Abstract
Chloroplasts are intracellular semiautonomous organelles central to photosynthesis and are essential for plant growth and yield. The significance of the function of chloroplast-related genes in response to climate change has not been well studied in crops. In the present study, the initial focus was on genes that were predicted to be located in the chloroplast genome in rice, a model crop plant, with genes either preferentially expressed in the leaf or ubiquitously expressed in all organs. The characteristics were analyzed by Gene Ontology (GO) enrichment and MapMan functional classification tools. It was then identified that 110 GO terms (45 for leaf expression and 65 for ubiquitous expression) and 1,695 genes mapped to MapMan overviews were strongly associated with chloroplasts. In particular, the MapMan cellular response overview revealed a close association between heat stress response and chloroplast-related genes in rice. Moreover, features of these genes in response to abiotic stress were analyzed using a large-scale publicly available transcript dataset. Consequently, the expression of 215 genes was found to be upregulated in response to high temperature stress. Conversely, genes that responded to other stresses were extremely limited. In other words, chloroplast-related genes were found to affect abiotic stress response mainly through high temperature response, with little effect on response to drought and salinity stress. These results suggest that genes involved in diurnal rhythm in the leaves participate in the reaction to recognize temperature changes in the environment. Furthermore, the predicted protein–protein interaction network analysis associated with high temperature stress is expected to provide a very important basis for the study of molecular mechanisms by which chloroplasts will respond to future climate changes.
1. Introduction
Chloroplasts are cellular organelles in which photosynthesis occurs, and they are found in cyanobacteria, algae, and higher plants [1]. Chloroplasts have a double-celled composite membrane and an extensively folded thylakoid membrane. The chloroplast is divided into two compartments, the soluble stroma and lumen, with the latter enclosed by the thylakoid membrane, which carries chlorophylls and other pigments [2]. Chloroplasts produce the energy needed to sustain life through photosynthesis and oxygen-release processes. The key function of chloroplasts is photosynthesis. However, they also affect the physiology and development of plants by their involvement in the synthesis of amino acids, nucleotides, fatty acids, phytohormones, and vitamins [3]. Furthermore, metabolites synthesized in chloroplasts protect plants from environmental (abiotic) and biotic stresses, including heat, cold, drought, salt, light, and pathogens [4, 5]. Chloroplasts have genomes containing approximately 120 genes, which encode key proteins involved in metabolic processes in green plants [6–8].
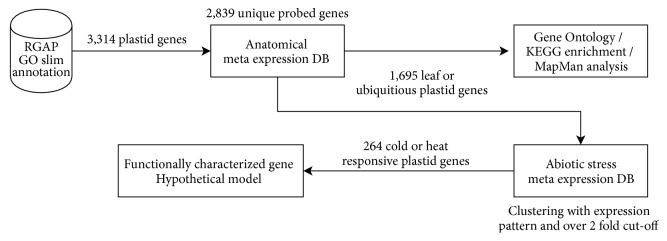
The role of chloroplasts in plant abiotic stress responses continues to be highlighted. The development of high-throughput sequencing technologies has made possible many advances in plant genetics and genomics. In this study, we tried a new approach through transcriptome analysis of chloroplast-related genes. Initially, we selected 3,314 plastid-related genes via Rice Genome Annotation Project (RGAP) Gene Ontology (GO) slim annotation. Then, we used the anatomical meta-expression database and, as a result, identified 1,695 leaf-preferred or ubiquitously expressed plastid-related genes. Subsequently, GO enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and MapMan analyses of these 1,695 genes were performed and significant functional groups were identified. Of the 1,695 leaf-preferred or ubiquitously expressed plastid-related genes, meta-expression analysis under abiotic stress conditions identified 264 cold or heat stress-responsive plastid-related genes. The analysis of functionally characterized plastid-related genes emphasized the significance of our candidate genes for crosstalk between chloroplast development and heat stress. Ultimately, the hypothetical network model will provide a valuable backbone for future studies (Figure 1).
Figure 1.
Workflow diagram summarizing analysis process about rice plastid-related genes. The workflow illustrates the entire analysis process in this study. First of all, we retrieved 4,707 plastid transcripts from GO slim annotation at the RGAP database. Then we removed unannotated and duplicated information to collect 3,314 plastid genes. By querying these genes to our meta-expression data source, we obtained 2,839 genes that only have the most highly expressed probe. We clustered these intensity values with KMC algorithm as 20 clusters. As a result, we identified 1,695 leaf-preferred or ubiquitously expressed genes for further analysis. With these two sets of genes, we performed functional characterization like GO enrichment, KEGG enrichment, and MapMan analysis to characterize their functions. In addition to these analyses, we queried the 1,695 genes to an abiotic stress expression database (DB) to identify stress-responsive plastid genes. As a result, we clustered 264 cold or heat stress-responsive plastid genes and conducted a literature search. Altogether, we constructed a hypothetical protein–protein interaction model of stress-related plastid genes.
2. Materials and Methods
2.1. Integration of Plastid Genes from Public Data Source
We obtained information on plastid-related genes from the GO slim annotation data at the Rice Genome Annotation Project web database (RGAP) [9]. We first selected 3,314 plastid-related genes in the cellular component subontology from the data. Then, we removed duplicated loci because of their transcript isoforms. Finally, we collected 3,314 plastid-related genes for further analysis.
2.2. Collection of Microarray Data
We collected transcriptomics data to analyze anatomical and abiotic expression patterns of 1,696 plastid-related genes, for which we used the data source described in our previous report [10]. Detailed information is as below. For analysis of anatomical expression profiles, we integrated anatomical data from the rice oligonucleotide array database ROAD [11]. For the abiotic stress database, we retrieved 13 expression dataset series, GSE16108, GSE18930, GSE21651, GSE23211, GSE24048, GSE25176, GSE26280, GSE28209, GSE31077, GSE33204, GSE37940, GSE38023, and GSE6901 from the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds), and one data series, E-MEXP-2401, from the European Bioinformatics Institute (EMBL-EBI) ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/).
2.3. Clustering of Transcriptome Data
MeV (Multiple Experiment Viewer) is a widely used program to visualize transcriptome data and to perform statistical analysis [12]. We used MeV software (version 4.9.0) to visualize our microarray data. For analysis of transcriptome data, we applied a k-means clustering (KMC) algorithm with Euclidean distance metric embedded in MeV with the same method as that used for the identification of late-pollen-preferred genes in rice [13]. In the case of the abiotic microarray data, we clustered 1,695 leaf-preferred or ubiquitously expressed plastid genes with the same KMC algorithm and matrix. In addition, we selected only genes with an average log2-fold-change value (treatment/control) in a cluster greater than 1(log2value) and a p-value of one-way analysis of variance (ANOVA) of less than 0.05 to ensure clear correlation. We used the row.oneway.anova function in the HybridMTest package in R to perform a one-way ANOVA test and used Illustrator software (Adobe Illustrator CS6) to present heatmap images [14].
2.4. GO Enrichment Analysis
GO enrichment is commonly used to interpret functional roles of large-scale transcriptomics data [15]. In the current study, we used the ROAD database to find GO terminology for each cluster (http://ricephylogenomics-khu.org/ROAD_old/analysis/go_enrichment.shtml, temporary homepage for updating). To perform GO enrichment analysis, we applied the following criteria: query number of >2, hyper p-value of <0.05, and fold-enrichment value (query number/query expected number) of >2, by referencing previous reports about GO enrichment analysis [13, 16]. We selected significant GO terms and integrated cluster information from transcriptomic data analysis with each selected GO term. Finally, we visualized these data via R Studio (version 1.1.453) and ggplot2 R package (version 3.0.0) [17].
2.5. KEGG Enrichment Analysis
We performed KEGG enrichment analysis using R Studio and the clusterProfiler package [18]. To use the enrichKEGG function in this package, we used input data consisting of cluster information and rice annotation project database ID. In addition, we chose data for organism code and filtered out results by applying adjusted p-value cut-offs of less than 0.05, as in other studies [19, 20]. For visualization of the results, we used dotplot function in the package and modified the figure with the ggplot2 package (version 3.0.0).
2.6. MapMan Analysis
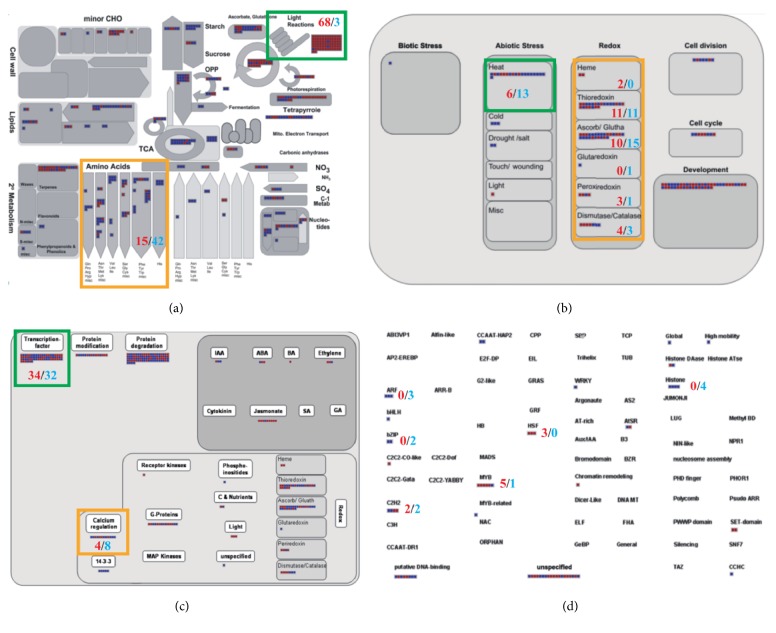
To obtain a systemic view of the 1,695 leaf-preferred and ubiquitously expressed plastid-related genes within rice, we performed MapMan analysis as previously described [21]. In detail, we conducted the functional characterization of the genes by uploading the locus list with cluster information to MapMan software (v3.6.0 RC1) [22]. Among the various functional classifications, we analyzed the metabolism, cellular response, regulation, and transcription factor overviews in detail (Figure 3).
Figure 3.
Functional classification of the leaf-preferred or ubiquitously expressed plastid genes via MapMan analysis. MapMan analysis of the two clusters for functional classification within various biological processes. (a) Metabolism overview, (b) cellular response overview, (c) regulation overview, and (d) transcription factor overview. Red and blue squares indicate members of leaf-preferred and ubiquitously expressed clusters, respectively. In addition to squares, red and blue digits show number of squares. Green box and orange box in metabolism, cellular response, and regulation overview highlight areas that are discussed in the Results.
2.7. Analysis of Functionally Characterized Genes via Literature Search
To identify the previously characterized functional roles of the 264 plastid-related genes associated with heat or cold stresses, we used the funRiceGenes database for functionally characterizing rice genes (https://funricegenes.github.io/) [23]. In this database, information on 3,148 functionally characterized genes is available. As in our previous study [24], we parsed functional roles for the 264 genes and gene clusters, using meta-expression data. Subsequently, we summarized the data in Table 1, using Excel software.
Table 1.
Summary of functionally characterized plastid genes associated with cold and abiotic stress.
| Categorya | Tissueb | Abioticc | Locus_ID | Gene symbol | Keywordd | Titlee |
|---|---|---|---|---|---|---|
| BAf | Ubi | Heat | LOC_Os06g50300 | Hsp90|rHsp90|OSGrp94 | abiotic stress | rHsp90 gene expression in response to several environmental stresses in rice (Oryza sativa L.) |
| BA | Leaf | Heat | LOC_Os08g29110 | OsTRXZ | cold stress | TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. |
| BA | Ubi | Heat | LOC_Os04g32950 | OsCNX | drought tolerance | Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum |
| BA | Leaf | Heat | LOC_Os03g19380 | OsCP12 | drought tolerance | The rice RING E3 ligase, OsCTR1, inhibits trafficking to the chloroplasts of OsCP12 and OsRP1, and its over-expression confers drought tolerance in Arabidopsis |
| BA | Ubi | Heat | LOC_Os07g34570 | OsDR8 | disease resistance | Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation |
|
| ||||||
| MTg | Leaf | Heat | LOC_Os03g45400 | OsNUS1|V1 | leaf | A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice |
| MT | Leaf | Heat | LOC_Os12g37610 | TCD11 | leaf | The rice TCD11 encoding plastid ribosomal protein S6 is essential for chloroplast development at low temperature |
| MT | Leaf | Heat | LOC_Os05g34040 | TCD5|CSV1|TSV | leaf | Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. |
| MT | Leaf | Heat | LOC_Os03g29810 | VYL | leaf | A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants |
| MT | Leaf | Heat | LOC_Os04g51280 | WSP1 | leaf | The RNA editing factor WSP1 is essential for chloroplast development in rice. |
| MT | Leaf | Heat | LOC_Os06g02580 | ZN | leaf | ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development |
| MT | Leaf | Heat | LOC_Os02g33610 | GRY79 | seedling | GRY79 encoding a putative metallo-β-lactamase-trihelix chimera is involved in chloroplast development at early seedling stage of rice. |
| MT | Leaf | Heat | LOC_Os12g17910 | OsCPn60alpha1|OsCPn60a1 | seedling | OsCpn60alpha1, encoding the plastid chaperonin 60alpha subunit, is essential for folding of rbcL |
| MT | Ubi | Heat | LOC_Os11g09280 | OsPDIL1;1 | seedling | Formation of protein disulfide bonds catalyzed by OsPDIL1;1 is mediated by microRNA5144-3p in rice. |
| MT | Ubi | Heat | LOC_Os06g41990 | SPP | root | A rice stromal processing peptidase regulates chloroplast and root development |
| MT | Leaf | Heat | LOC_Os05g48200 | OsNADH-GOGAT2 | spikelet number | Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice |
| MT | Ubi | Heat | LOC_Os06g08770 | OIP30 | pollen | OIP30, a RuvB-like DNA helicase 2, is a potential substrate for the pollen-predominant OsCPK25/26 in rice |
|
| ||||||
| PTh | Ubi | Heat | LOC_Os03g19510 | OsClpP5 | chloroplast | An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice |
| PT | Ubi | Heat | LOC_Os01g16040 | OsPAPST1 | chloroplast | Identification of a dual-targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice |
| PT | Ubi | Heat | LOC_Os03g60910 | OspTAC2 | chloroplast | OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. |
| PT | Ubi | Heat | LOC_Os07g06940 | WP1|OsValRS2 | chloroplast | WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. |
| PT | Leaf | Heat | LOC_Os03g40550 | OsFLN2|HSA1 | chloroplast | FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. |
| PT | Ubi | Heat | LOC_Os07g08340 | OsPK2 | grain | OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. |
| PT | Ubi | Heat | LOC_Os01g52500 | NADP-ME2|OsNADP-ME2 | growth | Expression of an NADP-malic enzyme gene in rice (Oryza sativa L.) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance |
| PT | Leaf | Heat | LOC_Os07g43810 | RNP29 | phosphate | Identification and characterization of chloroplast casein kinase II from Oryzasativa (rice). |
| PT | Ubi | Heat | LOC_Os03g55090 | Pho1 | starch | Mutation of the plastidial alpha-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm |
a Agronomic traits associated with functionally characterized genes out of candidate genes in this study.
b Anatomical cluster information of the gene.
c Abiotic cluster information of the gene.
d Key trait of characterized role of the gene.
e Title of research paper of the gene.
f Biotic or abiotic stress related trait.
g Morphological trait.
h Physiological trait.
2.8. Analysis of a Predicted Protein–Protein Interaction Network
Using the Rice Interactions Viewer tool (http://bar.utoronto.ca/interactions/cgi-bin/rice_interactions_viewer.cgi), we generated a hypothetical protein–protein interaction network, involving transcription factors (TFs), redox reactions, and functionally characterized genes. The network was edited with the Cytoscape tool (3.2.0 version) [25].
3. Results
3.1. Integration of 3,314 Plastid-Related Genes from GO Slim Annotation at the Rice Reference Database, RGAP
We applied GO slim annotation at a rice reference database, RGAP. GO provides controlled information on each gene, which classifies the genes into three categories, namely, biological process, molecular function, and cellular component [26]. We retrieved 4,707 plastid-related transcripts with the cellular component information of GO. Then, we selected only chromosome-annotated genes without duplication. As a result, we selected 3,314 genes for further analysis.
3.2. Anatomical Dissection of Chloroplast-Related Genes via Meta-Expression Analysis
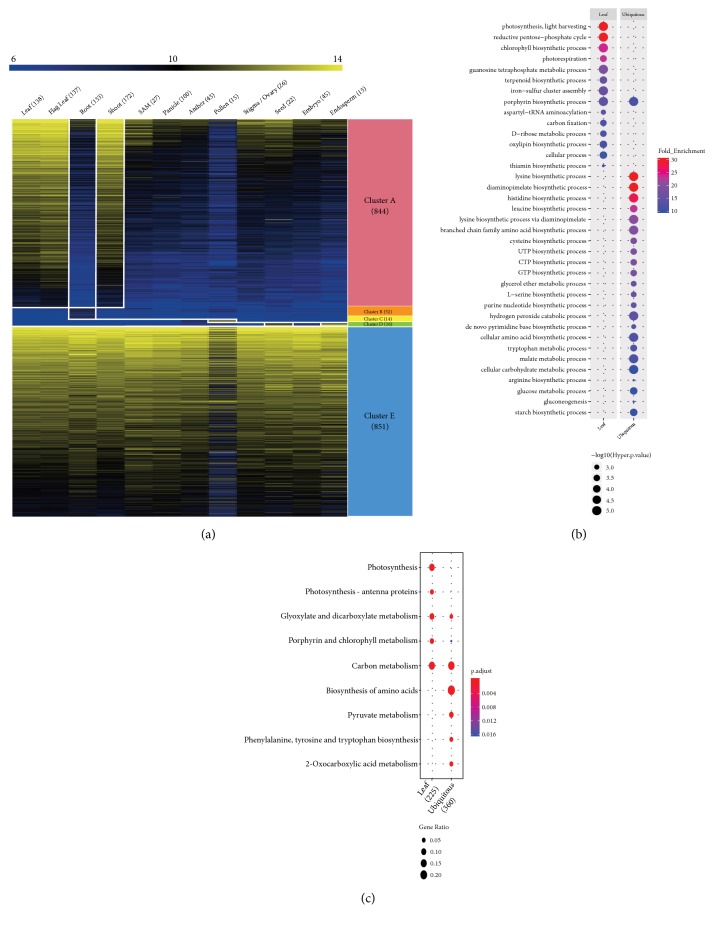
To assess the functional roles of 3,314 plastid-related genes, we used meta-anatomical expression profiles consisting of 983 rice Affymetrix array anatomical sample data [10]. By applying clustering analysis based on the Euclidian distance algorithm, we grouped 2,839 genes with probes on the Affymetrix array into 20 anatomical clusters (Figure S1). Based on this analysis, we identified plastid-related genes, with respect to organ-selective expression in leaves or ubiquitous expression pattern. For example, cluster A, with 844 genes, had preferential expression in leaves and cluster E, with 851 genes, exhibited an ubiquitous expression pattern, which was related to housekeeping functions (Figure 2(a)). In addition, we found that clusters B and C were closely associated with roots (52 genes) and pollen (14 genes), respectively, and cluster D with seed (16 genes).
Figure 2.
Meta-expression profile and functional analysis of the 1,695 leaf-preferred or ubiquitously expressed genes. We performed meta-expression analysis with large-scale microarray dataset for elucidating tissue-specific patterned plastid genes. In addition, we performed GO and KEGG enrichment analysis to identify functional roles for two clusters, leaf-preferred cluster A and ubiquitous cluster E. (a) Heatmap analysis of plastid-related genes and identification of five clusters. We performed KMC clustering into 20 clusters using Euclidean distance matric and selected 10 clusters on the basis of tissue-specific expression patterns. Among these clusters, we selected two major clusters, A (leaf-preferred genes) and E (ubiquitous genes) for further functional enrichment analysis. Digits under or beside each clusters indicate number of the genes that were classified into each cluster. (b) GO enrichment analysis of 1,695 leaf-preferred and ubiquitous expressed genes. To reveal characteristics of each cluster, we conducted GO enrichment analysis and visualized the result with ggplot2 package. GO terms were classified according to biological process GO terms. Dot color indicates fold-enrichment value (blue color is 2-fold, which is the minimum cut-off to select significant fold-enrichment value, and red color indicates higher fold-enrichment value greater than two), and dot size indicates statistical significance (-log10(hyper p-values) are used, with higher values having greater significance). (c) KEGG enrichment analysis of two clusters, A and E. Enriched KEGG pathway indicated with dot size representing the ratio of selected genes to total genes in the pathway and dot color illustrating adjusted p-value. The numbers below clusters indicate the number of mapped genes to selected KEGG pathways.
3.3. Functional Enrichment Analysis of Plastid-Related Genes with Leaf-Preferred or Ubiquitous Expression Using Gene Ontology and KEGG
We focused on plastid-related genes in two clusters associated with the leaf or all organs (ubiquitously expressed genes) through meta-expression analysis. We then performed a functional-group enrichment analysis for each of the two anatomical clusters. To determine significant functional groups associated with the two anatomical clusters, we performed GO and KEGG enrichment analyses (Figures 2(b) and 2(c)). In all, 45 GO terms in the biological process category were highly overrepresented in the leaf gene list (cluster A), with p-values of <0.05 and (log2)-fold-enrichment values of >2, as we previously reported [27]. They included biological processes related to photosynthesis, namely, light harvesting (36.3-fold enrichment, GO:0009765), reductive pentose-phosphate cycle (30.9, GO:0019253), chlorophyll biosynthetic process (23.9, GO:0015995), photorespiration (23.2, GO:0009853), guanosine tetraphosphate metabolic process (19.3, GO:0015969), terpenoid biosynthetic process (15.4, GO:0016114), iron-sulfur cluster assembly (15.4, GO:0016226), aspartyl-tRNA aminoacylation (14.5, GO:0006422), porphyrin biosynthetic process (14.5, GO:0006779), D-ribose metabolic process (11.9, GO:0006014), carbon fixation (11.9, GO:0015977), oxylipin biosynthetic process (11.4, GO:0031408), cellular process (10.7, GO:0009987), and thiamin biosynthetic process (10.5, GO:0009228) (Figure 2(b)). The 31 GO terms with fold-enrichment values less than 10 are shown in Figure S2. Similar to the results from GO enrichment analysis, KEGG enrichment also showed that photosynthesis, photosynthesis−antenna proteins, glyoxylate and dicarboxylate metabolism, porphyrin and chlorophyll metabolism, and carbon metabolism were enriched in leaves (Figure 2(c)). These results suggest that plastid-related genes with high expression levels in leaves are closely related to photosynthesis, as would be expected.
Next, we performed GO enrichment and KEGG enrichment analyses on ubiquitously expressed genes. As a result, we found that 65 GO terms were enriched in cluster E: lysine biosynthetic process (30.8-fold enrichment, GO:0009085), diaminopimelate biosynthetic process (30.8, GO:0019877), histidine biosynthetic process (27.4, GO:0000105), leucine biosynthetic process (23.0, GO:0009098), lysine biosynthetic process via diaminopimelate (20.5, GO:0009089), branched chain family amino acid biosynthetic process (19.6, GO:0009082), GTP biosynthetic process (18.5, GO:0006183), UTP biosynthetic process (18.5, GO:0006228), CTP biosynthetic process (18.5, GO:0006241), cysteine biosynthetic process (18.5, GO:0019344), purine nucleotide biosynthetic process (15.3, GO:0006164), L-serine biosynthetic process (15.3, GO:0006564), glycerol ether metabolic process (15.3, GO:0006662), hydrogen peroxide catabolic process (14.2, GO:0042744), de novo pyrimidine base biosynthetic process (13.2, GO:0006207), cellular amino acid biosynthetic process (12.5, GO:0008652), tryptophan metabolic process (12.3, GO:0006568), malate metabolic process (12.0, GO:0006108), porphyrin biosynthetic process (11.5, GO:0006779), glucose metabolic process (10.3, GO:0006006), gluconeogenesis (10.3, GO:0006094), arginine biosynthetic process (10.3, GO:0006526), starch biosynthetic process (10.3, GO:0019252), and cellular carbohydrate metabolic process (10.3, GO:0044262) (Figure 2(b)). The 41 GO terms with fold-enrichment values less than 10 are shown in Figure S2. Consistent with the GO enrichment analysis results, KEGG enrichment also showed that amino acid biosynthesis, including that of phenylalanine, tyrosine, and tryptophan, carbon metabolism, pyruvate metabolism, and 2-oxocarboxylic acid metabolism were enriched in all plants (Figure 2(c)). These results suggest that plastid-related genes with ubiquitous expression are closely related to basic metabolic processes.
3.4. Analysis of Various MapMan Overviews Associated with Plastid-Related Genes with Leaf-Preferred or Ubiquitous Expression
The MapMan program is an effective tool for visualizing diverse overviews associated with high-throughput transcriptome data [28]. We uploaded plastid-related locus IDs for the 844 upregulated genes in leaves (Figure 3, red squares) and the 851 ubiquitously expressed genes (Figure 3, blue squares). In the metabolism overview, we observed that genes with increased expression in leaves were mainly found to be involved in light reactions (71 elements, 68 for leaf-preferred expression/3 for ubiquitous expression, i.e., 68/3), whereas those with ubiquitous expression were found to be largely involved in amino acid metabolism (57 elements, 15/42) (Figure 3(a), green and orange box). Most of the light reactions were found to be associated with leaf-preferred genes, whereas amino acid metabolism was mainly identified in ubiquitously expressed genes. Interestingly, these results are consistent with the findings of the GO enrichment and KEGG enrichment analyses.
In the cellular response overview, heat stress within abiotic stress response was closely associated with both leaf-preferred or ubiquitously expressed genes (19 elements, 6/13) (Figure 3(b), green box). Many antioxidant proteins such as thioredoxin, ascorbate, glutathione, and peroxiredoxin and catalases were found in the redox cellular response (61 elements, 30/31) (Figure 3(b), orange box), with leaf-preferred or ubiquitously expressed genes being identified in similar numbers in heat stress and redox responses. Independent of organ specificity, these results suggest that plastid-related genes are closely related to heat stress and utilize proteins catalyzing redox reactions to effectively remove superoxide and H2O2 produced under stressful conditions.
Finally, we identified leaf-preferred or ubiquitously expressed genes associated with TFs (66 elements, 34/32) and calcium regulation (12 elements, 4/8) in the regulation overview (Figure 3(c), green and orange box). As a result of detailed examination of TFs, we found six (5/1) myeloblastosis (MYB) oncogenes, four (0/4) histones, four (2/2) Cys2His2 (C2H2) zinc fingers, three (0/3) auxin-response factors (ARFs), three (3/0) heat shock TFs (HSFs), and two (0/2) basic leucine zipper (bZIP) TFs (Figure 3(d)). Most of the MYB and HSFs were found in leaf-preferred genes, and histone, ARFs, and bZIP TFs were mainly identified in ubiquitously expressed genes. We have identified various TFs and calcium regulation elements in Figures 3(c) and 3(d). These results suggest that Ca2+ is involved in the activation of genes encoding HSFs and heat shock proteins (HSPs) during heat stress.
3.5. Abiotic Stress Dissection of Plastid-Related Genes via Meta-Expression Analysis
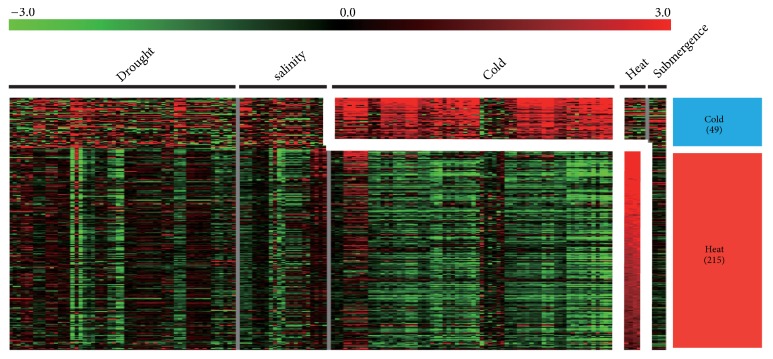
In the previous section, we used MapMan analysis to deduce that plastid-related genes were closely associated with heat stress response. To test this hypothesis, we conducted meta-expression analysis under abiotic stress conditions, such as drought, salinity, cold, heat, or submergence, for 1,695 genes (844 genes with preferential expression in leaves and 851 genes with ubiquitous expression), using meta-expression data (Figure S3). As a result, 215 genes were associated with heat stress and 49 genes with cold stress (Figure 4). However, we did not identify candidate genes showing differential expression patterns clearly associated with drought, salinity, or submergence. These results suggest that plastid-related genes respond mainly to changes in external temperature rather than to other stresses.
Figure 4.
Heatmap analysis of cold- and heat-responsive plastid genes which show leaf-preferred or ubiquitous expression patterns. Heatmap of cold or heat stress-responsive plastid-related genes. Similarly to the anatomy clustering, we used KMC algorithm with Euclidean distance matric to cluster abiotic-responsive genes. To define stress responsiveness, we applied criteria that were greater than average 1 log2-fold change (2-fold) in each stress and p-value less than 0.05 in one-way ANOVA test. As a result, we identified 264 cold or heat stress-responsive plastid genes.
3.6. Evaluation of Candidate Genes Associated with Plastid-Related Genes Using Rice Genes with Known Functions
To evaluate the functional significance of the 215 genes with increased expression under heat stress, we searched the literature to determine what functions of heat stress-responsive plastid genes have been identified in previous studies [23]. Of six genes found in that database (Table 1), five have been linked to various biotic and abiotic stress responses in rice. They include heat shock protein 90 [29] for environmental stresses; OsTRXZ [30] for cold stress; OsCNX [31], and OsCP12 [32] for drought tolerance; and OsDR8 [33] for disease resistance. Moreover, 12 of the genes are associated with morphological traits: OsNUS1 [34], TCD11 [35], TCD5 [36], VYL [37], WSP1 [38], and ZN [39] with leaf development; GRY79 [40], OsCPn60a1 [41], and OsPDIL1;1 [42] with seedling development; SPP [43] with root development; OsNADH-GOGAT2 [44] with spikelet number; and OIP30 [45] with pollen trait. In addition, nine genes are associated with physiological traits: OsClpP5 [46], OsPAPST1 [47], OspTAC2 [48], OsValRS2 [49], and OsFLN2 [50] with chloroplasts; OsPK2 [51] with grain; OsNADP-ME2 [52] with growth; RNP29 [53] for phosphate; and Pho1 [54] for starch.
As expected, most of the heat stress-responsive genes having functions involved in leaf or seedling development were associated with morphological traits, whereas most of the genes associated with physiological traits were related to chloroplasts. Interestingly, in genes associated with abiotic stress responses, more were reported to have functions related to drought, high salinity, and cold stress than with heat stress. For example, the 90 kDa heat shock protein (rHsp90) accumulates after exposure to abiotic stresses such as high salinity, desiccation, and high pH as well as high temperature, and tobacco transgenic plants overexpressing rHsp90 exhibit increased tolerance to salinity [29]. In addition, temperature-sensitive virescent (tsv) showed defective chloroplasts and decreased chlorophyll content under cold stress. Interestingly, TSV, interacting with OsTRXZ (a subunit of plastid-encoded RNA polymerase (PEP) in chloroplasts), enhanced OsTRXZ stability at low temperatures. These results suggest that plastid-related genes are also associated with various abiotic stresses.
3.7. Analyses of Predicted Protein–Protein Interactions Associated with Chloroplast-Related Genes
Regulatory genes are primary targets when investigating diverse stress responses and developmental processes. Understanding the regulatory relationship between them can lead to a new strategy for the manipulation of chloroplasts to improve plant tolerance to heat stress. To expand our knowledge of this mechanism, we utilized the Rice Interactions Viewer to generate a hypothetical protein–protein interaction network associated with the 215 upregulated genes associated with heat stress response [55]. We then refined the network by using genes in the following four categories as the query: 29 plastid-related genes with elevated expression in heat stress (orange circles, Figure 5), 24 TFs (green circles), six functionally characterized genes (purple circles), and six redox proteins (blue circles).
Figure 5.
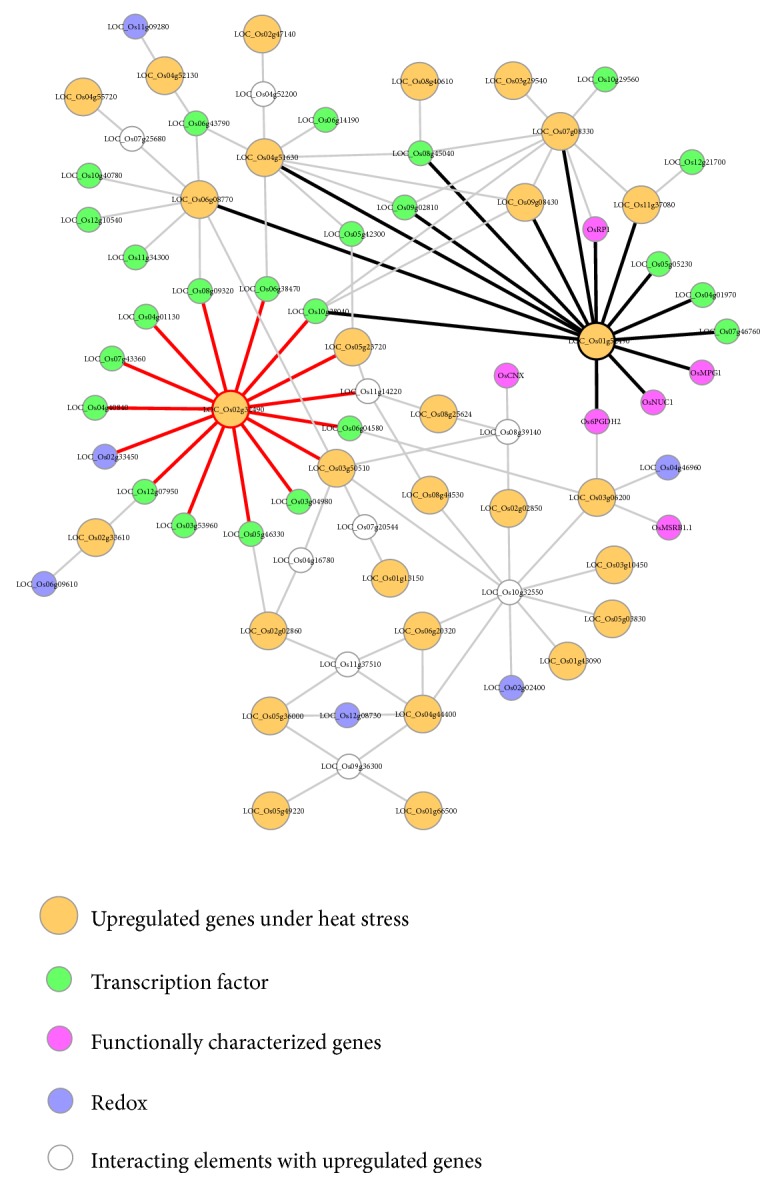
Construction of regulatory network associated with genes upregulated under high temperature. Using Rice Interaction Viewer and Cytoscape tools, we queried the predicted protein–protein interaction network associated with 29 upregulated genes under heat stress (orange circles), 24 transcription factors (green circles), six functionally characterized genes (purple circles), and six redox proteins (blue circles).
We found two interesting genes in this network. LOC_Os02g32490, encoding the AMP-binding enzyme, is predicted to interact with 11 TFs and one involved in the redox reaction (red lines, Figure 5). Another gene, LOC_Os01g52490, encoding a 40S ribosomal protein, is likely to be associated with four functionally characterized genes and six TFs (black lines, Figure 5). The four functionally characterized genes are all associated with response to abiotic stress. For example, OsCTR1, encoding the RING Ub E3 ligase, interacts with two chloroplast-localized proteins (OsCP12 and OsRP1) and is involved in drought tolerance [32]. The transcript level of Os6PGDH2, encoding 6-phosphogluconate dehydrogenase (6PGDH), increases under drought, cold, and high salinity conditions and in response to abscisic acid treatments, under which conditions 6PGDH activity also increases [56]. Finally, the shorter Nucleolin1 gene (OsNUC1-S) reduces oxidative stress during high salinity treatment [57].
4. Discussion
4.1. Chloroplasts Maintain Reactive Oxygen Species (ROS) Homeostasis through Redox Enzymes Such as Superoxide Dismutase (SOD) and the Ascorbate-Glutathione (ASC-GSH) Cycle during Heat Stress
We found that chloroplast-related genes were associated with heat stress under abiotic stress conditions by MapMan analysis (Figure 3(b), green box). In addition, several antioxidants such as thioredoxin, ascorbate, glutathione, peroxiredoxin, and catalases, which are known to play an important role in scavenging ROS, have also been found (Figure 3(b), orange box). ROS are chemically reactive species and products of aerobic metabolism [58]. ROS are mainly produced by chloroplasts, mitochondria, and peroxides and are scavenged by the antioxidant mechanisms [59]. The balance between ROS generation and ROS scavenging is disturbed by various abiotic stresses like extreme temperatures, high salinity, drought, and heavy metals [5]. The plant has developed two efficient antioxidant mechanisms to maintain the ROS homeostasis of cells [5]: (i) antioxidant enzymes, like superoxide dismutase (SOD), ascorbate peroxidase (APX), catalase (CAT), guaiacol peroxidase (GPX), glutathione reductase (GR), dehydroascorbate reductase (DHAR), and monodehydroascorbate reductase (MDHAR); (ii) nonenzymatic antioxidants, such as ascorbic acid (AA), reduced glutathione (GSH), α-tocopherol, carotenoids, and flavonoids [58].
The chloroplast utilizes a well-organized thylakoid membrane system to efficiently capture light [60]. In thylakoids, photosystem I (PSI) and photosystem II (PSII) play a key role in the light harvesting system and are the major sources of ROS production. However, abiotic stresses such as drought, high salinity, and temperature extremes induce the formation of O•−2 in the photosystem through the Mehler reaction. Subsequently, a membrane-bound Cu/Zn SOD at the PSI converts O•−2 into H2O2 [58], which is then converted to H2O via the ascorbate-glutathione (ASC-GSH) cycle [61]. One of the integral components of the ASC-GSH cycle, APX, reduces H2O2 to H2O and DHA, using AA as a reducing agent in the chloroplast [62]. Another key component, GR, is a flavoprotein oxidoreductase, mainly found in chloroplasts. GR uses NADPH to reduce GSSG (glutathione, oxidized form) to GSH, with GSH reacting with and quenching detrimental ROS species such as 1O2 and OH• [63].
This ROS scavenging by antioxidant enzymes or nonenzymatic antioxidants protects plants from heat-induced oxidative stress. For example, Zea mays plants showed greater expression of enzymatic antioxidants, such as CAT, APX, and GR, compared with O. sativa and maintained significant levels of nonenzymatic antioxidants such as AA and GSH at high temperatures (45°C for day/40°C for night). These results suggest that Z. mays will be better able to cope with oxidative damage by heat stress through the ROS- scavenging process than its fellow grass, O. sativa [64]. Furthermore, heat-acclimated turf grass was found to maintain low ROS levels by increasing the synthesis of AA and GSH at high temperatures [65]. These results suggest that the antioxidant defense mechanism plays an important role in heat stress tolerance [66]. We have identified various enzymatic or nonenzymatic antioxidants in chloroplast-related genes through MapMan analysis (Figure 3(b)), and these candidate genes can be used as a major route to develop crops tolerant to abiotic stress, including heat stress.
4.2. Transcriptional Activity in the Nucleus Is Partially Regulated by Signals Derived from the Plastids
Chloroplast retrograde signaling refers to a communication pathway in which transcriptional activities in the nucleus are partially regulated by signals derived from plastids [67]. In general, chloroplasts of plants are predicted to be descendants of ancient photosynthetic bacteria and have circular genomes for their transcription and translation machinery [68]. The first discovery of retrograde signaling was reported in barley. Two barley chloroplast ribosome-deficient mutants caused downregulation of nuclear-encoded plastid proteins due to a defect in plastid function [69]. After that, studies on the function of retrograde signaling were conducted by coordinating chlorophyll biosynthesis with the expression of nuclear genes in some plants [70, 71]. Conversely, genome-uncoupled mutants, in which communication between the chloroplast and the nucleus was disrupted, expressed nuclear-encoded photosynthetic genes despite defective chloroplast physiology or inhibited biogenesis [70].
In chloroplast retrograde signaling, heat shock proteins (HSPs) play an important role in heat stress [4]. Interestingly, we identified a number of TFs, including heat shock transcription factors (HSFs), in Figure 3(d). Heat shock genes (HSGs), encoding HSPs, are upregulated during heat stress. HSPs play a role as chaperones to prevent denaturation of intracellular proteins and to preserve stability through protein folding [72]. Under heat stress conditions, plant HSP expression is rapidly activated by specific HSFs binding to the conserved sequences of the heat shock elements in the promoters of heat-responsive genes [73, 74]. For example, H2O2 in rice increased tolerance to oxidative stress by inducing the expression of chloroplast-localized small HSPs [75]. In addition, high H2O2 in Arabidopsis has been shown to induce activation of some chaperones, HSPs, and HSFs at the mRNA levels [76]. Until now, the retrograde signals have been perceived only in the cytosol, and the mechanism of communication with the nucleus remains largely unknown. We found several HSFs and other TFs in chloroplast-related genes through MapMan analysis. These might be potential candidate TFs to mediate plastid-nucleus signaling.
5. Conclusions
Chloroplasts are sensitive to environmental changes and have developed a complex network of plastid signals to protect plants from environmental stresses. ROS is a by-product of aerobic metabolism and acts as a marker under environmental stress. Chloroplast retrograde regulation is essential for coordinating gene expression, including that involving photosynthesis, in both the nucleus and the chloroplast. To cope with environmental stress, retrograde signals derived from chloroplasts must be delivered rapidly by cytosolic messengers or by distinct signal cascade pathways to the nucleus. H2O2, the HSP-associated complex, and some TFs have been suggested as possible retrograde signaling molecules, but more research remains to be conducted to determine how they function [4]. We performed functional dissection of chloroplast-related genes via diverse meta-expression data analysis based on microarray data and functional classification analyses. This work might provide new insights into the role of chloroplasts in producing crop plants with enhanced abiotic stress tolerance.
Acknowledgments
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01325901 and PJ01366401 to Ki-Hong Jung), Basic Science Research Program from the National Research Foundation (NRF) of Korea (NRF- 2018R1A4A1025158 to Ki-Hong Jung), and a Global Ph.D. Fellowship Program supported by the NRF (NRF-2018H1A2A1060336 to Woo-Jong Hong). We thank Prof. Gynheung An for providing valuable advice at all times.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
We have no conflicts of interest to declare.
Authors' Contributions
Yo-Han Yoo and Woo-Jong Hong contributed equally to this research.
Supplementary Materials
Table S1. List of 3,314 plastid-localized genes. Table S2. Anatomical expression data and cluster information of 1,695 plastid-related genes. Table S3. Abiotic stress expression data and cluster information of 264 plastid-related genes.
Figure S1. KMC clustering analysis for anatomical expression data of 3,314 genes using the Affymetrix array.
Figure S2. Detailed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of 1,695 leaf-preferred or ubiquitously expressed genes.
Figure S3. Abiotic stress clusters for 1,695 genes using Affymetrix array.
References
- 1.Serrano I., Audran C., Rivas S. Chloroplasts at work during plant innate immunity. Journal of Experimental Botany. 2016;67(13):3845–3854. doi: 10.1093/jxb/erw088. [DOI] [PubMed] [Google Scholar]
- 2.Vella F. Biochemical Education. 2003. The cell. A molecular approach. [Google Scholar]
- 3.Daniell H., Lin C. S., Yu M., Chang W. J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biology. 2016;17:p. 134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun A.-Z., Guo F.-Q. Chloroplast retrograde regulation of heat stress responses in plants. Frontiers in Plant Science. 2016;7:p. 398. doi: 10.3389/fpls.2016.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das K., Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Frontiers in Environmental Science. 2014;2(53):13. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- 6.Daniell H., Chan H., Pasoreck E. K. Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annual Review of Genetics. 2016;50(1):595–618. doi: 10.1146/annurev-genet-120215-035349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen J. F., de Paula W. B., Puthiyaveetil S., Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends in Plant Science. 2011;16(12):645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Bobik K., Burch-Smith T. M. Chloroplast signaling within, between and beyond cells. Frontiers in Plant Science. 2015;6(781) doi: 10.3389/fpls.2015.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang S., Zhu W., Hamilton J., et al. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Research. 2007;35(supplement 1):D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandran A. K. N., Yoo Y.-H., Cao P., et al. Updated Rice Kinase Database RKD 2.0: enabling transcriptome and functional analysis of rice kinase genes. Rice. 2016;9(1) doi: 10.1186/s12284-016-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao P., Jung K.-H., Choi D., Hwang D., Zhu J., Ronald P. C. The rice Oligonucleotide array database: an atlas of rice gene expression. Rice. 2012;5(17) doi: 10.1186/1939-8433-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeed A., Sharov V., White J., et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 13.Moon S., Oo M. M., Kim B., et al. Genome-wide analyses of late pollen-preferred genes conserved in various rice cultivars and functional identification of a gene involved in the key processes of late pollen development. Rice. 2018;11(28) doi: 10.1186/s12284-018-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pounds S., Fofana D. HybridMTest: Hybrid Multiple Testing. R package version 1.10.0, 2011.
- 15.Khatri P., Drǎghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung K., Dardick C., Bartley L. E., et al. Refinement of light-responsive transcript lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS ONE. 2008;3(10) doi: 10.1371/journal.pone.0003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginestet C. ggplot2: elegant graphics for data analysis. Journal of The Royal Statistical Society Series A-Statistics in Society. 2011 [Google Scholar]
- 18.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012;16(5) doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z., Liu Y., Li L., et al. Whole transcriptome sequencing of Pseudomonas syringae pv. actinidiae-infected kiwifruit plants reveals species-specific interaction between long non-coding RNA and coding genes. Scientific Reports. 2017;7, article no. 4910 doi: 10.1038/s41598-017-05377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L. Plasminogen: a potential target gene for dietary supplements and biomarker of the early stage of obesity by fatigue mice. Biomedical Research (India) 2017;28(10):4299–4304. [Google Scholar]
- 21.Hong W., Yoo Y., Park S., et al. Genome-wide identification and extensive analysis of rice-endosperm preferred genes using reference expression database. Journal of Plant Biology. 2017;60(3):249–258. doi: 10.1007/s12374-016-0552-z. [DOI] [Google Scholar]
- 22.Thimm O., Bläsing O., Gibon Y., et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37(6):914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 23.Yao W., Li G., Yu Y., Ouyang Y. FunRiceGenes dataset for comprehensive understanding and application of rice functional genes. GigaScience. 2018;7(1):1–9. doi: 10.1093/gigascience/gix119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong W., Jung K. Comparative analysis of flanking sequence tags of T-DNA/transposon insertional mutants and genetic variations of fast-neutron treated mutants in rice. Journal of Plant Biology. 2018;61(2):80–84. doi: 10.1007/s12374-017-0425-0. [DOI] [Google Scholar]
- 25.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo Y.-H., Nalini Chandran A. K., Park J.-C., et al. OsPhyB-mediating novel regulatory pathway for drought tolerance in rice root identified by a global RNA-seq transcriptome analysis of rice genes in response to water deficiencies. Frontiers in Plant Science. 2017;8(580) doi: 10.3389/fpls.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoo Y., Choi H., Jung K. Genome-wide identification and analysis of genes associated with lysigenous aerenchyma formation in rice roots. Journal of Plant Biology. 2015;58(2):117–127. doi: 10.1007/s12374-014-0486-2. [DOI] [Google Scholar]
- 29.Liu D., Zhang X., Cheng Y., Takano T., Liu S. rHsp90 gene expression in response to several environmental stresses in rice (Oryza sativa L.) Plant Physiology and Biochemistry. 2006;44(5-6):380–386. doi: 10.1016/j.plaphy.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Sun J., Zheng T., Yu J., et al. TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. New Phytologist. 2017;215(1):240–255. doi: 10.1111/nph.14482. [DOI] [PubMed] [Google Scholar]
- 31.Sarwat M., Naqvi A. R. Heterologous expression of rice calnexin (OsCNX) confers drought tolerance in Nicotiana tabacum. Molecular Biology Reports. 2013;40(9):5451–5464. doi: 10.1007/s11033-013-2643-y. [DOI] [PubMed] [Google Scholar]
- 32.Lim S. D., Lee C., Jang C. S. The rice RING E3 ligase, OsCTR1, inhibits trafficking to the chloroplasts of OsCP12 and OsRP1, and its overexpression confers drought tolerance in Arabidopsis. Plant, Cell & Environment. 2014;37(5):1097–1113. doi: 10.1111/pce.12219. [DOI] [PubMed] [Google Scholar]
- 33.Wang G., Ding X., Yuan M., et al. Dual function of rice OsDR8 gene in disease resistance and thiamine accumulation. Plant Molecular Biology. 2006;60(3):437–449. doi: 10.1007/s11103-005-4770-x. [DOI] [PubMed] [Google Scholar]
- 34.Kusumi K., Mizutani A., Nishimura M., Iba K. A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. The Plant Journal. 1997;12(6):1241–1250. doi: 10.1046/j.1365-313x.1997.12061241.x. [DOI] [Google Scholar]
- 35.Wang W., Zheng K., Gong X., et al. The rice TCD11 encoding plastid ribosomal protein S6 is essential for chloroplast development at low temperature. Journal of Plant Sciences. 2017;259:1–11. doi: 10.1016/j.plantsci.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Zhang J., Shi X., et al. Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. Journal of Experimental Botany. 2016;67(17):5187–5202. doi: 10.1093/jxb/erw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong H., Fei G.-L., Wu C.-Y., et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiology. 2013;162(4):1867–1880. doi: 10.1104/pp.113.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Cui X., Wang Y., Wu J., Gu X., Lu T. The RNA editing factor WSP1 is essential for chloroplast development in rice. Molecular Plant. 2017;10(1):86–98. doi: 10.1016/j.molp.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Pandeya D., Nath K., et al. ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. The Plant Journal. 2010;62(4):713–725. doi: 10.1111/j.1365-313X.2010.04183.x. [DOI] [PubMed] [Google Scholar]
- 40.Wan C., Li C., Ma X., et al. GRY79 encoding a putative metallo-β-lactamase-trihelix chimera is involved in chloroplast development at early seedling stage of rice. Plant Cell Reports. 2015;34(8):1353–1363. doi: 10.1007/s00299-015-1792-y. [DOI] [PubMed] [Google Scholar]
- 41.Kim S., Yang J., An G. OsCpn60α1, encoding the plastid chaperonin 60α subunit, is essential for folding of rbcL. Molecules and Cells. 2013;35(5):402–409. doi: 10.1007/s10059-013-2337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia K., Zeng X., Jiao Z., et al. Formation of protein disulfide bonds catalyzed by OsPDIL1;1 is mediated by MicroRNA5144-3p in rice. Plant & Cell Physiology (PCP) 2018;59(2):331–342. doi: 10.1093/pcp/pcx189. [DOI] [PubMed] [Google Scholar]
- 43.Yue R., Wang X., Chen J., et al. A rice stromal processing peptidase regulates chloroplast and root development. Plant & Cell Physiology (PCP) 2010;51(3):475–485. doi: 10.1093/pcp/pcq012. [DOI] [PubMed] [Google Scholar]
- 44.Tamura W., Kojima S., Toyokawa A., et al. Disruption of a novel NADH-glutamate synthase2 gene caused marked reduction in spikelet number of rice. Frontiers in Plant Science. 2011;2(57) doi: 10.3389/fpls.2011.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C., Chen W., Lin L., Lee C., Tseng T., Leu W. OIP30, a RuvB-Like DNA Helicase 2, is a potential substrate for the pollen-predominant OsCPK25/26 in rice. Plant & Cell Physiology (PCP) 2011;52(9):1641–1656. doi: 10.1093/pcp/pcr094. [DOI] [PubMed] [Google Scholar]
- 46.Tsugane K., Maekawa M., Takagi K., et al. An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. The Plant Journal. 2006;45(1):46–57. doi: 10.1111/j.1365-313X.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- 47.Xu J., Yang J., Wu Z., et al. Identification of a dual-targeted protein belonging to the mitochondrial carrier family that is required for early leaf development in rice. Plant Physiology. 2013;161(4):2036–2048. doi: 10.1104/pp.112.210831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang D., Liu H., Zhai G., Wang L., Shao J., Tao Y. OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. Journal of Genetics and Genomics. 2016;43(10):601–608. doi: 10.1016/j.jgg.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Wang C., Zheng M., et al. WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiology. 2016;170(4):2110–2123. doi: 10.1104/pp.15.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L., Zhang S., Qiu Z., et al. FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. Journal of Integrative Plant Biology. 2018;60(2):94–111. doi: 10.1111/jipb.12631. [DOI] [PubMed] [Google Scholar]
- 51.Cai Y., Li S., Jiao G., et al. OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnology Journal. 2018;16(11):1878–1891. doi: 10.1111/pbi.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S., Cheng Y., Zhang X., et al. Expression of an NADP-malic enzyme gene in rice (Oryza sativa. L) is induced by environmental stresses; over-expression of the gene in Arabidopsis confers salt and osmotic stress tolerance. Plant Molecular Biology. 2007;64(1-2):49–58. doi: 10.1007/s11103-007-9133-3. [DOI] [PubMed] [Google Scholar]
- 53.Lu Q., Ding S., Reiland S., et al. Identification and characterization of chloroplast casein kinase II from Oryza sativa (rice) Journal of Experimental Botany. 2015;66(1):175–187. doi: 10.1093/jxb/eru405. [DOI] [PubMed] [Google Scholar]
- 54.Satoh H., Shibahara K., Tokunaga T., et al. Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. The Plant Cell. 2008;20(7):1833–1849. doi: 10.1105/tpc.107.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ho C.-L., Wu Y., Shen H.-B., Provart N. J., Geisler M. A predicted protein interactome for rice. Rice. 2012;5(1):p. 15. doi: 10.1186/1939-8433-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou F.-Y., Huang J., Yu S.-L., Zhang H.-S. The 6-phosphogluconate dehydrogenase genes are responsive to abiotic stresses in rice. Journal of Integrative Plant Biology. 2007;49(5):655–663. doi: 10.1111/j.1744-7909.2007.00460.x. [DOI] [Google Scholar]
- 57.Sripinyowanich S., Chamnanmanoontham N., Udomchalothorn T., et al. Overexpression of a partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Journal of Plant Sciences. 2013;213:67–78. doi: 10.1016/j.plantsci.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 58.Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment. 2010;33(4):453–467. doi: 10.1111/j.1365-3040.2009.02041.x. [DOI] [PubMed] [Google Scholar]
- 59.Foyer C. H., Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. The Plant Cell. 2005;17(7):1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfannschmidt T. Chloroplast redox signals: how photosynthesis controls its own genes. Trends in Plant Science. 2003;8(1):33–41. doi: 10.1016/S1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 61.Foyer C. H., Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011;155(1):2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma P., Dubey R. Ascorbate peroxidase from rice seedlings: properties of enzyme isoforms, effects of stresses and protective roles of osmolytes. Journal of Plant Sciences. 2004;167(3):541–550. doi: 10.1016/j.plantsci.2004.04.028. [DOI] [Google Scholar]
- 63.Asada K. The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- 64.Kumar S., Gupta D., Nayyar H. Comparative response of maize and rice genotypes to heat stress: status of oxidative stress and antioxidants. Acta Physiologiae Plantarum. 2012;34(1):75–86. doi: 10.1007/s11738-011-0806-9. [DOI] [Google Scholar]
- 65.Xu S., Li J., Zhang X., Wei H., Cui L. Effects of heat acclimation pretreatment on changes of membrane lipid peroxidation, antioxidant metabolites, and ultrastructure of chloroplasts in two cool-season turfgrass species under heat stress. Environmental and Experimental Botany. 2006;56(3):274–285. doi: 10.1016/j.envexpbot.2005.03.002. [DOI] [Google Scholar]
- 66.Hasanuzzaman M., Nahar K., Alam M. M., Roychowdhury R., Fujita M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. International Journal of Molecular Sciences. 2013;14(5):9643–9684. doi: 10.3390/ijms14059643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pogson B. J., Woo N. S., Förster B., Small I. D. Plastid signalling to the nucleus and beyond. Trends in Plant Science. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 68.Sugiura M., Hirose T., Sugita M. Evolution and mechanism of translation in chloroplasts. Annual Review of Genetics. 1998;32(1):437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 69.Bradbeer J. W., Atkinson Y. E., Börner T., Hagemann R. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature. 1979;279(5716):816–817. doi: 10.1038/279816a0. [DOI] [Google Scholar]
- 70.Susek R. E., Ausubel F. M., Chory J. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74(5):787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- 71.Sullivan J. A., Gray J. C. Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. The Plant Cell. 1999;11(5):901–910. doi: 10.1105/tpc.11.5.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baniwal S. K., Bharti K., Chan K. Y., et al. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. Journal of Biosciences. 2004;29(4):471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- 73.Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K.-D. Complexity of the heat stress response in plants. Current Opinion in Plant Biology. 2007;10(3):310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 74.Nover L., Scharf K. D., Gagliardi D., Vergne P., Czarnecka-Verner E., Gurley W. B. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress & Chaperones. 1996;1(4):215–223. doi: 10.1379/1466-1268(1996)001<0215:THWCAP<2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee B., Won S., Lee H., et al. Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene. 2000;245(2):283–290. doi: 10.1016/S0378-1119(00)00043-3. [DOI] [PubMed] [Google Scholar]
- 76.Desikan R. Regulation of the arabidopsis transcriptome by oxidative stress. Plant Physiology. 2002;127(1):159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of 3,314 plastid-localized genes. Table S2. Anatomical expression data and cluster information of 1,695 plastid-related genes. Table S3. Abiotic stress expression data and cluster information of 264 plastid-related genes.
Figure S1. KMC clustering analysis for anatomical expression data of 3,314 genes using the Affymetrix array.
Figure S2. Detailed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of 1,695 leaf-preferred or ubiquitously expressed genes.
Figure S3. Abiotic stress clusters for 1,695 genes using Affymetrix array.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.