Abstract
Mammalian target of rapamycin complex 1 (mTORC1), a protein complex containing the serine/threonine kinase mTOR, integrates various growth stimulating signals. mTORC1 is expressed in intestinal epithelial cells (IECs), whereas the physiological roles of this protein complex in homeostasis of IECs remain virtually unknown. We here generated mice, in which tuberous sclerosis complex 2 (Tsc2), a negative regulator of mTORC1, was specifically ablated in IECs (Tsc2 CKO mice). Ablation of Tsc2 enhanced the phosphorylation of mTORC1 downstream molecules such as ribosomal S6 protein and 4E-BP1 in IECs. Tsc2 CKO mice manifested the enhanced proliferative activity of IECs in intestinal crypts as well as the promoted migration of these cells along the crypt-villus axis. The mutant mice also manifested the increased apoptotic rate of IECs as well as the increased ectopic Paneth cells, which are one of the major differentiated IECs. In addition, in vitro study showed that ablation of Tsc2 promoted the development of intestinal organoids without epidermal growth factor, while mTORC1 inhibitor, rapamycin, diminished this phenotype. Our results thus suggest that Tsc2-mTORC1 signaling regulates the proliferation, migration, and positioning of IECs, and thereby contributes to the proper regulation of intestinal homeostasis.
Keywords: Mammalian target of rapamycin complex 1, Tuberous sclerosis complex 2, Small intestine, Intestinal epithelial cells, Intestinal homeostasis
Intestinal epithelial cells (IECs) of the small intestine are regenerated continuously from intestinal stem cells (ISCs) that reside in a region near the base of intestinal crypt [1,2]. ISCs that give rise to IECs generate proliferating progeny, known as transient amplifying (TA) cells. TA cells migrate out of the stem cell niche in the crypt, divide rapidly, and initiate differentiation into the various cell lineages, including absorptive enterocytes, mucin-secreting goblet cells, peptide hormone–secreting enteroendocrine cells, and antimicrobial peptide–producing Paneth cells. Absorptive enterocytes, goblet cells, and enteroendocrine cells mature and migrate up the crypt toward the tip of intestinal villi. Paneth cells travel down to the base of the crypt. In particular, absorptive enterocytes are expelled into the gut lumen after they have reached at the tip of the villi. The continuous production of new IECs from each crypt is thus balanced by the elimination of older cells at the tip of the villi, resulting in a rapid turnover of IECs.
Mammalian target of rapamycin (mTOR) may be involved in regulating the turnover of IECs along the crypt-villus axis [3]. mTOR is a serine/threonine kinase that forms two distinct multiprotein complexes called mTOR complex (mTORC) 1 and mTORC2. mTOR is known as a sensor of growth factors, nutrients, energy, oxidation state and stress, and coordinates cell growth, survival and autophagy [4]. The activity of mTORC1 is inactivated by tuberous sclerosis complex (Tsc) 1/2 in the basal state. In response to extracellular stimuli, Tsc1/2 is inactivated by phosphorylation and then mTORC1 is activated. Previous studies have suggested that intestinal tumorigenesis driven by the loss of Apc activity requires mTORC1 [5,6], whereas the physiological roles of mTORC1 in IECs remain unclear.
In this study, we generated the mice, in which Tsc2 was specifically ablated in IECs (Tsc2 CKO mice) to highlight the role of mTORC1 activity in IECs. We here clarify the physiological roles of Tsc2 and mTORC1 in homeostasis of IECs.
MATERIALS AND METHODS
Antibodies and reagents
A mouse monoclonal antibody (mAb) to β-catenin (# 610153) was obtained from BD Biosciences (San Diego, CA). A mouse mAb to β-tubulin (# T4026) was obtained from Sigma-Aldrich (St. Louis, MO). A rat mAb to bromodeoxyuridine (BrdU) (# ab6326) was obtained from Abcam (Cambridge, MA). Rabbit polyclonal antibodies (pAbs) to Ki67 (# AM11168PU-S) were obtained from Acris (Herford, Germany). Rabbit pAbs to mucin 2 (# sc-15334) were obtained from Santa Cruz Biotechnology. Rabbit pAbs to lysozyme (# A0099) were obtained from Dako (Glostrup, Denmark). Rabbit pAbs to cleaved caspase-3 (# 9661S), to S6 (# 2217S), to phosphorylated S6 (# 4858S), to Tsc2 (# 4308S), to 4E-BP1 (# 2855S), and to phosphorylated 4E-BP1 (# 9452S) were obtained from Cell Signaling Technology (Beverly, MA). Secondary antibodies labeled with Alexa488 or Cy3 for immunofluorescence analysis were obtained from ThermoFisher (Waltham, MA) and Jackson ImmunoResearch (West Grove, PA), respectively. Horseradish peroxidase–conjugated goat secondary antibodies for immunoblot analysis were obtained from Jackson ImmunoResearch. BrdU was from Sigma-Aldrich. Mayer’s hemalum solution was obtained from Merck KGaA (Darmstadt, Germany), and eosin was obtained from Wako (Osaka, Japan).
Mice
To generate Tsc2fl/+;villin-cre mice, villin-cre mice were crossed with Tsc2fl/fl mice [7]. The resulting Tsc2fl/+;villin-cre offspring were crossed with Tsc2fl/fl mice to obtain Tsc2fl/fl;villin-cre (Tsc2 CKO) mice and Tsc2fl/fl (control) mice. These mice were maintained at the Institute for Experimental Animals at Kobe University Graduate School of Medicine under the specific pathogen–free (SPF) condition. This study was approved by the Institutional Animal Care and Use Committee of Kobe University (Permit Number: P170707, P150506, P120508-R2), and all animal experiments were performed according to Kobe University Animal Experimentation Regulations. All efforts were made to minimize suffering.
Detection of deleted and floxed alleles of Tsc2 by PCR
Several tissues of adult mice were washed with ice-cold phosphate-buffered saline (PBS), incubate overnight at 56°C in lysis buffer (100 mM Tris-HCl [pH 8.5], 5 mM EDTA, 0.2% sodium dodecyl sulfate [SDS], 200 mM NaCl, 50 μg/ml proteinase K), and centrifuged at 17,500 × g for 20 min at room temperature. The resulting supernatant was then subjected to isopropanolol precipitation of genomic DNA. The floxed allele (237 bp product) of Tsc2 was identified by PCR with the sense primer (5′-CAGCCTTGCCTGTATCTATG-3′) and antisense primer (5′-GGTGTTGGAACTGAGCAGAT-3′). The deleted allele (282 bp product) of Tsc2 was identified by PCR with the sense primer (5′-AATGCCCTAAGTGCAACCTG-3′) and antisense primer (5′-GGTGTTGGAACTGAGCAGAT-3′).
Histology and immunofluorescence analysis
For histological analysis, the small intestine was removed and immediately fixed with 4% paraformaldehyde in PBS for >12 h at room temperature. Parafin-embedded sections with a thickness of 6 μm were then prepared and stained with hematoxylin-eosin. For immunofluorescence analysis, the small intestine was removed and fixed immediately for 3 h at room temperature with 4% paraformaldehyde in PBS. The small intestine was then transferred to 30% (wt/vol) sucrose solutions in PBS overnight, embedded in optimal cutting temperature (OCT) compound (Sakura, Tokyo, Japan), and rapidly frozen in liquid nitrogen. Frozen sections with a thickness of 5 μm were prepared with a cryostat. The sections were then mounted on glass slides, air-dried, and subjected to immunofluorescence analysis with primary antibodies and fluorescent dye–labeled secondary antibodies as described previously [8,9]. Fluorescence images were obtained with a fluorescence microscope (BX51; Olympus, Tokyo, Japan).
BrdU incorporation assay
Mice were injected intraperitoneally with BrdU (10 mg per kilogram of body weight). After 2, 24, 48 or 72 h, frozen sections were prepared as described above for immunofluorescence analysis. The sections were incubated for 30 min at 65°C with 0.025 M HCl, washed with 0.1 M borate buffer (pH 8.5), and incubated at room temperature first for 2 h with mAbs to BrdU and to β-catenin. The sections were then stained for 1 h with fluorescent dye–labeled secondary antibodies. Fluorescence images were obtained with a fluorescence microscope (BX51, Olympus). IEC migration distance, which was defined as the distance from the crypt base to the BrdU-positive cells that had migrated the farthest, was measured with the use of ImageJ software (NIH).
Isolation of mouse IECs
Mouse IECs were isolated as previously described [10] but with a slight modification. In brief, the small intestine was washed with PBS, cut into small pieces, and washed three times with Hanks’ balanced salt solution (HBSS) containing 1% fetal bovine serum and 25 mM HEPES-NaOH (pH 7.5). They were then incubated three times on a rolling platform for 15 min at room temperature in HBSS containing 5 mM EDTA and 25 mM HEPES-NaOH (pH 7.5). After removing debris, IECs were isolated by centrifugation at 250 × g for 10 min at 4°C. Isolated IECs were washed three times with PBS.
Immunoblot analysis
Isolated IECs were lysed with lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM NaF, 1 mM sodium vanadate, 1% protease inhibitor cocktail (Nacalai Tesque)]. The lysates were then centrifuged at 17,500 × g for 15 min at 4°C. The resulting supernatants were subjected to immunoblot analysis as previously described [9,10].
Reverse transcription (RT) and real-time PCR analysis
Isolation of total RNA and quantitative RT-PCR analysis were performed as described previously [11], with minor modifications. In brief, total RNA was prepared from isolated IECs with the use of Sepasol RNA I (Nacalai Tesque) and an RNeasy Mini Kit (Qiagen). The first-strand cDNA was synthesized from portions (0.8 μg) of the RNA with the use of a QuantiTect Reverse Transcription Kit (Qiagen). The cDNA fragments of interest were amplified by PCR with the use of Fast Start SYBR Green Master (Roche, Penzberg, Germany) and a LightCycler 480 instrument (Roche). The amplification was analyzed with the use of LightCycler 480 software (Roche). The abundance of each target mRNA was normalized by that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. Primer sequences (forward and reverse, respectively) were as follows: GAPDH, 5′-AGGTCGGTGTGAACGGATTTG-3′ and 5′-TGTAGACCATGTAGTTGAGGTCA-3′; Hes1, 5′-GGACAAACCAAAGACGGCCTCTGAGCACAG-3′ and 5′-TGCCGGGAGCTATCTTTCTTAAGTGCATCC-3′; Atoh1, 5′-GCTGGTAAGGAGAAGCGGCTGTG-3′ and 5′-TGTACCCCATTCACCTGTTTGC-3′.
Intestinal organoid culture
Intestinal organoid culture was performed as previously described [12]. In brief, crypts were isolated from the small intestine by incubation for 30 min at 4°C in PBS containing 5 mM EDTA. The isolated crypts were mixed with Matrigel (BD Biosciences) and transferred to 48-well plates. After polymerization of the Matrigel, advanced Dulbecco’s modified Eagle’s medium–F12 (Invitrogen), which was supplemented with penicillin-streptomycin (100 U/ml) (Invitrogen), 10 mM HEPES (Invitrogen), 1× GlutaMAX (Invitrogen), 1× N2 (Invitrogen), 1× B27 (Invitrogen), 1.25 mM N-acetylcysteine (Sigma-Aldrich), 10% R-spondin1–Fc–conditioned medium, EGF (50 ng/ml) (Peprotech, Rocky Hill, NJ), and Noggin (100 ng/ml) (Peprotech), was overlaid on the gel. When the organoids were cultured in the medium without EGF, they were treated with or without 1 μM Rapamycin (LC Laboratorie, Woburn, MA). The organoids were maintained in an incubator (37°C, 5% CO2) for 3 or 4 days.
Quantification of organoid area and bud formation in intestinal organoids
Intestinal organoids were imaged with an Axiovert 200 microscope (Carl Zeiss, Oberkochen, Germany). The organoid area was measured by encircling the periphery of each organoid with the use of ImageJ software (NIH). Any protrusions were defined as buds, and their number per organoid was counted.
Statistical analysis
Quantitative data are presented as means ± standard error (SE). Data were analyzed with Student’s t test or by analysis of variance (ANOVA) followed by Tukey’s test, as appropriate. A P value of <0.05 was considered statistically significant. Analysis was performed with the use of GraphPad Prism software version 6.0 (GraphPad, San Diego, CA).
RESULTS
Tsc2 ablation induces the hyperactivation of mTORC1 in IECs
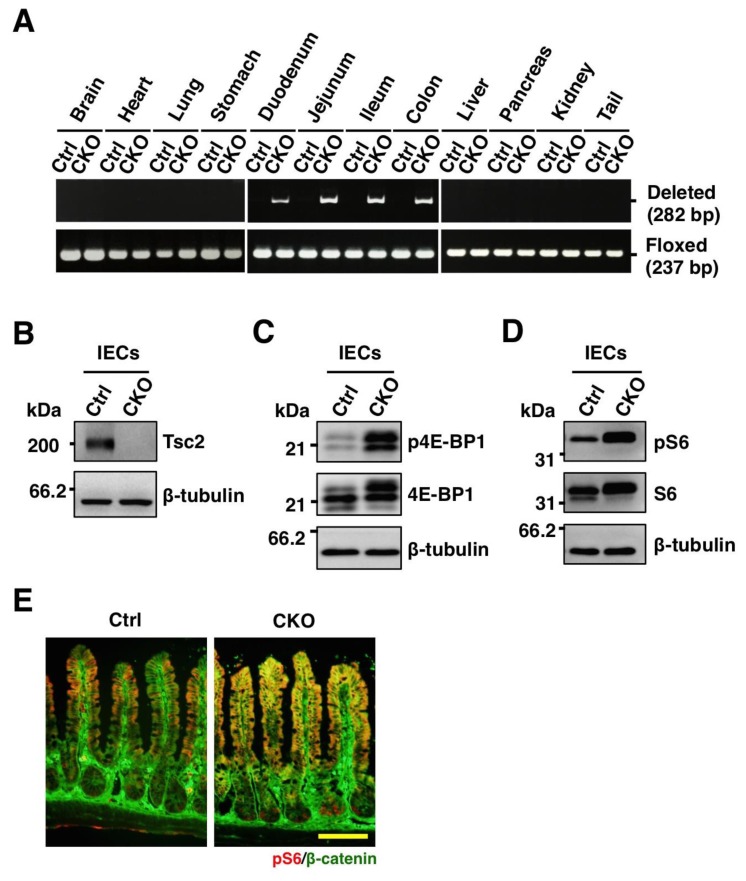
To investigate the role of mTORC1 in IECs of the small intestine, we crossed mice homozygous for a floxed Tsc2 allele (Tsc2fl/fl) with villin-cre transgenic mice [13]. The specific deletion of Tsc2 allels in adult Tsc2fl/fl;villin-cre (Tsc2 CKO) mice was determined by PCR analysis of genomic DNA isolated from several tissues including intestine. Consistent with the results of previous studies with the villin-cre transgene [13], the deletion of Tsc2 alleles was detected in the duodenum, jejunum, ileum, and colon of Tsc2 CKO mice, but not in any other organ (Figure 1A). The abundance of Tsc2 protein in IECs isolated from the small intestine of Tsc2 CKO mice was greatly reduced compared with that for control mice (Figure 1B). These results thus indicated that the villin-cre transgene directs the specific deletion of the Tsc2 gene in IECs of the mouse small intestine.
Figure 1.
Generation of IEC-specific Tsc2 CKO mice. (A) Genomic DNA extracted from the indicated organs of adult control (Ctrl) or Tsc2 CKO (CKO) mice were subjected to PCR analysis with primers specific for deleted or floxed alleles of Tsc2. (B) Lysates of IECs from the small intestine of control or Tsc2 CKO mice were subjected to immunoblot analysis with antibodies to Tsc2 and β-tubulin. (C) Lysates of IECs from the small intestine of control and Tsc2 CKO mice were subjected to immunoblot analysis with antibodies to phospho-4E-BP1 (p4E-BP1), 4E-BP1, and β-tubulin. (D) Lysates of IECs from the small intestine of control and Tsc2 CKO mice were subjected to immunoblot analysis with antibodies to phospho-S6 ribosomal protein (pS6), S6 ribosomal protein (S6), and β-tubulin. (E) Frozen sections of the small intestine from control or Tsc2 CKO mice were immunostained with antibodies to pS6 (red) and to β-catenin (green). Scale bar, 100 μm.
Given that Tsc2 has been known to suppress mTORC1 activity [4], we next examined whether the loss of Tsc2 activates mTORC1 signaling in IECs. Immunoblot analysis showed that the amounts of 4E-BP1 and ribosomal S6 protein, both of which are downstream proteins of mTORC1, were markedly increased in IECs from the small intestine of Tsc2 CKO mice (Figure 1C and 1D). The phosphorylation levels of these proteins were also increased in IECs from the small intestine of Tsc2 CKO mice (Figure 1C and 1D). We next subjected sections of the small intestine to immunohistofluorescence staining with antibodies to phosphorylated S6. Whereas staining for phosphorylated S6 was relatively weak throughout the intestinal crypts and villi of control mice, it was prominent in villi and crypts of Tsc2 CKO mice (Figure 1E). These results thus indicated that Tsc2 ablation resulted in hyperactivation of mTORC1 in IECs of mouse small intestine.
Promotion of the proliferative activity and turnover of IECs in Tsc2 CKO mice
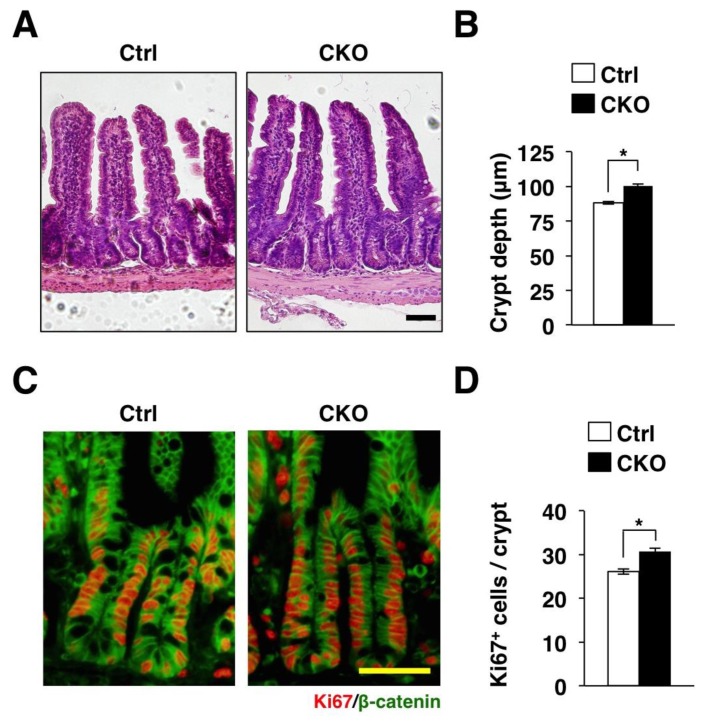
We next histologically analysed the phenotype of the small intestine in Tsc2 CKO mice by hematoxylin-eosin staining (Figure 2A). Ablation of Tsc2 did not affect the number of villus and crypt (data not shown). Ablation of Tsc2 also did not affect the length of individual villus (data not shown). In contrast, the depth of crypts in the small intestine was increased in Tsc2 CKO mice (Figure 2B). The intestinal crypt is the region where intestinal stem cells (ISCs) generate proliferating progeny, known as TA cells, that migrate out of the stem cell niche, cease to proliferate, and initiate differentiation into the various cell lineages for renewal of the villus [14,15]. Immunostaining for Ki67, a marker for cell proliferation [16], showed that the number of Ki67-positive IECs was markedly increased in the small intestinal crypts of Tsc2 CKO mice (Figure 2C and 2D). Thus, proliferative IECs are likely increased in Tsc2 CKO mice.
Figure 2.
Promotion of the proliferative activity of IECs in Tsc2 CKO mice. (A) Hematoxylin-eosin staining of paraffin-embedded sections of the small intestine from control or Tsc2 CKO mice. Scale bar, 50 μm. (B) The crypt depth was determined from sections similar to those in (A). Data are means ± SE for 90 crypts of three mice. *, P < 0.05. (C) Frozen sections of the small intestine from control or Tsc2 CKO mice were immunostained with antibodies to Ki67 (red) and to β-catenin (green). Scale bar, 50 μm. (D) The number of Ki67-positive cells per crypt was determined from sections similar to those in (C). Data are means ± SE for 90 crypts of three mice. *, P < 0.05.
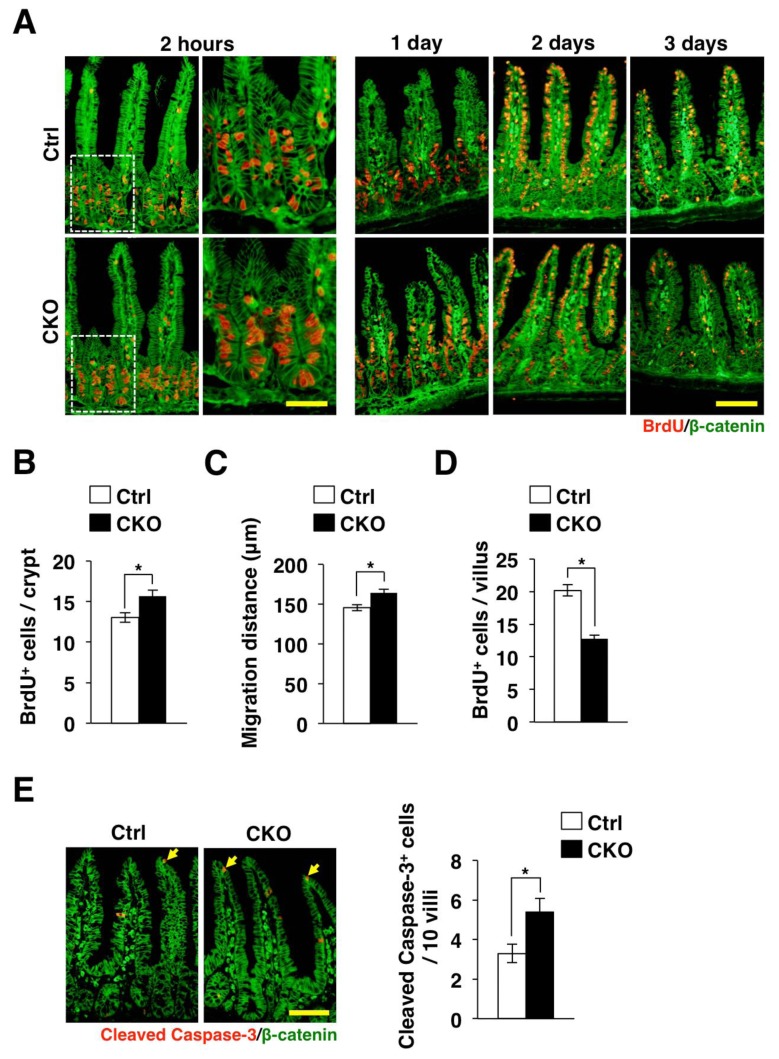
We also examined the incorporation of bromodeoxyuridine (BrdU) into IECs as well as the turnover of BrdU-labeled IECs in Tsc2 CKO mice. Consistent with the increase of Ki67-positive IECs in Tsc2 CKO mice (Figure 2C and 2D), the number of BrdU-positive IECs in crypts of the small intestine was markedly increased for Tsc2 CKO mice compared with control mice at 2 h after BrdU injection (Figure 3A and 3B). In addition, we found that, at 1 day after BrdU injection, whereas most BrdU-positive IECs remained in a position immediately above or within crypt areas of the small intestine in control mice, most such cells in the small intestine of Tsc2 CKO mice had migrated to the middle region of villi (Figure 3A). Quantitative analysis confirmed that the migration distance of BrdU-labeled IECs in the small intestine was markedly increased for Tsc2 CKO (Figure 3C). BrdU-positive IECs of both control and Tsc2 CKO mice reached at the top of villi at 2 days after BrdU injection (Figure 3A). At 3 days after BrdU injection, BrdU-positive IECs in villi of the small intestine in Tsc2 CKO mice was greatly reduced compared with that apparent for control mice (Figure 3A and 3D). These results thus confirmed that loss of Tsc2 results in promotion of the proliferative activity and migration of IECs in the small intestine.
Figure 3.
Promotion of the turnover of IECs in Tsc2 CKO mice. (A) Frozen sections of the small intestine from control or Tsc2 CKO mice were prepared at 2 h or 1, 2, or 3 days after BrdU injection and were immunostained with antibodies to BrdU (red) and to β-catenin (green). The boxed areas in the left panels for the sections prepared at 2 h after BrdU injection are shown at higher magnification in the right panels. Scale bars, 50 μm (higher magnification) or 100 μm (lower magnification). (B) The number of BrdU-positive cells per crypt at 2 h after BrdU injection was determined from sections similar to those in (A). Data are means ± SE for 90 crypts of three mice. *, P < 0.05. (C) The migration distance for BrdU-positive cells at 1 day after BrdU injection was determined from sections similar to those in (A). Data are means ± SE for 90 villi from three mice. *, P < 0.05. (D) The number of BrdU-positive cells per villus at 3 days after BrdU injection was determined from sections similar to those in (A). Data are means ± SE for 90 villi of three mice. *, P < 0.05. (E) Frozen sections of the small intestine from control or Tsc2 CKO mice were immunostained with antibodies to cleaved caspase-3 (red) and to β-catenin (green). Representative images are shown in the left panels. Scale bar, 100 μm. The number of cleaved caspase-3-positive cells per 10 villi in such sections was also determined. Data are means ± SE for 100 villi of three mice. *, P < 0.05.
IECs are expelled into the gut lumen after they have migrated to the tip of villi. This elimination of IECs is thought to be triggered, at least in part, by spontaneous apoptosis [17]. To investigate whether the accelerated migration of IECs in the small intestine of Tsc2 CKO mice might be attributable to an increased frequency of apoptosis, we performed immunostaining with antibodies to the cleaved form of caspase-3. We found that the number of cleaved caspase-3–positive IECs in the small intestine was significantly increased for Tsc2 CKO mice compared with control mice (Figure 3E). Together, above results suggested that the turnover of mature IECs was markedly accelerated in the small intestine of Tsc2 CKO mice.
Regulation of Paneth cell clustering by Tsc2
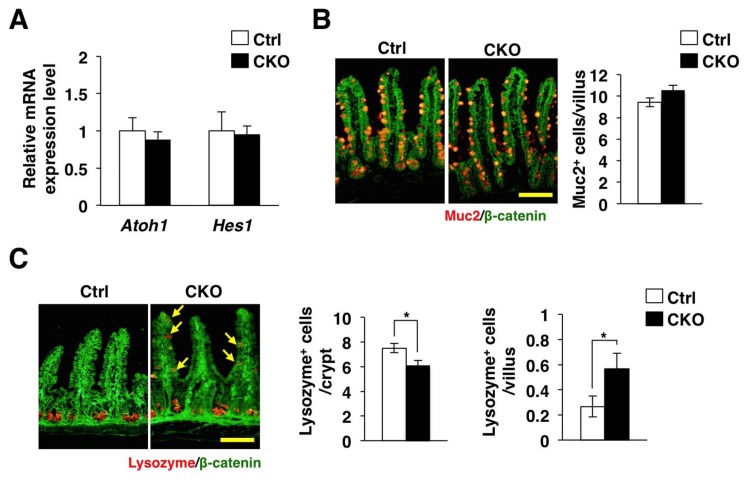
We next investigated whether IEC-specific ablation of Tsc2 affects the differentiation of IECs. The Notch signaling pathway is thought to regulate the absorptive versus secretory fate decision in the intestinal epithelium [15,18,19]. Activation of this signaling pathway induces expression of Hes1, which in turn prevents expression of a factor, known as Atoh1, that promotes the development of secretory cells including goblet and Paneth cells [20]. In addition, previous study showed that systemic over-expression of a dominant negative Tsc2 allele reduced the number of goblet cells or Paneth cells through Notch/Hes1 signaling [21]. We found that ablation of Tsc2 had no effect on the expression of Hes1 and Atoh1, however (Figure 4A). We also found that ablation of Tsc2 unaffected the number of Mucin 2–positive goblet cells in the small intestine (Figure 4B). In contrast, the number of Paneth cells, which were stained with antibodies to lysozyme, was significantly reduced at the base of crypts, whereas the number of these cells in the villus was significantly increased in the small intestine of Tsc2 CKO mice (Figure 4C). These results thus suggest that Tsc2 controls the correct clustering of Paneth cells in the small intestine.
Figure 4.
Abnormal clustering of Paneth cells in Tsc2 CKO mice. (A) Quantitative RT-PCR analysis of Atoh1 and Hes1 in isolated IECs from the small intestine of control or Tsc2 CKO mice. The amount of each mRNA was normalized by that of GAPDH mRNA and then expressed relative to the normalized value for control mice. Data are means ± SE representative of four mice. (B) Frozen section of the small intestine from control or Tsc2 CKO mice were immunostained with antibodies to mucin 2 (Muc2) (red) and to β-catenin (green). Representative images are shown in the left panels. Scale bar, 100 μm. The number of Muc-2 positive cells per villus was also determined from such sections. Data are means ± SE for 180 villi of six mice. (C) Frozen section of the small intestine from control and Tsc2 CKO mice were immunostained with antibodies to lysozyme (red) and to β-catenin (green). Representative images are shown in the left panels. Arrows indicate cells positive for lysozyme in the villi. Scale bar, 100 μm. The number of lysozyme positive cells per crypt was determined from such sections. The number of lysozyme positive cells per villus was also determined. Data are means ± SE for 90 crypts or villi of three mice. *, P < 0.05.
Promotion of the development of intestinal organoids by IEC-specific Tsc2 ablation
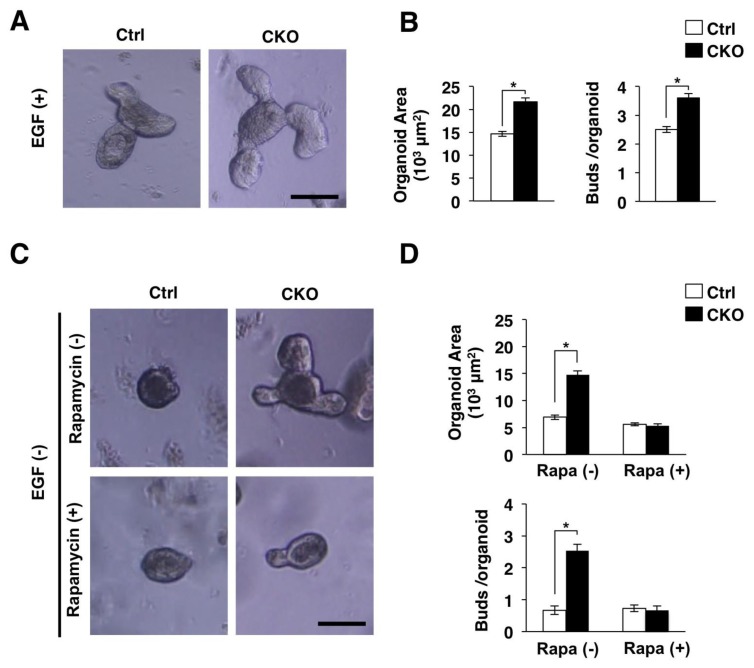
We next examined the development of mouse intestinal organoids from Tsc2 CKO mice to clarify the detailed effect of Tsc2 ablation on IECs. The intestinal organoid is a model of three-dimensional “mini-guts”, and they are thought to mimic in vivo proliferation and differentiation of IECs [12]. We found that the development of intestinal organoids from Tsc2 CKO mice was markedly promoted compared with that for control mice (Figure 5A and 5B). Epidermal growth factor (EGF) is essential for normal development of intestinal organoids [12]. However, we also found that ablation of Tsc2 promoted organoid development without EGF (Figure 5C and 5D). Thus, Tsc2 ablation is likely to promote IEC proliferation in an EGF independent manner. We also examined the effects of an mTORC1 inhibitor, rapamycin, on the development of mouse intestinal organoids to clarify whether the promotion of organoid development by Tsc2 ablation is attributable to the direct activation of mTORC1 on IECs. We found that the promotion of organoid development by Tsc2 ablation was diminished by rapamycin treatment (Figure 5C and 5D). Together, these results suggested that the increased proliferative activity in Tsc2-deficient organoids was attributable to hyperactivation of mTORC1.
Figure 5.
Promotion of the development of intestinal organoids by IEC-specific Tsc2 ablation. (A) Isolated crypts were cultured in the Matrigel with EGF for 3 days as described in Materials and Methods. Scale bar 100 μm. (B) Areas of intestinal organoids cultured as for (A) were determined. The number of buds per organoid was also determined. Data are means ± SE for a total of 90 organoids per group in three separate experiments. *, P < 0.05. (C) Intestinal organoids cultured without EGF were stimulated with (+) or without (−) Rapamycin for 4 days. Scale bar 100 μm. (D) Areas of intestinal organoids cultured as for (C) were determined. The number of buds per organoid was also determined. Data are means ± SE for a total of 90 organoids per group in three separate experiments. *, P < 0.05.
DISCUSSION
We have here shown that the proliferative activity of IECs as well as the migration of mature IECs along the crypt-villus axis were markedly promoted in the small intestine of Tsc2 CKO mice. We also confirmed that ablation of Tsc2 enhanced the phosphorylation of mTORC1 downstream molecules such as ribosomal S6 protein and 4E-BP1 in IECs. mTORC1 activity has been known to coordinate cell proliferation through 4E-BP1 [22]. Thus, excessive mTORC1 activity is likely to promote the proliferation of IECs in the small intestinal crypt, and then promote the migration and turnover of mature IECs.
Differentiation of secretory IECs such as Paneth cells and goblet cells is controlled by Notch signaling [20]. Previous study demonstrated that systemic over-expression of a dominant negative Tsc2 allele in mice (Tsc2-RGΔ TG mice) activated Notch signaling, which in turn inhibited the development of goblet cells or Paneth cells in the intestinal mucosa [21]. In contrast, we found that IEC-specific ablation of Tsc2 did not affect Notch signaling. Consistently, the number of goblet cells was normal in Tsc2 CKO mice. By using IEC-specific mTOR or Raptor CKO mice, another group demonstrated that mTORC1 promoted terminal maturation of absorptive enterocytes and secretory IECs, but not their specification [23]. Thus, differences in phenotypes between Tsc2-RGΔ TG mice and our Tsc2 CKO mice likely reflect differences in the particular cells reducing mTORC1 activity. We also found that ablation of Tsc2 disturbed Paneth cell clustering at the base of crypts. This phenotype resembles to mice, in which Tsc1 is ablated in IECs [24]. Taken together, excessive mTORC1 activity in IECs does not likely affect the differentiation to secretory IECs, whereas it disturbs Paneth cell clustering at the base of crypts.
We finally demonstrated that ablation of Tsc2 promoted the development of intestinal organoids. The intestinal organoid is a model of three-dimensional “mini-guts” with crypt-villus domains that contain all the mature IECs [12]. EGF is thought to serve as a major driver of TA cell and IEC proliferation through activation of the Ras–MAPK signaling pathway [12,14]. Indeed, EGF is an essential component in the standard culture medium for the development of intestinal organoids [12]. We found that ablation of Tsc2 promoted organoid development without EGF, however. In contrast, mTORC1 inhibitor, rapamycin, diminished this phenotype. Thus, excessive mTORC1 signaling likely promotes proliferation of IECs in an EGF independent manner.
In conclusion, the present study showed that Tsc2-mTORC1 signaling regulates the proliferation, migration, and positioning of IECs. Recent studies suggest that the down-regulation of mTORC1 activity maintains stem cell pool [25,26]. Thus, excessive mTORC1 activity may convert ISCs to proliferating IECs such as TA cells, and thereby promotes the turnover of IECs. Further investigation is certainly necessary to clarify the detailed molecular mechanisms by which Tsc2-mTORC1 signaling regulates homeostasis of IECs.
ACKNOWLEDGEMENTS
We thank Dr. C. J. Kuo (Stanford University) for providing HEK293T cells expressing R-spondin1–Fc as well as Dr. T. Sato (Keio University) for technical supports. This work was supported by a Grant-in-Aid for Challenging Exploratory Research (16K15219 to T. Matozaki) and a Grant-in-Aid for Scientific Research (C) (16K08586 to T. Kotani) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This work was also supported by the Uehara Memorial Foundation (to T. Kotani) and Takeda Science Foundation (to T. Kotani). None of the authors has any conflicts of interest or any financial ties to disclose.
REFERENCES
- 1.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H, van den Born M, Danenberg E, van den Brink S, Korving J, Abo A, Peters PJ, Wright N, Poulsom R, Clevers H. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang H, Xiong X, Wang X, Yin Y. Mammalian target of rapamycin signaling pathway changes with intestinal epithelial cells renewal along crypt-villus axis. Cell Physiol Biochem. 2016;39:751–759. doi: 10.1159/000445665. [DOI] [PubMed] [Google Scholar]
- 4.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujishita T, Aoki K, Lane HA, Aoki M, Taketo MM. Inhibition of the mTORC1 pathway suppresses intestinal polyp formation and reduces mortality in ApcΔ716 mice. Proc Natl Acad Sci U S A. 2008;105:13544–13549. doi: 10.1073/pnas.0800041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, Dudek KM, Casey HA, Scopelliti A, Cordero JB, Vidal M, Pende M, Ryazanov AG, Sonenberg N, Meyuhas O, Hall MN, Bushell M, Willis AE, Sansom OJ. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigeyama Y, Kobayashi T, Kido Y, Hashimoto N, Asahara S, Matsuda T, Takeda A, Inoue T, Shibutani Y, Koyanagi M, Uchida T, Inoue M, Hino O, Kasuga M, Noda T. Biphasic response of pancreatic β-cell mass to ablation of tuberous sclerosis complex 2 in mice. Mol Cell Biol. 2008;28:2971–2979. doi: 10.1128/MCB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita H, Kotani T, Park JH, Murata Y, Okazawa H, Ohnishi H, Ku Y, Matozaki T. Role of the protein tyrosine phosphatase Shp2 in homeostasis of the intestinal epithelium. PLoS One. 2014;9:e92904. doi: 10.1371/journal.pone.0092904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadakata H, Okazawa H, Sato T, Supriatna Y, Ohnishi H, Kusakari S, Murata Y, Ito T, Nishiyama U, Minegishi T, Harada A, Matozaki T. SAP-1 is a microvillus-specific protein tyrosine phosphatase that modulates intestinal tumorigenesis. Genes Cells. 2009;14:295–308. doi: 10.1111/j.1365-2443.2008.01270.x. [DOI] [PubMed] [Google Scholar]
- 10.Murata Y, Mori M, Kotani T, Supriatna Y, Okazawa H, Kusakari S, Saito Y, Ohnishi H, Matozaki T. Tyrosine phosphorylation of R3 subtype receptor-type protein tyrosine phosphatases and their complex formations with Grb2 or Fyn. Genes Cells. 2010;15:513–524. doi: 10.1111/j.1365-2443.2010.01398.x. [DOI] [PubMed] [Google Scholar]
- 11.Murata Y, Kotani T, Supriatna Y, Kitamura Y, Imada S, Kawahara K, Nishio M, Daniwijaya EW, Sadakata H, Kusakari S, Mori M, Kanazawa Y, Saito Y, Okawa K, Takeda-Morishita M, Okazawa H, Ohnishi H, Azuma T, Suzuki A, Matozaki T. Protein tyrosine phosphatase SAP-1 protects against colitis through regulation of CEACAM20 in the intestinal epithelium. Proc Natl Acad Sci U S A. 2015;112:E4264–4271. doi: 10.1073/pnas.1510167112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 13.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 14.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 15.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Holt PR, Moss SF, Kapetanakis AM, Petrotos A, Wang S. Is Ki-67 a better proliferative marker in the colon than proliferating cell nuclear antigen? Cancer Epidemiol Biomarkers Prev. 1997;6:131–135. [PubMed] [Google Scholar]
- 17.Hall PA, Coates PJ, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 18.Sancho R, Cremona CA, Behrens A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 2015;16:571–581. doi: 10.15252/embr.201540188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 20.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Rychahou P, Wang Q, Weiss HL, Evers BM. TSC2/mTORC1 signaling controls Paneth and goblet cell differentiation in the intestinal epithelium. Cell Death Dis. 2015;6:e1631. doi: 10.1038/cddis.2014.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, Kozma SC, Thomas G, Sonenberg N. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–1176. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampson LL, Davis AK, Grogg MW, Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barron L, Sun RC, Aladegbami B, Erwin CR, Warner BW, Guo J. Intestinal epithelial-specific mTORC1 activation enhances intestinal adaptation after small bowel resection. Cell Mol Gastroenterol Hepatol. 2017;3:231–244. doi: 10.1016/j.jcmgh.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, Nielsen GP, Mino-Kenudson M, Zukerberg LR, Bhan AK, Deshpande V, Sabatini DM. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haller S, Kapuria S, Riley RR, O’Leary MN, Schreiber KH, Andersen JK, Melov S, Que J, Rando TA, Rock J, Kennedy BK, Rodgers JT, Jasper H. mTORC1 activation during repeated regeneration impairs somatic stem cell maintenance. Cell Stem Cell. 2017;21:806–818. doi: 10.1016/j.stem.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]