaces, and are known potential pathobionts, particularly associated with oral infections75. Prevotella spp. have been demonstrated to promote pro-inflammatory Th17-mediated immune responses75,76; thus, a decreased abundance of this bacterial family in the presence of GI helminth infections could be linked to the known immune-regulatory properties of these parasites77. Indeed, a reduction in Th17-inducing segmented filamentous bacteria has been previously reported in association with Nippostrongylus brasiliensis infection in a rodent model34, indicating that this may be a common immune-modulatory mechanism in GI helminth infections. Nevertheless, in herbivores, Prevotellaceae also play a key role in the break-down of indigestible fibres78,79; moreover, in a study by Houlden et al.36, an observed increase in long chain fatty acids in the faeces of T. muris infected mice was attributed to inefficient breakdown of plant fibres as a result of reduced abundance of Prevotella and Parabacteroides. This dichotomy of function in Prevotella spp. has been well described80, and demonstrates that, whilst Prevotella are commensal bacteria with an important metabolic role, they can be pro-inflammatory in association with mucosal pathology. It is also worth reporting that, in contrast to our findings, a positive association between helminth infections and Prevotella spp. abundance was observed in the abomasum of 3 mo small ruminants colonised by the GI helminth H. contortus46. Whilst this apparent discrepancy may be linked to fundamental differences between the host:parasite systems under investigation and infection sites (stomach versus large intestine), this data calls for further investigations of the mechanisms governing the interactions between GI helminths and Prevotella spp. and their implications for the developing microbiome of young vertebrates.
Within the phylum Firmicutes, a number of families belonging to class Clostridia were significantly more abundant in the faecal microbiota of C-high when compared to C-low at D0, according to LEfSe (Mogibacteriaceae and Eubacteriaceae) and network (Dehalobacteriaceae and Clostridiacae) analyses. Clostridia have been frequently implicated in studies of helminth-microbiome interactions, with increased abundances being reported in the gut of mice experimentally infected with T. muris36,81,82, and in association with mixed GI helminth infections in humans82. Clostridia are known to exert immune-regulatory functions, since they produce SCFAs with anti-inflammatory properties, including butyrate83,84. Hence, the increased abundances of Mogibacteriaceae and other Clostridiaceae observed in the faecal microbiota of youngstock with heavy parasite burdens could represent a mechanism by which helminths suppress host immune responses, thus reducing pathology and facilitating the establishment of chronic infections83. However, Mogibacteriaceae are not butyrate producers85; in addition, metabolomic analysis of faecal samples collected in this study revealed lower levels of butyrate in faeces from C-high when compared to C-low prior to anthelmintic treatment. In accordance with this observation, a recent study investigating the faecal metabolome of human volunteers with chronic infections by Strongyloides stercoralis also detected lower levels of butyrate in samples from parasite-colonised individuals42. These data indicate that, whilst increases in SFCAs have been described previously in association with GI helminth infection83, this link may not be applicable to all host-parasite systems. Of note, Clostridia belonging to the family Mogibacteriaceae have also been associated with GI inflammation in periodontal disease86, and hence their increased abundance in animals with higher infection levels in this study may suggest a role for these bacteria in the onset of mucosal pathology. On the other hand, increased levels of Mogibacteriaceae have also been identified as a biomarker of health in studies comparing subjects with inflammatory bowel disease and irritable bowel disease, to healthy controls87–89. These apparent contradictions are analogous to those described above for Prevotella spp., and highlight the need for mechanistic studies to unravel the complex function of microbial species within the gut flora colonising different hosts, and in various disease states.
Amongst other bacterial taxa with well-known immune-modulatory functions, those belonging to the order Lactobacillales (phylum Firmicutes, class Bacilli), were also expanded in the faecal microbiota of C-high versus C-low at D0, and bacteria of the family Lactobacillaceae were significantly reduced following anthelmintic treatment in the former group. The existence of a mutualistic association between Lactobacillaceae and parasitic helminths has been hypothesised based on experimental evidence obtained from murine models of infection with the intestinal nematode Heligmosomoides polygyrus38,90. Indeed, in a key study by Reynolds et al.90, H. polygyrus infections were associated with significantly expanded populations of Lactobacillaceae in the gut of colonised mice; in addition, oral administration of L. taiwanensis prior to helminth exposure was followed by the onset of T regulatory cell mediated immune responses and significantly increased worm burdens91. Based on this knowledge, it is tempting to speculate that similar relationships may exist between cyathostomin parasites and equine hosts. Moreover, the increased abundance of bacterial taxa with immune-modulatory roles in animals with heavy burdens of helminth infections may alter the susceptibility of the equine hosts to colonisation by other, ‘opportunistic’ pathogens. Indeed, potential pathobionts such as Campylobacter jejuni and Pasteurellaceae were expanded in animals with high FEC, and reflected data collected in previous investigations of helminth-infected horses and pigs92,93. In humans and rodents, Lactobacillaceae are also generally considered to have positive health benefits for the host94; however, the beneficial effects of Lactobacillaceae to the health and homeostasis of the equine GI microbiota is yet to be demonstrated54,95. A recent review of data collected from equine gut microbiome studies has highlighted the roles of other bacterial taxa, such as Lachnospiraceae (phylum Firmicutes, class Clostridia), in supporting intestinal health54. In particular, increased abundances of OTUs assigned to the family Lachnospiraceae have been reported in the faecal microbiome of healthy horses when compared with animals with colitis51,68; furthermore, ponies with innate resistance to helminth infection, and consequently lower parasite burdens, have been shown to harbour larger populations of this bacterial family93. Accordingly, in our study, the genus Pseudobutyrivibrio (family Lachnospiraceae) was significantly more abundant in the faecal microbiome of C-low versus C-high prior to treatment. Together, this data points towards a detrimental effect of acute cyathostomin infections on the development and maturation of the equine gut microbiome, an effect which is highly likely to have repercussions on susceptibility to a range of infectious and non-infectious diseases1,27,93,96. In support of this hypothesis, faecal microbial richness (a measure of microbial alpha diversity and a proxy of gut ‘health’29,66,97–99) was significantly lower in samples from C-high when compared to C-low prior to treatment. Acute helminth infections have been frequently associated with decreased alpha diversity of microbial gut populations36,42,43 and attributed primarily to alterations of the GI microenvironment in response to mounting immune reactions against the invading parasites. Nevertheless, these data contrast with our previously published work in which we reported a trend towards increased alpha diversity in the gut microbiome of chronically helminth-infected adult horses40. Differences between the systems investigated (i.e. acutely versus chronically infected horses) may provide a possible explanation, whereby infections of immunologically naïve animals and subsequent pro-inflammatory responses are accompanied by a significant decrease in gut microbial diversity; the latter may be restored (or increased; cf.39,42,100) during chronic helminth infections, due to a synergic effect of host adaptive immune responses and immune-suppressive properties of helminth parasites101 that contributes to the dampening of local inflammation102.
Given the impact of helminth infections on the composition and diversity of the youngstock gut microbiome, we sought to investigate the associations between such changes and the composition of the faecal metabolome. A multivariate analyses of faecal metabolite levels in samples from C-high and C-low prior to and following ivermectin administration revealed only moderate differences between the faecal metabolic profiles of these two groups. A greater abundance of the SCFA isobutyrate, the amino acids leucine, lysine and phenylananine, and the products of carbohydrate breakdown glucose and trehalose were observed in samples from C-high versus C-low prior to ivermectin administration. Of note, increases in selected amino acid abundance have also been described in the faecal metabolome of both mice36 and humans infected by GI helminths42. This consistent observation may indicate a reduction in the absorption of the products of microbial metabolism, for example, as a consequence of ongoing intestinal inflammation caused by helminth colonisation. This hypothesis is supported by the lack of significant correlations between the abundance of specific gut bacteria and metabolites detected by PLS analysis, which might indicate that host-related factors, such as malabsorption, might have been responsible for the observed changes. In the context of livestock management, a reduction in amino acid absorption is likely to have considerable implications for animal performance and production. Thus, the relationships between metabolite production and absorption should be further investigated via metabolomics analyses of faeces and other biofluids (e.g. urine and blood) of equines and other livestock species infected by parasitic helminths103. Administration of anthelmintics to C-high did not result in significant alterations of concentrations of faecal metabolites detected before treatment, suggesting that any malabsorptive component of the observed increase in amino acids and glucose, did not resolve over the time-scale of the study. In contrast, concentrations of glucose, uracil, inosine and trehalose in faecal samples from C-low were significantly increased post-treatment. As these animals did not harbour significant parasite burdens prior to anthelmintic treatment, this finding supports a possible role of other environmental factors, such as grass quality and/or anthelmintic administration, on faecal metabolite and gut bacteria abundance over time.
Concluding remarks
Data from both bacterial 16S rRNA sequences and 1H-NMR analyses of faecal samples from equine youngstock with high versus low parasite burdens revealed a number of helminth-associated perturbations to GI microbial composition and metabolism, with likely repercussions on medium- to long-term host susceptibility to a range of infections and diseases which should be further investigated. In particular, we advocate for in-depth studies of the impact that early colonisation of young production animals by parasites (e.g. calves, lambs and piglets) exerts on host gut microbial composition and metabolism, given the likely severe economic impact of alterations of the gut homeostasis of these species. In addition, data from our study calls for further explorations of the role/s that parasite-associated changes in gut microbiota may play in the immunopathology of helminth infections in children from endemic areas, and their susceptibility to other parasitic, bacterial and viral infectious agents in areas of poor sanitation.
Methods
Ethics statement
This study was approved and carried out in strict accordance and compliance with the guidelines of the Institutional Ethical Review Committee, Department of Veterinary Medicine, University of Cambridge, UK (Ref. No. CR190). Written informed consent was obtained from the stud farm from which study samples were collected.
Study population and diagnostic procedures
A cohort of TB yearlings was recruited from a stud farm in eastern England, UK. The stud hosts ~50 yearlings each year, which are kept at pasture in groups of 2–8 across 486 hectares. Routine control of parasite infections in the stud relies on administration of targeted anthelmintic treatments (with ivermectin and fenbendazole), based on evaluation of individual parasite burdens (inferred by FEC, that consist in counting the number of parasite e.p.g. of faeces) at 2 monthly intervals. In addition, praziquantel is administered to each yearling three times a year for tapeworm control, whilst a single moxidectin treatment is administered in late November for control of cyathostomins. Samples used in this study were collected in April 2017; all yearlings had received ivermectin (0.2 mg/kg) and fenbendazole (10 mg/kg) in February 2017, and praziquantel (1.5 mg/kg) in March 2017. A total of 53 TB yearlings, between 12–16 months of age at the time of sampling, were screened for infection by cyathostomins. Briefly, duplicate faecal samples were collected from all horses on D0; aliquots of each sample were subjected to (i) FEC analysis using a centrifugal floatation technique sensitive to one e.p.g.104, and (ii) screening for infections with the common equine cestode Anoplocephala perfoliata using a double sugar flotation technique105, and (iii) larval culture and microscopic examination to screen for infections with large strongyle nematodes, e.g. Strongylus vulgaris. Horses were recruited in the study if they satisfied the following criteria: (i) FEC of ≥100 e.p.g. (=C-high) or ≤10 e.p.g. (C-low) in duplicate faecal samples at D0; (ii) negative for co-infections with other GI helminths (i.e. Strongylus spp., Parascaris equorum, Strongyloides westeri, A. perfoliata) and no history of or clinical signs of infection by Oxyuris equi; (iii) no antibiotic treatment for at least 2 months prior to sampling. Of the 53 horses screened, 23 matched these criteria, of which 9 were enrolled into the C-high group, and 14 into C-low (Supplementary Table S1). A power calculation was performed based on a bacterial 16S rRNA amplicon sequence dataset from a previous study conducted using samples from equines on the same farm40. A Wilcoxon-Mann-Whitney test for comparing OTU-specific abundances between two samples, as described by Mattiello et al.106 (https://fedematt.shinyapps.io/shinyMB), demonstrated that the current study with 23 samples had sufficient power (0.81) to detect an effect size of 2, in at least 20 OTUs between C-high and C-low.
Anthelmintic treatment and sampling
Individual, freshly voided, faecal samples were collected from the centre of the faecal mass from all C-high and C-low animals at D0. Immediately following sample collection, an anthelmintic treatment (Eqvalan: ivermectin 0.2 mg/kg) was administered to each animal. Sampling was repeated as above at D2 and D14. A 100 g aliquot of each faecal sample was snap frozen, transported to the laboratory and stored at −80 °C within 2 h of collection, prior to genomic DNA extraction, high-throughput sequencing of the bacterial 16S rRNA gene and metabolite extraction; the remainder was kept fresh and subjected to FEC analysis as described above.
DNA extractions and bacterial 16S rRNA gene Illumina sequencing
Previously published40,42 bacterial 16S rRNA high-throughput sequencing protocols and bioinformatics analyses of sequence data were adapted for this study. Briefly, genomic DNA was extracted from each faecal sample, as well as from five negative ‘blank’ (=no DNA) controls, using the PowerSoil® DNA Isolation Kit (Qiagen, Carlsbad, CA, USA), according to the manufacturers’ instructions. High-throughput sequencing of the V3-V4 hypervariable region of the bacterial 16S rRNA gene was performed by Eurofins Genomics on an Illumina MiSeq platform according to the standard protocols with minor adjustments. Briefly, the V3-V4 region was PCR-amplified using universal primers, that contained the adapter overhang nucleotide sequences for forward (TACGGGAGGCAGCAG) and reverse primers (CCAGGGTATCTAATCC). Amplicons were purified using AMPure XP beads (Beckman Coulter) and set up for the index PCR with Nextera XT index primers (Illumina). The indexed samples were purified using AMPure XP beads (Beckman Coulter) and quantified using the Fragment Analyzer Standard Sensitivity NGS Fragment Analysis Kit (Advanced Analytical) and equal quantities from each sample were pooled. The resulting pooled library was quantified using the Agilent DNA 7500 Kit (Agilent), and sequenced using the v3 chemistry (2 × 300 bp paired-end reads, Illumina).
Bioinformatics and statistical analyses of 16S rRNA sequencing data
Raw paired-end Illumina reads were trimmed for 16S rRNA gene primer sequences using Cutadapt (https://cutadapt.readthedocs.org/en/stable/) and sequence data were processed using the Quantitative Insights Into Microbial Ecology 2 (QIIME2-2018.4; https://qiime2.org) software suite107. Successfully joined sequences were quality filtered (Read cut-off: 17; 286 and 17; 255 for forward and reverse, respectively), dereplicated, chimeras identified, and paired-end reads merged in QIIME2 using DADA2108. A phylogenetic tree was generated for diversity analysis, followed by calculation of alpha and beta diversity metrics using the ‘core-metrics-phylogenetic command’ in QIIME2. Sequences were assigned to taxonomy using the feature classifier: Greengenes 13_8 99% OTUs full-length sequences. A feature table with the assigned taxonomy was exported from QIIME2 alongside a weighted UniFrac distance matrix for downstream biostatistical analysis. Statistical analyses were executed using the Calypso software109 (cgenome.net/calypso/); total sum scaling (TSS) normalisation was applied, followed by square route transformation to account for the non-normal distribution of taxonomic counts data. Microbial community profiles were ordinated using PCoA (Bray-Curtis distances) and supervised CCA including infection status and sample collection time point as explanatory variables. Differences in bacterial alpha diversity (richness, evenness and Shannon Index) between study groups (C-high and C-low at D0; as well as D0, D2 and D14 for each C-high and C-low) were evaluated based on rarefied data (read depth of 25,868) using ANOVA. Differences in beta diversity (weighted UniFrac distances) were measured using PERMDISP110. Differences in the abundances of individual microbial taxa between groups were assessed using the LEfSe workflow111, accounting for the paired nature of samples pre- and post-anthelmintic treatment. Furthermore, networks of correlation were constructed using the Calypso software109 to identify clusters of co-occurring bacteria based on their association with helminth infection status. Taxa and explanatory variables were represented as nodes, taxa abundance as node size, and edges represented positive associations, while nodes were coloured according to infection status. Taxa abundances were associated with infection status using Pearson’s correlation and nodes were then coloured based on the strength of this association. Networks were generated by first computing associations between taxa using Pearsons’s rho and the resulting pairwise correlations were converted into dissimilarities and then used to ordinate nodes in a two-dimensional plot by PCoA. Therefore, correlating nodes were located in close proximity and anti-correlating nodes were placed at distant locations in the network.
Metabolite extraction
Metabolites were extracted from 200 mg aliquots of each faecal sample using a methanol–chloroform–water (2:2:1) procedure, as described previously42. In particular, 600 μl of methanol–chloroform mix (2:1 v:v) were added, samples were homogenised using stainless steel beads and sonicated for 15 min at room temperature. 200 μl each of chloroform and water were added, samples were centrifuged and the separated aqueous and lipid phases were collected. The procedure was repeated twice, and the aqueous fraction from each extraction were pooled. The aqueous fraction was dried in a vacuum concentrator (Concentrator Plus, Eppendorf).
1H-NMR analysis of aqueous extracts
Protocols of 1H-NMR metabolite analysis have been described previously42. Briefly, the dried aqueous fractions were re-dissolved in 600 μl D2O, containing 0.2 mM sodium-3-(tri-methylsilyl)-2,2,3,3-tetradeuteriopropionate (TSP) (Cambridge Isotope Laboratories, MA, USA) as an internal standard and phosphate buffer (40 mM NaH2PO4/160 mM Na2HPO4). The samples were analysed using an AVANCE II + NMR spectrometer operating at 500.13 MHz for the 1H frequency and 125.721 MHz for the 13C frequency (Bruker, Germany) using a 5 mm TXI probe. The instrument is equipped with TopSpin 3.2. Spectra were collected using a solvent suppression pulse sequence based on a one-dimensional nuclear Overhauser effect spectroscopy (NOESY) pulse sequence to saturate the residual 1 H water signal (relaxation delay = 2 s, t1 increment = 3 us, mixing time = 150 ms, solvent pre-saturation applied during the relaxation time and the mixing time). One hundred and twenty-eight transients were collected into 16 K data points over a spectral width of 12 ppm at 27 °C. In addition, representative samples of each data set were also examined by two-dimensional Correlation Spectroscopy (COSY), using a standard pulse sequence (cosygpprqf) and 0.5 s water presaturation during relaxation delay, 8 kHz spectral width, 2048 data points, 32 scans per increment, 512 increments. Peaks were assigned using the COSY spectra in conjunction with reference to previous literature and databases and the Chenomx spectral database contained in Chenomx NMR Suite 7.7 (Chenomx, Alberta, Canada). 1D-NMR spectra were processed using TopSpin. Free induction decays were Fourier transformed following multiplication by a line broadening of 1 Hz, and referenced to TSP at 0.0 ppm. Spectra were phased and baseline corrected manually. The integrals of the different metabolites were obtained using Chenomx. Metabolites were normalised to faecal dry matter, total area and differential abundance of metabolites between samples from C-high and C-low, at D0 and D14. Faecal metabolite abundances from each sample were ordinated by PCoA according to infection status (C-high and C-low) and sample collection time point (D0 and D14). Predicted associations among metabolites identified in the faecal metabolome of each sample group were also identified by Pearson’s correlation heatmaps in Calypso109 (cgenome.net/calypso/). In particular, heatmaps were constructed to identify associations between metabolite abundance and infection status (i.e. C-high and C-low) and time point pre- and post-treatment (i.e. D0 and D14). Differences in individual metabolite abundance between groups were evaluated for statistical significance using ANOVA with FDR correction for multiple comparisons. In order to identify linear correlations between metabolites and bacterial OTUs identified in faecal samples, PLS analysis was performed on data from all samples at D0 (pre-treatment) in Simca-P v15, with metabolites as the dependent and OTU as the independent matrix, respectively.
Supplementary information
Acknowledgements
L.E.P. is the grateful recipient of a Horserace Betting Levy Board postdoctoral research fellowship. T.P.J. PhD scholarship is funded by the Biotechnology and Biological Sciences Research Council (UK). Research in the C.C. laboratory is funded by grants awarded by the Isaac Newton Trust/Wellcome Trust/University of Cambridge joint research scheme, the Isaac Newton Trust and the Royal Society (UK). The authors wish to thank the managers and employees of the UK stud farm for contributing to this study.
Author Contributions
L.E.P. and C.C. designed the research; L.E.P., R.A.M. and Ce.C. carried out the research and performed data processing and analyses. L.E.P. and C.C. drafted the manuscript text, with input from Ce.C. and J.L.G. The figures were prepared by T.P.J., with input from L.E.P. and C.C. All authors reviewed and approved the manuscript prior to submission.
Data Availability
The raw 16s rRNA and HNMR data, metadata and QIIME2 feature table are available at Mendeley Data (10.17632/95m8sfd3kt.1).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laura E. Peachey, Email: laura.peachey@bristol.ac.uk
Cinzia Cantacessi, Email: cc779@cam.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-47204-6.
References
- 1.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 4.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:852. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013;7:1069–79. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa MC, Stämpfli HR, Allen-Vercoe E, Weese JS. Development of the faecal microbiota in foals. Equine Vet. J. 2016;48:681–688. doi: 10.1111/evj.12532. [DOI] [PubMed] [Google Scholar]
- 8.Dong TS, Gupta A. Influence of early life, diet, and the environment on the microbiome. Clin. Gastroenterol. Hepatol. 2018;17:231–242. doi: 10.1016/j.cgh.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adami AJ, et al. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr. Res. 2018;84:426–434. doi: 10.1038/s41390-018-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iizumi T, Battaglia T, Ruiz V2, Perez Perez GI. Gut microbiome and antibiotics. Arch. Med. Res. 2017;48:727–734. doi: 10.1016/j.arcmed.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, et al. Marked response in microbial community and metabolism in the ileum and cecum of suckling piglets after early antibiotics exposure. Front. Microbiol. 2018;9:1166. doi: 10.3389/fmicb.2018.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sylvia KE, Deyoe JE, Demas GE. Early-life sickness may predispose Siberian hamsters to behavioral changes following alterations of the gut microbiome in adulthood. Brain Behav. Immun. 2018;73:571–583. doi: 10.1016/j.bbi.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schokker D, et al. Long-lasting effects of early-life antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PLoS One. 2015;10:e0116523. doi: 10.1371/journal.pone.0116523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astbury S, et al. High fructose intake during pregnancy in rats influences the maternal microbiome and gut development in the offspring. Front. Genet. 2018;9:203. doi: 10.3389/fgene.2018.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berer K, et al. Dietary non-fermentable fiber prevents autoimmune neurological disease by changing gut metabolic and immune status. Sci. Rep. 2018;8:10431. doi: 10.1038/s41598-018-28839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8:143–171. doi: 10.1080/19490976.2016.1278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, et al. Early-life food nutrition, microbiota maturation and immune development shape life-long health. Crit. Rev. Food. Sci. Nutr. 2018;59:S30–S38. doi: 10.1080/10408398.2018.1485628. [DOI] [PubMed] [Google Scholar]
- 18.Francino MP. Birth mode-related differences in gut microbiota colonization and immune system development. Ann. Nutr. Metab. 2018;73:12–16. doi: 10.1159/000490842. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67:1614–1625. doi: 10.1136/gutjnl-2018-315988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmermann P, Curtis N. Factors influencing the intestinal microbiome during the first year of life. Pediatr. Infect. Dis. J. 2018;37:e315–e335. doi: 10.1097/INF.0000000000002103. [DOI] [PubMed] [Google Scholar]
- 21.Valentine G, Chu DM, Stewart CJ, Aagaard KM. Relationships between perinatal interventions, maternal-infant microbiomes, and neonatal outcomes. Clin. Perinatol. 2018;45:339–355. doi: 10.1016/j.clp.2018.01.00. [DOI] [PubMed] [Google Scholar]
- 22.Lundgren SN, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson RC, et al. Maternal omega-3 fatty acids regulate offspring obesity through persistent modulation of gut microbiota. Microbiome. 2018;6:95. doi: 10.1186/s40168-018-0476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrahamsson TR, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy. 2014;44:842–50. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 25.Sozanska B. Microbiome in the primary prevention of allergic diseases and bronchial asthma. Allergol. Immunopathol. (Madr). 2018;47:79–84. doi: 10.1016/j.aller.2018.03.005,. [DOI] [PubMed] [Google Scholar]
- 26.Abrahamsson Thomas R., Jakobsson Hedvig E., Andersson Anders F., Björkstén Bengt, Engstrand Lars, Jenmalm Maria C. Low diversity of the gut microbiota in infants with atopic eczema. Journal of Allergy and Clinical Immunology. 2012;129(2):434-440.e2. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Knutie SA, Wilkinson CL, Kohl KD, Rohr JR. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 2017;8:86. doi: 10.1038/s41467-017-00119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernstein CN, Burchill C, Targownik LE, Singh H, Roos LL. Events within the first year of life, but not the neonatal period, affect risk for later development of inflammatory bowel diseases. Gastroenterology. 2019;156:2190–2197. doi: 10.1053/j.gastro.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ismail IH, et al. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr. Allergy Immunol. 2012;23:674–81. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 30.Korpela K, de Vos WM. Early life colonization of the human gut: microbes matter everywhere. Curr. Opin. Microbiol. 2018;44:70–78. doi: 10.1016/j.mib.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Broadhurst MJ, et al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012;8:e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cattadori IM, et al. Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS One. 2016;11:e0159770. doi: 10.1371/journal.pone.0159770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glendinning L, Nausch N, Free A, Taylor DW, Mutapi F. The microbiota and helminths: sharing the same niche in the human host. Parasitology. 2014;141:1255–71. doi: 10.1017/S0031182014000699. [DOI] [PubMed] [Google Scholar]
- 34.Fricke WF, et al. Type 2 immunity-dependent reduction of segmented filamentous bacteria in mice infected with the helminthic parasite Nippostrongylus brasiliensis. Microbiome. 2015;3:40. doi: 10.1186/s40168-015-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes KS, et al. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391–4. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houlden A, et al. Chronic Trichuris muris infection in C57BL/6 mice causes significant changes in host microbiota and metabolome: Effects reversed by pathogen clearance. PLoS One. 2015;10:e0125945. doi: 10.1371/journal.pone.0125945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20140295. doi: 10.1098/rstb.2014.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dea-Ayuela MA, Rama-Iniguez S, Bolas-Fernandez F. Enhanced susceptibility to Trichuris muris infection of B10Br mice treated with the probiotic Lactobacillus casei. Int. Immunopharmacol. 2008;8:28–35. doi: 10.1016/j.intimp.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Lee SC, et al. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl. Trop. Dis. 2014;8:e2880. doi: 10.1371/journal.pntd.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peachey LE, et al. The relationships between faecal egg counts and gut microbial composition in UK Thoroughbreds infected by cyathostomins. Int. J. Parasitol. 2018;48:403–412. doi: 10.1016/j.ijpara.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenkins TP, et al. Schistosoma mansoni infection is associated with quantitative and qualitative modifications of the mammalian intestinal microbiota. Sci. Rep. 2018;8:12072. doi: 10.1038/s41598-018-30412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkins TP, et al. A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area. Sci. Rep. 2018;8:15651. doi: 10.1038/s41598-018-33937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm JB, et al. Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of Lactobacilli. PLoS One. 2015;10:e0125495. doi: 10.1371/journal.pone.0125495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNeilly TN, Nisbet AJ. Immune modulation by helminth parasites of ruminants: implications for vaccine development and host immune competence. Parasite. 2014;21:51. doi: 10.1051/parasite/2014051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridler A. Disease threats to sheep associated with intensification of pastoral farming. N. Z. Vet. J. 2008;56:270–273. doi: 10.1080/00480169.2008.36846. [DOI] [PubMed] [Google Scholar]
- 46.Li RW, et al. The effect of helminth infection on the microbial composition and structure of the caprine abomasal microbiome. Sci. Rep. 2016;6:20606. doi: 10.1038/srep20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li RW, Wu S, Li W, Huang Y, Gasbarre LC. Metagenome plasticity of the bovine abomasal microbiota in immune animals in response to Ostertagia ostertagi infection. PLoS One. 2011;6:e24417. doi: 10.1371/journal.pone.0024417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El-Ashram S, et al. Microbial community and ovine host response varies with early and late stages of Haemonchus contortus infection. Vet. Res. Commun. 2017;41:263–277. doi: 10.1007/s11259-017-9698-5. [DOI] [PubMed] [Google Scholar]
- 49.Almeida ML, et al. Intense exercise and aerobic conditioning associated with chromium or L-carnitine supplementation modified the fecal microbiota of fillies. PLoS One. 2016;11:e0167108. doi: 10.1371/journal.pone.0167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blackmore TM, et al. Strong stability and host specific bacterial community in faeces of ponies. PLoS One. 2013;8:e75079. doi: 10.1371/journal.pone.0075079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa MC, et al. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS One. 2012;7:e41484. doi: 10.1371/journal.pone.0041484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costa MC, et al. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015;205:74–80. doi: 10.1016/j.tvjl.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Costa MC, et al. Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Vet. Res. 2015;11:19. doi: 10.1186/s12917-015-0335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costa MC, Weese JS. Understanding the intestinal microbiome in health and disease. Vet. Clin. North. Am. Equine. Pract. 2018;34:1–12. doi: 10.1016/j.cveq.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Costa MC, Weese JS. The equine intestinal microbiome. Anim. Health. Res. Rev. 2012;13:121–128. doi: 10.1017/S1466252312000035. [DOI] [PubMed] [Google Scholar]
- 56.Daly K, et al. Alterations in microbiota and fermentation products in equine large intestine in response to dietary variation and intestinal disease. Br. J. Nutr. 2012;107:989–995. doi: 10.1017/S0007114511003825. [DOI] [PubMed] [Google Scholar]
- 57.Dougal K, et al. Characterisation of the faecal bacterial community in adult and elderly horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS One. 2014;9:e87424. doi: 10.1371/journal.pone.0087424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dougal K, et al. A comparison of the microbiome and the metabolome of different regions of the equine hindgut. FEMS Microbiol. Ecol. 2012;82:642–52. doi: 10.1111/j.1574-6941.2012.01441.x. [DOI] [PubMed] [Google Scholar]
- 59.Ericsson AC, Johnson PJ, Lopes MA, Perry SC, Lanter HR. A Microbiological map of the healthy equine gastrointestinal tract. PLoS One. 2016;11:e0166523. doi: 10.1371/journal.pone.0166523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandes KA, et al. Faecal microbiota of forage-fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS One. 2014;9:e112846. doi: 10.1371/journal.pone.0112846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leng J, et al. Exploration of the fecal microbiota and biomarker discovery in equine grass sickness. J. Proteome Res. 2018;17:1120–1128. doi: 10.1021/acs.jproteome.7b00784. [DOI] [PubMed] [Google Scholar]
- 62.Mach N, et al. The effects of weaning methods on gut microbiota composition and horse physiology. Front. Physiol. 2017;8:535. doi: 10.3389/fphys.2017.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metcalf JL, et al. Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci. Rep. 2017;7:15497. doi: 10.1038/s41598-017-15375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Proudman CJ, et al. Characterisation of the faecal metabolome and microbiome of Thoroughbred racehorses. Equine Vet. J. 2015;47:580–6. doi: 10.1111/evj.12324. [DOI] [PubMed] [Google Scholar]
- 65.Schoster A, Mosing M, Jalali M, Staempfli HR, Weese JS. Effects of transport, fasting and anaesthesia on the faecal microbiota of healthy adult horses. Equine Vet. J. 2016;48:595–602. doi: 10.1111/evj.12479. [DOI] [PubMed] [Google Scholar]
- 66.Schoster A, Staempfli HR, Guardabassi LG, Jalali M, Weese JS. Comparison of the fecal bacterial microbiota of healthy and diarrheic foals at two and four weeks of life. BMC Vet. Res. 2017;13:144. doi: 10.1186/s12917-017-1064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart HL, et al. Characterization of the fecal microbiota of healthy horses. Am. J. Vet. Res. 2018;79:811–819. doi: 10.2460/ajvr.79.8.811. [DOI] [PubMed] [Google Scholar]
- 68.Weese JS, et al. Changes in the faecal microbiota of mares precede the development of post partum colic. Equine Vet. J. 2015;47:641–649. doi: 10.1111/evj.12361. [DOI] [PubMed] [Google Scholar]
- 69.Zhao Y, et al. Comparison of fecal microbiota of mongolian and thoroughbred horses by high-throughput sequencing of the V4 region of the 16S rRNA gene. Asian-Australas J. Anim. Sci. 2016;29:1345–1352. doi: 10.5713/ajas.15.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matthews JB. Facing the threat of equine parasitic disease. Equine Vet. J. 2011;43:126–32. doi: 10.1111/j.2042-3306.2010.00356.x. [DOI] [PubMed] [Google Scholar]
- 71.Love S, Murphy D, Mellor D. Pathogenicity of cyathostome infection. Veterinary Parasitology. 1999;85(2-3):113–122. doi: 10.1016/S0304-4017(99)00092-8. [DOI] [PubMed] [Google Scholar]
- 72.Willing B, et al. Changes in faecal bacteria associated with concentrate and forage-only diets fed to horses in training. Equine Vet. J. 2009;41:908–914. doi: 10.2746/042516409X447806. [DOI] [PubMed] [Google Scholar]
- 73.Warzecha CM, et al. Influence of short-term dietary starch inclusion on the equine cecal microbiome. J. Anim. Sci. 2017;95:5077–5090. doi: 10.2527/jas2017.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martinez I, et al. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. Elife. 2018;7:e36521. doi: 10.7554/eLife.36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Aquino SG, et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014;192:4103–4011. doi: 10.4049/jimmunol.1301970. [DOI] [PubMed] [Google Scholar]
- 76.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brosschot TP, Reynolds LA. The impact of a helminth-modified microbiome on host immunity. Mucosal Immunol. 2018;11:1039–1046. doi: 10.1038/s41385-018-0008-5. [DOI] [PubMed] [Google Scholar]
- 78.Thomas F, Hehemann J, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Front. Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 80.Zhang XS, et al. Antibiotic-induced acceleration of type 1 diabetes alters maturation of innate intestinal immunity. Elife. 2018;7:e37816. doi: 10.7554/eLife.37816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White EC, et al. Manipulation of host and parasite microbiotas: Survival strategies during chronic nematode infection. Sci. Adv. 2018;4:eaap7399. doi: 10.1126/sciadv.aap7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ramanan D, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaiss MM, et al. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity. 2015;43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 85.Nakazawa F, et al. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2000;50:Pt, 679–88. doi: 10.1099/00207713-50-2-679. [DOI] [PubMed] [Google Scholar]
- 86.Lourenvarsigmao TGB, Spencer SJ, Alm EJ, Colombo APV. Defining the gut microbiota in individuals with periodontal diseases: an exploratory study. J. Oral. Microbiol. 2018;10:1487741. doi: 10.1080/20002297.2018.1487741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heitkemper MM, et al. Stool and urine trefoil factor 3 levels: associations with symptoms, intestinal permeability, and microbial diversity in irritable bowel syndrome. Benef. Microbes. 2018;9:345–355. doi: 10.3920/BM2017.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hollister, E. B. et al. Relationships of microbiome markers with extraintestinal, psychological distress and gastrointestinal symptoms, and quality of life in women with irritable bowel syndrome. J. Clin. Gastroenterol. ePub ahead of print, 10.1097/MCG.000000000000110 (2018). [DOI] [PMC free article] [PubMed]
- 89.Altomare, A. et al. Gut mucosal-associated microbiota better discloses Inflammatory Bowel Disease differential patterns than faecal microbiota. Dig. Liver. Dis51, 648–656, doi:0.1016/j.dld.2018.11.021 (2018). [DOI] [PubMed]
- 90.Reynolds LA, et al. Commensal-pathogen interactions in the intestinal tract: Lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes. 2014;5:522–532. doi: 10.4161/gmic.32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor SL, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J. Allergy. Clin. Immunol. 2018;141:94–103 e15. doi: 10.1016/j.jaci.2017.03.044. [DOI] [PubMed] [Google Scholar]
- 92.Wu S, et al. Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS One. 2012;7:e35470. doi: 10.1371/journal.pone.0035470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clark A, et al. Strongyle infection and gut microbiota: Profiling of resistant and susceptible horses over a grazing season. Front. Physiol. 2018;9:272. doi: 10.3389/fphys.2018.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wells JM. Immunomodulatory mechanisms of Lactobacilli. Microb. Cell Fact. 2011;10(Suppl 1):S17. doi: 10.1186/1475-2859-10-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schoster A, Weese JS, Guardabassi L. Probiotic use in horses - what is the evidence for their clinical efficacy? J. Vet. Intern. Med. 2014;28:1640–52. doi: 10.1111/jvim.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knutie SA, et al. Early-life diet affects host microbiota and later-life defenses against parasites in frogs. Integr. Comp. Biol. 2017;57:732–742. doi: 10.1093/icb/icx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 2007;13:675–683. doi: 10.1002/ibd.20101. [DOI] [PubMed] [Google Scholar]
- 98.Ott SJ, Schreiber S. Reduced microbial diversity in inflammatory bowel diseases. Gut. 2006;55:1207. [PMC free article] [PubMed] [Google Scholar]
- 99.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Giacomin P, et al. Experimental hookworm infection and escalating gluten challenges are associated with increased microbial richness in celiac subjects. Sci. Rep. 2015;5:13797. doi: 10.1038/srep13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McSorley HJ, Maizels RM. Helminth infections and host immune regulation. Clin. Microbiol. Rev. 2012;25:585–608. doi: 10.1128/CMR.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peachey LE, Jenkins TP, Cantacessi C. This gut ain’t big enough for both of us. or is it? Helminth-microbiota interactions in veterinary species. Trends Parasitol. 2017;33:619–632. doi: 10.1016/j.pt.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 103.Goldansaz SA, et al. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS One. 2017;12:e0177675. doi: 10.1371/journal.pone.0177675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Christie, M. & Jackson, F. Specific identification of strongyle eggs in small samples of sheep faeces. Res. Vet. Sci. 32, 113–117, doi: 0.1016/S0034-5288(18)32448-2 (1982). [PubMed]
- 105.Rehbein S, Lindner T, Visser M, Winter R. Evaluation of a double centrifugation technique for the detection of Anoplocephala eggs in horse faeces. J. Helminthol. 2011;85:409–14. doi: 10.1017/S0022149X10000751. [DOI] [PubMed] [Google Scholar]
- 106.Mattiello F, et al. A web application for sample size and power calculation in case-control microbiome studies. Bioinformatics. 2016;32:2038–40. doi: 10.1093/bioinformatics/btw099. [DOI] [PubMed] [Google Scholar]
- 107.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zakrzewski M, et al. Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 2006;9:683–93. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 111.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw 16s rRNA and HNMR data, metadata and QIIME2 feature table are available at Mendeley Data (10.17632/95m8sfd3kt.1).