Abstract
Infection-related cancer comprises one-sixth of the global cancer burden. Oncoviruses can directly or indirectly contribute to tumorigenesis. Ubiquitination is a dynamic and reversible posttranslational modification that participates in almost all cellular processes. Hijacking of the ubiquitin system by viruses continues to emerge as a central theme around the viral life cycle. Deubiquitinating enzymes (DUBs) maintain ubiquitin homeostasis by removing ubiquitin modifications from target proteins, thereby altering protein function, stability, and signaling pathways, as well as acting as key mediators between the virus and its host. In this review, we focus on the multiple functions of DUBs in RIG-I-like receptors (RLRs) and stimulator of interferon genes (STING)-mediated antiviral signaling pathways, oncoviruses regulation of NF-κB activation, oncoviral life cycle, and the potential of DUB inhibitors as therapeutic strategies.
1. Introduction
About 15-16% of cancer cases are attributable to infection [1]. Viral infection is one of the main risk factors for the development of infection-related cancers. Currently, the known oncogenic viruses include Epstein-Barr virus (EBV) [2–4], Kaposi sarcoma herpes virus (KSHV) [5], human T-cell lymphotropic virus type 1 (HTLV-1) [1, 6], hepatitis B virus (HBV), hepatitis C virus (HCV), human papillomavirus (HPV), and human immunodeficiency virus type 1 (HIV-1) [7]. EBV, also known as human herpes virus 4, was the first virus to be associated with human malignancy. EBV is a double-stranded DNA virus. EBV infects approximately 95% of the world's population, which is the most common and persistent viral infection in humans. HTLV was the first human retrovirus to be identified. About 3–5% of HTLV-1-infected individuals develop adult T-cell leukemia/lymphoma (ATL), which is an aggressive and lethal malignancy with few effective therapeutic options [8]. Hepatocellular cancer (HCC) is the fifth most prevalent malignant tumor and the third leading cause of cancer-related deaths. HCC is a highly lethal cancer and is mainly associated with chronic HBV and HCV infections with about 80% of HCC caused by HBV and HCV infections [9]. Around 5% of global human cancers are caused by HPV [10]. HIV infection increases cancer risk mostly by immunosuppression and chronic immune activation [7] (Table 1).
Table 1.
Viral caused cancer types.
| Virus type | Cancer-related virus | Cancer types | Mechanisms | Ref. |
|---|---|---|---|---|
| RNA virus | HIV-1 | Lymphomas (most EBV-positive), KSHV-caused Kaposi sarcoma, and HPV-associated cervical and Anogenital carcinomas | indirect | [12] |
| HTLV-1 | Adult T-cell leukemia/lymphoma (ATL) | direct | [6, 11] | |
| HCV | Hepatocellular cancer, Non-Hodgkin lymphoma (especially B-cell lymphoma) | indirect | [14] | |
| DNA virus | HBV | Hepatocellular cancer | indirect | [14] |
| HPV | Cervix, Anal, Vulvar, and Penile cancers, and a subset of head and neck squamous cell carcinomas | direct | [1] | |
| KSHV | Kaposi sarcoma, primary effusion lymphoma | direct | [10] | |
| EBV | Nasopharyngeal carcinoma, Gastric cancer, Non-Hodgkin lymphomas (nhls), and Burkitt lymphoma, Nature killer/T-cell lyphoma | direct | [7–9] |
The fate and function of most proteins depend on posttranslational modifications [11]. Ubiquitin is a posttranslational modifier and a key regulatory molecule participating in various cellular activities. Aberrant ubiquitin system activity is linked to many diseases, including cancer [12], infection [13, 14], and neurodegeneration [15]. All viruses need host machinery to maintain infection and replication. Therefore, oncoviruses rely on the ubiquitin system at many levels, and even hijack the ubiquitin system to satisfy their survival needs. Ubiquitination is dynamic and it can be reversed by deubiquitinating enzymes (deubiquitinases or DUBs). This explains why DUBs are the main regulators in the interactions between the virus and its host. Some viruses even encode viral deubiquitinating enzymes to affect multiple host cell processes. However, relevant research findings are very limited. Thus far, identifying and taking full advantage of viral-related DUBs is a continuing challenge [13]. Here, we review current knowledge from both the host and viral points of view, discussing how the DUBs are involved in the viral life cycle and how oncoviruses avoid or utilize the DUBs to satisfy their survival needs.
2. General Functions of DUBs
DUBs maintain ubiquitin system homeostasis by cleaving polyubiquitin chains or completely removing ubiquitin chains from ubiquitinated proteins and then generating and recycling free ubiquitin [16]. Deubiquitination has important functions in regulating the ubiquitin-dependent pathways, including cell cycle regulation, cell death, protein degradation, protein function, gene expression, and signal transduction [17]. Thus far, about 100 DUBs have been identified in six different families and are classified into two categories (Table 2) [18, 19]. Imbalances in DUBs activities are involved in multiple diseases, including cancer, inflammation, neurological disorders, and microbial infections [17]. DUBs, such as A20, OTULIN, and CYLD, mediate NF-κB and cell death to maintain optimal signal transduction and immune homeostasis [20]. Compared with normal cells, cancer cells need elevated synthesis of growth-promoting proteins and protein-degradation capacity to satisfy uncontrolled mitosis. Much research has focused on studying their function and substrates to elucidate the role of DUBs in specific diseases. Abnormal expression of DUBs-encoding genes has been detected in human cancers. A mutant tumor suppressor gene CYLD has been identified in familial cylindromatosis and CYLD is downregulated in multiple cancer types [21]. Hajek, et. al have identified a distinct subset of HPV-associated head and neck squamous cell carcinomas that have TRAF3/CYLD mutations [22]. Multiple oncoviruses utilize these DUBs to edit ubiquitin chains and alter ubiquitin signaling, contributing to virus infection, replication, and pathogenesis. To date, vaccines against HBV and HPV have already begun to decrease the incidence of cancers attributed to these oncoviruses. However, other oncoviruses have no existing vaccines. In addition to prevention by vaccines, targeting the interplay between oncoviruses and their host might give rise to effective and inexpensive treatment strategies with minimal toxicity.
Table 2.
DUBs classification.
| Categories | Families | DUBs |
|---|---|---|
| Cysteine proteases | USP | USP 1-8, USP 9X, USP 9Y, USP 10-16, USP 17 L1, USP 17 L2, USP 18-26, USP 27X, USP 28-54, USP L1, CYLD |
| UCH | UCH L1, UCH L3, UCH L5, BAP1 | |
| MJD | ATXN3, ATXN3L, JOSD1, JOSD2 | |
| OTU | OTUB1, OTUB2, OTUD1, OTUD3, OTUD4, OTUD5, OTUD6A, OTUD6B, OTUD7A, OTUD7B, A2O, HIN1L, VCPIP1, TRABID, YOD1 | |
| MINDY | FAM63A, FAM63B, FAM188A, FAM188B | |
| Metalloproteases | JAMM | AMSH, AMSH-LP, BRCC36, COPS5, COPS6, EIF3F, EIF3H, MPND, MYSM1, PSMD7, PSMD14, PRPF8 |
Six classes of DUBs in the human genome are classified into two categories, cysteine proteases, and metalloproteases. Five classes are cysteine proteases: USP, ubiquitin-specific proteases; UCH, ubiquitin carboxyl-terminal hydrolases; MJD, Machado-Joseph disease protein domain proteases; OTU, ovarian-tumor proteases; MINDY, motif interacting with Ub-containing DUB family. One class is metalloproteases: JAMM, JAMM/MPN domain-associated metallopeptidases.
3. DUBs Participate in Antiviral Innate Responses
As the first line of host defense against viral infection, host pattern recognition receptors (PRRs), including RLRs, toll-like receptors (TLRs), and cytosolic dsDNA sensors (such as STING), recognize viral nucleic acids inducing innate immune responses, resulting in the production of type I interferons (IFNs) and proinflammatory cytokines [23, 24]. Using or bypassing host immune signaling is important for viruses to successfully establish infection. A thorough understanding of the molecular mechanisms between virus-related deubiquitination and antiviral innate immunity signaling is necessary for the control of infectious diseases and for developing therapeutic targets.
3.1. DUBs Are Involved in RLRs-Mediated Innate Immunity against RNA Oncoviruses
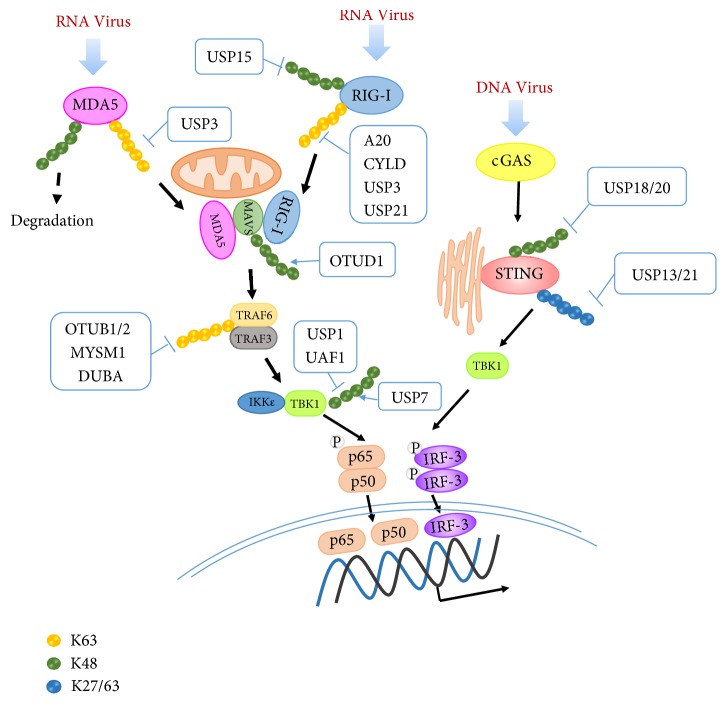
RNA viruses are mainly recognized by RLRs. RLRs recognize viral RNAs through the RNA helicase domain (RLD), and then interact with the mitochondrial antiviral signaling protein, MAVS [25]. The RLRs include retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which belong to a family of cytosolic host RNA helicases that recognize distinct nonself RNA signatures and trigger innate immune responses against several RNA viral infections. After recognition of viral RNA through the RNA helicase domain (RLD), RIG-I or MDA5 binds to MAVS. The K63-linked polyubiquitination of these adaptors is essential for signal activation. On the other hand, DUBs have also been shown to regulate antiviral innate immunity. Some DUBs negatively regulate the innate immune system to guard against excessive self-destructive immune responses and thus play a critical role in maintaining the balance of the immune system. USP21 [26], USP3 [27], and CYLD [28] negative regulate RIG-I and MDA5 activation by binding to and removing K63-linked polyubiquitin chains. The deubiquitinases OTUB1/2 [29, 30] and MYSM1 [31] inhibit K63-linked ubiquitination of TRAF3/6 and negatively regulate IFNs production. OTUD1 can also remove K48-linked ubiquitination from Smurf1, which targets MAVS for K48-linked ubiquitination and degradation, contributing to the degradation of MAVS [25]. Zhang et al. found that RNA viral infection can utilize the OTUD1-Smurf1 axis through the NF-κB signaling pathway to promote downregulation of the MAVS, TRAF3, and TRAF6 proteins and IFNs production [32]. In addition to the DUBs mentioned above, the host also uses positive regulation of DUBs against viral infection. USP15 reduces the K48-linked ubiquitination of TRIM25 (targeting RIG-I K63-linked ubiquitination and activation) leading to its stabilization [33] and promoting RIG-I activation. USP25 clears virus-triggered K48-linked ubiquitination, promoting the stability of TRAF3 and TRAF6 [34] and positively regulating RNA virus-triggered innate immune responses. USP1 and UAF1 bind to TBK1, remove its K48-linked polyubiquitination, and reverses the degradation process of TBK1. This USP1–UAF1 complex enhances TLR3/4 and RIG-I–induced IFN regulatory factor 3 (IRF3) activation and subsequent IFN-β secretion [35]. These studies indicate that DUBs play a critical role in regulating the virus-triggered RIG-I-like pathway and IFNs production, which are crucial for RNA viruses to establish efficient infection at an early stage (Figure 1).
Figure 1.
DUBs participate in antiviral innate immunity. During virus infection, K63-linked polyubiquitination of RLRs promotes their interaction with MAVS and signal transmission. USP15 inhibits K48-ubiquitination of RNA sensor RIG-I to inhibit RIG-I degradation; A20, CYLD, USP3, and USP21 inhibit K63-ubiquitination of RIG-I to negatively regulate RIG-I activation. USP3 inhibits K63 ubiquitination of MDA5 to inhibit its activation. RIG-I and MDA5 bind to and activate MAVS. Activated MAVS works as a scaffold to recruit various TRAFs, leading to TBK1/IƘB kinase Ɛ (IKK-Ɛ)-mediated phosphorylation and nuclear translocation of IRF3 and IRF7, and production of IFNs and OTUD1 stabilizes MAVS by removing K48-ubiquitination. Deubiquitinases OTUB1/2, MYSM1, and DUBA inhibit K63-linked ubiquitination of TRAF3 or TRAF6 and negatively regulate IFNs production. HSV infection can recruit USP21 to deubiquitinate the K27/63-linked polyubiquitin chain on STING. USP13 removes K27-linked polyubiquitin chains from STING and thereby impairs the recruitment of TBK1 to reduce the antiviral immune response against DNA viruses. USP18 recruits USP20 in an enzymatic activity-independent manner and facilitates USP20 to remove K33- and K48-linked ubiquitin chains from STING, thereby preventing degradation of STING caused by DNA virus infection. USP7 interacts with TRIM27 and removes its K48-linked polyubiquitination, promoting the degradation of TBK1. USP1 and UAF1 inhibit K48 polyubiquitin chains to stabilize TBK1 contributing to IFNs production.
3.2. DUBs Are Involved in STING-Mediated Innate Immunity against DNA Oncoviruses
Host cells express multiple cytosolic DNA sensors to recognize exogenous viral nucleic acids, such as DAI, DDX41, IFI16, and cyclic GMP-AMP synthase (cGAS). These sensors trigger signaling pathways and activate the adaptor protein stimulator of IFN genes (STING; also known as MITA) to induce the expression of type I IFN [36]. STING is a key adaptor protein for most DNA sensing pathways. Ubiquitination of STING caused by viral infection plays critical roles in virus-triggered signaling [37]. K27- or K63-linked ubiquitination mediated by various E3 ubiquitin ligases, such as TRIM32, AMFR, and INSIG1 [38, 39], is essential for full activation of STING. Double-stranded DNA viruses, such as EBV, use ubiquinase TRIM29 to ubiquitinate and degrade STING, suppressing host innate immunity that leads to the persistence of DNA viral infections [40]. HSV infection can recruit USP21 to STING through p38-mediated phosphorylation of USP21 at Ser538. USP21 deubiquitinates the K27/63-linked polyubiquitin chain on STING, thereby leading to reduced production of type I IFNs [41]. During HTLV-1 and HBV infection, Tax and HBV polymerases decrease the K63-linked ubiquitination of STING and disrupt the interactions between STING and TBK1, which leads to loss of STING function and subsequent impairment of IRF3 activation, IFN-induction, and an antiviral response [42, 43]. In addition, USP13 removes K27-linked polyubiquitin chains from STING and then decreases the antiviral immune response against DNA viruses by disrupting the recruitment of TBK1 [44]. To inhibit DNA viral infection, USP18 recruits USP20 in an enzymatic activity-independent manner and facilitates USP20 to remove K33- and K48-linked ubiquitin chains from STING, thereby preventing degradation of STING caused by DNA viral infection [45] (Figure 1). HPV upregulates UCHL1 to clear K63-linked ubiquitin chains from TRAF3, resulting in a lower amount of the downstream signaling complex TRAF3-TBK-1 to suppress the type I IFN pathway [46]. Further research is still needed to find and clarify the functions of DUBs during viral infection. More information will help control infectious diseases and facilitate the development of clinical antiviral therapies.
4. DUBs Regulate Oncovirus Infection and Activation in an NF-κB-Dependent Manner
RLR-, TLR-, and STING-induced innate immune response contribute to activation of NF-κB. NF-κB signaling plays an essential role in immune regulation and its role has been explored in almost all aspects of cellular activity. To achieve successful infection, oncoviruses have developed mechanisms to hijack the NF-κB pathway. Multiple DUBs are key regulators of NF-κB signaling. Several DUBs, such as CYLD and A20, have been extensively studied in the negative regulation of NF-κB signaling. During the viral infection stage, HCV stimulation upregulates A20/ABIN1 expression, thereby suppressing NF-κB activity and leading to inefficient M1 macrophage polarization to promote HCV infection [58]. EBV deubiquitinating Enzyme (v-DUB) BPLF1 inhibits TLR signaling through both MyD88- and TRIF-dependent pathways by removing ubiquitin chains from signaling intermediates, such as TRAF6, NEMO, and IκBα [59, 60]. This leads to reduced NF-κB activation and proinflammatory cytokine production in response to EBV and contributes to virus infectivity. During the infection stage, oncoviruses upregulate NF-κB inhibitory DUBs or encode viral DUBs disrupting secretion of antiviral cytokines and interfering with the innate antiviral immune responses by inhibiting NF-κB activation.
NF-κB activation also plays an important role in virus reactivation, replication, and virus-mediated cell transformation. HIV inhibits CYLD to facilitate the NF-κB pathway, playing an important role in HIV reactivation from latency [61]. HTLV-1- encoded Tax inactivates the NF-κB negative regulators, A20 and CYLD, which allows chronic NF-κB activation in HTLV-1-transformed cells [62]. USP20 deubiquitinates TRAF6 and Tax, thus suppressing interleukin 1β (IL-1β)- and Tax-induced NF-κB activation, suggesting USP20 as a key negative regulator of Tax-induced NF-κB signaling [63]. The HPV-encoded E6 protein targets CYLD, resulting in ubiquitination and proteasomal degradation of CYLD to induce NF-κB activation [64]. In keratinocytes, HPV infection inhibits CYLD expression, resulting in enhanced K63-linked polyubiquitination and nuclear translocation of BCL-3, which leads to activation of the NF-κB signaling pathway [65, 66]. Mutation of CYLD in HPV-positive head and neck squamous cell carcinomas (HNSCCs) leads to the activation of NF-κB signaling and maintenance of episomal HPV in tumors. In KSHV-infected primary effusion lymphoma cell lines, KSHV-encoded viral FLICE inhibitory protein (vFLIP) K13 can induce NF-κB activation, which upregulates A20 expression. A20 interacts with K13 and blocks K13-induced excessive NF-κB activation in a negative feedback manner [67, 68]. The regulation of NF-κB signaling by oncoviruses is not only important for the viral life cycle, but also contributes to the development of malignant tumors. Focusing on the role of DUBs in viral biology and NF-κB may contribute to infection-related cancer prevention and treatment.
5. Oncoviruses Use Host DUBs or Encode v-DUBs to Facilitate Viral Infection and Replication
5.1. EBV
EBV-encoded latent membrane protein 1 (LMP1) is an important tumorigenic protein. Our previous studies have shown that LMP1 rescues p53-induced cell cycle arrest and apoptosis by promoting K63-linked ubiquitination of p53. LMP1 also inhibits cell necroptosis by modulating RIPK1/3(receptor interacting protein kinase 1/3) ubiquitination [69, 70]. LMP1 can also induce the expression of UCH-L1 and it may contribute to viral transformation and the progression of lymphoid malignancies [71, 72]. EBV nuclear antigen 1 (EBNA1) plays important roles in promoting EBV genome replication and persistence, and EBV latent gene expression. EBNA1 interacts with USP7, which is also known as herpes virus associated ubiquitin-specific protease (HAUSP). The EBNA1 and USP7 interaction can promote cell survival and contribute to EBNA1 functions at the EBV oriP and inhibit p53-mediated antiviral responses [73]. The EBV nuclear antigen 3 (EBNA3) family targets and interacts with USP46/USP12 deubiquitination complexes. The complex exhibits DUB activity and contributes to EBNA3-mediated lymphoblastoid cell growth [74]. Besides utilizing host DUBs, EBV can also encode the viral deubiquitinating enzyme, BPLF1, which is an immune evasion gene product that can suppress antiviral immune responses during primary infection [47]. BPLF1 is expressed during the late phase of lytic EBV infection and is incorporated into viral particles. It can eliminate K63- and/or K48-linked ubiquitin chains and act as an active DUB during the productive lytic cycle and EBV infection [48] (Table 3).
Table 3.
Oncoviruses encoded v-DUBs.
5.2. KSHV
KSHV-encoded viral interferon regulatory factor 1 (vIRF1) can bind to USP7 and decrease the deubiquitinase activity of USP7 for stabilizing p53, thereby disrupting the p53 signaling pathway [73]. Latency-associated nuclear antigen (LANA) induces the expression of UCH-L1, which might lead to viral transformation and the progression of lymphoid malignancies [71]. KSHV encoded tegument protein ORF64, which has deubiquitinase activity can inhibit the ubiquitination of RIG-I and suppress RIG-I-mediated IFN signaling. It is necessary for KSHV infection [49] (Table 3).
5.3. HPV
E6 and E7 are the main oncoproteins encoded by HPV. USP11 and USP15 can greatly increase the steady state level of HPV-16 E6 and E7 by reducing their ubiquitination and degradation, thereby increasing the oncogenic potential of HPV [75, 76].
5.4. HIV
HIV-1 Tat is encoded at an early stage after infection and is in charge of enhancing viral production. USP7 and USP47 stabilize the HIV-1 Tat protein by removing its K48 polyubiquitination chain [77]. The stabilization of Tat leads to enhanced HIV-1 gene expression, facilitates virus spread, and also reduces immune recognition in HIV-1- expressing cells [78].
5.5. HCV
HCV encodes the core protein and nonstructural (NS) proteins NS3 and NS5A and promotes oncogenic transformation, replication, and virus assembly [9]. Studies show that NS5A binds to the ovarian tumor protein, deubiquitinase 7B (OTUD7B) and enhances OTUD7B DUB activity, which may contribute to viral replication and infection [50].
Oncoviruses utilize host DUBs to stabilize viral proteins, which increases the oncogenic potential of oncoviruses. Oncogenic viral products disturb host cell signaling pathways by enhancing the level of specific DUBs or DUB activity to promote viral genome replication and persistence. One DUB exhibited an opposite role in different oncoviruses, which indicates that if a DUB is used as an antiviral target, the potential effect on other viruses must be considered. Further studies are still needed to describe the detailed mechanisms between DUBs and oncoviruses.
6. DUB Inhibitors (DIs) as Potential Therapeutic Strategies
Inhibition of proteasome deubiquitinating activity is a new cancer therapy. Most DIs are small molecule compounds, exerting their function by suppressing DUB activity. The ubiquitin-specific proteases (USPs) are the largest and the most diverse DUB family and gene mutations, altered activity, or abnormal expression of USPs has been linked to multiple cancer types. USPs attractive are therapeutic targets and interest is growing in the development of enzyme selective or specific chemical inhibitors as antiviral and anticancer agents. The USP7-specific small molecule inhibitors, HBX41, 108, and P5091, induce apoptosis by stabilizing p53 in multiple myeloma cells resistant to conventional bortezomib therapies [55]. b-AP15 inhibits USP14 and UCHL5 and was shown to inhibit tumor growth in multiple solid tumor mouse models and attenuated tumor invasion in acute myelogenous leukemia in in vivo models [56]. WPI130 targets USP5, USP9X, and USP14 and inhibits viral progeny production of several RNA viruses, induces apoptosis, and suppresses growth of breast cancer cells [53, 57]. The USP1 inhibitors, GW7647 and ML323, attenuate growth of leukemic cells, non-small-cell lung cancer cells, and osteosarcoma cells [52, 54]. In light of these findings, DIs could be significant as potential therapeutic modalities in the treatment of multiple cancers. Given the multiple functions of DUBs in viral infection, developing inhibitors targeting the functional activities of virus-associated DUBs or virus-encoded DUBs might contribute to the reduction of oncovirus infections and could be used in infection-related cancers as accessory treatments (Table 4).
Table 4.
Chemical DUB inhibitors.
| DUB Inhibitors(DIs) | target | Cancer types | reference |
|---|---|---|---|
| HBX 41,108 | USP5, 7, 8 and UCH-L3 | myeloma | [50] |
| HBX -19,818 | USP7 | colon carcinoma | [51] |
| HBX-28,258 | USP7 | colon carcinoma | [51] |
| P5091 | USP7 | myeloma | [50] |
| P22077 | USP7 | - | [52] |
| GW7674 | USP1 | non-small cell lung cancer | [53, 54] |
| ML323 | USP1 and some DUBs | non-small cell lung cancer and osteosarcoma | [53, 54] |
| b-AP15 (VLX1500) |
UCHL5, USP14 and some DUBs | nonspecific | [55] |
| WPI 130 | USP5/USP9x/USP14/UCHL1/UCHL5 | breast cancer | [56, 57] |
| PR-619 | broad-range DUB inhibitor | - | [52] |
7. Conclusions and Perspectives
DUBs are central component in the ubiquitin signaling system to modulate proteostasis and have been shown to participate in all aspects of the viral life cycle. To escape from host immune responses, hijacking of the ubiquitin system by viruses continues to emerge as a central theme around virus infection and replication. In this review, we summarized recent studies focusing on the role of deubiquitinases in antiviral immune responses, modulation of the NF-κB pathway, as well as on RNA and DNA oncovirus infection, replication, and pathogenesis. However, the detailed mechanisms between viruses, host, and DUBs are still not clear. As for the potential use of DIs as therapeutic strategies against cancer, many have been identified but none have been used clinically. As a new cancer therapy target, many challenges remain to be addressed for further understanding of DUBs function in order to develop compounds that inhibit or induce their activity to control the pathogenesis of oncoviruses.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81430064, 81602402, and 81672705) and the College Students' Innovation Project of Central South University (2018zzts234 and 2018zzts230).
Conflicts of Interest
The authors have no conflicts of interest.
References
- 1.Plummer M., de Martel C., Vignat J., Ferlay J., Bray F., Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. The Lancet Global Health. 2016;4(9):e609–e616. doi: 10.1016/S2214-109X(16)30143-7. [DOI] [PubMed] [Google Scholar]
- 2.Cao Y. EBV based cancer prevention and therapy in nasopharyngeal carcinoma. Nature Partner Journals Precision Oncology. 2017;1(1):p. 10. doi: 10.1038/s41698-017-0018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass A. J., Thorsson V., Shmulevich I., et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suarez F., Lecuit M. Infection-associated non-Hodgkin lymphomas. Clinical Microbiology and Infection. 2015;21(11):991–997. doi: 10.1016/j.cmi.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 5.Moore P. S., Chang Y. The conundrum of causality in tumor virology: The cases of KSHV and MCV. Seminars in Cancer Biology. 2014;26:4–12. doi: 10.1016/j.semcancer.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: A Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 7.Carbone A., Vaccher E., Gloghini A., et al. Diagnosis and management of lymphomas and other cancers in HIV-infected patients. Nature Reviews Clinical Oncology. 2014;11(4):223–238. doi: 10.1038/nrclinonc.2014.31. [DOI] [PubMed] [Google Scholar]
- 8.Giam C., Semmes O. HTLV-1 infection and adult T-cell leukemia/lymphoma—a tale of two proteins: tax and HBZ. Viruses. 2016;8(6):p. 161. doi: 10.3390/v8060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arzumanyan A., Reis H. M. G. P. V., Feitelson M. A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nature Reviews Cancer. 2013;13(2):123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 10.Casper C., Fitzmaurice C. Infection-related cancers: prioritising an important and eliminable contributor to the global cancer burden. The Lancet Global Health. 2016;4(9):e580–e581. doi: 10.1016/S2214-109X(16)30169-3. [DOI] [PubMed] [Google Scholar]
- 11.Liu X., Shi F., Li Y., et al. Post-translational modifications as key regulators of TNF-induced necroptosis. Cell Death & Disease. 2016;7(7):e2293–e2293. doi: 10.1038/cddis.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeller D., Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 13.Isaacson M. K., Ploegh H. L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host & Microbe. 2009;5(6):559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Castejon G., Edelmann M. J. Deubiquitinases: novel therapeutic targets in immune surveillance? Mediators of Inflammation. 2016;2016:13. doi: 10.1155/2016/3481371.3481371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciechanover A., Kwon Y. T. Degradation of misfolded proteins in neurodegenerative diseases: therapeutic targets and strategies. Experimental & Molecular Medicine. 2015;47(3, article e147) doi: 10.1038/emm.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farshi P., Deshmukh R. R., Nwankwo J. O., et al. Deubiquitinases (DUBs) and DUB inhibitors: a patent review. Expert Opinion on Therapeutic Patents. 2015;25(10):1191–1208. doi: 10.1517/13543776.2015.1056737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfoh R., Lacdao I. K., Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocrine-Related Cancer. 2015;22(1):T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Arcy P., Wang X., Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacology & Therapeutics. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Abdul Rehman S., Kristariyanto Y., Choi S., et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Molecular Cell. 2016;63(1):146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lork M., Verhelst K., Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: So similar, yet so different. Cell Death & Differentiation. 2017;24(7):1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolaou K., Tsagaratou A., Eftychi C., Kollias G., Mosialos G., Talianidis I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21(6):738–750. doi: 10.1016/j.ccr.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 22.Hajek M., Sewell A., Kaech S., Burtness B., Yarbrough W. G., Issaeva N. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer. 2017;123(10):1778–1790. doi: 10.1002/cncr.30570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Wu J., Du F., Chen X., Chen Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbalat R., Ewald S. E., Mouchess M. L., Barton G. M. Nucleic acid recognition by the innate immune system. Annual Review of Immunology. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 25.Quicke K. M., Diamond M. S., Suthar M. S. Negative regulators of the RIG-I-like receptor signaling pathway. European Journal of Immunology. 2017;47(4):615–628. doi: 10.1002/eji.201646484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Y., Mao R., Yu Y., et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. The Journal of Experimental Medicine. 2014;211(2):313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J., Song Y., Li Y., et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Research. 2014;24(4):400–416. doi: 10.1038/cr.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman C. S., O'Donnell M. A., Legarda-Addison D., et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO Reports. 2008;9(9):930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S., Zheng H., Mao A., et al. Regulation of Virus-triggered Signaling by OTUB1- and OTUB2-mediated Deubiquitination of TRAF3 and TRAF6. The Journal of Biological Chemistry. 2010;285(7):4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y., Xu R., Zheng X., Thiel V. HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathogens. 2014;10(4) doi: 10.1371/journal.ppat.1004041.e1004041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panda S., Nilsson J. A., Gekara N. O. Deubiquitinase MYSM1 regulates innate immunity through inactivation of TRAF3 and TRAF6 complexes. Immunity. 2015;43(4):647–659. doi: 10.1016/j.immuni.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L., Liu J., Qian L., et al. Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathogens. 2018;14(5) doi: 10.1371/journal.ppat.1007067.e1007067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauli E., Chan Y. K., Davis M. E., et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Science Signaling. 2014;7(307):p. ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin D., Zhang M., Zhang M.-X., et al. Erratum: Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6 (Proceedings of the National Academy of Sciences of the United States of America (2015) 112 (11324-11329) DOI: 10.1073/pnas.1509968112) Proceedings of the National Acadamy of Sciences of the United States of America. 2015;112(43):11324–11329. doi: 10.1073/pnas.1519363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z., Song H., Jia M., et al. USP1–UAF1 deubiquitinase complex stabilizes TBK1 and enhances antiviral responses. The Journal of Experimental Medicine. 2017;214(12):3553–3563. doi: 10.1084/jem.20170180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paludan S., Bowie A. Immune Sensing of DNA. Immunity. 2013;38(5):870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Q., Sun L., Chen Z. J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nature Immunology. 2016;17(10):1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 38.Wang Q., Liu X., Cui Y., et al. The E3 Ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity. 2014;41(6):919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Hu M., Wang Y., Shu H. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. The Journal of Biological Chemistry. 2012;287(34):28646–28655. doi: 10.1074/jbc.m112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing J., Zhang A., Zhang H., et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nature Communications. 2017;8(1):p. 945. doi: 10.1038/s41467-017-00101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y., Wang L., Jin J., et al. p38 inhibition provides anti–DNA virus immunity by regulation of USP21 phosphorylation and STING activation. The Journal of Experimental Medicine. 2017;214(4):991–1010. doi: 10.1084/jem.20161387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Li J., Chen J., et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. Journal of Virology. 2015;89(4):2287–2300. doi: 10.1128/JVI.02760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Yang S., Liu L., Wang H., Yang B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Research. 2017;232:13–21. doi: 10.1016/j.virusres.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 44.Sun H., Zhang Q., Jing Y., et al. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nature Communications. 2017;8(1) doi: 10.1038/ncomms15534.15534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M., Zhang M., Zhang Q., et al. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Research. 2016;26(12):1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karim R., Tummers B., Meyers C., et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte's innate immune response. PLoS Pathogens. 2013;9(5) doi: 10.1371/journal.ppat.1003384.e1003384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenzie J., Lopez-Giraldez F., Delecluse H., Walsh A., El-Guindy A., Ling P. D. The epstein-barr virus immunoevasins BCRF1 and BPLF1 are expressed by a mechanism independent of the canonical late pre-initiation complex. PLoS Pathogens. 2016;12(11) doi: 10.1371/journal.ppat.1006008.e1006008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehurst C. B., Ning S., Bentz G. L., et al. The epstein-barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. Journal of Virology. 2009;83(9):4345–4353. doi: 10.1128/JVI.02195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inn K., Lee S., Rathbun J. Y., et al. Inhibition of RIG-I-mediated signaling by kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. Journal of Virology. 2011;85(20):10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sianipar I. R., Matsui C., Minami N., et al. Physical and functional interaction between hepatitis C virus NS5A protein and ovarian tumor protein deubiquitinase 7B. Microbiology and Immunology. 2015;59(8):466–476. doi: 10.1111/1348-0421.12278. [DOI] [PubMed] [Google Scholar]
- 51.Altun M., Kramer H. B., Willems L. I., et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chemistry & Biology. 2011;18(11):1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Liang Q., Dexheimer T. S., Zhang P., et al. A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses. Nature Chemical Biology. 2014;10(4):298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pal A., Donato N. J. Ubiquitin-specific proteases as therapeutic targets for the treatment of breast cancer. Breast Cancer Research. 2014;16(5):p. 461. doi: 10.1186/s13058-014-0461-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J., Dexheimer T. S., Ai Y., et al. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chemistry & Biology. 2011;18(11):1390–1400. doi: 10.1016/j.chembiol.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colland F., Formstecher E., Jacq X., et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular Cancer Therapeutics. 2009;8(8):2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 56.D'Arcy P., Brnjic S., Olofsson M. H., et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature Medicine. 2011;17(12):1636–1640. doi: 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 57.Perry J. W., Ahmed M., Chang K., et al. Antiviral activity of a small molecule deubiquitinase inhibitor occurs via induction of the unfolded protein response. PLoS Pathogens. 2012;8(7) doi: 10.1371/journal.ppat.1002783.e1002783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan C., Zhang Y., Zhou Y., et al. Up-regulation of A20/ABIN1 contributes to inefficient M1 macrophage polarization during Hepatitis C virus infection. Virology Journal. 2015;12(1) doi: 10.1186/s12985-015-0379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Gent M., Braem S. G., de Jong A., et al. Epstein-barr virus large tegument protein BPLF1 contributes to innate immune evasion through interference with toll-like receptor signaling. PLoS Pathogens. 2014;10(2) doi: 10.1371/journal.ppat.1003960.e1003960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehurst C. B., Vaziri C., Shackelford J., Pagano J. S. Epstein-barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase recruitment to DNA damage sites. Journal of Virology. 2012;86(15):8097–8106. doi: 10.1128/JVI.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Manganaro L., Pache L., Herrmann T., et al. Tumor suppressor cylindromatosis (CYLD) controls HIV transcription in an NF-B-dependent manner. Journal of Virology. 2014;88(13):7528–7540. doi: 10.1128/JVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pujari R., Hunte R., Thomas R., et al. Human T-cell leukemia virus type 1 (HTLV-1) tax requires CADM1/TSLC1 for inactivation of the NF-κB inhibitor A20 and constitutive NF-κB signaling. PLoS Pathogens. 2015;11(3) doi: 10.1371/journal.ppat.1004721.e1004721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasunaga J., Lin F. C., Lu X., Jeang K. T. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-B signaling. Journal of Virology. 2011;85(13):6212–6219. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An J., Mo D., Liu H., et al. Inactivation of the CYLD deubiquitinase by HPV E6 mediates hypoxia-induced NF-κB activation. Cancer Cell. 2008;14(5):394–407. doi: 10.1016/j.ccr.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shostak K., Zhang X., Hubert P., et al. NF-κB-induced KIAA1199 promotes survival through EGFR signalling. Nature Communications. 2014;5(1) doi: 10.1038/ncomms6232.5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shostak K., Chariot A. EGFR and NF-κB: partners in cancer. Trends in Molecular Medicine. 2015;21(6):385–393. doi: 10.1016/j.molmed.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 67.Matta H., Gopalakrishnan R., Punj V., Yi H., Suo Y., Chaudhary P. M. A20 is induced by kaposi sarcoma-associated herpesvirus-encoded viral FLICE inhibitory protein (vFLIP) K13 and blocks K13-induced nuclear factor-κB in a negative feedback manner. The Journal of Biological Chemistry. 2011;286(24):21555–21564. doi: 10.1074/jbc.M111.224048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakakibara S., Espigol-Frigole G., Gasperini P., Uldrick T. S., Yarchoan R., Tosato G. A20/TNFAIP3 inhibits NF-κB activation induced by the Kaposi’s sarcoma-associated herpesvirus vFLIP oncoprotein. Oncogene. 2013;32(10):1223–1232. doi: 10.1038/onc.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li L., Li W., Xiao L., et al. Viral oncoprotein LMP1 disrupts p53-induced cell cycle arrest and apoptosis through modulating K63-linked ubiquitination of p53. Cell Cycle. 2014;11(12):2327–2336. doi: 10.4161/cc.20771. [DOI] [PubMed] [Google Scholar]
- 70.Liu X., Li Y., Peng S., et al. Epstein-Barr virus encoded latent membrane protein 1 suppresses necroptosis through targeting RIPK1/3 ubiquitination. Cell Death & Disease. 2018;9(2) doi: 10.1038/s41419-017-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bentz G. L., Bheda-Malge A., Wang L., Shackelford J., Damania B., Pagano J. S. KSHV LANA and EBV LMP1 induce the expression of UCH-L1 following viral transformation. Virology. 2014;448:293–302. doi: 10.1016/j.virol.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L., Tao Q., Jin H., et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clinical Cancer Research. 2010;16(11):2949–2958. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- 73.Chavoshi S., Egorova O., Lacdao I. K., Farhadi S., Sheng Y., Saridakis V. Identification of kaposi sarcoma herpesvirus (KSHV) vIRF1 protein as a novel interaction partner of human deubiquitinase USP7. The Journal of Biological Chemistry. 2016;291(12):6281–6291. doi: 10.1074/jbc.m115.710632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohashi M., Holthaus A. M., Calderwood M. A., et al. The EBNA3 family of epstein-barr virus nuclear proteins associates with the USP46/USP12 deubiquitination complexes to regulate lymphoblastoid cell line growth. PLoS Pathogens. 2015;11(4) doi: 10.1371/journal.ppat.1004822.e1004822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin C., Chang H., Yu W. C. USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. The Journal of Biological Chemistry. 2008;283(23):15681–15688. doi: 10.1074/jbc.M708278200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vos R. M., Altreuter J., White E. A., Howley P. M. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. Journal of Virology. 2009;83(17):8885–8892. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ali A., Raja R., Farooqui S. R., Ahmad S., Banerjea A. C. USP7 deubiquitinase controls HIV-1 production by stabilizing Tat protein. Biochemical Journal. 2017;474(10):1653–1668. doi: 10.1042/BCJ20160304. [DOI] [PubMed] [Google Scholar]
- 78.Setz C., Friedrich M., Rauch P., et al. Inhibitors of deubiquitinating enzymes block HIV-1 replication and augment the presentation of gag-derived MHC-I epitopes. Viruses. 2017;9(8):p. 222. doi: 10.3390/v9080222. [DOI] [PMC free article] [PubMed] [Google Scholar]