Abstract
Infant gut-associated bifidobacteria has a metabolic pathway that specifically utilizes lacto-N-biose I (Gal-β1,3-GlcNAc) and galacto-N-biose (Gal-β1,3-GalNAc) from human milk and mucin glycans. UDP-glucose 4-epimerase (GalE) from Bifidobacterium longum (bGalE) catalyzes epimerization reactions of UDP-Gal into UDP-Glc and UDP-GalNAc into UDP-GlcNAc with the same level of activity that is required to send galacto-hexoses into glycolysis. Here, we determined the crystal structures of bGalE in three ternary complex forms: NAD+/UDP, NAD+/UDP-GlcNAc, and NAD+/UDP-Glc. The broad specificity of bGalE was explained by structural features of the binding pocket for the N-acetyl or C2 hydroxy group of the substrate. Asn200 is located in a pocket of the C2 group, and its side chain adopts different conformations in the complex structures with UDP-Glc and UDP-GlcNAc. On the other side, Cys299 forms a large pocket for the C5 sugar ring atom. The flexible C2 pocket and the large C5 pocket of bGalE are suitable for accommodating both the hydroxy and N-acetyl groups of the substrate during sugar ring rotation in the catalytic cycle. The substrate specificity and active site structure of bGalE were distinct from those of Esherichia coli GalE but similar to those of human GalE.
Subject terms: Enzyme mechanisms, Bacteria, X-ray crystallography
Introduction
Bifidobacteria are anaerobic gram-positive bacteria belonging to the genus Bifidobacterium and are usually found in the gastrointestinal tract of humans and animals1. It is known that they play a significant role in human health by acting with various transporters, glycosidases, and metabolic enzymes2,3. Several species of Bifidobacterium, e.g., Bifidobacterium longum subsp. longum (B. longum), Bifidobacterium bifidum, Bifidobacterium breve, and Bifidobacterium longum subsp. infantis, are the predominant intestinal bacteria in breastfed infants4–6. Recent studies revealed that the infant gut-associated bifidobacteria has specialized metabolic pathways for utilizing oligosaccharides contained in human milk with a considerably large amount (up to 12–24 g/L), suggesting a symbiotic relationship between humans and these microorganisms7–9. The human milk oligosaccharides (HMOs) consist of more than 240 different kinds of molecular species with a degree of polymerization of more than three10. The structural unit of lacto-N-biose I (Gal-β1,3-GlcNAc, LNB) is predominantly present in HMOs but not in milk of other mammals11, and LNB has been shown to be the bifidus factor, which promotes growth of infant gut-associated bifidobacteria12,13. The metabolic pathway dedicated for utilization of LNB also utilizes galacto-N-biose (Gal-β1,3-GalNAc, GNB) that is present in glycoconjugates, such as mucin glycoproteins14,15. The gene cluster for the GNB/LNB pathway in bifidobacteria is composed of a specific ABC transporter and four intracellular enzymes, GNB/LNB phosphorylase (GLNBP, EC 2.4.1.211; locus tag = BLLJ_1623), N-acetylhexosamine 1-kinase (NahK, EC 2.7.1.162; BLLJ_1622), UDP-glucose—hexose-1-phosphate uridylyltransferase (GalT, EC 2.7.7.12; BLLJ_1621), and UDP-glucose 4-epimerase (GalE, EC 5.1.3.2; BLLJ_1620). Of note, here we use the accepted name of GalE recommended by Nomenclature Committee of IUBMB-IUPAC, although the other name “UDP-galactose 4-epimerase” has been generally used in the structural biology community until recently. We previously reported crystal structures of the solute-binding protein of the GNB/LNB-specific transporter16, GLNBP17, and NahK18. Here, we focused on structural features of GalE from B. longum JCM1217 (bGalE).
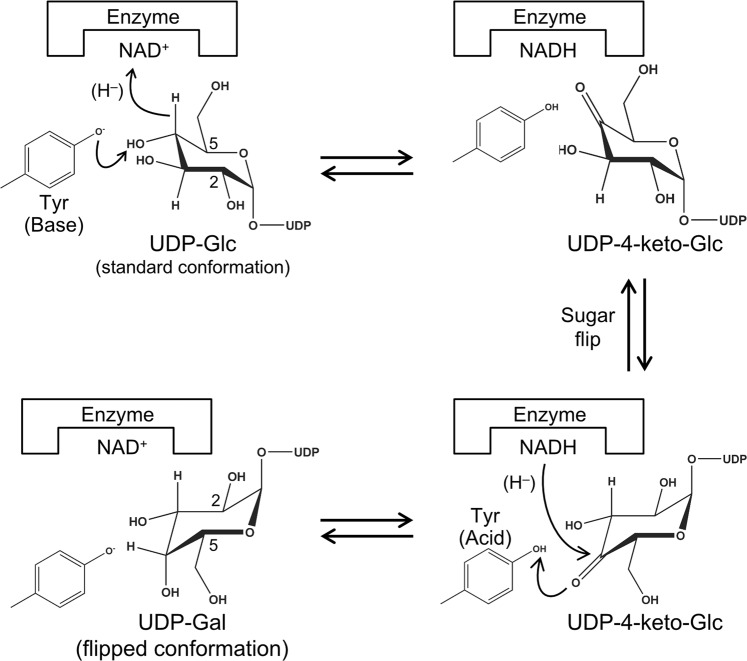
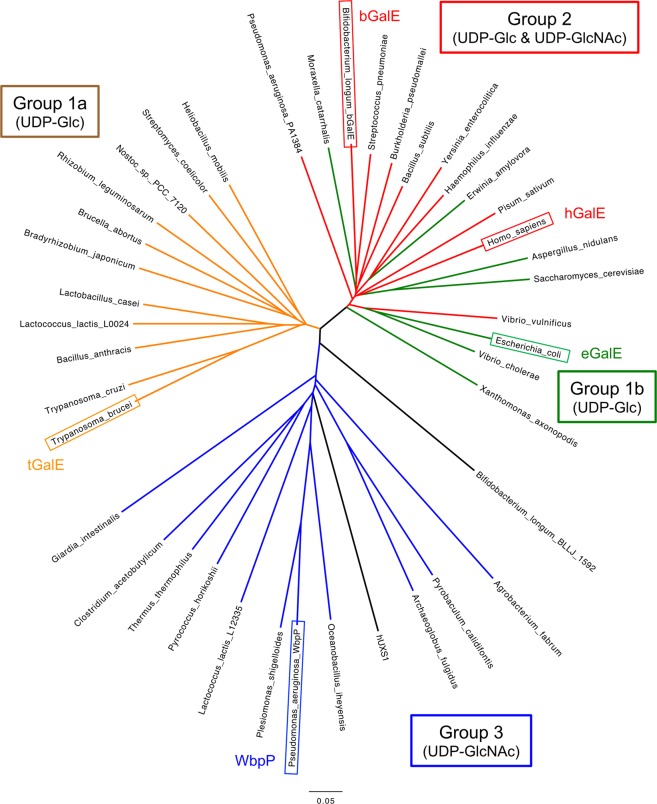
GalE catalyzes NAD+-dependent oxidoreductive interconversion of gluco- and galacto-hexoses (C4-epimerization) linked to UDP (Fig. 1) and generally plays a key role in the metabolism of galactose in various organisms19–21. Crystal structures of GalEs from Escherichia coli (eGalE)22–27, Trypanosoma brucei (tGalE)28, human (hGalE)29–31, Pseudomonas aeruginosa (WbpP)32, Pyrobaculum calidifontis33, Aspergillus nidulans34, and several others have been reported. Ishiyama et al. classified GalE enzymes into three major groups based on their substrate specificity32. Group 1 enzymes preferentially catalyze the epimerization between UDP-Glc and UDP-Gal (eGalE and tGalE), group 2 enzymes do not show a preference for either UDP-Glc/UDP-Gal or UDP-GlcNAc/UDP-GalNAc (hGalE), and group 3 enzymes preferentially catalyze the epimerization between UDP-GlcNAc and UDP-GalNAc (WbpP) (Table 1). Group 1 enzymes are further divided into two subfamilies (1a and 1b) due to their separation in the phylogenetic tree (Fig. 2). bGalE catalyzes epimerization reactions of UDP-Gal into UDP-Glc and UDP-GalNAc into UDP-GlcNAc with the same level of activity and is categorized into group 215. Amino acid sequence identities between bGalE and the representative of each group are as follows: hGalE (group 2) = 54.0%, eGalE (group 1b) = 54.3%, tGalE (group 1a) = 37.5%, and WbpP (group 3) = 24.3%. GalEs are classified as SDR1E family of a large short-chain dehydrogenases/reductase (SDR) superfamily, which includes oxidoreductases, epimerases, and lyases35. In this study, we describe the crystal structures of bGalE and compare them with GalEs from other organisms.
Figure 1.
Proposed reaction mechanism of GalE. The catalytic base/acid tyrosine residue of bGalE is Tyr150. The NAD+ cofactor is tightly bound to the protein as a prosthetic group and facilitates catalysis via hydride transfer.
Table 1.
Enzyme activities of GalE enzymes.
| UDP-Glc | UDP-Gal | UDP-GlcNAc | UDP-GalNAc | Note | Ref. | |
|---|---|---|---|---|---|---|
| bGalE | — | 158 | 147 | — | Specific activity (U/mg) at 1 mM UDP-sugar | 15 |
| hGalE | — | 33.8 | — | 26 | Specific activity (U/mg) at 0.41 mM UDP-Gal or 0.66 mM UDP-GalNAc | 27 |
|
hGalE (C307Y) |
1.21 | 0.02 | Relative activity compared with the wild type enzyme at 2 mM UDP-Gal or 1.89 mM UDP-GalNAc | 43 | ||
| eGalE | — | 23.9 | — | 0.003 | Specific activity (U/mg) at 0.41 mM UDP-Gal or 0.66 mM UDP-GalNAc | 27 |
|
eGalE (Y299C) |
— | 5.1 | — | 0.69 | ||
| tGalE | — | 5.9 | — | — | Specific activity (U/mg) at 20 mM UDP-Gal | 54 |
| WbpP | 0.124 | 0.188 | 120 | 271 | kcat (min−1). Km values were similar for all substrates (197~251 uM) | 55 |
—, Not determined.
Figure 2.
Phylogenetic tree of GalEs and UDP-xylose synthase. Source organisms of biochemically and structurally characterized GalE enzymes and those of uncharacterized protein sequences are shown. The group classification and substrate specificity prediction are performed according to the method described in Ishiyama et al.32. Human UDP-xylose synthase 1 (hUXS1) is also shown. Bar, 5% sequence divergence.
Results and Discussion
Overall structure
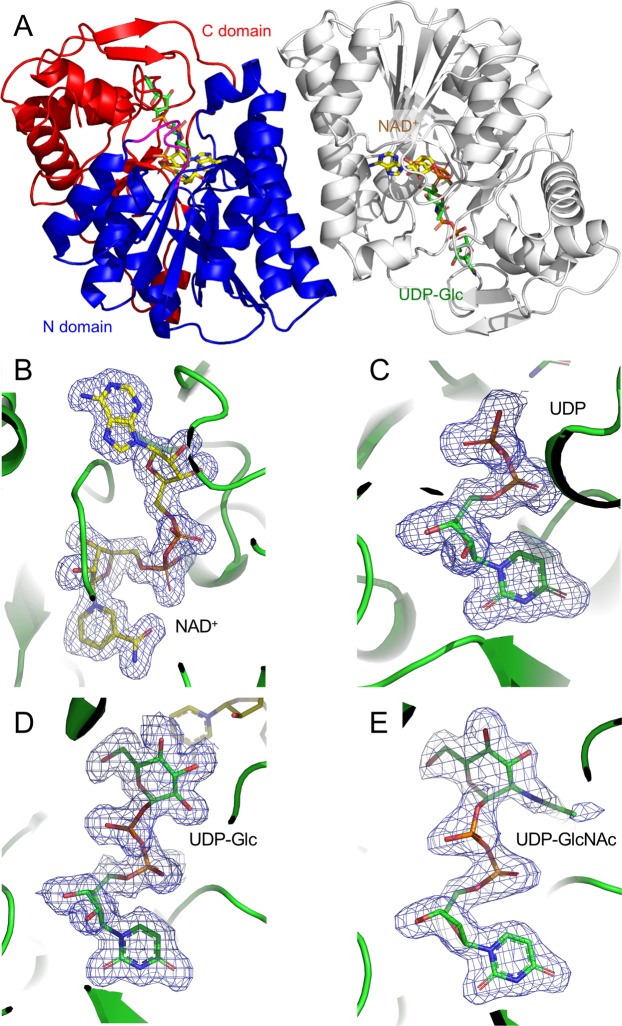
The molecular masses of the purified recombinant bGalE protein (348 amino acids) as deduced from the amino acid sequence, estimated by SDS-PAGE and gel filtration chromatography were 38.3, 38, and 68 kDa, respectively, suggesting that this protein is dimeric in solution. Crystals of bGalE belong to the P6522 space group and contain one monomer per asymmetric unit. Since the recombinant bGalE protein contains one NAD+ molecule per monomer as a noncovalently bound prosthetic group, we prepared ternary complex form crystals with UDP, UDP-Glc, or UDP-GlcNAc by cocrystallization and determined their crystal structures at 1.8–2.0 Å resolution (Table 2). A plausible dimer assembly formed by a crystallographic 2-fold rotation symmetry is present in the crystal packing (Fig. 3A). A molecular interface analysis using PISA36 indicates that the whole surface area, buried area, and ΔGint value (the solvation free energy gain upon formation of the assembly) are 23,790 Å2, 6,340 Å2, and –44.7 kcal/mol, respectively. The dimer interface consists of a bundle of 4 α-helices and contains 8 hydrogen bonds and 4 salt bridges. bGalE has a typical fold of SDR superfamily35. The overall monomer protein structure can be roughly divided into an N-terminal NAD+ binding domain (N domain, residues 1–177 and 236–262) and a C-terminal UDP-sugar binding domain (C domain, residues 178–235 and 263–340). The N domain adopts a typical Rossmann fold composed of a seven-stranded parallel β-sheet flanked by nine α-helices. The C domain possesses an α/β motif consisting of four α-helices and four β-strands. The three ternary complex structures have virtually the same main chain structures because the root mean square deviation (RMSD) values for the Cα atoms (no distance cutoff) between them are less than 0.18 Å. The main chain structure of bGalE is similar to those of bacterial and eukaryotic GalEs. Cα RMSD values (distance cutoff = 2.0 Å) of the UDP-GlcNAc complex with hGalE (PDB ID 1HZJ, chain B), eGalE (PDB ID 1XEL), tGalE (PDB iD 2CNB, chain A), and WbpP (PDB ID 1SB8) are 0.69 Å (306 a.a.), 0.88 Å (284 a.a.), 0.72 Å (239 a.a.), and 1.41 Å (231 a.a.), respectively.
Table 2.
Data collection and refinement statistics.
| UDP + NAD+ | UDP-GlcNAc + NAD+ | UDP-Glc + NAD+ | |
|---|---|---|---|
| Data collectiona | |||
| Beamline | BL17A | BL5A | NW12A |
| Wavelength | 0.979 | 1.000 | 1.000 |
| Space group | P6522 | P6522 | P6522 |
| Unit cell (Å) | a = b = 70.1, c = 322.2 | a = b = 69.4, c = 322.0 | a = b = 70.4, c = 322.6 |
| Resolution (Å)a |
48.50–1.80 (1.84–1.80) |
48.13–2.00 (2.05–2.00) |
48.62–1.80 (1.84–1.80) |
| Total reflections | 952,581 (55,646) | 551,727 (41,132) | 800,942 (45,652) |
| Unique reflections | 45,059 (2,591) | 32,430 (2,323) | 44,487 (2,504) |
| Completenessb | 100.0 (100.0) | 100.0 (100.0) | 98.8 (97.7) |
| Multiplicityb | 21.1 (21.5) | 17.0 (17.7) | 18.0 (18.2) |
| Mean I/σ(I)b | 9.3 (4.0) | 18.1 (3.7) | 24.9 (6.9) |
| Rmergeb | 0.253 (0.947) | 0.133 (0.855) | 0.100 (0.552) |
| CC1/2b | 0.983 (0.961) | 0.993 (0.974) | 0.999 (0.974) |
| Refinementc | |||
| Resolution | 40.17–1.80 | 34.75–2.00 | 36.78–1.80 |
| No. of reflections | 42,417 | 30,845 | 42,233 |
| R factor/Rfree (%) | 14.9 (18.0) | 18.1 (22.1) | 16.1 (19.0) |
| No. of atoms | 3,033 | 2,898 | 2,990 |
| No. of solvents | 1 (UDP), 1 (NAD+), 1 (Mg2+), 375 (water) | 1 (UDP-GlcNAc), 1 (NAD+), 2 (Mg2+), 211 (water) | 1 (UDP-Glc), 1 (NAD+), 1 (Mg2+), 322 (water) |
| RMSD from ideal values | |||
| Bond lengths (Å) | 0.014 | 0.012 | 0.015 |
| Bond angles (°) | 1.73 | 1.66 | 1.77 |
| Ramachandran plot (%) | |||
| Favored | 99.1 | 98.8 | 99.1 |
| Allowed | 0.6 | 0.9 | 0.6 |
| Outlier | 0.3 | 0.3 | 0.3 |
| PDB ID | 6K0G | 6K0H | 6K0I |
aCalculated using XDS.
bValues in parentheses are for the highest resolution shell.
cThe structures were refined using diffraction datasets processed by HKL2000.
Figure 3.
Crystal structure of bGalE. (A) Overall structure of the biological dimer assembly. One protomer is shown in blue (N domain, residues 1–177 and 236–262) and red (C domain, residues 178–235 and 263–340), and the other symmetry-related protomer is shown in gray. mFo-DFc omit map of NAD+ (B) and UDP (C) in the NAD+ + UDP complex, UDP-Glc (D), and UDP-GlcNAc (E) are shown with a contour level of 3.0σ. NAD+ and UDP or UDP-sugar molecules are shown with yellow and green sticks, respectively.
NAD+ binding site
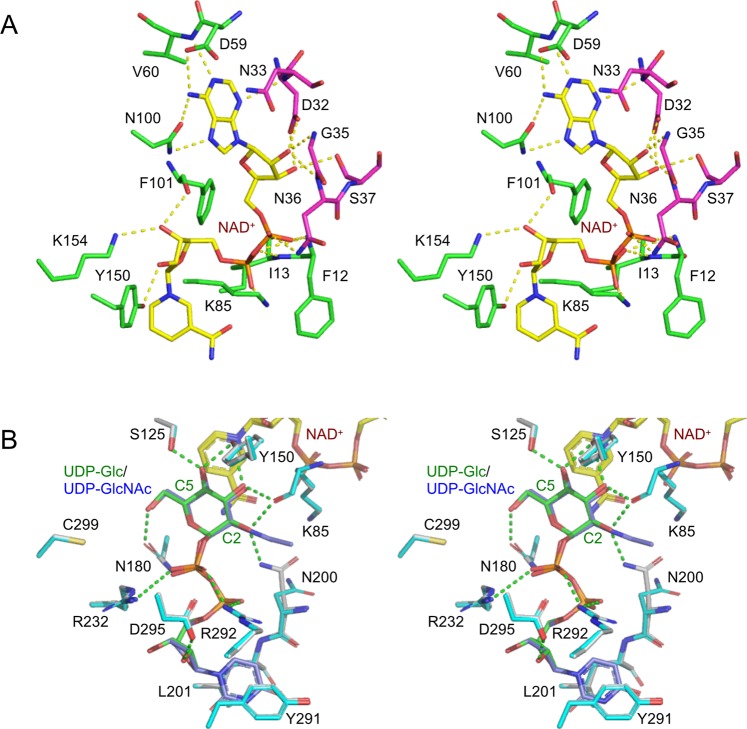
The electron density of the NAD+ molecule bound in the N domain was clearly observed in all three complex structures. Figure 3B shows the mFo-DFc omit map of NAD+ in the UDP complex. The nicotinamide ribose and adenine ribose moieties are in syn-C2′-endo and anti-C2′-endo conformations, respectively. The ribose conformations of NAD+ are identical to those observed in eGalE37 and other GalE enzymes studied so far. Figure 4A shows interactions of the NAD+ molecule with bGalE. Residues recognizing the NAD+ cofactor are basically conserved in GalEs. In bGalE, the nicotinamide ribose moiety interacts with Phe101, Lys154, and Tyr150, the pyrophosphate moiety interacts with Phe12, Ile13, and Lys85, and the adenosine base moiety interacts with Asn33, Asp59, Val60, and Asn100. A loop region (Asp32–Ser37, magenta in Fig. 4A) forms several direct hydrogen bonds with the base, ribose, and phosphate groups of the adenosine moiety.
Figure 4.
Stereoview of the cofactor and substrate binding sites of bGalE. (A) NAD+ binding site of the UDP complex structure. (B) Superimposition of the complex structures with UDP-Glc (white and green for protein and UDP-Glc) and UDP-GlcNAc (cyan and blue for protein and UDP-GlcNAc). NAD+ and hydrogen bonds are shown as yellow sticks and dotted lines, respectively.
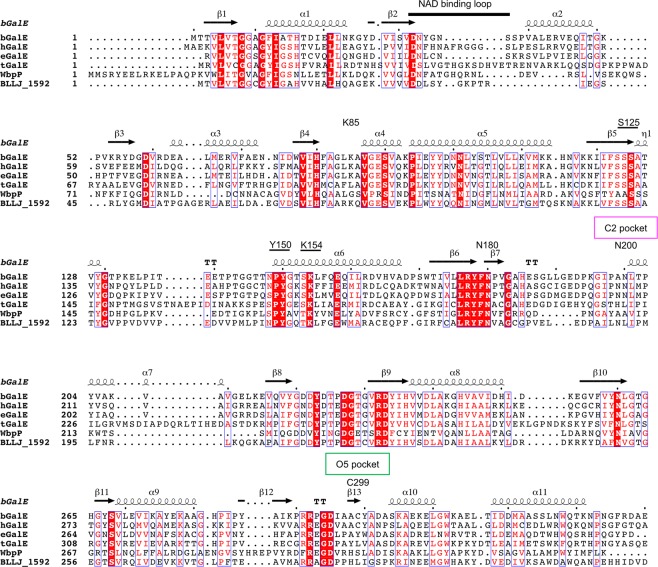
The cofactor NAD+ was intrinsically bound to bGalE without any supplementation throughout experimental processes in all complexes. It was reported that when NAD+ was removed from the GalE proteins, the proteins were irreversibly denatured37. The loop region of Asp32–Ser37 in the N domain interacts with the adenosine moiety of NAD+ (magenta in Fig. 3A). The corresponding region (residues 32–43) in P. calidifontis GalE was defined as the “NAD-binding loop”33. The NAD-binding loop is present in all GalE homologs (Fig. 5). In contrast, l-threonine dehydrogenases (l-ThrDHs), which share similar structural fold with GalEs, lack this loop and easily release NAD+ from the protein38. Sakuraba et al. indicated that the NAD-binding loop of GalEs plays a key role in preventing the release of the catalytically relevant cofactor and contributing to the protein stability of P. calidifontis GalE33.
Figure 5.
Amino acid sequence alignment of GalEs. Representatives of group 2 (bGalE and hGalE), group 1b (eGalE), group 1a (tGalE), group 3 (WbpP), and a paralog of bGalE (BLLJ_1592) are shown. The catalytic triad residues are underlined.
UDP-sugar binding site
As shown in Fig. 3C–E, the electron densities of UDP, UDP-Glc, and UDP-GlcNAc were clearly observed. Interactions of UDP-Glc and UDP-GlcNAc with bGalE are shown in Fig. 4B. Residues recognizing the UDP moiety are basically conserved in GalEs. In bGalE, the UDP moiety is recognized by stacking and hydrophobic interactions by Tyr291 and Leu201, hydrogen bonds by Asp295 and Asn180, and salt bridges by Arg292 and Arg232. These interactions are identically present in the UDP complex (data not shown). The hydroxy groups of the Glc moiety are recognized by hydrogen bonds from the main chain of Lys85 (O2 and O3) and the side chain atoms of Asn200 (O2), Tyr150 (O3 and O4), Ser125 (O4), and Asn180 (O6). The GlcNAc moiety is similarly recognized, but a significant displacement of the side chain of Asn200 was observed. In the UDP-GlcNAc complex, the side chain of Asn200 adopts a distinct conformation (designated as “swing out”) from that in the UDP-Glc complex (“swing in”) to accommodate the 2-N-acetyl group. This phenomenon was also reported for the corresponding residue (Asn207) in the UDP-GlcNAc complex of hGalE30 (discussed below).
The glucopyranose rings of the UDP-sugars in bGalE are both in the “standard” conformation (Fig. 1), in which the H4 atom faces the pro-S side of the NAD+ nicotinamide. The distances between the C4 atom of the sugar ring and the C4 atom of the nicotinamide ring are 3.4 Å and 3.5 Å in the UDP-Glc and UDP-GlcNAc complex structures, respectively. In the proposed reaction mechanism of GalE, a hydride transfer is thought to occur between these carbon atoms, leading to formation of a 4-ketopyranose intermediate and NADH (Fig. 1)37. It has been reported so far that the distance between the C4–C4 atoms in a productive mode is within the range of 3.0 ~ 3.7 Å22,26,29,30,34,39. Tyr150 forms a direct hydrogen bond (2.6 Å) with the O4 atom in both the UDP-Glc and UDP-GlcNAc complex structures. The corresponding residue of hGalE (Tyr157) is the catalytic base residue (proton acceptor) from the O4 hydroxy in the first reaction step (Fig. 1)29. In the subsequent reaction steps, the sugar moiety of the substrate is assumed to flip its sugar ring, and the nicotinamide ring of the reduced cofactor (NADH) transfers back the hydride from the si-face to the opposite face of the C4-keto-intermediate to produce an epimer19. In the latter step, the catalytic tyrosine residue acts as a general acid (proton donor) to the O4 hydroxy group of the sugar moiety. A comprehensive study using X-ray crystallography, in situ NMR, and steady-state and stopped-flow kinetics revealed a detailed reaction mechanism of human UDP-xylose synthase 1 (hUXS1; EC 4.1.1.35; other name: UDP-glucuronic acid decarboxylase), which also belongs to the SDR1E family of SDR superfamily40. In addition to the decarboxylation reaction that forms xylose from glucuronic acid, the proposed reaction mechanism involves base/acid catalysis by Tyr147, which is located at the corresponding position (near the C4 atom of the sugar moiety) to Tyr150 of bGalE.
A completely conserved SYK catalytic triad motif has been recognized in SDR superfamily enzymes35. In bGalE, Ser125, Tyr150, and Lys154 correspond to the triad (Fig. 5). Lys154 contributes to the binding of nicotinamide ribose, while Ser125 and Tyr150 form direct hydrogen bonds with the O3 and O4 hydroxy groups of the Glc or GlcNAc (Fig. 4), respectively. These residues appear to support precise transfer of hydride and proton, which is the most important step of GalE catalysis. For eGalE, site-directed mutagenesis studies indicated the catalytic importance of the triad residues (Ser124, Tyr149, and Lys153)41,42. Structural analyses on mutants of the Ser and Tyr residues were also reported24,26. Tyr149 and Ser124 are in position to participate in the acid-base catalysis required to drive hydride transfer and synergistically contribute to the catalysis by coordinating the reactive O4 hydroxy group20. Asn180, which recognizes the O6 hydroxy of the sugar moiety, is also completely conserved (Fig. 5). The other three residues involved in forming the sugar binding pocket (Lys85, Asn200, and Cys299) exhibit variety in the three GalE groups (1, 2, and 3) and prescribe their substrate specificity.
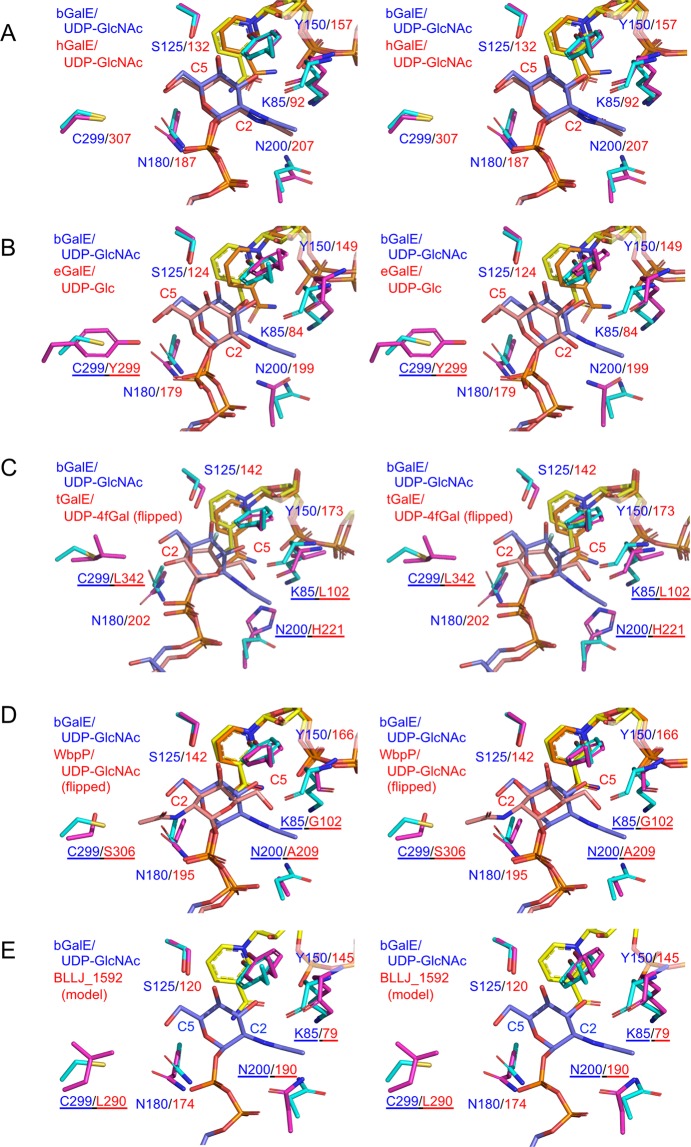
Figure 6 shows superimpositions of the active site of bGalE + UDP-GlcNAc and representative enzymes in group 2 (hGalE), group 1b (eGalE), group 1a (tGalE), and group 3 (WbpP), presenting both the standard (bGalE + UDP-GlcNAc, hGalE + UDP-GlcNAc, and bGalE + UDP-Glc) and the flipped (tGalE + UDP-4-deoxy-4-fluoro-α-galactose (UDP-4fGal) and WbpP + UDP-GlcNAc) conformations of the sugar moiety. The UDP-GlcNAc complex structures of bGalE and hGalE (both in group 2) superimpose very well, and the residues forming the sugar binding site are completely conserved (Figs 5 and 6A). It is noteworthy that most UDP sugars observed in the crystal structures of GalE enzymes in groups 1 and 2 were in the standard conformation. For eGalE and tGalE in group 1, the flipped conformation structure was obtained only by using the S124A/Y149F double mutant26 or by using a substrate analog UDP-4fGal39. On the other hand, for a group 3 enzyme WbpP, the wild-type enzyme bound the natural substrate UDP-GlcNAc in the flipped conformation32.
Figure 6.
Stereoview of the UDP-sugar binding site in GalEs. Superimposition of bGalE + UDP-GlcNAc (cyan, blue, and yellow for protein, UDP-sugar, and NAD+) and other GalE enzymes (magenta, pink, and orange for protein, UDP-sugar, and NAD+) are shown. (A) hGalE + UDP-GlcNAc (PDB ID: 1HZJ chain B), (B) eGalE + UDP-Glc (PDB ID: 1XEL), (C) tGalE + UDP-4fGal (PDB ID: 2CNB chain A), (D) WbpP + UDP-GlcNAc (PDB ID: 1SB8), and (E) a homology model of BLLJ_1592.
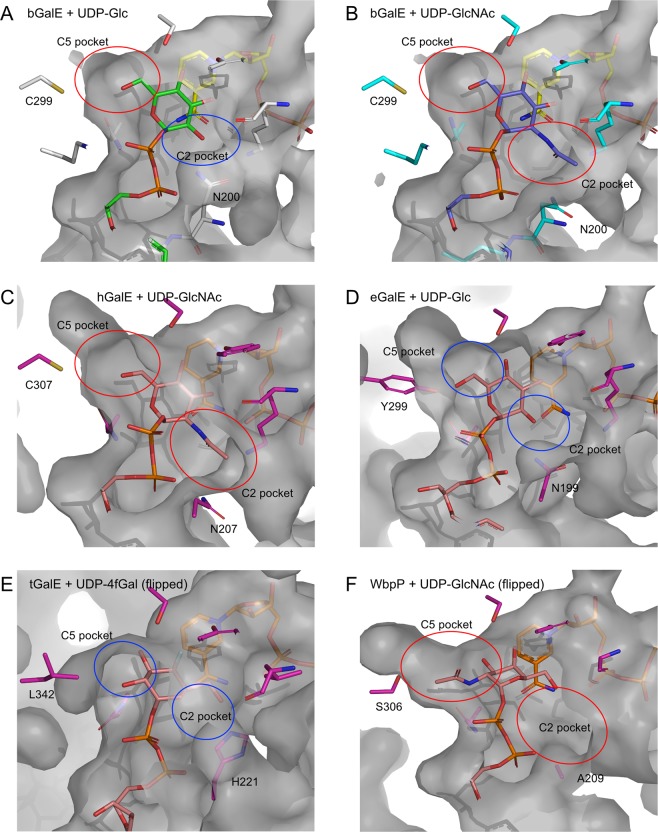
Ishiyama et al. proposed a conceptual model of the GalE active site surrounded by a hexagon-shaped box (six walls) and used it for discussion of the relationship between sequence (active site structure) and substrate specificity32. Here, we simplify the concept and focus on two pockets (walls in the original concept) responsible for the substrate specificity between UDP-Glc/Gal and UDP-GlcNAc/GalNAc. As shown in Fig. 7A,B, Asn200 and Cys299 form pockets for the groups connected to the C2 and C5 sugar ring atoms, respectively, in the “standard” conformation. Therefore, we hereafter designate these structural regions as C2 pocket and C5 pocket. These two pockets can discriminate the size of the C2 hydroxy or N-acetyl groups during the sugar group rotation in the catalytic cycle of the GalE reaction.
Figure 7.
Surface presentation of the UDP-sugar binding site in GalEs. (A) bGalE + UDP-Glc, (B) bGalE + UDP-GlcNAc, (C) hGalE + UDP-GlcNAc, (D) eGalE + UDP-Glc (E) tGalE + UDP-4fGal, and (F) WbpP + UDP-GlcNAc are shown. Color codes and PDB IDs are as described in Figs 4 and 6. The C5 and C2 pockets are shown as red or blue circles for large or small pockets, respectively. The probe radius for surface calculation was 1.4 Å.
It has been shown that the size difference of the C5 pocket is the primary factor for the substrate specificity of GalEs32. Group 2 and 3 enzymes (bGalE, hGalE, and WbpP) have large C5 pocket with small Cys or Ser residue at this position whereas group 1 enzymes (eGalE and tGalE) have small C5 pocket with a bulky Tyr or Leu residue (Fig. 7). Although the crystal structure of eGalE Y299C mutant complexed with UDP-GlcNAc was reported, the sugar moiety was bound in a displaced nonproductive mode, and the distance between the C4 atom of GlcNAc and the C4 atom of nicotinamide ring was 9.4 Å27. The Y299C mutant of eGalE showed 5-fold reduction in the activity toward UDP-Gal, while it exhibited a 230-fold increase in activity toward UDP-GalNAc (Table 1). For hGalE, C307Y mutation resulted in significant loss of activity toward UDP-GalNAc, while the activity toward UDP-Gal was not affected (Table 1)43.
In the C2 pocket, the size and flexibility of the amino acid side chain contributed to accommodation of the N-acetyl group of the substrate. Similar to the case of bGalE observed in the present work, hGalE (group 2) adopts “swing in” and “swing out” conformations in the UDP-Glc and UDP-GlcNAc complex structures, respectively29,30. eGalE (group 1b) also has Asn199 residue at this position (Fig. 7D). However, in the crystal structure of the wild-type eGalE enzyme, the sugar moiety of UDP-GlcNAc was disordered27, suggesting limited flexibility of the Asn199 side chain. We further investigated the protein structures around the Asn residue and C2 pocket of the group 1b and 2 enzymes, but no significant structural feature that contribute to the flexibility difference between them was found (data not shown). The corresponding residues at the C2 pocket are large His221 in tGalE (group 1a specific for UDP-Glc) and small Ala209 in WbpP (group 3 specific for UDP-GlcNAc/GalNAc) (Fig. 7E,F). Therefore, the C2 pocket size is likely to contribute to the substrate specificity in these groups. In addition to the steric hindrance, a hydrogen-bonded solvent (water) network near Ala209 in WbpP is involved in the recognition of the N-acetyl group of the substrate32.
Three-dimensional structures of GalEs from a wide range of organisms are now available in the Protein Data Bank due to structural genomics projects and studies from several groups. Such efforts and advance in protein homology modeling have enabled more precise prediction of GalE specificities than the pioneering work by Ishiyama et al.32. We previously investigated biochemical characteristics of a paralog GalE enzyme from the same organism, B. longum JCM1217 (BLLJ_1592 corresponding to BL1671 of B. longum NCC2705)44, whose gene is not accompanied by other galactose metabolic genes. Although we did not measure the activity for UDP-GlcNAc/GalNAc, the specific activity of BLLJ_1592 for UDP-Gal (260 U/mg) was higher than that of bGalE (140 U/mg). Interestingly, BLLJ_1592 was suggested to be inhibited by GalNAc more severely than bGalE because the enzymatic GNB production using BLLJ_1592 under high GalNAc concentration (600 mM) was less efficient than that using bGalE. A phylogenetic tree analysis indicated that BLLJ_1592 is distantly located from any group (Fig. 2), and multiple amino acid sequence alignment could not predict its substrate specificity because of the low sequence homology around the C2 pocket-forming region (Fig. 5). We built a reliable homology model of BLLJ_1592 (GQME = 0.72 and QMEAN = −2.34) using the SWISS-MODEL server45 with an unpublished structure of GalE from Burkholderia pseudomallei (PDB ID: 3ENK, sequence identity = 42.3%) as a template. As shown in Fig. 6E, the C2 and C5 pocket of BLLJ_1592 are formed by Asn190 and Leu290, respectively. The inhibition by GalNAc suggests that the side chain of Asn190 is flexible and can accommodate the N-acetyl side chain. Therefore, BLLJ_1592 was predicted to have group 1-like specificity (low activity for UDP-GlcNAc/GalNAc) due to the large side chain of Leu290 in the C5 pocket.
Concluding remarks
In this study, we revealed the structural basis for the broad substrate specificity of a bacterial enzyme that is required for efficient catabolism of HMO and mucin O-glycans. Interestingly, the structure of GalE from the human symbiont (B. longum) was more similar to that of its host animal (H. sapiens) compared to the enzyme from a gut microbe living in the same niche (E. coli). GalEs in group 1b and 2 are closely located in the phylogenetic tree (Fig. 2), and a small number of amino acid mutations in the active site (especially Cys or Tyr residue at the C5 pocket) can change the substrate specificity. Gal and GalNAc sugars are utilized by wide range of organisms from microbes to animals for various biological events such as breast feeding (lactose and HMO) and utilization of glycoproteins and glycoconjugates. To metabolize and utilize galacto-type sugars in relationship with other organisms (symbiosis or pathogenesis), organisms may be taking advantage of the GalE enzymes’ characteristics, such as the specificity being prone to change. Interestingly, hUXS1, which catalyzes the considerably different reaction from GalEs40, is located within the clade of group 3 GalEs in a phylogenetic tree (Fig. 2). The UDP-xylose synthase plays a key role in the glycosaminoglycan synthesis on the protein core of extracellular matrix proteoglycans of mammals. Moreover, a study using kinetic isotope effects reveled a more complicated reaction mechanism of UDP-D-apiose/UDP-D-xylose synthase, which is related to hUXS1. In addition to the NAD+-dependent oxidation and base catalysis by Tyr, the UDP-D-apiose/UDP-D-xylose synthase reaction involves decarboxylation, retro-aldol sugar ring opening, rearrangement, and ring contraction46. The unusually high catalytic potential of SDR superfamily enzymes cannot be estimated from the phylogenetic analysis alone but careful inspection based on three-dimensional protein structures is required.
Methods
Protein production and purification
A gene encoding C-terminally His6-tagged GalE (pET-30-lnpD, residues 1–340) was cloned from the genomic DNA of B. longum JCM1217 as previously described15. The expression plasmid was introduced into E. coli BL21 CodonPlus (DE3)-RIL (Stratagene, La Jolla, CA) for protein expression. The transformant was cultivated at 37 °C in Luria-Bertani medium containing 100 mg/L kanamycin and 20 mg/L chloramphenicol until the absorbance at 600 nm reached 0.6. Protein expression was induced by adding 0.1 mM isopropyl-β-D-thiogalactopyranoside to the medium. The culture was incubated for an additional 20 h at 25 °C. Expressed cells were harvested by centrifugation and suspended in 50 mM Tris-HCl (pH 7.0) and 0.1 mM phenylmethylsulfonyl fluoride. The cells were disrupted via sonication, and the supernatant was purified by sequential column chromatography. Ni-affinity chromatography was conducted using a HisTrap FF crude column (GE Healthcare, Fairfield, CT) with two steps of 20 mM and 250 mM imidazole in 50 mM Tris-HCl (pH 7.0). Gel-filtration chromatography was conducted using a Superdex 200 pg 16/60 column (GE Healthcare) equilibrated with 50 mM Tris-HCl (pH 7.0) and 150 mM NaCl at a flow rate of 1 mL/min. The purified protein concentrations were determined by a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA) with bovine serum albumin as the standard.
Crystallography
Crystals of bGalE were grown at 4 °C using the sitting drop vapor diffusion method by mixing 1.0 µL of a 7 mg/ml protein solution with an equal volume of a reservoir solution containing 30% (v/v) PEG400, 0.2 M MgCl, and 0.1 M HEPES-NaOH (pH 7.5). Crystals grew in 2 weeks. Each complex was obtained by cocrystallisation with a ligand (10 mM UDP, 10 mM UDP-Gal, or 10 mM UDP-GlcNac). Crystals were flash-cooled at 100 K in a stream of nitrogen gas. The diffraction data sets were collected using synchrotron radiation at beamlines BL5A, BL17A, and AR-NW12A of the Photon Factory at the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan). The bGalE crystals were long pillar-shaped, and we used helical scan mode for the data collection to reduce radiation damage. The data sets were processed using the HKL200047 and XDS48. The initial phase was determined by the molecular replacement method using MOLREP49. Manual model rebuilding was achieved using Coot50. Crystallographic refinement was performed using Refmac551. Babinet’s bulk solvent type scaling was applied, and TLS (Translation/Libration/Screw) refinement was not used. We used UDP-Gal for the soaking experiment but the resultant electron density map indicated that the protein was complexed with UDP-Glucose, suggesting that the catalytic conversion was occurred in crystallo. The refined crystal structures were validated using MolProbity (http://molprobity.biochem.duke.edu) server52. Molecular graphic images and amino acid sequence alignment were prepared using PyMOL (Schrödinger, LLC, New York, NY) and ESPript53.
Accession codes
The coordinates and structure factors of UDP + NAD+, UDP-GlcNAc + NAD+, and UDP-Glc + NAD+ have been deposited in the Protein Data Bank under accession codes 6K0G, 6K0H, and 6K0I, respectively.
Acknowledgements
We thank the staff of the Photon Factory for the X-ray data collection and Dr. Takayoshi Wakagi for helpful discussion. This work was supported by Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry (25010 A) and in part by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Agency for Medical Research and Development (AMED).
Author Contributions
S.F. and M.K. conceived and designed the experiments. Y.-W.N. conducted the experiments. M.N. constructed the expression plasmid. Y.-W.N., S.F. and T.A. analyzed the results. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Picard C, et al. Review article: Bifidobacteria as probiotic. Aliment. Pharmacol. Ther. 2005;22:495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 2.Ventura M, Turroni F, Motherway MOC, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends in Microbiology. 2012;20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Tojo R, et al. Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World Journal of Gastroenterology. 2014;20:15163–15176. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakata S, et al. Culture-independent analysis of fecal microbiota in infants, with special reference to Bifidobacterium species. FEMS Microbiol. Lett. 2005;243:417–423. doi: 10.1016/j.femsle.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tannock GW, et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl. Environ. Microbiol. 2013;79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fushinobu S. Unique Sugar Metabolic Pathways of Bifidobacteria. Biosci. Biotechnol. Biochem. 2010;74:2374–2384. doi: 10.1271/bbb.100494. [DOI] [PubMed] [Google Scholar]
- 8.Pacheco AR, Barile D, Underwood MA, Mills DA. The Impact of the Milk Glycobiome on the Neonate Gut Microbiota. Annu. Rev. Anim. Biosci. 2014;3:419–445. doi: 10.1146/annurev-animal-022114-111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama T. Host-derived glycans serve as selected nutrients for the gut microbe: Human milk oligosaccharides and bifidobacteria. Bioscience, Biotechnology and Biochemistry. 2016;80:621–632. doi: 10.1080/09168451.2015.1132153. [DOI] [PubMed] [Google Scholar]
- 10.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: Structural, Functional, and Metabolic Aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 11.Urashima T, Hirabayashi J, Sato S, Kobata A. Human Milk Oligosaccharides as Essential Tools for Basic and Application Studies on Galectins. Trends Glycosci. Glycotechnol. 2018;30:SE51–SE65. doi: 10.4052/tigg.1734.1SE. [DOI] [Google Scholar]
- 12.Kiyohara M, et al. Prebiotic effect of lacto-N-biose I on bifidobacterial growth. Biosci. Biotechnol. Biochem. 2009;73:1175–1179. doi: 10.1271/bbb.80697. [DOI] [PubMed] [Google Scholar]
- 13.Xiao J-Z, et al. Distribution of In Vitro Fermentation Ability of Lacto-N-Biose I, a Major Building Block of Human Milk Oligosaccharides, in Bifidobacterial Strains. Appl. Environ. Microbiol. 2010;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitaoka M, Tian J, Nishimoto M. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl. Environ. Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimoto M, Kitaoka M. Identification of N-Acetylhexosamine 1-Kinase in the Complete Lacto-N-Biose I/Galacto-N-Biose Metabolic Pathway in Bifidobacterium longum. Appl. Environ. Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki R, et al. Structural and thermodynamic analyses of solute-binding Protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J. Biol. Chem. 2008;283:13165–73. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 17.Hidaka M, et al. The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J. Biol. Chem. 2009;284:7273–83. doi: 10.1074/jbc.M808525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato M, et al. Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase. Biochim. Biophys. Acta - Proteins Proteomics. 2015;1854:333–340. doi: 10.1016/j.bbapap.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Holden HM, Rayment I, Thoden JB. Structure and Function of Enzymes of the Leloir Pathway for Galactose Metabolism. J. Biol. Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 20.Frey PA, Hegeman AD. Chemical and stereochemical actions of UDP-galactose 4-epimerase. Acc. Chem. Res. 2013;46:1417–1426. doi: 10.1021/ar300246k. [DOI] [PubMed] [Google Scholar]
- 21.Beerens K, Soetaert W, Desmet T. UDP-hexose 4-epimerases: a view on structure, mechanism and substrate specificity. Carbohydr. Res. 2015;414:8–14. doi: 10.1016/j.carres.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Thoden JB, Frey PA, Holden HM. Molecular Structure of the NADH/UDP-glucose Abortive Complex of UDP-galactose 4-Epimerase from Escherichia coli: Implications for the Catalytic Mechanism. Biochemistry. 1996;35:5137–5144. doi: 10.1021/bi9601114. [DOI] [PubMed] [Google Scholar]
- 23.Thoden JB, Frey PA, Holden HM. High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol. Protein Sci. 1996;5:2149–2161. doi: 10.1002/pro.5560051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoden JB, Gulick AM, Holden HM. Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10685–10695. doi: 10.1021/bi9704313. [DOI] [PubMed] [Google Scholar]
- 25.Thoden JB, et al. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:6294–6304. doi: 10.1021/bi970025j. [DOI] [PubMed] [Google Scholar]
- 26.Thoden JB, Holden HM. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1998;37:11469–11477. doi: 10.1021/bi9808969. [DOI] [PubMed] [Google Scholar]
- 27.Thoden JB, Henderson JM, Fridovich-Keil JL, Holden HM. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J. Biol. Chem. 2002;277:27528–27534. doi: 10.1074/jbc.M204413200. [DOI] [PubMed] [Google Scholar]
- 28.Shaw MP, et al. High-resolution crystal structure of Trypanosoma brucei UDP-galactose 4′-epimerase: A potential target for structure-based development of novel trypanocides. Mol. Biochem. Parasitol. 2003;126:173–180. doi: 10.1016/S0166-6851(02)00243-8. [DOI] [PubMed] [Google Scholar]
- 29.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 30.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-Epimerase. J. Biol. Chem. 2002;276:15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 31.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94M-substituted UDP-galactose 4-epimerase. J. Biol. Chem. 2001;276:20617–20623. doi: 10.1074/jbc.M101304200. [DOI] [PubMed] [Google Scholar]
- 32.Ishiyama N, Creuzenet C, Lam JS, Berghuis AM. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: Substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 2004;279:22635–22642. doi: 10.1074/jbc.M401642200. [DOI] [PubMed] [Google Scholar]
- 33.Sakuraba H, Kawai T, Yoneda K, Ohshima T. Crystal structure of UDP-galactose 4-epimerase from the hyperthermophilic archaeon Pyrobaculum calidifontis. Arch. Biochem. Biophys. 2011;512:126–134. doi: 10.1016/j.abb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Dalrymple, S. A., Ko, J., Sheoran, I., Kaminskyj, S. G. W. & Sanders, D. A. R. Elucidation of Substrate Specificity in Aspergillus nidulans UDP-Galactose-4-Epimerase. PLoS One8 (2013). [DOI] [PMC free article] [PubMed]
- 35.Persson B, Kallberg Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs) Chemico-Biological Interactions. 2013;202:111–115. doi: 10.1016/j.cbi.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Thoden JB, Frey PA, Holden HM. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 38.Yoneda K, Sakuraba H, Muraoka I, Oikawa T, Ohshima T. Crystal structure of UDP-galactose 4-epimerase-like l -threonine dehydrogenase belonging to the intermediate short-chain dehydrogenase-reductase superfamily. FEBS J. 2010;277:5124–5132. doi: 10.1111/j.1742-4658.2010.07916.x. [DOI] [PubMed] [Google Scholar]
- 39.Alphey MS, et al. Trypanosoma brucei UDP-galactose-4′-epimerase in ternary complex with NAD+ and the substrate analogue UDP-4-deoxy-4-fluoro-α-D- galactose. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:829–834. doi: 10.1107/S1744309106028740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eixelsberger Thomas, Sykora Sabine, Egger Sigrid, Brunsteiner Michael, Kavanagh Kathryn L., Oppermann Udo, Brecker Lothar, Nidetzky Bernd. Structure and Mechanism of Human UDP-xylose Synthase. Journal of Biological Chemistry. 2012;287(37):31349–31358. doi: 10.1074/jbc.M112.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, et al. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4- epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 42.Swanson Barbara A., Frey Perry A. Identification of lysine 153 as a functionally important residue in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1993;32(48):13231–13236. doi: 10.1021/bi00211a035. [DOI] [PubMed] [Google Scholar]
- 43.Schulz JM, et al. Determinants of function and substrate specificity in human UDP-galactose 4′-epimerase. J. Biol. Chem. 2004;279:32796–32803. doi: 10.1074/jbc.M405005200. [DOI] [PubMed] [Google Scholar]
- 44.Nishimoto M, Kitaoka M. One-pot enzymatic production of β-d-galactopyranosyl-(1→3)-2-acetamido-2-deoxy-d-galactose (galacto-N-biose) from sucrose and 2-acetamido-2-deoxy-d-galactose (N-acetylgalactosamine) Carbohydr. Res. 2009;344:2573–2576. doi: 10.1016/j.carres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 45.Biasini Marco, Bienert Stefan, Waterhouse Andrew, Arnold Konstantin, Studer Gabriel, Schmidt Tobias, Kiefer Florian, Cassarino Tiziano Gallo, Bertoni Martino, Bordoli Lorenza, Schwede Torsten. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research. 2014;42(W1):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eixelsberger Thomas, Horvat Doroteja, Gutmann Alexander, Weber Hansjörg, Nidetzky Bernd. Isotope Probing of the UDP-Apiose/UDP-Xylose Synthase Reaction: Evidence of a Mechanism via a Coupled Oxidation and Aldol Cleavage. Angewandte Chemie International Edition. 2017;56(9):2503–2507. doi: 10.1002/anie.201609288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed]
- 49.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Chen Vincent B., Arendall W. Bryan, Headd Jeffrey J., Keedy Daniel A., Immormino Robert M., Kapral Gary J., Murray Laura W., Richardson Jane S., Richardson David C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D Biological Crystallography. 2009;66(1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42 (2014). [DOI] [PMC free article] [PubMed]
- 54.Roper JR, Guther MLS, Milne KG, Ferguson MAJ. Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2002;99:5884–9. doi: 10.1073/pnas.092669999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Creuzenet C, Belanger M, Wakarchuk WW, Lam JS. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 2000;275:19060–19067. doi: 10.1074/jbc.M001171200. [DOI] [PubMed] [Google Scholar]