Abstract
The plant innate immune system has two major branches, the pathogen-triggered immunity and the effector-triggered immunity (ETI). The effectors are molecules released by plant attackers to evade host immunity. In addition to the foreign intruders, plants possess endogenous instigators produced in response to general cellular injury termed as damage-associated molecular patterns (DAMPs). In plants, DAMPs or alarmins are released by damaged, stressed, or dying cells following abiotic stress such as radiation, oxidative and drought stresses. In turn, a cascade of downstream signaling events is initiated leading to the upregulation of defense or response-related genes. In the present study, we have investigated more thoroughly the conservation status of the molecular mechanisms implicated in the danger signaling primarily in plants. Towards this direction, we have performed in silico phylogenetic and structural analyses of the associated biomolecules in taxonomically diverse plant species. On the basis of our results, the defense mechanisms appear to be largely conserved within the plant kingdom. Of note, the sequence and/or function of several components of these mechanisms was found to be conserved in animals, as well. At the same time, the molecules involved in plant defense were found to form a dense protein-protein interaction (PPi) network, suggesting a crosstalk between the various defense mechanisms to a variety of stresses, like oxidative stress.
1. Introduction
Plants lack a global immune surveillance system, but they have acquired through evolution a highly effective innate immune response. There are currently three major types of elicitors of immune response in plants: (a) the “nonself” foreign molecule microbial/pathogen-associated molecular patterns (MAMPs/PAMPs) recognized by surface-localized pattern recognition receptors (PRRs), (b) signals produced by herbivores, nematodes, or parasitic plants, and (c) their effectors released by the attackers in order to counteract host defense [1]. Based on the principle on the wide variety of all these patterns inducing immune response, a unifying converging theory has emerged that points towards the concept of host perception of “danger” independently of its origin [2, 3].
DAMPs as death-, danger-, or damage-associated molecular patterns are of biological origin and are considered the major immunogenic mediators released passively by damaged, stressed, or dying cells, including tumor cells targeted by oxidative injury, radiation, or chemotherapy [4]. Endogenous or self signals are of two kinds, the primary DAMPS (e.g., cell wall fragments) and the secondary signals produced in response to danger, like processed protein fragments, such as phytocytokines, similar to animal cytokines. A third class that has evolved lately and is not well-defined includes the abiotic danger signals, like nanomaterials [5]. DAMPs are host cell-derived, as opposed to exogenously derived (nonself) MAMPs and PAMPs [6]. In a seminal study on this field by Matzinger [7], it was suggested that the immune system does not actually distinguish between self and nonself but rather detects “danger” through a series of positive and negative signals derived from damaged or stressed tissues mediated by DAMPs [7]. In plants, DAMPs, similar to animals, are perceived by pattern recognition receptors (PRRs), such as plasma membrane-localized receptors, thereby leading to a cascade of events including cytoplasmic Ca2+ elevation, depolarization of the cell membrane, production of reactive oxygen species (ROS), the transient phosphorylation of mitogen-activated protein kinases (MAPKs), and the transcriptional upregulation of defense or response-related genes [6, 8, 9]. In humans, oxidative stress, through the production of ROS, activates components of the MAPK-mediated signaling pathway including ERK, JNK, and p38 MAPKs; this can have both prosurvival and proapoptotic effects [10]. Although in yeast and mammals the main mechanisms triggering MAPK signaling activation under stress conditions have been studied extensively, in plants they remain largely unexplored [11].
In plant cells, there is emerging evidence of the connection between DAMPs and the DNA damage response (DDR), as nucleic acid recognition represents a fundamental step in host defense. Plants have been documented to perceive both extracellular DNA and RNA [12–14]. Toll-like receptors (TLRs) play a central role in the preferential recognition and binding of extracellular DNA in animals [15–19]. Despite the fact that TLR homologs have not been identified in plants, extracellular self-DNA (sDNA) was shown to act as a DAMP in plants. It has been reported that sDNA can trigger ROS- and MAPK-dependent signaling cascades [20], alter the CpG DNA methylation status (hypomethylation), and elicit defense-related responses [21]. Moreover, DNA damage and alteration of the primary chromatin structure were shown to induce the expression of defense-related genes [22, 23]. Elucidation of the plant-specific receptors that recognize extracellular sDNA as DAMPs could advance our knowledge on sDNA-dependent danger signaling in plant cells.
Plants have evolved mechanisms of innate immunity against detrimental pathogenic microorganisms and herbivorous animals. As mentioned above, DAMPs or alarmins, i.e., biomolecules released by stressed cells, share similarities between plants and animals in different aspects. For example, the recently discovered Arabidopsis HMGB3 is the counterpart of the pivotal animal DAMP HMGB1 [6]. Recent studies highlight the role of plants as unique biological models to monitor various types of exogenous (environmental) stress. More specifically, green leaf volatiles (GLVs) can act as airborne infochemicals that regulate the expression of defense response-related plant genes as shown in Arabidopsis thaliana [24]. In addition, medicinal plants and their derivatives can mitigate nephrotoxicity and anticancer drug side effects due to their intrinsic antioxidant and anti-inflammatory properties (reviewed in [25]). It would be necessary for the better understanding of the complex response network of various organisms to abiotic stresses to highlight the key genes and the interactions of this network to inflammatory and immune response networks. DAMPs are regarded as the link between these networks. Therefore, the analytical description of the molecular mechanisms and regulatory pathways that orchestrate plant responses to abiotic stresses and, particularly, oxidative stress, cell death, and DDR is crucial for the application of this knowledge to more complicated organisms.
Plants' immune response against intruders is regulated mainly by two antagonistic defense signaling pathways: (i) the salicylic acid- (SA-) mediated signal transduction pathway elicited by biotrophic and hemiobiotrophic pathogens and (ii) the octadecanoid signaling pathway with the key hormone jasmonic acid (JA) induced by heterotrophic pathogens and herbivores [1]. Of note, the oxidized lipids that participate in octadecanoid signaling are the plant functional equivalents of the mammalian oxidized phospholipids [26].
To date, there is still a knowledge gap concerning the evolutionary origin of the different DAMP-dependent signaling cascades identified in the plant kingdom and the way this valuable knowledge can be projected to more complex organisms, especially animal cells. We believe that an evolutionary perspective could provide useful insights into the origin of DAMP-mediated mechanisms and their role across the different levels of biological organization and complexity. In this study, we have made an effort to assess the conservation of the defense mechanisms by conducting phylogenetic and structural analyses of the molecules implicated in different stages of danger signaling in 11 vascular plant species that represent diverse taxonomic divisions: Arabidopsis thaliana (thale cress), Zea mays, Oryza sativa (rice), Hordeum vulgare (barley), Medicago truncatula (barrel medic), Nicotiana tabacum (tobacco), Populus trichocarpa (cottonwood), Solanum lycopersicum (tomato), Prunus persica (peach), Vitis vinifera (wine grape), and Pisum sativum (pea).
2. Methods
2.1. Sequence Dataset and Homology Searching
The bibliographic database PubMed/MEDLINE (https://www.ncbi.nlm.nih.gov/pubmed) was thoroughly searched using relevant keywords such as “damage-associated molecular patterns”, “danger signals”, “alarmins”, “danger-associated molecular patterns”, “DAMPs”, “endogenous danger signaling”, “plants”, and “viridiplantae”. The names and/or accession numbers of the characterized proteins reported in the articles were used to retrieve their corresponding sequences from the publicly accessible sequence databases UniProtKB [27] and NCBI's GenBank [28]. To identify more orthologous protein sequences, the known sequences were used as a query in reciprocal BLASTp and tBLASTn [29] searches (cutoff E − value ≤ 1.0E − 9) of the genomes of 11 vascular plant species, representing diverse taxonomic divisions (Figure S1). The canonical or longest transcripts were selected. Any partial or ambiguous sequences were not included in the subsequent steps of the study.
2.2. Phylogenetic Analyses
The amino acid sequences of the corresponding proteins (Table S1) were aligned with MAFFT, version 3.7 [30]. Phylogenetic analyses were performed by employing a maximum likelihood (ML) method. To this end, the resulting multiple alignments were provided as input to PhyML v.3.0 [31], which optimizes a distance-based starting tree (BioNJ) [32] by employing a heuristic algorithm. The expected number of amino acid substitutions per site was estimated with the JTT model [33]. Protein sequences distantly related to those under study were used as outgroups. The robustness of the reconstructed phylogenetic trees was evaluated by bootstrapping (200 bootstrap pseudoreplicates). Trees were illustrated using Dendroscope [34].
2.3. Pairwise Distance Estimation
Pairwise distances between protein sequences were computed using the software package MEGA, version 7.0 [35].
2.4. Protein Domain Organization
The protein sequences under study were searched against the protein signature databases SMART v.8.0 [36] and CDD v.3.16 [37] in order to determine the boundaries of their constituent domains.
2.5. Sequence Motifs
The amino acid sequences corresponding to the catalytic kinase domain of the orthologous proteins DORN1, MAPK3, MAPK6, PEPR1/2, SERK3/4/5, SR160, and WAKL (Table S1) were aligned using MAFFT v.3.7 [30] and edited with Utopia's CINEMA alignment editor [38]. Ungapped sequence motifs were extracted from the alignment and submitted to WebLogo 3.5.0 [39] with default parameters, to create consensus sequences.
2.6. Tertiary Structure Analysis
The plant MAPK3 and MAPK6 sequences under study were aligned using the PROMALS3D multiple sequence alignment program, which incorporates evolutionary and tertiary structural information to improve alignment accuracy [40, 41]. The degree of conservation of amino acid residues of the homologous plant MAPK3 and MAPK6 proteins was estimated with the usage of the ConSurf program [42]. To this end, the resulting multiple sequence alignment of the MAPK3 and MAPK6 amino acid sequences was provided as input to ConSurf to map the conservation grades of the amino acid residues onto the resolved tertiary structure of the Arabidopsis thaliana MAPK6 (PDB ID: 5CI6 [43]). The protein structure was displayed using the PyMOL molecular graphics program (http://www.pymol.org/).
2.7. Structural Modeling of Protein-Protein Interactions
The tertiary structure of the extracellular LRR domain of the A. thaliana PEPR2 was predicted by homology modeling. The X-ray crystal structure of the Arabidopsis PEPR1LRR (PDB ID: 5GR8, chain A [44]) was used as a template to model PEPR2LRR using the I-TASSER server [45]. The quality of the final modeled protein structures was evaluated with Procheck [46].
In order to assemble the trimeric PEPR2LRR-AtPEP1-BAK1 complex, the following steps were carried out: LRR domains (italicized) present in the PEPR1LRR-AtPEP1 (PDB ID: 5GR8) and FLS2LRR-FLG22-BAK1 (PDB ID: 4MN8) complexes and the PEPR2LRR homology model were structurally superimposed by using the alignment algorithm FATCAT [47]. Following this superimposition, the coordinates of FLS2LRR-FLG22-BAK1 and PEPR1LRR were deleted. As a result of this procedure, a crude model of the PEPR2LRR-AtPEP1-BAK1 complex was obtained. This crude model was refined with the water refinement step of the HADDOCK webserver [48]. During refinement, the interaction between PEPR2LRR and AtPEP1 was used as a restraint, with the following restraint definition: (i) assign (resi 438 and segid A) (resi 23 and segid C) 3.00 3.00 0.05 and (ii) assign (resi 392 and segid A) (resi 23 and segid C) 3.00 3.00 0.05. All of the interface statistics were carried out through the CoCoMaps tool (https://www.molnac.unisa.it/BioTools/cocomaps/).
2.8. Functional Association Network
The associations among the plant proteins implicated in danger signaling, in this study, were investigated using STRING v11 [49], a database of either experimental or predicted, direct or indirect, protein-protein interactions; a relatively high confidence interaction score (0.7) was selected. Intermediate nodes connecting the input nodes were also predicted, with a maximum number of 5 interactors. To this end, the Arabidopsis thaliana proteins WAK1, MAPK3, MAPK6, DORN1, PEPR1, PEPR2, SR160, HMGB1, HMGB2, HMGB3, SERK3, and SERK4 were used as input to STRING to generate an interaction network.
3. Results and Discussion
3.1. Classes of DAMPs
The diverse DAMPs that have been identified in plants are arbitrarily grouped, in this study, in the following classes.
3.1.1. Cell Wall-Derived DAMPs
Oligogalacturonide (OG) fragments are released from the degradation of the plant cell wall constituent homogalacturonan (HGA) by pathogen-encoded polygalacturonases. OG fragments trigger a danger signaling cascade involving ROS production, increase in cytoplasmic Ca2+ concentration, activation of MAPK3 and MAPK6, upregulation of resistance genes, and activation of components of the SA, JA, and ethylene pathways [50–52]. The wall-associated kinase 1 (WAK1) was suggested to perceive OGs [53, 54]. Besides OGs, Claverie et al. suggest that xyloglucans (Xh), components of the cell wall hemicellulose, act as DAMPs and induce defense responses similar to OGs [55].
Likewise, in animals, glycosaminoglycan hyaluronan fragments are released during damage and are implicated in wound repair and regeneration. These fragments are recognized by the Toll-like receptors TLR2 and TLR4, leading to activation of the inflammatory gene expression [56]. Of note, hyaluronan fragments and MAPK activation were found to mediate ROS-induced upregulation of the defense-related gene MUC5AC [57].
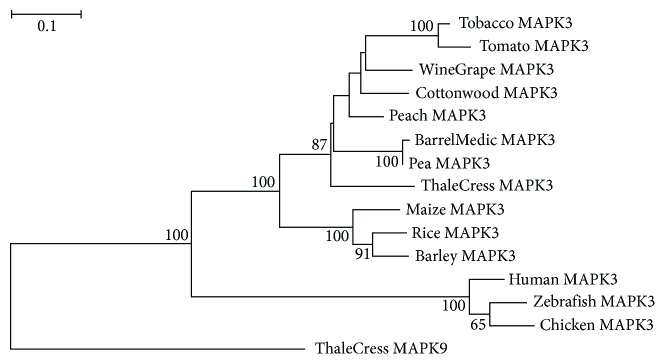
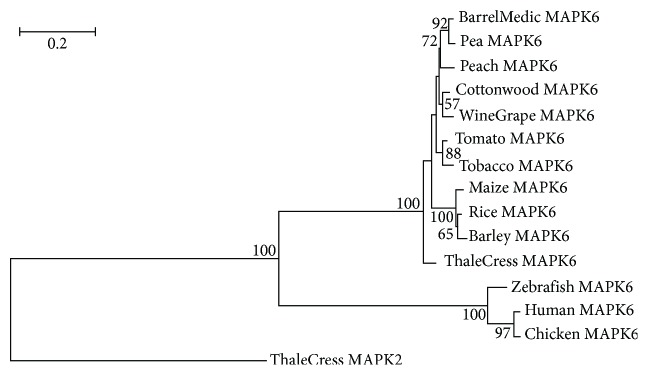
The viridiplantae MAPK3 and MAPK6 proteins (Figure 1) have orthologs in metazoa which are also implicated in danger signaling [58]. The plant MAPK3 and MAPK6 have rather evolved independently from their animal orthologs as they form distinct monophyletic groups with bootstrap values of 100, and they are connected to the corresponding metazoan clade with a long branch which reflects long evolutionary distance (Figures 2 and 3). The monocotyledonous plant MAPK3 and MAPK6 sequences form well-separated clades in their respective phylogenetic trees, leading to the suggestion that these sequences have probably appeared after the monocot-eudicot divergence about 200 million years ago (MYA) [59]. The tomato and tobacco MAPK3 and MAPK6 sequences cluster together with high statistical confidence, indicating that these sequences emerged after the members of the order solanales branched off from the other vascular plants (Figures 2 and 3).
Figure 1.
Phylogenetic distribution of the proteins implicated in plant defense.
Figure 2.
ML-based tree of MAPK3 protein sequences. The branch lengths are proportional to the evolutionary distance. Bootstrap support values ≥ 50% are shown at the nodes. The scale bar at the upper left denotes the length of amino acid replacements per position. The sequence ThaleCress MAPK9 was used as outgroup.
Figure 3.
Tree of MAPK6 protein sequences. The sequence ThaleCress MAPK9 was used as outgroup. The conventions are the same as in Figure 2.
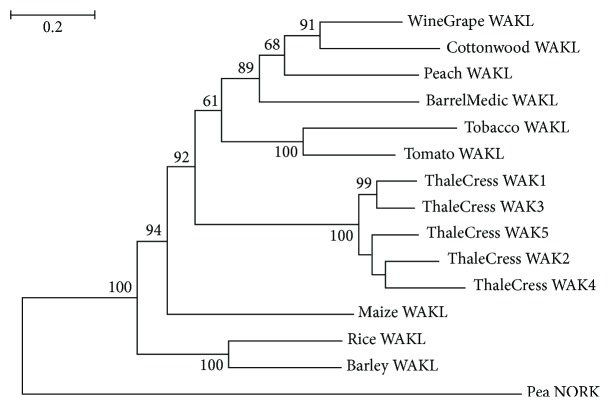
A cluster of five highly similar WAK (WAK1-5) genes are present in A. thaliana (thale cress) [60]. The Arabidopsis WAK1-5 protein sequences form a separate highly supported clade (Figure 4). Based on sequence database searches, a single copy of WAK was found in the other plant species (Figure 1). WAK homologs were not detected in the pea; this is probably due to incomplete annotation of the Pisum sativum genome. The encoded protein was arbitrarily referred to as WAKL, where “L” stands for “like,” by virtue of sequence similarity to the thale cress WAK1-5 (Figure 4). A primordial WAK gene might have undergone a series of duplications in A. thaliana, thereby giving rise to fıve WAK paralogs. However, any fully sequenced and annotated genomes of other members of the order brassicales are not currently available, apart from the one of the model plant organism A. thaliana, which would allow us to examine whether the WAK gene expansion is species- or order-specific. Based on extensive bibliographic searches, no experimental evidence was found about A. thaliana WAK2-5 acting as PRRs. WAK1 has apparently acquired a more specialized function during the course of evolution. WAK proteins are likely restricted to vascular plants, since orthologous WAK sequences were not found in metazoa. The role of these WAKL proteins in the perception of OGs in their host plant organisms remains to be investigated experimentally.
Figure 4.
Tree of WAKL protein sequences. The sequence Pea NORK (nodulation receptor kinase) was used as outgroup. The conventions are the same as in Figure 2.
3.1.2. Extracellular ATP
Extracellular ATP (eATP), released from stressed or damaged cells into the extracellular milieu, constitutes a class of danger signals both in plants and animals. In plants, eATP plays an important signaling role in plant cells by participating in several processes including cell development, viability, and stress responses [61, 62]. It is recognized by the plant-specific PRR, DORN1 (DOes not Respond to Nucleotides 1), which is a membrane lectin receptor kinase [63]. DORN1 is required for the ATP-mediated intracellular influx of Ca2+, formation of ROS, phosphorylation of MAPK3 and MAPK6, and stimulation of the expression of defense genes [64, 65]. Tripathi and colleagues demonstrated that eATP elicits defense responses via the JA signaling pathway [66].
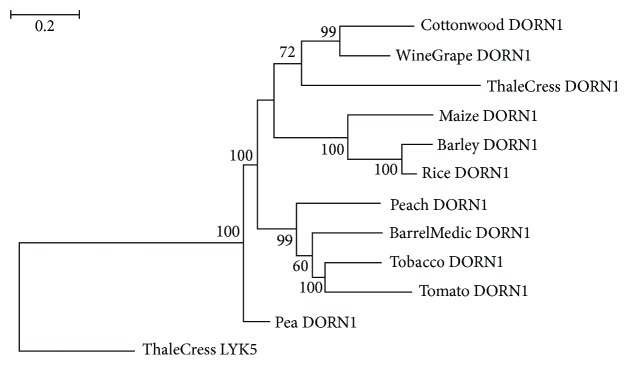
Putative DORN1 homologous sequences were detected in all plant species under investigation (Figure 1). The cereal plants as well as the solanales DORN1 sequences form their own monophyletic group with bootstrap values of 100, suggesting class- and order-specific evolution of DORN1 (Figure 5). DORN homologous sequences were found exclusively in plants, wherein might be implicated in the recognition of eATP.
Figure 5.
Tree of DORN1 protein sequences. The sequence ThaleCress LYK5 was used as outgroup. The conventions are the same as in Figure 2.
In animals, eATP was shown to mediate oxidative stress response [67, 68] and to be involved in inflammatory responses [64, 69]. Two types of P2 purinoceptors, the ligand-gated cation channel P2X receptors and the metabotropic (G-protein-coupled) P2Y receptors [70], are activated by eATP and are involved in the eATP-induced increase of cytosolic Ca2+ levels [71].
3.1.3. Polypeptide-Based DAMPs
A wide range of polypeptide-based DAMPs derived from larger precursors can act as robust inducers of plant defense responses [72, 73]. In Arabidopsis thaliana, the 23-amino acid PEP1 (Plant Elicitor Peptide) is processed from a 92-amino acid precursor upon damage, JA, and ethylene [74]. Based on homology searches, only one ortholog of AtPEP1 was detected in the fellow plants, that is, ZmPEP1, in maize. This protein is considered a DAMP, as well, given that it acts as an endogenous elicitor able to regulate the pathogenesis-related gene through sucrose-mediated signaling [75]. In A. thaliana, PEP1 is perceived by two PRRs, PEPR1 (PEPtide Receptor 1) and PEPR2, leading to ROS production and upregulation of defensin-like genes [76]. Of note, the only PEP orthologs were found in two species belonging to divergent taxonomic groups, eudicots (A. thaliana) and monocots (Zea mays), and not any taxonomically related species (such as fellow eudicots). This suggests that even distantly related organisms respond to similar stimuli with similar mechanisms.
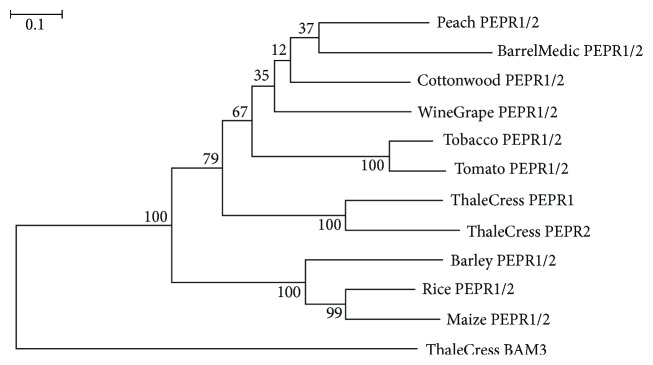
PEPs' cognate receptors, however, were found in all plants under study (except pea) (Figure 1). Thale cress harbors a total of seven PEPs (PEP1-PEP7) and two PEPR paralogs, the encoded proteins of which share 67% identity. In the inferred phylogenetic tree, the AtPEPR1 and AtPEPR2 protein sequences form a highly monophyletic branch with bootstrap support 100 (Figure 6). However, a single copy of PEPR was detected in the other plant species; the corresponding protein of which is arbitrarily called PEPR1/2 due to its amino acid sequence similarity to both AtPEPR1 and AtPEPR2. The cereal plants form a separate highly supported clade, leading to the suggestion that PEPR1/2 genes evolved after the monocots-dicots split (Figure 6). PEPRs are also kingdom-restricted, since PEPR orthologs were detected exclusively in vascular plants.
Figure 6.
Tree of PEPR1/2 protein sequences. The sequence ThaleCress BAM3 was used as outgroup. The conventions are the same as in Figure 2.
AtPEPs exhibit functional similarity to the 18-amino acid peptide systemin, which is processed from the 200-amino acid hormone prosystemin (SYST) in the tomato upon wounding [77]. Prosystemin was found only in the tomato and not in tobacco which is also member of the same solanales order (Figure 1). However, two hydroxyproline-rich systemin (HSY) peptides, designated HSYA and HSYB, cleaved from a larger preprotein, were identified in tobacco (Figure 1) which do not bear any sequence similarity to systemin [77]. Both SYST and HSY induce a JA-mediated signaling pathway that leads to the activation of defense-related genes [77, 78].
Of particular note, as in the case of AtPEPRs, systemin cognate receptors (SR160) were detected in all plant species under investigation (Figure 1). It would be intriguing to suggest that PEPR and SR160 genes were probably propagated through vertical gene transfer in plants, and the peptides that specifically bind to them evolved later in certain species in order to carry out species-specific functions.
Protein fragments, such as fibronectin fragments (FN-fs), are the equivalents of the plant polypeptide-based DAMPs in mammals, and they are perceived by Toll-like receptors (TLRs), like TLR4 [79]. Of note, FN-fs were found to stimulate ROS production in human articular chondrocytes [80].
PEPR1/2 and SR160 are transmembrane (TM) proteins comprised of LRR (Leucine-Rich Repeats) motifs, one TM domain, and one catalytic kinase domain in the cytoplasmic region. PEPRs resemble the mammalian TLRs in terms that they share LRR motifs in their amino-termini, suggesting that the extracellular LRR domain is used for DAMP perception both in plants and mammals [76, 81].
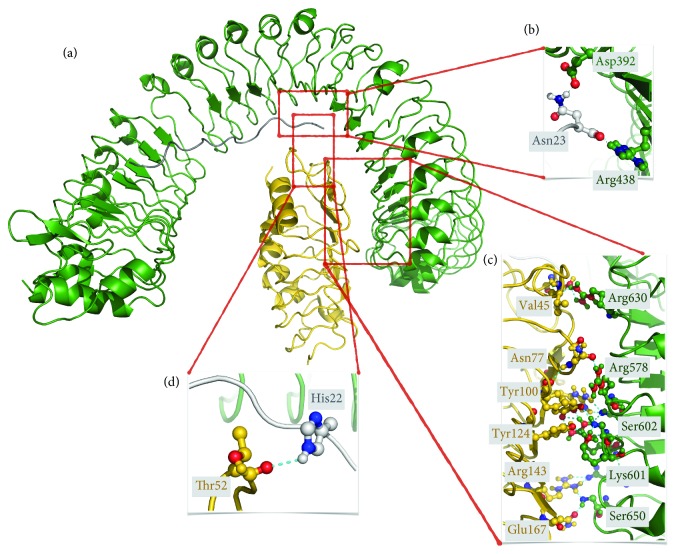
In this study, we aimed to get an evolutionary glimpse into the binding pattern of the A. thaliana PEPR2LRR to AtPEP1. The interaction between PEPR2LRR-AtPEP1 (Figure 7) was also modeled by using the crystal structure of PEPR1LRR-AtPEP1 (PDB ID: 5GR8) as a template. According to 5GR8, there are 16 hydrogen bonds formed between PEPR1LRR and its AtPEP1 peptide [44]. In our PEPR2LRR-AtPEP1 model, this number increases to 17. Two hydrogen bonds, among all, were found to be strictly conserved, i.e., the ones between TYR395-GLN21 and ASP441-ASN23 across the PEPR1LRR-AtPEP1 interface and TYR346-GLN21 and ASP392-ASN23 across the PEPR2LRR-AtPEP1 interface. This reflects the importance of these stabilizing interactions within the PEPR family. The extent of the PEPR1LRR-AtPEP1 and PEPR2LRR-AtPEP1 interfaces appear to be conserved, where 66 amino acids interact over an area of 1033 Å2 in the case of PEPR1LRR-AtPEP1, and 65 amino acids interact through a 1046 Å2-sized interface. PEPR2LRR-BAK1 and its template FLS2LRR-BAK1 (PDB ID: 4MN8) entail a similar number of amino acids across their interfaces (57 for PEPR2LRR-BAK1 and 56 for FLS2LRR-BAK1) (Figure 7). Although this is the case, PEPR2LRR-BAK1 makes more polar interactions (through 12 hydrogen bonds) compared to FLS2LRR-BAK1 (entails 5 hydrogen bonds only), indicating the polar character of the PEPR2LRR surface and a possibly higher affinity between PEPR2LRR-BAK1. Of particular interest, in this study, the structurally important residues TYR395 and ASP441 in AtPEPR2LRR were also found to be conserved in the fellow plant PEPR1/2 primary amino acid sequences, based on multiple sequence alignment. This leads to the suggestion that the Zea mays PEPR1/2LRR could possibly bind to its corresponding ZmPEP1.
Figure 7.
(a) Structural model of the Arabidopsis thaliana PEPR2LRR (green)-AtPEP1 (gray)-BAK1 (gold) complex. Important polar interactions formed across the PEPR2LRR and AtPEP1, PEPR2LRR and BAK1, AtPEP1 and BAK1 interfaces are depicted in (b), (c), and (d), respectively. Polar interactors are represented in ball-and-sticks, with oxygen atoms colored in red and nitrogens in blue.
3.1.4. HMGB3
A relatively novel class of plant DAMPs, the Arabidopsis thaliana HMGB3 (High-Mobility Group Box 3), was shown to induce plant innate immunity, including activation of MAPK3/4/6 and defense-related genes, as well as increased resistance to necrotrophic pathogens. This response is mediated by the LRR-PRR BAK1 (Brassinosteroid insensitive 1-Associated Kinase 1)/SERK3 (Somatic Embryogenesis Receptor-like Kinase 3) and BKK1 (BAK1-like 1)/SERK4 [82].
The mammalian counterpart of the plant HMGB3, HMGB1, is released upon tissue wounding and inflammation [83], as well as response to oxidative [84] and ionizing radiation-induced stress [85]. Release of HMGB1 into the extracellular milieu is associated with the occurrence of severe stress that may have a negative effect on tissue function and homeostasis [85]. HMGB1 is a chromosomal scaffold protein involved in chromatin remodeling induced in response to DNA damage accumulation. It promotes nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR) pathways, facilitating DNA protein kinase (DNA-PK) activity [86]; similarly to the catalytic subunit of DNA-PK, DNA-PKcs, HMGB1 binds CpG oligonucleotides, inducing the TLR9-mediated inflammatory response [87]. Based on protein domain analysis, the human HMGB1 harbors two HMG boxes, BoxA and BoxB, whereas the A. thaliana HMGB proteins have only one HMG box. In the plant species under study, single HMGB homologs were detected and were found to be related to Arabidopsis HMGB1/2/3. This is because the A. thaliana proteins HMGB1, HMGB2, and HMGB3 share a high degree of sequence identity/similarity. HMGB1/2/3-related sequences were detected in all plant species (Figure S1). This finding might represent an interesting starting point to explore more in depth the correlation between DAMPs and DDR also in plants and to provide clues about the degree of conservation, at the trans-kingdom level, of such molecular processes.
Genes encoding SERKs have been identified in the genomes of vascular plants, both monocots and dicots [88]. Furthermore, Sasaki et al. [89] reported the presence of SERK homologs in the less evolved plant liverwort Marchantia polymorpha and even in the unicellular green alga Closterium ehrenbergii. SERKs are ancient, essential genes, conserved during speciation. Multiple SERK genes are found in dicots, monocots, and nonvascular plants. This indicates that SERK genes are at least 450 million years old and were present before the split of nonvascular and vascular plants [90]. Putative SERK-related proteins were detected in the species under investigation (except pea) (Figure 1).
3.2. Conserved Structural Features of Plant Proteins Implicated in Danger Signaling
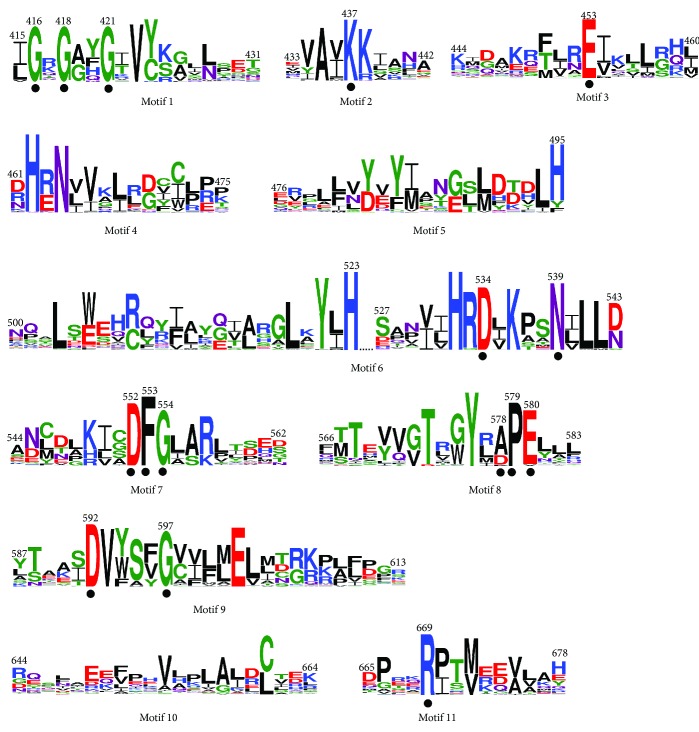
In the present study, the 11 conserved catalytic sequence motifs reported by Hanks et al. [91] were identified in the kinase domain of the plant proteins DORN1, MAPK3, MAPK6, PEPR1/2, SERK3/4/5, SR160, and WAKL investigated in this study (Figure 8). In motif 1, the consensus signature GxGxxG of phosphate binding, conserved across nucleotide binding proteins [92, 93], was detected. The invariant lysine (K) in motif 2 is essential in protein kinase activity as it participates actively in the phosphate transfer reaction [92]. The invariant aspartate (D) and asparagine (N) amino acids in motif 6, as well as the stretch of [94] Asp (D), Phe (F), and Gly (G) residues in motif 7, are actively involved in ATP binding. In motif 8, the highly conserved Glu (E) and invariant Pro (P) were found to be flanked by a conserved alanine (A) residue in [91]. However, in the plant kinase domain, at the same position, Asp (D) was found to be the most frequent residue instead. Phosphorylation of these residues was associated with enhanced catalytic activity in a number of protein kinases [95, 96]. Moreover, several highly conserved amino acids with an unknown role identified by Hanks et al. [91] were also found to be conserved in plant kinases, including Asp (D) in motif 3, Asp (D) and Gly (G) in motif 9, and Arg (R) in motif 11.
Figure 8.
Sequence logos of the conserved protein motifs identified in the catalytic domain of protein kinases. The first and last amino acid residues of each motif are numbered according to A. thaliana WAK1. The conserved/invariant amino acid residues critical for the protein kinase activity are denoted by dots. The height of each letter depicts the relative frequency of the corresponding amino at that position, and the letters are ordered in such a way that the most frequent one is on the top.
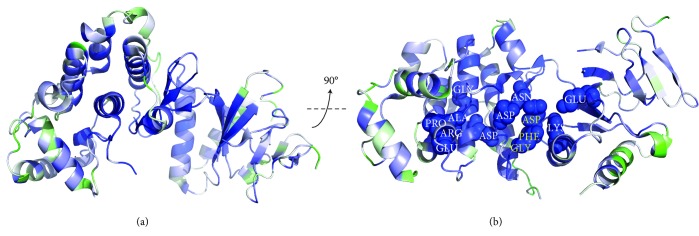
The degree of conservation of the amino acid residues critical for kinase activity is depicted in the three-dimensional structure of A. thaliana MAPK6 [43] (Figure 9). A total 13 out of the 16 amino acids found to be conserved/invariant in the primary structures of the kinase domain of plant proteins (Figure 8) are also shown to be conserved in the tertiary structure of this domain (Figure 9).
Figure 9.
(a) Tertiary structure of the Arabidopsis thaliana MAPK6 (PDB ID: 5CI6), where blue corresponds to the most highly conserved region and green to the least one. The conserved DFG (207-209) motif is highlighted in yellow ((b), at the center of the molecule). The other important conserved residues can be listed as (from left to right) GLY251, ALA 231, PRO232, GLU233, ARG336, ASP246, ASP189, ASN194, GLU110, and LYS92. The numbering is given as in 5CI6.
3.3. Functional Interactome
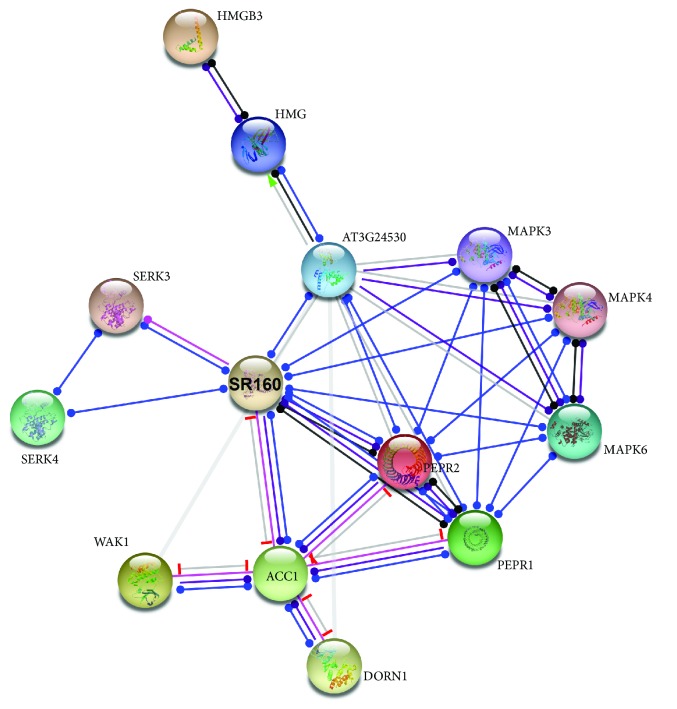
The input A. thaliana molecules involved in plant defense appear to form a dense network (Figure 10), leading to the suggestion that the individual pathways (e.g., DORN1-mediated pathway, etc.) interact into a rather complex plant endogenous danger signal transduction network. For example, the DAMP receptors DORN1, WAK1, PEPR1/2, and SR160 appear to be connected through the multifunctional enzyme ACC1 (acetyl-CoA carboxylase 1), while SERK3/4 interacts directly with SR160. The ubiquitous acetyl-CoA carboxylase is a pivotal enzyme in the synthesis of fatty acids in both eukaryotes and prokaryotes. Both plastid and cytosolic ACC1/2 are nuclear-encoded multidomain enzymes of eukaryotic origin and have central roles in the control of cell division and differentiation and most importantly in many biosynthetic and homeostatic mechanisms [97]. ACC1 catalyzes the key reaction in the biosynthesis of very-long-chain fatty acids giving rise to cuticular waxes, suberin, and sphingolipids, and it is connected to several abiotic and biotic stress response networks [98]. Of note, cuticular waxes play active roles in both local and systemic resistance during plant-pathogen interaction since the plant cuticle is part of the first step of defense pattern-triggered immunity that includes DAMPs [99]. The predicted interaction between ACC1 and the several DAMP receptors might reflect some aspects, still uncovered, of the mechanisms underlying the plant resistance or susceptibility to pathogens. It has been suggested that decreased levels of specific cutin monomers or wax components contribute to the decreased expression of virulence factors or may act as receptors triggering the defense-associated signaling pathway [100]. Further investigation towards elucidating the functional or physical associations between the master regulator of fatty acids ACC1 and the DAMP receptors would provide us with a more detailed picture of the ACC1-DAMP interactome in plants.
Figure 10.
Functional network of the Arabidopsis thaliana molecules involved in DAMP-mediated danger signaling. The nodes represent proteins, and the lines denote edges.
4. Conclusions
It has been only about two decades when the significance of DAMPs for the survival and homeostasis of multicellular organisms under stress conditions has been highlighted. A wide range of molecules have been recognized as DAMPs, and related patterns have been deciphered, whereas a DAMP-like behavior has been evidenced for other molecules. According to Rubartelli [101], the acute phase protein serum amyloid A (SAA) can be considered as a DAMP-like molecule because it is able to prime glial cells and activate the inflammasome in the absence of a pathogen-derived priming stimulus. This response is observed following brain injury and allows to induce the inflammatory response under completely sterile conditions [101]. In plants, a DAMP-like behavior has been described for methanol, a volatile byproduct of pectin methylesterases, rapidly released from the plant cell wall in response to pathogens [102]. Differently from other DAMPs, methanol has no elicitor activity; however, its solvent properties could affect the cell wall and plasma membrane-bound proteins and plasma membrane integrity, resulting in damaged-self signal [102]. These findings further support the apparent complexity of the DAMPs-related processes and functions. Related to more than twenty DAMPs discovered in animals to date, relatively few have been recognized in plants [6].
In our study, we have focused on the well-characterized plant DAMPs. We have performed comprehensive phylogenetics and protein structural analyses in order to identify “true” orthologs between and within the plant and animal kingdom. Our findings support the notion that all efforts to elucidate the pathway(s) through which innate immunity is triggered could likely uncover additional signaling components that are shared by all kingdoms, from plants to humans. The “ancestral” mechanisms of defense are conserved across kingdoms. Several key molecular “players” of those mechanisms appear to be largely conserved (e.g., eATP, calcium, and MAPKs). Some plant proteins involved in endogenous danger signaling were found to have sequence homologs in animals (e.g., MAPKs), whereas most of them were detected exclusively in plants. Both in plants and animals, selective pressure has been likely exerted to maintain the functional integrity of the defense mechanisms by recruiting or coopting into the existing molecular pathway novel players. These players may have no apparent sequence/structural similarity but, through convergent evolution, have rather acquired similar functions, as in the case of DORN1 and P2 purinoreceptors, in plants and animals, respectively.
These findings could provide the foundation for expanding the current knowledge on endogenous danger signaling molecules in plants and their role in adaptive defense mechanisms such as oxidative stress and other types of endogenous or exogenous stress. Of importance, based on the finding of this study, it would be tempting to suggest that plants could serve as basal model systems to study the individual signal transduction pathways implicated in danger signaling and further extrapolate this information to more complex organisms like mammals.
Acknowledgments
A.P. and E.K. would like to extend their thanks to Izmir Biomedicine and Genome Center (IBG), Izmir, Turkey, for its support in implementing the project. Dr. E. Karaca acknowledges partial support from Alexander von Humboldt Foundation Return Fellowship. Dr. A.G. Georgakilas acknowledges funding from the DAAD Grant “DNA Damage and Repair and Their Relevance to Carcinogenesis” (No. 57339330). Dr. A. Balestrazzi acknowledges research supported by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022)-Dept. of Biology and Biotechnology “L. Spallanzani,” Università degli Studi di Pavia (to A.B.).
Contributor Information
Athanasia Pavlopoulou, Email: athanasia.pavlopoulou@deu.edu.tr.
Alexandros G. Georgakilas, Email: alexg@mail.ntua.gr.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
A.P. and A.G.G. designed and supervised the study. A.P. and E.K. performed data analyses. A.P., E.K., A.B., and A.G.G. wrote the manuscript. All authors reviewed and approved the final manuscript.
Supplementary Materials
Figure S1: a cladogram extracted from NCBI's Taxonomy Database [103] depicting the evolutionary relationships among plant species. Table S1: accession numbers of the protein sequences investigated in the present study.
References
- 1.Campos M. L., Kang J. H., Howe G. A. Jasmonate-triggered plant immunity. Journal of Chemical Ecology. 2014;40(7):657–675. doi: 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296(5566):301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 3.Land W. G. Damage-Associated Molecular Patterns in Human Diseases: Volume 1: Injury-Induced Innate Immune Responses. Springer; 2018. [DOI] [Google Scholar]
- 4.Garg A. D., Galluzzi L., Apetoh L., et al. Molecular and translational classifications of DAMPs in immunogenic cell death. Frontiers in Immunology. 2015;6:p. 588. doi: 10.3389/fimmu.2015.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gust A. A., Pruitt R., Nurnberger T. Sensing danger: key to activating plant immunity. Trends in Plant Science. 2017;22(9):779–791. doi: 10.1016/j.tplants.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Choi H. W., Klessig D. F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biology. 2016;16(1):p. 232. doi: 10.1186/s12870-016-0921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matzinger P. Tolerance, danger, and the extended family. Annual Review of Immunology. 1994;12(1):991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 8.Heil M., Land W. G. Danger signals – damaged-self recognition across the tree of life. Frontiers in Plant Science. 2014;5:p. 578. doi: 10.3389/fpls.2014.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lorenzo G., Ferrari S., Cervone F., Okun E. Extracellular damps in plants and mammals: immunity, tissue damage and repair. Trends in Immunology. 2018;39(11):937–950. doi: 10.1016/j.it.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Son Y., Cheong Y. K., Kim N. H., Chung H. T., Kang D. G., Pae H. O. Mitogen-activated protein kinases and reactive oxygen species: how can ros activate MAPK pathways? Journal of Signal Transduction. 2011;2011:6. doi: 10.1155/2011/792639.792639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalmi S. K., Sinha A. K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Frontiers in Plant Science. 2015;6:p. 769. doi: 10.3389/fpls.2015.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat A., Ryu C. M. Plant perceptions of extracellular DNA and RNA. Molecular Plant. 2016;9(7):956–958. doi: 10.1016/j.molp.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Gallucci S., Maffei M. E. DNA sensing across the tree of life. Trends in Immunology. 2017;38(10):719–732. doi: 10.1016/j.it.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Heil M., Vega-Muñoz I. Chapter Five - Nucleic acid sensing in mammals and plants: facts and caveats. International Review of Cell and Molecular Biology. 2019;345:225–285. doi: 10.1016/bs.ircmb.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Hardarson H. S., Baker J. S., Yang Z., et al. Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. American Journal of Physiology-Heart and Circulatory Physiology. 2007;292(1):H251–H258. doi: 10.1152/ajpheart.00398.2006. [DOI] [PubMed] [Google Scholar]
- 16.Krug A., French A. R., Barchet W., et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21(1):107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Lund J. M., Alexopoulou L., Sato A., et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(15):5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Means T. K., Latz E., Hayashi F., Murali M. R., Golenbock D. T., Luster A. D. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. Journal of Clinical Investigation. 2005;115(2):407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Brann T. W., Zhou M., et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. The Journal of Immunology. 2011;186(8):4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duran-Flores D., Heil M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain, Behavior, and Immunity. 2018;72:78–88. doi: 10.1016/j.bbi.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Vega-Muñoz I., Feregrino-Pérez A. A., Torres-Pacheco I., Guevara-González R. G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Functional Plant Biology. 2018;45(10):1065–1072. doi: 10.1071/FP18011. [DOI] [PubMed] [Google Scholar]
- 22.Hadwiger L. A., Tanaka K. Non-host resistance: DNA damage is associated with SA signaling for induction of PR genes and contributes to the growth suppression of a pea pathogen on pea endocarp tissue. Frontiers in Plant Science. 2017;8:p. 446. doi: 10.3389/fpls.2017.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadwiger L. A., Tanaka K. DNA damage and chromatin conformation changes confer nonhost resistance: a hypothesis based on effects of anti-cancer agents on plant defense responses. Frontiers in Plant Science. 2018;9, article 1056 doi: 10.3389/fpls.2018.01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamauchi Y., Matsuda A., Matsuura N., Mizutani M., Sugimoto Y. Transcriptome analysis of Arabidopsis thaliana treated with green leaf volatiles: possible role of green leaf volatiles as self-made damage-associated molecular patterns. Journal of Pesticide Science. 2018;43(3):207–213. doi: 10.1584/jpestics.D18-020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidari-Soreshjani S., Asadi-Samani M., Yang Q., Saeedi-Boroujeni A. Phytotherapy of nephrotoxicity-induced by cancer drugs: an updated review. Journal of Nephropathology. 2017;6(3):254–263. doi: 10.15171/jnp.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieterse C. M. J., Leon-Reyes A., van der Ent S., van Wees S. C. M. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5(5):308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 27.The UniProt Consortium. Uniprot: The universal protein knowledgebase. Nucleic Acids Research. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson D. A., Cavanaugh M., Clark K., et al. Genbank. Nucleic Acids Research. 2018;46(D1):D41–D47. doi: 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K., Rozewicki J., Yamada K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guindon S., Lethiec F., Duroux P., Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Research. 2005;33(Web Server):W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gascuel O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14(7):685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- 33.Jones D. T., Taylor W. R., Thornton J. M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 34.Huson D. H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Systematic Biology. 2012;61(6):1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I., Bork P. 20 years of the smart protein domain annotation resource. Nucleic Acids Research. 2018;46(D1):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A., Bryant S. H. CD-search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32(Web Server):W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parry-Smith D. J., Payne A. W., Michie A. D., Attwood T. K. Cinema—a novel colour interactive editor for multiple alignments. Gene. 1998;221(1):GC57–GC63. doi: 10.1016/s0378-1119(97)00650-1. [DOI] [PubMed] [Google Scholar]
- 39.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. Weblogo: a sequence logo generator. Genome Research. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei J., Grishin N. V. Promals3d: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. In: Russell D., editor. Multiple Sequence Alignment Methods. Vol. 1079. Totowa, NJ, USA: Humana Press; 2014. pp. 263–271. (Methods in molecular biology (Methods and Protocols)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei J., Kim B. H., Grishin N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Research. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashkenazy H., Abadi S., Martz E., et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research. 2016;44(W1):W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B., Qin X., Wu J., et al. Analysis of crystal structure of Arabidopsis MPK6 and generation of its mutants with higher activity. Scientific Reports. 2016;6(1, article 25646) doi: 10.1038/srep25646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J., Han Z., Sun Y., Zhang H., Gong X., Chai J. Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Research. 2015;25(1):110–120. doi: 10.1038/cr.2014.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research. 2015;43(W1):W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laskowski R. A., Rullmann J. C., MacArthur M. W., Kaptein R., Thornton J. M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR. 1996;8(4):477–486. doi: 10.1007/bf00228148. [DOI] [PubMed] [Google Scholar]
- 47.Ye Y., Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Research. 2004;32(Web Server):W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonvin A. M. J. J., Karaca E., Kastritis P. L., Rodrigues J. P. G. L. M. Defining distance restraints in haddock. Nature Protocols. 2018;13(7):p. 1503. doi: 10.1038/s41596-018-0017-6. [DOI] [PubMed] [Google Scholar]
- 49.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Denoux C., Galletti R., Mammarella N., et al. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant. 2008;1(3):423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Droillard M. J., Thibivilliers S., Cazale A. C., Barbier-Brygoo H., Lauriere C. Protein kinases induced by osmotic stresses and elicitor molecules in tobacco cell suspensions: two crossroad map kinases and one osmoregulation-specific protein kinase. FEBS Letters. 2000;474(2-3):217–222. doi: 10.1016/S0014-5793(00)01611-2. [DOI] [PubMed] [Google Scholar]
- 52.Galletti R., Ferrari S., De Lorenzo G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea . Plant Physiology. 2011;157(2):804–814. doi: 10.1104/pp.111.174003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brutus A., Sicilia F., Macone A., Cervone F., de Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decreux A., Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant and Cell Physiology. 2005;46(2):268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- 55.Claverie J., Balacey S., Lemaître-Guillier C., et al. The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana . Frontiers in Plant Science. 2018;9, article 1725 doi: 10.3389/fpls.2018.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang D., Liang J., Noble P. W. Hyaluronan in tissue injury and repair. Annual Review of Cell and Developmental Biology. 2007;23(1):435–461. doi: 10.1146/annurev.cellbio.23.090506.123337. [DOI] [PubMed] [Google Scholar]
- 57.Casalino-Matsuda S. M., Monzon M. E., Day A. J., Forteza R. M. Hyaluronan fragments/CD44 mediate oxidative stress–induced MUC5B up-regulation in airway epithelium. American Journal of Respiratory Cell and Molecular Biology. 2009;40(3):277–285. doi: 10.1165/rcmb.2008-0073OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kyriakis J. M., Avruch J. Mammalian mapk signal transduction pathways activated by stress and inflammation: a 10-year update. Physiological Reviews. 2012;92(2):689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 59.Wolfe K. H., Gouy M., Yang Y. W., Sharp P. M., Li W. H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(16):6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He Z. H., Cheeseman I., He D., Kohorn B. D. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of arabidopsis. Plant Molecular Biology. 1999;39(6):1189–1196. doi: 10.1023/A:1006197318246. [DOI] [PubMed] [Google Scholar]
- 61.Cao Y., Tanaka K., Nguyen C. T., Stacey G. Extracellular ATP is a central signaling molecule in plant stress responses. Current Opinion in Plant Biology. 2014;20:82–87. doi: 10.1016/j.pbi.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka K., Choi J., Cao Y., Stacey G. Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Frontiers in Plant Science. 2014;5:p. 446. doi: 10.3389/fpls.2014.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi J., Tanaka K., Cao Y., et al. Identification of a plant receptor for extracellular ATP. Science. 2014;343(6168):290–294. doi: 10.1126/science.343.6168.290. [DOI] [PubMed] [Google Scholar]
- 64.Burnstock G., Boeynaems J. M. Purinergic signalling and immune cells. Purinergic Signalling. 2014;10(4):529–564. doi: 10.1007/s11302-014-9427-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi J., Tanaka K., Liang Y., Cao Y., Lee S. Y., Stacey G. Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis . Biochemical Journal. 2014;463(3):429–437. doi: 10.1042/BJ20140666. [DOI] [PubMed] [Google Scholar]
- 66.Tripathi D., Zhang T., Koo A. J., Stacey G., Tanaka K. Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiology. 2018;176(1):511–523. doi: 10.1104/pp.17.01477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad S., Ahmad A., Ghosh M., Leslie C. C., White C. W. Extracellular ATP-mediated signaling for survival in hyperoxia-induced oxidative stress. Journal of Biological Chemistry. 2004;279(16):16317–16325. doi: 10.1074/jbc.M313890200. [DOI] [PubMed] [Google Scholar]
- 68.Cruz C. M., Rinna A., Forman H. J., Ventura A. L. M., Persechini P. M., Ojcius D. M. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. Journal of Biological Chemistry. 2007;282(5):2871–2879. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoshida K., Ito M., Matsuoka I. Divergent regulatory roles of extracellular ATP in the degranulation response of mouse bone marrow-derived mast cells. International Immunopharmacology. 2017;43:99–107. doi: 10.1016/j.intimp.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 70.Burnstock G., Kennedy C. Is there a basis for distinguishing two types of P2-purinoceptor? General Pharmacology: The Vascular System. 1985;16(5):433–440. doi: 10.1016/0306-3623(85)90001-1. [DOI] [PubMed] [Google Scholar]
- 71.Wareham K. J., Seward E. P. P2X7 receptors induce degranulation in human mast cells. Purinergic Signalling. 2016;12(2):235–246. doi: 10.1007/s11302-016-9497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Albert M. Peptides as triggers of plant defence. Journal of Experimental Botany. 2013;64(17):5269–5279. doi: 10.1093/jxb/ert275. [DOI] [PubMed] [Google Scholar]
- 73.Marmiroli N., Maestri E. Plant peptides in defense and signaling. Peptides. 2014;56:30–44. doi: 10.1016/j.peptides.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 74.Huffaker A., Pearce G., Ryan C. A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(26):10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huffaker A., Dafoe N. J., Schmelz E. A. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiology. 2011;155(3):1325–1338. doi: 10.1104/pp.110.166710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krol E., Mentzel T., Chinchilla D., et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. Journal of Biological Chemistry. 2010;285(18):13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pearce G., Strydom D., Johnson S., Ryan C. A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science. 1991;253(5022):895–897. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 78.Pearce G., Ryan C. A. Systemic signaling in tomato plants for defense against herbivores: Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. Journal of Biological Chemistry. 2003;278(32):30044–30050. doi: 10.1074/jbc.M304159200. [DOI] [PubMed] [Google Scholar]
- 79.Okamura Y., Watari M., Jerud E. S., et al. The extra domain A of fibronectin activates Toll-like receptor 4. Journal of Biological Chemistry. 2001;276(13):10229–10233. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 80.Del Carlo M., Schwartz D., Erickson E. A., Loeser R. F. Endogenous production of reactive oxygen species is required for stimulation of human articular chondrocyte matrix metalloproteinase production by fibronectin fragments. Free Radical Biology and Medicine. 2007;42(9):1350–1358. doi: 10.1016/j.freeradbiomed.2007.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsushima N., Tanaka T., Enkhbayar P., et al. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007;8(1):p. 124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi H. W., Manohar M., Manosalva P., Tian M., Moreau M., Klessig D. F. Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid. PLoS Pathogens. 2016;12(3, article e1005518) doi: 10.1371/journal.ppat.1005518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lotze M. T., Tracey K. J. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nature Reviews Immunology. 2005;5(4):331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 84.Tang D., Kang R., Zeh H. J., III, Lotze M. T. High-mobility group box 1, oxidative stress, and disease. Antioxidants & Redox Signaling. 2011;14(7):1315–1335. doi: 10.1089/ars.2010.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mavragani I. V., Laskaratou D. A., Frey B., et al. Key mechanisms involved in ionizing radiation-induced systemic effects. A current review. Toxicology Research. 2016;5(1):12–33. doi: 10.1039/C5TX00222B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lange S. S., Vasquez K. M. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Molecular Carcinogenesis. 2009;48(7):571–580. doi: 10.1002/mc.20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ivanov S., Dragoi A. M., Wang X., et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 2007;110(6):1970–1981. doi: 10.1182/blood-2006-09-044776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.aan den Toorn M., Albrecht C., de Vries S. On the origin of SERKs: bioinformatics analysis of the somatic embryogenesis receptor kinases. Molecular Plant. 2015;8(5):762–782. doi: 10.1016/j.molp.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 89.Sasaki G., Katoh K., Hirose N., et al. Multiple receptor-like kinase cDNAs from liverwort Marchantia polymorpha and two charophycean green algae, Closterium ehrenbergii and Nitella axillaris: extensive gene duplications and gene shufflings in the early evolution of streptophytes. Gene. 2007;401(1-2):135–144. doi: 10.1016/j.gene.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 90.Bowman J. L., Floyd S. K., Sakakibara K. Green genes—comparative genomics of the green branch of life. Cell. 2007;129(2):229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 91.Hanks S. K., Quinn A. M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 92.Carrera A. C., Alexandrov K., Roberts T. M. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring atp. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(2):442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng Z., Goncearenco A., Berezovsky I. N. Nucleotide binding database NBDB – a collection of sequence motifs with specific protein-ligand interactions. Nucleic Acids Research. 2016;44(D1):D301–D307. doi: 10.1093/nar/gkv1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Denhez F., Galibert F., Heimann B., et al. An invariant asparagine residue belonging to a highly conserved domain in all protein kinases is instrumental in the protein kinase activity of the v-mil gene product. Oncogene Research. 1988;3(2):199–205. [PubMed] [Google Scholar]
- 95.Ellis L., Clauser E., Morgan D. O., Edery M., Roth R. A., Rutter W. J. Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell. 1986;45(5):721–732. doi: 10.1016/0092-8674(86)90786-5. [DOI] [PubMed] [Google Scholar]
- 96.Weinmaster G., Zoller M. J., Smith M., Hinze E., Pawson T. Mutagenesis of Fujinami sarcoma virus: evidence that tyrosine phosphorylation of P130gag-fps modulates its biological activity. Cell. 1984;37(2):559–568. doi: 10.1016/0092-8674(84)90386-6. [DOI] [PubMed] [Google Scholar]
- 97.Faure J. D., Vittorioso P., Santoni V., et al. The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development. 1998;125(5):909–918. doi: 10.1242/dev.125.5.909. [DOI] [PubMed] [Google Scholar]
- 98.Lu S., Zhao H., Parsons E. P., et al. The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiology. 2011;157(3):1079–1092. doi: 10.1104/pp.111.185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aragon W., Reina-Pinto J. J., Serrano M. The intimate talk between plants and microorganisms at the leaf surface. Journal of Experimental Botany. 2017;68(19):5339–5350. doi: 10.1093/jxb/erx327. [DOI] [PubMed] [Google Scholar]
- 100.Ziv C., Zhao Z., Gao Y. G., Xia Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Frontiers in Plant Science. 2018;9, article 1088 doi: 10.3389/fpls.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rubartelli A. DAMP-mediated activation of NLRP3-inflammasome in brain sterile inflammation: the fine line between healing and neurodegeneration. Frontiers in Immunology. 2014;5:p. 99. doi: 10.3389/fimmu.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hann C. T., Bequette C. J., Dombrowski J. E., Stratmann J. W. Methanol and ethanol modulate responses to danger- and microbe-associated molecular patterns. Frontiers in Plant Science. 2014;5:p. 550. doi: 10.3389/fpls.2014.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sayers E. W., Barrett T., Benson D. A., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2012;40(D1):D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: a cladogram extracted from NCBI's Taxonomy Database [103] depicting the evolutionary relationships among plant species. Table S1: accession numbers of the protein sequences investigated in the present study.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.