Abstract
HIV integrates its provirus into the host genome and establishes latent infection. Antiretroviral therapy (ART) can control HIV viremia, but cannot eradicate or cure the virus. Approaches targeting host epigenetic machinery to repress HIV, leading to an aviremic state free of ART, are needed. Bromodomain and extraterminal (BET) family protein BRD4 is an epigenetic reader involved in HIV transcriptional regulation. Using structure-guided drug design, we identified a small molecule (ZL0580) that induced epigenetic suppression of HIV via BRD4. We showed that ZL0580 induced HIV suppression in multiple in vitro and ex vivo cell models. Combination treatment of cells of aviremic HIV-infected individuals with ART and ZL0580 revealed that ZL0580 accelerated HIV suppression during ART and delayed viral rebound after ART cessation. Mechanistically different from the BET/BRD4 pan-inhibitor JQ1, which nonselectively binds to BD1 and BD2 domains of all BET proteins, ZL0580 selectively bound to BD1 domain of BRD4. We further demonstrate that ZL0580 induced HIV suppression by inhibiting Tat transactivation and transcription elongation as well as by inducing repressive chromatin structure at the HIV promoter. Our findings establish a proof of concept for modulation of BRD4 to epigenetically suppress HIV and provide a promising chemical scaffold for the development of probes and/or therapeutic agents for HIV epigenetic silencing.
Keywords: AIDS/HIV
Keywords: Drug therapy, Epigenetics
Introduction
HIV infection continues to cause a global pandemic. Antiretroviral therapy (ART) can effectively suppress active HIV replication and has improved survival and quality of life of HIV-infected individuals. However, because HIV integrates its provirus into the host genome and establishes stable latent infection, ART fails to eradicate the virus (1–3). Discontinuation of ART almost inevitably results in reemergence of viral replication (4–6). Even under ART, a low level of residual HIV replication was believed to occur in some individuals (7–9), although this observation remains controversial (10, 11). Intensification of ART does not appear to reduce residual viral expression, which is thought to be a driver of inflammation and HIV-associated complications (12). Drug resistance is also a public health issue with the scale-up of ART (13). Due to the limitations of ART, approaches directed toward host mechanisms to suppress HIV (to minimize potential emergence of drug-resistant strains) are needed. These approaches can be complimentary to ART and further suppress residual viremia with the hope of eventually inducing HIV remission free of ART.
HIV gene expression is regulated by host cell epigenetic and transcriptional mechanisms (14, 15). Various approaches targeting these mechanisms to suppress integrated HIV have been reported. Small antisense RNAs targeted to the HIV promoter were shown to induce HIV transcriptional suppression and/or silencing (16–20). Chemical inhibition of Tat, a key viral protein in HIV transcription, by an inhibitor (dCA) can suppress Tat transactivation and induce a state of epigenetic repression in the HIV promoter (21, 22). In vivo administration of dCA can reduce residual HIV viremia and delay viral rebound in ART-suppressed, HIV-infected humanized mice (23). Despite substantial efforts to understand mechanisms for epigenetic regulation of HIV, effective approaches, and especially those utilizing small molecules and targeting host epigenetic factors to silence HIV, are limited.
The bromodomain and extraterminal (BET) family proteins, including BRD4, are a group of epigenetic factors characterized by 2 N-terminal bromodomains (BDs) that bind acetylated histones in chromatin (24, 25). As an epigenetic reader, BRD4 is functionally versatile and interacts with a variety of partnering proteins to regulate gene expression (26–29). Accumulating evidence has suggested that BRD4 plays an important role in HIV transcriptional regulation (29–32). It was shown that BRD4 can suppress HIV transcription elongation by competing with Tat for cellular p-TEFb/CDK9 (30–32). Targeted modulation of BET/BRD4 by pan-BET inhibitor JQ1 (33) relieves the competition between BRD4 and Tat and therefore reactivates latent HIV (30, 31). Via its BDs, BRD4 can be recruited to HIV LTR through interactions with acetyl-histone H3 (AcH3) and AcH4. It was recently reported that differential interactions of BRD4 with AcH4 and AcH3 in the HIV promoter are associated with different effects on HIV transcription and latency establishment (34). Together, these studies support the functional versatility of BRD4 in regulation of target gene expression, depending on the partnering proteins it interacts with, and indicate that BRD4 and its associated epigenetic machinery can be potentially modulated to exert positive or negative effects on HIV proviral transcription.
Given the established role of BRD4 in HIV transcription, we were interested in discovering approaches that can modulate this pathway to induce HIV transcriptional activation or repression. Using structure-guided drug design, we have synthesized multiple libraries of BRD4 modulators (35). Screening these compounds led to identification of a small molecule, named ZL0580, that is more selective to the BD1 domain of BRD4 and induces a functional impact on HIV transcription distinct from that of JQ1. We found that, unlike JQ1, ZL0580 induced epigenetic suppression of HIV in multiple in vitro and ex vivo models. Gene KO and overexpression analysis confirmed that BRD4 played a predominant role and is specifically required in ZL0580-induced HIV suppression. We further showed that ZL0580 induced HIV suppression by inhibiting Tat transactivation and transcription elongation as well as by inducing a more repressive chromatin structure at the HIV LTR.
Results
Discovery of a lead small molecule (ZL0580) that suppresses HIV.
We have synthesized a batch class of small molecules designed to modulate BRD4 (35) and screened these compound libraries for their activities on HIV transcription using the HIV latently infected J-Lat cells (full-length 10.6) (36). Our initial goal was to identify new BRD4 modulators that are superior to JQ1 in activating latent HIV and, therefore, will serve as more potent latency reversing agents (LRA). Cells were treated with individual compounds, carrier (DMSO; negative control [NC]), JQ1, or PMA (positive control) singly for 24 hours. Activation of latent HIV was measured based on detection of GFP expression by flow cytometry. In one compound library (62 compounds; called C1-C62), we identified 3 compounds (ZL0454, ZL0482, and ZL0519) that modestly activate HIV; however, their potency is either weaker than or comparable to that of JQ1 (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI120633DS1).
Intriguingly, in an HIV suppression model in which J-Lat cells were stimulated with PMA to activate HIV and treated with individual compounds, we identified one lead compound (C48: ZL0580) that is distinct from JQ1, but suppresses PMA-induced HIV activation (Figure 1A). The chemical structure of ZL0580 is shown in Figure 1B, with its design and synthesis detailed in the Supplemental Methods. Further analysis showed that ZL0580 suppresses PMA-induced HIV activation in a dose-dependent manner (Figure 1, C and D). As a control, JQ1 alone activates HIV and synergistically enhances PMA-stimulated HIV activation (Figure 1, C and D). To determine whether HIV suppression by ZL0580 occurs at the transcriptional level, we quantified HIV mRNAs and showed that ZL0580 reduces both Gag and 3′ LTR RNA levels, while JQ1 enhances their levels (Figure 1E), supporting the idea that ZL0580 induces HIV transcriptional suppression. Kinetic analysis showed that single ZL0580 treatment (10 μM) suppresses both PMA-stimulated and basal HIV transcription through day 14 after treatment (Figure 1, F and G). In J-Lat cells, basal HIV transcription under resting conditions is readily detectable by quantitative PCR (qPCR) (Cq values shown in Supplemental Figure 2). In addition, we treated resting J-Lat cells with a single dose of ZL0580 for 3 days, followed by reactivation by LRAs. ZL0580 pretreatment rendered J-Lat cells more resistant to HIV reactivation by suberoylanilide hydroxamic acid (SAHA) or prostratin (Figure 1H), indicating that ZL0580 may induce epigenetic reprogramming of HIV LTR.
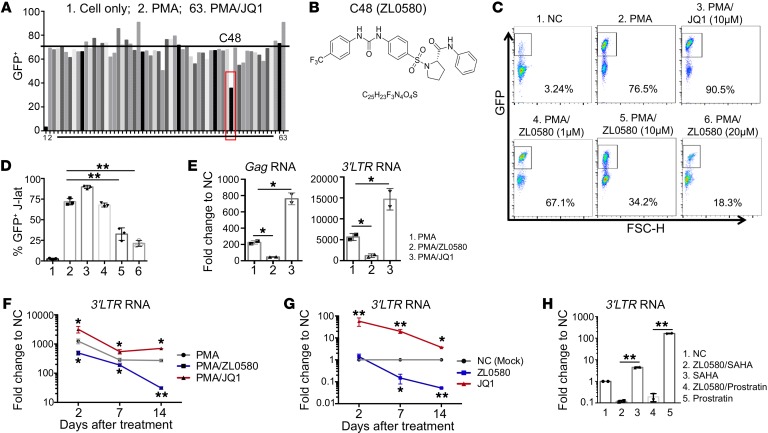
Figure 1. Discovery of a small molecule suppressing HIV in J-Lat cells.
(A) Screening of compounds (C1–C62) designed as new BRD4 modulators in J-Lat cells (10.6). Cells were stimulated with PMA (1 μg/mL) to activate HIV and treated with individual compounds (10 μM) for 24 hours (PMA/C1–C62). Cell only (NC), PMA, and PMA/JQ1 (10 μM) were included as controls (labeled as 1, 2, and 63). HIV activation was measured by flow cytometry (GFP+%). (B) Chemical structure of ZL0580. (C and D) Dose-dependent suppression of PMA-induced HIV activation by ZL0580. Cells were treated with PMA and ZL0580 (0 μM, 1 μM, 10 μM, 20 μM) for 24 hours. NC or PMA/JQ1 (10 μM) was included as a control. Representative FACS plots for GFP expression (C) and cumulative data for percentage of GFP+ in J-Lat cells of 3 experimental repeats (D) (mean ± SD) are shown. (E) Comparison of HIV transcription. HIV RNAs (Gag and 3′ LTR) were quantified by qPCR in cells 24 hours after treatment. Results are shown as fold change relative to NC. (F and G) Kinetics of ZL0580-induced HIV suppression in PMA-activated (F) or resting (G) J-Lat cells. Cells were treated as indicated for 24 hours. HIV 3′ LTR RNA was quantified on days 2, 7, and 14 after treatment. Data are shown as fold change relative to NC for each time point. Asterisks denote comparison of PMA/ZL0580 or PMA/JQ1 with PMA (F) or comparison of ZL0580 or JQ1 with NC (G). Error bars in E–G represent SD of PCR duplicate. (H) Unstimulated J-Lat cells were treated with NC or ZL0580 (10 μM), followed by stimulation with SAHA or prostratin 3 days after treatment. HIV reactivation was measured based on 3′ LTR RNA, and results are shown as fold change relative to NC. All experiments were repeated at least 3 times. *P < 0.05; **P < 0.005, 1-way ANOVA (D) and paired Student’s t test (E–H).
We examined toxic effects of ZL0580 on J-Lat cells by treating them with a wide range of ZL0580 (0–80 μM) for various lengths of time (1 and 3 days), followed by LIVE/DEAD aqua blue staining and flow cytometric analysis for cell viability. ZL0580 did not cause significant cell death at concentrations below 40 μM (Supplemental Figure 3A). In HIV-suppression kinetic analysis (Figure 1, F and G), treatment of J-Lat cells with ZL0580 (10 μM) also did not cause significant cell death on days 2, 7, and 14 compared with NC in both PMA-activated and unstimulated cells (Supplemental Figure 3B). These data indicate that the observed effect of ZL0580 is independent of cell toxicity.
In this compound library, in addition to ZL0580, we noted that 2 other compounds (ZL0506 and ZL0549) could also modestly suppress HIV (Supplemental Figure 4). In contrast, the 3 compounds described above (ZL0482, ZL0454, and ZL0519) that could activate latent HIV manifested effects similar to those of JQ1 and synergistically enhanced PMA-induced HIV activation (Supplemental Figure 4C). These data suggest that, instead of being a pan-assay interference compound (37), the HIV-suppressive effect of ZL0580 in J-Lat cells is specific.
ZL0580 suppresses HIV in in vitro HIV-infected human CD4+ T cells. To determine whether ZL0580 can suppress HIV in a more relevant system, we used human CD4+ T cells that were infected with HIV in vitro. Normal PBMCs were stimulated with phytohemagglutinin (PHA) (typically more efficient in activating T cells than PMA for longer-term culture) for 2 days, followed by HIV infection (US-1 strain) in the presence of ZL0580 (0–8 μM) or a control compound, ZL0454 (0–8 μM). ZL0454 was selected from the same library, since ZL0454 did not suppress HIV in J-Lat cells (Supplemental Figure 1 and Supplemental Figure 4C). HIV infection in CD4+ T cells was assessed 3 days after viral exposure by flow cytometry based on intracellular p24 expression. Consistent with the results in J-Lat cells, ZL0580 could also suppress HIV in activated human CD4+ T cells in a dose-dependent manner (Figure 2, A and B). Of note, the potency of ZL0580 to suppress HIV in primary CD4+ T cells appeared to be stronger than that in J-Lat cells, since single ZL0580 treatment (8 μM) led to almost complete loss of productive HIV infection in CD4+ T cells (Figure 2, A and B). As a control, treatment with ZL0454 did not suppress HIV in CD4+ T cells (Figure 2, A and B). To ensure that ZL0580 itself did not affect HIV infection of PBMCs, we quantified cell-associated HIV DNA and found that ZL0580 treatment did not significantly alter HIV DNA levels (Figure 2C), but reduced HIV transcription in PBMCs (Figure 2D).
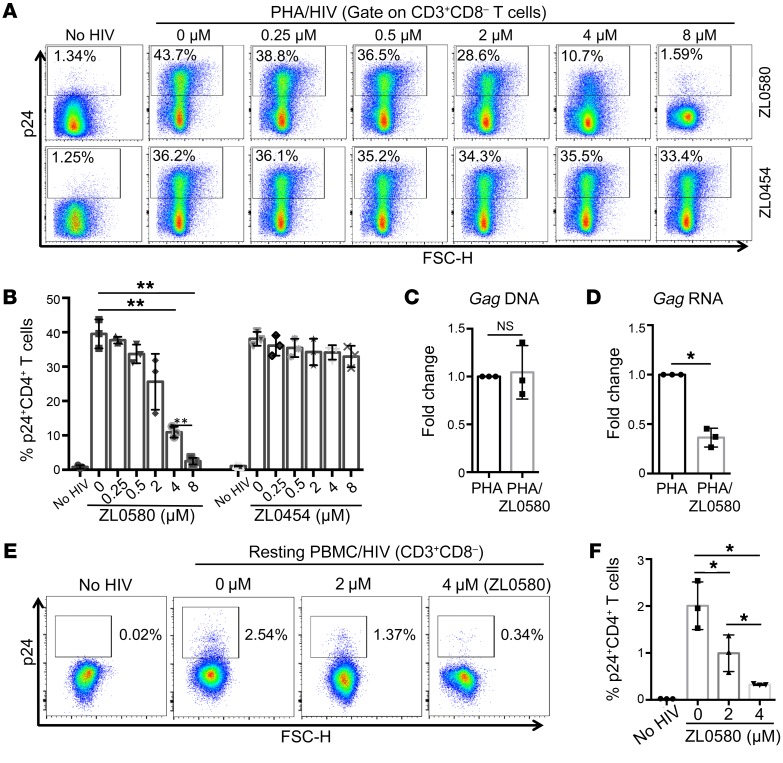
Figure 2. HIV suppression by ZL0580 in in vitro HIV-infected human CD4+ T cells.
(A) HIV infection of PHA-activated CD4+ T cells in normal PBMCs. PBMCs (n = 3) were stimulated with PHA (1 μg/mL) for 2 days, followed by infection with R5 HIV (US-1) in the absence or presence of ZL0580 (top) or ZL0454 (bottom) at various concentrations, as indicated. Three days after viral exposure, HIV infection in CD4+ T cells was measured by flow cytometry based on intracellular p24 staining. Representative FACS plots are shown. (B) Comparison of percentage of p24+CD4+ T cells in PBMCs. (C and D) Quantification of HIV DNA (C) and Gag RNA (D) in PBMCs following different treatments by qPCR. Data are shown as fold change of PHA/ZL0580 (4 μM) relative to PHA alone. (E) Representative FACS plots showing HIV infection (intracellular p24) of unactivated CD4+ T cells in PBMCs on day 6 after treatment. (F) Comparison of p24+ percentage in unactivated CD4+ T cells in PBMCs. (B–D and F) Mean ± SD from 3 PBMC donors. *P < 0.05; **P < 0.01, 1-way ANOVA (B and F) and paired Student’s t test (C and D).
We also evaluated the impact of ZL0580 on HIV in unactivated (resting) CD4+ T cells. Normal PBMCs were not stimulated, but were directly infected with HIV by spinoculation. Twenty-four hours after HIV infection, cells were extensively washed and treated with ZL0580 (2 μM and 4 μM) or not treated (NC). As expected, compared with activated cells, HIV replication kinetics in unactivated CD4+ T cells was slower. However, we were able to detect low but significant levels of HIV replication (intracellular p24 expression) on day 6 after HIV inoculation (%p24+, 2.54%) (Figure 2E). Of importance, ZL0580 also dose dependently suppressed HIV in unactivated CD4+ T cells (Figure 2, E and F).
We assessed PBMC viability and found that it was comparable between NC and ZL0580 treatment (at 8 μM) (Supplemental Figure 5A), indicating that the HIV-suppressive effect of ZL0580 in human CD4+ T cells was not due to overt cell toxicity. Potential toxic effects of ZL0580 on PBMCs were evaluated with a wider range of ZL0580 concentrations (0–80 μM). Similarly to what occurred with J-Lat cells, ZL0580 did not cause significant PBMC death at concentrations below 40 μM (Supplemental Figure 5B).
We further examined the effects of ZL0580 on T cell phenotypes and activation markers associated with HIV infection and replication. ZL0580 did not significantly alter the expression of HIV entry receptors (CD4 and CCR5) on CD4+ T cells (Supplemental Figure 6, A and B). Expression of activation markers (CD25, CD38, and HLA-DR) is also comparable between NC and ZL0580 treatment for both activated and unactivated T cells (Supplemental Figure 6, C and D). To more broadly assess the impact of ZL0580 on T cells, we examined the expression of an array of genes in PHA-activated and resting PBMCs following ZL0580 treatment or no treatment. A total of 17 genes closely associated with T cell phenotypes and functions were chosen, including cytokines and chemokines, transcription factors, lineage differentiation factors, innate HIV restriction factors, and transcript elongation factors (Supplemental Figure 6, E and F). The data showed that ZL0580 did not significantly alter the expression of these genes in PBMCs (Supplemental Figure 6, E and F), indicating that ZL0580 did not induce global changes in human T cells.
ZL0580 suppresses HIV transcription ex vivo in PBMCs of viremic HIV-infected participants.
We next assessed HIV-suppressive activity of ZL0580 in PBMCs of HIV-infected individuals ex vivo. At this point, our goal was determining whether ZL0580 manifested any suppressive effect on induced HIV transcription. Therefore, we chose PBMCs of viremic HIV-infected participants (RV21 cohort) (38) (Supplemental Table 2), which enabled HIV transcription to be more readily detectable due to higher levels of viral transcription. PBMCs were activated by PHA in the absence or presence of ZL0580 treatment (8 μM) for 2 days, followed by measurement of HIV transcription by quantifying gag mRNA. Highly consistent with the results in J-Lat cells and human CD4+ T cells infected in vitro, ZL0580 could also induce fairly potent suppression of induced HIV transcription ex vivo in PBMCs of viremic HIV-infected individuals (Figure 3). While the potency of suppression varied among different participants, ZL0580 induced significant suppression in 7 out 8 PBMCs examined (Figure 3).
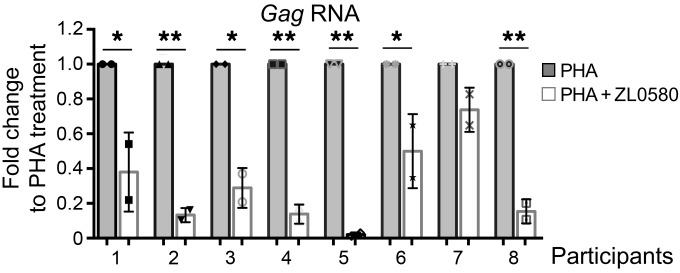
Figure 3. ZL0580 suppresses HIV transcription ex vivo in PBMCs of viremic HIV-infected individuals.
PBMCs of RV21 participants (n = 8) were activated by PHA and treated with ZL0580 (8 μM) or not treated. Two days after treatment, cell-associated HIV Gag RNA was quantified by qPCR. For each PBMC, data are shown as fold change of PHA+ZL0580-treated cells relative to PHA-treated cells. PCR was conducted in duplicate, and error bars represent SD of PCR replicate for each PBMC. *P < 0.05; **P < 0.005, paired Student’s t test.
ZL0580 suppresses HIV in PBMCs of aviremic HIV-infected participants.
To explore the impact of ZL0580 on latent HIV, PBMCs of ART-suppressed, aviremic HIV-infected participants in the RV254 cohort (39) (Supplemental Table 3) were examined in 2 different models. First, PBMCs (n = 5; 6 months after ART) were stimulated with anti-CD3/CD28 to activate latent HIV and to induce CD4+ T cell expansion. An advantage of this model is that expanded T cells can be cultured for more than 3 to 4 weeks with good viability and allow durability analysis (22). Cells were treated with ART alone (efavirenz, zidovudine, raltegravir), ART plus ZL0580 (2 μM), or mock treatment (NC) in IL-2–containing medium. ART was initiated at the beginning to control spreading infections. Medium was replaced every 3 days (maintaining the same drugs), and HIV production was measured by ultrasensitive nested PCR (22). We showed that this PCR method can sensitively detect less than 10 HIV copies (Supplemental Figure 7). As shown in Figure 4A, while HIV production kinetics varied among the 5 PBMCs, ZL0580 treatment promoted HIV suppression during ART and delayed viral rebound after ART cessation (Figure 4A). Quantitative analysis was performed to evaluate the impact of ZL0580 on promoting HIV suppression (before treatment cessation) and on viral rebound (after treatment cessation) by comparing the length of time (days) and AUC between ART alone and ART plus ZL0580. For HIV suppression, ART alone required 15 ± 3.3 days to induce full suppression, whereas ART plus ZL0580 required 12 ± 3.6 days (P < 0.05) (Figure 4B). Comparison of AUC before treatment cessation also revealed modest, but significant, differences between ART and ART plus ZL0580 (Figure 4B). Notably, the impact of ZL0580 on viral rebound was more significant (Figure 4C). For ART alone, viral rebound was quickly detected within 3 to 6 days in all 5 PBMCs after treatment removal, while ART plus ZL0580 led to marked delays in viral rebound in 3 out of 5 PBMCs (Figure 4A). Similar quantitative analysis showed that days to viral rebound in ART alone and ART plus ZL0580 treatments were 2.4 ± 1.3 and 15 ± 6.1, respectively (P < 0.005) (Figure 4C). Comparison of AUC (after treatment cessation) also revealed significant differences between the 2 groups (P < 0.05) (Figure 4C). Through the experiments, we closely monitored cell viability and observed that, while PBMC viability decreased over time, ZL0580 treatment did not cause significant toxicity to these cells compared with ART alone or NC (Supplemental Figure 8). Experiments were terminated when total cell viability dropped below 30% (around 27 to 33 days after initial treatment).
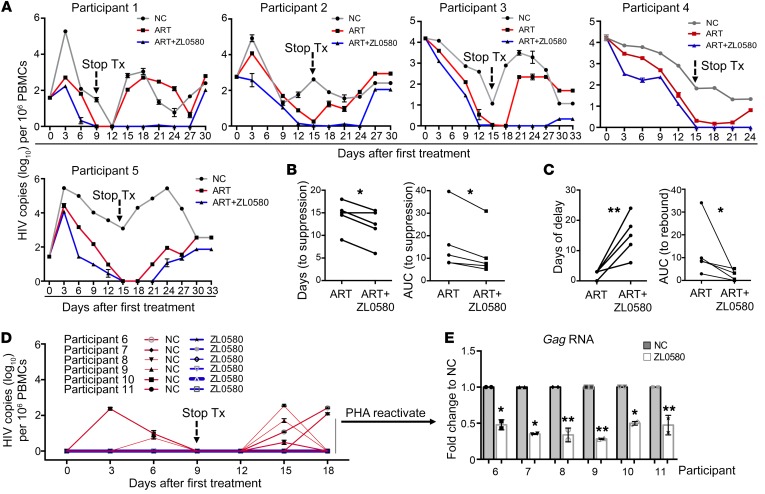
Figure 4. Suppressive effect of ZL0580 on HIV in PBMCs of ART-suppressed, aviremic HIV-infected participants.
(A) PBMCs of aviremic RV254 participants (n = 5) were stimulated with anti-CD3/CD28 and cultured in IL-2–containing medium to induce latent HIV activation and CD4+ T cell expansion. Cells were treated with ART alone, ART+ZL0580 (2.5 μM), or were mock treated (NC). HIV release in supernatants was quantified by the 2-step nested qPCR. Following full HIV suppression, treatments were stopped and viral RNA copies were continuously monitored every 3 days. Data are shown as HIV copies (log10) per 106 PBMCs. (B and C) Quantitative analysis of the effect of ZL0580 on promoting HIV suppression during ART (B) and on viral rebound following ART cessation (C). Comparison of length of time (days) and AUC prior to treatment cessation (B) and after treatment cessation up until first viral rebound (C) between ART and ART+ZL0580 for the 5 PBMCs. AUC for each PBMC was quantified using Prism. (D) HIV production by unactivated RV254 PBMCs (n = 6). PBMCs were directly treated with ZL0580 (2.0 μM) or not treated (NC) on days 0, 3, and 6 (treatment stopped on day 9). HIV production in supernatants was measured once every 3 days as indicated. (E) After day 18, PBMCs were stimulated with PHA to reactivate latent HIV. HIV transcriptional reactivation was measured by quantifying Gag RNA in cells. The data are shown as fold change of ZL0580 treatment relative to NC for each PBMC. (A, D, and E) PCR was conducted in duplicate, and error bars show PCR replicate SD. *P < 0.05; **P < 0.005, (B, C, and E), paired Student’s t test.
Second, PBMCs of aviremic participants (n = 6) were not activated, but were directly treated with ZL0580 (2 μM) or not treated (without ART). Treatment was given every 3 days for a total of 3 times (day 0, 3, 6) and stopped on day 9 (Figure 4D). This model aimed to determine whether ZL0580 could inhibit spontaneous HIV production and suppress LRA-stimulated latent HIV reactivation. Of interest, while low levels of spontaneous HIV production were readily detectable in some PBMCs of the NC group (1 PBMC on day 3, 2 on day 6, 4 on day 15, and 2 on day 18), ZL0580 treatment inhibited spontaneous HIV production in all 6 PBMCs (Figure 4D). Cell culture was continuously monitored to day 18, and cells remained in good condition. Cells were then stimulated with PHA to reactivate latent HIV. We found that treatment of these PBMCs with ZL0580 led to inhibition of PHA-activated HIV transcription compared with NC (Figure 4E). This finding is consistent with the result in J-Lat cells (Figure 1H) and indicates that ZL0580 displays a repressive effect on latent HIV in aviremic PBMCs.
ZL0580 induces HIV suppression via BRD4.
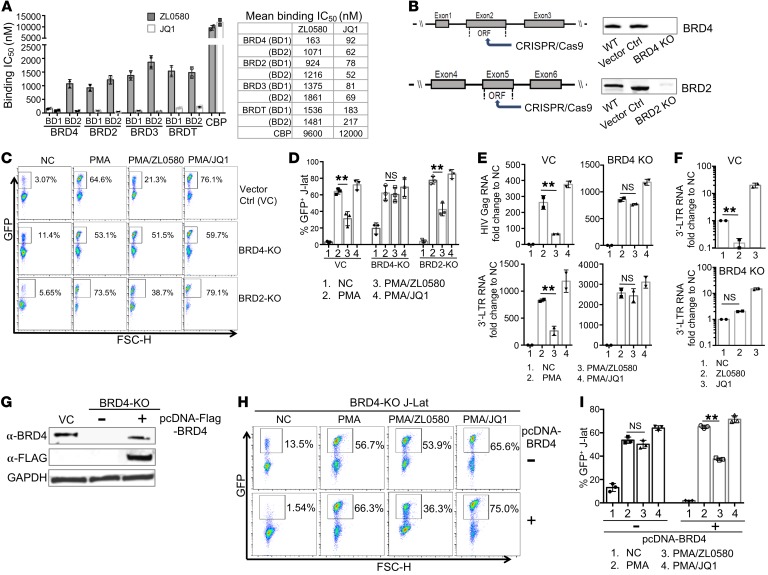
To understand mechanisms underlying ZL0580-induced HIV suppression, we explored roles of BET proteins. BET proteins consist of BRD2, BRD3, BRD4, and BRDT (25), among which BRD4 and BRD2 were shown to participate in HIV transcriptional regulation (30, 32, 40). First, we measured binding affinity of ZL0580 to BDs of BET as compared with JQ1 using the time-resolved fluorescence energy transfer (TR-FRET) assay and found that JQ1 nonselectively bound to both BD1 and BD2 all 4 BET proteins (Figure 5A), consistent with the previously described role of JQ1 as a pan-BET inhibitor (33). In contrast, ZL0580 more selectively bound to the BRD4 BD1 domain (IC50 = 163 nM) instead of BD2 (IC50 = 1071 nM). ZL0580 also displayed approximately 6- to 11-fold selectivity for BRD4 over other BET proteins (IC50 ranged from approximately 0.9 to 1.9 μM) as well as over a non-BET BD protein, CBP (IC50 >10 μM) (Figure 5A). We also assessed binding activity of ZL0580 to a broader panel of nonspecific cellular proteins and showed that ZL0580 also manifested no or weak binding activity toward these nonspecific targets (Supplemental Table 1).
Figure 5. ZL0580 selectively binds to BRD4 (BD1), and BRD4 is functionally required for ZL0580-induced HIV suppression.
(A) In vitro binding of ZL0580 or JQ1 to BD1 and BD2 of BET proteins measured by TR-FRET (IC50, nM). Error bars represent SD of assay replicates. (B) CRISPR/Cas9 KO of BRD4 and BRD2 in J-Lat cells. (C and D) BRD4 KO abrogates ZL5080-induced HIV suppression. VC, BRD4-KO, or BRD2-KO cells were treated as indicated. Representative FACS plots for GFP expression (C) and cumulative data comparing GFP+ percentage in VC and KO J-Lat cells from 3 independent experiments (mean ± SD) (D) are shown. (E) Expression of Gag and 3′ LTR RNAs in PMA-activated, VC, or BRD4-KO cells after treatment (24 hours). (F) Expression of 3′ LTR RNA in unstimulated, VC, or BRD4-KO J-Lat cells after different treatments (on day 7). (G) Exogenous BRD4 expression in BRD4-KO J-Lat. KO cells were nucleofected with pcDNA-FLAG-BRD4 plasmid (+) or not treated (–). BRD4 expression was measured by Western blot using anti-BRD4 and anti-FLAG antibody (day 4 after nucleofection). VC was included as a control. (H and I) Effect of BRD4 overexpression on ZL0580-induced HIV suppression. BRD4-KO cells without (top) or with BRD4 overexpression (bottom) were treated as indicated. Representative FACS plots for GFP expression (H) and cumulative data comparing GFP+ percentage of J-Lat cells among different treatments (I) are shown. In this figure, error bars show SD of experimental replicates (D and I) and of PCR duplicate (E and F). Experiments were repeated at least 3 times. **P < 0.005, paired Student’s t test (D–F, I).
To explore the potential binding mode of ZL0580 to BRD4 as compared with that of JQ1, we conducted docking analysis for ZL5080 binding to BRD4 BD1 and BD2 domains using the determined BRD4/JQ1 cocrystal structure (33) and showed that ZL0580 formed strong interactions with BD1 compared with BD2 of BRD4 (Supplemental Figure 9, A and B). Overlay analysis by superimposing the ZL0580 docked pose with the BRD4 (BD1)/(+)-JQ1 complex structure (33) (Supplemental Figure 9C) indicated that ZL0580 binds to BRD4 BD1 with adequate access into its acetyl-lysine (KAc) binding pocket, but manifests notable differences from JQ1. ZL0580 has a partial scaffold extended to an additional region; the proline sulfonamide fragment of ZL0580 overlaps with the crystallographically determined JQ1-binding mode, while the phenylurea sulfonamide moiety extends to the region between the WPF shelf and the ZA channel (Supplemental Figure 9C). These data imply that ZL0580 can interact with BRD4, but manifests a binding mode distinct from JQ1.
To directly examine functional roles of BRD4 and BRD2 in ZL0580-induced HIV suppression, we used CRISPR/Cas9 to generate stable BRD4- and BRD2-KO J-Lat cell lines (Figure 5B and Supplemental Figure 10). Efficient KO of BRD4 and BRD2 in J-Lat cells was confirmed by Western blotting (Figure 5B). We also showed that other BET proteins (BRD2, BRD3, BRDT) were normally expressed in the BRD4-KO J-Lat cells, indicating that CRISPR/Cas9 did not induce broad off-target effects (Supplemental Figure 11). Intriguingly, deletion of BRD4, but not BRD2, largely abrogated ZL0580-induced HIV suppression (Figure 5, C and D), supporting the idea that BRD4 is functionally required in this process. Another interesting observation is that BRD4 KO led to enhanced basal HIV activation in resting J-Lat cells (% GFP+ in NC, 11.4%) as compared with the vector control (VC) J-Lat cells (% GFP+, 3.07) (Figure 5C), indicating that BRD4 may repress basal HIV transcription as well.
In addition to detection of GFP, we measured HIV transcription in these J-Lat cells to further verify the functional role of BRD4 (Figure 5E). Consistent with the GFP results, ZL0580 can substantially inhibit Gag and 3′ LTR RNA expression in PMA-activated, VC J-Lat cells, which was largely abrogated in the BRD4-KO cells (Figure 5E). A similar pattern was observed in the unstimulated/resting J-Lat cells: while ZL0580 induced inhibition of basal HIV transcription (3′ LTR RNA on day 7) as compared with NC and JQ1 treatment (Figure 5F). Such inhibitory effects were abrogated to a large extent when BRD4 was knocked out (Figure 5F). Here, HIV transcription on day 7 after treatment was chosen for comparison based on the earlier data showing that ZL0580 induces potent suppression of basal HIV transcription in unstimulated J-Lat cells on that day (Figure 1G).
To further confirm that BRD4, instead of other BD-containing proteins (due to possible CRISPR/Cas9-induced off-target effects), is required in this process, exogenous BRD4 was overexpressed in the BRD4-KO cells by nucleofecting the cells with BRD4-encoding plasmid (pcDNA5-FLAG-BRD4; Addgene) (41). Efficient BRD4 expression was detected in the KO cells on day 4 after nucleofection (Figure 5G). Using an optimized protocol, nucleofection did not cause overt toxic effects to J-Lat cells. BRD4 overexpression was confirmed with both anti-BRD4 and anti-FLAG antibodies (Figure 5G). We showed that, while ZL0580 failed to induce HIV suppression in BRD4-KO cells (Figure 5, H and I), exogenous BRD4 expression restored to a large extent the ability of ZL0580 to suppress HIV in these cells (Figure 5, H and I). These data provide additional evidence that ZL0580 induces HIV suppression via BRD4.
ZL0580 inhibits Tat transactivation and key factors in HIV transcription elongation.
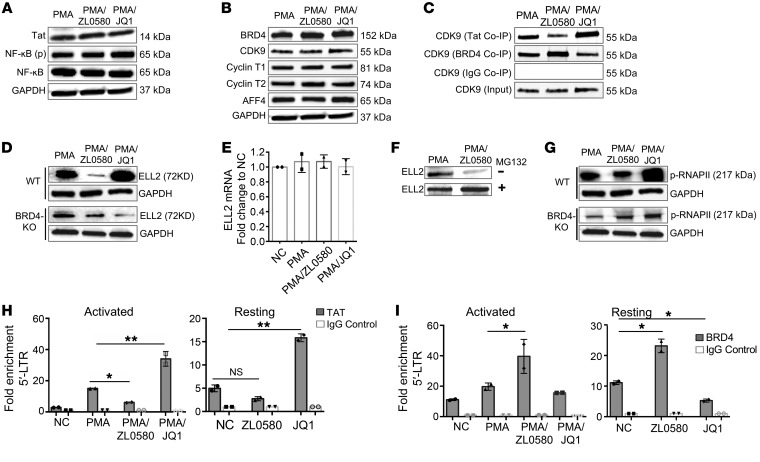
After demonstrating a functional requirement of BRD4 in ZL0580-induced HIV suppression, we next explored downstream biochemical mechanisms. We showed earlier (Figure 1) that ZL0580 can suppress HIV in activated J-Lat cells at 24 hours after treatment. Based on these data, we first measured Tat protein expression to explore the stage or stages of HIV transcription inhibited by ZL0580. As compared with PMA alone or PMA+JQ1, treatment with PMA+ZL0580 did not significantly (or slightly) reduce Tat protein (Figure 6A), indicating that ZL0580 may suppress HIV involving a stage or stages after Tat synthesis, at least at the 24-hour time point after treatment. Consistent with this result, expression of NF-κB, which is important for HIV transcriptional initiation, was not affected by ZL0580 either (Figure 6A).
Figure 6. ZL0580 inhibits Tat transactivation and key factors in HIV transcription elongation.
(A and B) Western blot measurement of Tat and NF-κB (A) and cellular proteins involved in transcription elongation (B) in WT J-Lat cells 24 hours after treatment. (C) Co-IP analysis for binding of CDK9 to Tat or BRD4 in WT J-Lat cells 24 hours after treatment. Control IgG Co-IP and input CDK9 were used as controls. Total/input CDK9 blots in panels (B and C) represent the same experiment. (D) ELL2 protein expression in WT and BRD4-KO J-Lat cells 24 hours after treatment. (E) ELL2 mRNA expression by qPCR in WT J-Lat cells 24 hours after treatment. (F) Effect of protease inhibition by MG132 on ELL2 protein levels in WT J-Lat cells. Cells were pretreated with proteasome inhibitor MG-132 for 6 hours (bottom) or not treated (top), followed by treatment with PMA or PMA+ZL0580 (10 μM) for 18 hours. ELL2 protein was measured by Western blot. (G) Phosphorylated RNAPII (Ser 2 CTD) in WT (top) and BRD4-KO (bottom) J-Lat cells after different treatments. Loading control GAPDH in panel (D and G) represents the same experiment. (H and I) ChIP-qPCR analysis for recruitment of Tat (H) or BRD4 (I) to HIV 5′ LTR in PMA-activated or unstimulated J-Lat cells 24 hours after treatment. ChIP using control IgG was included for normalization. qPCR was conducted to quantify the precipitated HIV 5′ LTR region. Data were normalized to NC. Error bars (E, H, and I) show SD of qPCR replicate. All experiments were independently conducted at least 3 times. *P < 0.05; **P < 0.005, 1-way ANOVA (H and I). ND, nondetectable.
Next, we explored mechanisms involved in Tat transactivation and transcription elongation. An established role of BRD4 is recruitment of cellular super elongation complex (SEC) (e.g., p-TEFb/CDK9) to target the gene promoter, stimulating RNA Pol II (RNAPII) activation and transcription elongation (42). In HIV-infected cells, BRD4 competes with Tat for cellular reservoirs of CDK9 and negatively regulates HIV transcription elongation (30–32). First, we examined major proteins involved in HIV transcription elongation, including BRD4, CDK9, cyclin T1/T2, and AFF4. Neither ZL0580 (PMA+ZL0580) nor JQ1 (PMA+JQ1) treatment altered expression of these proteins (Figure 6B). However, Co-IP analysis of protein-protein interactions led to interesting findings. Tat Co-IP showed that, compared with PMA alone, ZL0580 treatment (PMA+ZL0580) reduced CDK9 binding to Tat, whereas JQ1 enhanced CDK9 binding to Tat (Figure 6C). This is not simply due to differential input Tat levels, since they were comparable among different treatments (Figure 6A). In contrast, BRD4 Co-IP revealed an opposing result: compared with PMA alone, ZL0580 induced enhanced CDK9 binding to BRD4, whereas JQ1 decreased CDK9 binding to BRD4 (Figure 6C).
Like CDK9, ELL2 is another catalytic factor of SEC and is implicated in HIV transcriptional regulation (43). When measuring cellular proteins in these treated cells, we made an interesting observation, that ZL0580 and JQ1 differentially regulated ELL2 protein. Compared with PMA alone, ZL0580 substantially reduced ELL2 protein, whereas JQ1 increased its levels (Figure 6D). Such distinct effects on ELL2 by ZL0580 and JQ1 were abrogated, to a large extent, in BRD4-KO cells (Figure 6D), supporting a functional role of BRD4 in mediating the regulatory effects of ZL0580 and JQ1 on HIV transcription. Further, we identified that ZL0580 inhibited ELL2 by reducing its protein stability, since ELL2 mRNA transcription was not altered by ZL0580 (Figure 6E) and proteasome inhibition (by MG132) almost completely recovered ELL2 in ZL0580-treated cells (Figure 6F). These data indicate that ZL0580 inhibits ELL2 via mechanisms involving posttranslational degradation (44). Among the many proteins examined, only ELL2 was inhibited by ZL0580, indicating that this effect of ZL0580 on cellular proteins is likely selective for ELL2.
RNAPII activation is a rate-limiting step in transcription elongation (30, 31). Stimulation of RNAPII activation (phosphorylation of CTD Ser2 of RNAII) can be mediated by Tat-recruited SEC. We found that, compared with PMA alone, ZL0580 reduced RNAII activation, whereas JQ1 potentiated its activation (Figure 6G); notably, such differential effects on RNAPII activation were also abrogated to some extent when BRD4 was knocked out (Figure 6G). This finding is consistent with the above observations of CDK9 binding to Tat or BRD4 (Figure 6C) and the effects on ELL2 protein levels by ZL0580 and JQ1 (Figure 6, D–F), providing evidence that, in contrast with from JQ1, ZL0580 induces inhibition of key factors in HIV transcription elongation.
To directly examine the impact of ZL0580 on Tat transactivation, we measured recruitment of Tat to the HIV promoter (a small region overlapping with HIV transcription start site [TSS]) (31) by ChIP-qPCR and observed that, while JQ1 enhanced binding of Tat to the HIV promoter, ZL0580 reduced Tat recruitment to the HIV promoter (Figure 6H). As a control, no Tat recruitment to the GAPDH promoter was detected (Supplemental Figure 12). A similar pattern for Tat binding to the HIV promoter was also observed in unstimulated J-Lat cells (Figure 6H). These data support the idea that inhibition of Tat transactivation may represent a mechanism by which ZL0580 induces HIV suppression. Moreover, we measured BRD4 binding to the HIV promoter in these treated cells. Of interest, BRD4 displayed a binding pattern distinct from that of Tat in both activated and resting J-Lat cells: compared with no compound control, ZL0580 promoted BRD4 binding to the HIV promoter, whereas JQ1 reduced this binding (Figure 6I), indicating that BRD4 may completely block or reduce Tat transactivation following ZL0580 treatment.
ZL0580 induces a more repressive chromatin structure in HIV LTR.
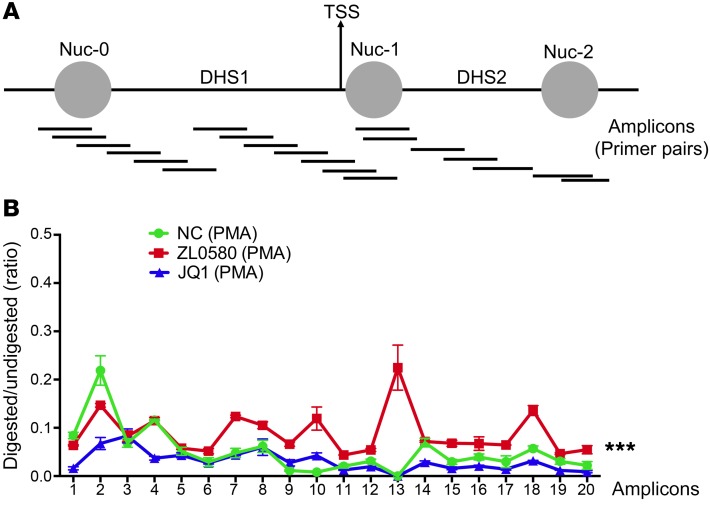
Nucleosome organization and structure in HIV LTR correlates with HIV proviral transcription (45). Positioning of a nucleosome (nuc-1) downstream of HIV TSS restricts HIV transcription (Figure 7A) (45). To this end, we have observed that, in addition to induced HIV transcription in the activated cells, ZL0580 also induced suppression of basal and latent HIV in unstimulated cells, in which HIV underwent low-level transcription and Tat protein was considered low. We therefore speculate that mechanisms beyond Tat-mediated transcription may also play a role in ZL0580-induced HIV suppression. We examined effects of ZL0580 on nucleosome organization and DNA accessibility in HIV LTR, using high-resolution MNase nucleosomal mapping (45). J-Lat cells were pretreated with ZL0580, JQ1, or not treated (NC) for 24 hours, followed by PMA activation for 24 hours. Chromatin isolated from cells was divided into undigested and MNase-digested samples. Digested and undigested DNA samples were probed with 20 separate sets of overlapping primers to amplify different regions along the HIV LTR (Figure 7A) (45). The principle is that DNA within nucleosomes would be protected (at least partially) from MNase digestion, whereas nucleosome-free and linker DNAs would be cleaved. Accordingly, the ratio for the PCR product in digested DNA to that of undigested control can be used to assess nucleosomal occupancy and DNA accessibility. We found that compared with the no-compound treatment control (NC), treatment with ZL0580 led to enhanced nucleosomal DNA protection in the majority of amplicon regions (6–20), especially in the amplicon 13 that covers Nuc-1 immediately downstream of TSS (Figure 7B). In contrast, compared with NC, treatment of J-Lat cells with JQ1 reduced nucleosomal DNA protection in several amplicon regions, although the effect of JQ1 appeared to be only modest (Figure 7B). These data indicate that JQ1 may cause a less repressive nucleosomal structure in the HIV LTR, consistent with a recent report (29). These data together suggest that ZL0580 induces chromatin remodeling and causes a more repressive nucleosomal structure in the HIV LTR.
Figure 7. Analysis of chromatin structure in HIV LTR by high-resolution MNase nucleosomal mapping.
(A) Schematic diagram illustrating PCR amplicons at the HIV LTR covering 40–902 nucleotides that correspond to Nuc-0, DNase hypersensitive site 1 (DHS1), Nuc-1, DHS2, and Nuc-2. PCR product sizes are 100 ± 10 bp approximately 30 bp apart from each other. (B) Profile of chromatin structure in the HIV LTR in J-Lat cells after different treatments. Cells were either untreated (NC) or treated with ZL0580 (10 μM) or JQ1 (10 μM) for 24 hours, followed by activation with PMA for 24 hours. Chromatin profile was determined by calculating the ratio (y axis) for the amount of PCR product in the MNase-digested DNA relative to that of the undigested control DNA for each primer pair. The x axis shows corresponding PCR amplicon with bp units with 0 as the start of LTR Nuc-0. The MNase assay was independently repeated twice. Error bars show SD of PCR replicates. ***P < 0.001 denotes statistical comparison among NC, ZL0580, and JQ1 by 1-way ANOVA.
Discussion
Epigenetic silencing of the chromatinized HIV provirus is considered a potential “block-and-lock” approach for HIV functional cure (46). The present study provides evidence supporting a proof of concept that BRD4 and its associated epigenetic machinery can be modulated to repress integrated HIV. We present a small molecule (ZL0580) that more selectively binds to BRD4 as compared with JQ1 and induces epigenetic suppression of HIV. Our data show that ZL0580 can suppress both induced and basal HIV transcription and that ZL0580 cotreatment with ART accelerates HIV suppression and delays viral rebound ex vivo. Mechanistically, ZL0580 induces HIV suppression by inhibiting Tat-mediated transcription elongation and inducing a repressive chromatin structure at the HIV promoter.
Modulation of the target protein or pathway by different regulatory agents (e.g., agonist and antagonist) to induce distinct functional outcomes has been reported in previous studies. In this study, we present several lines of evidence, especially gene KO and overexpression analysis (Figure 5), supporting the idea that ZL0580 induces HIV epigenetic suppression via BRD4. In an effort to understand the molecular basis for how ZL0580 and JQ1 may engage BET/BRD4 to differentially regulate HIV, our data identified several key differences in the interaction of these 2 molecules with BET/BRD4: (a) binding assay shows that JQ1 nonselectively binds to all 4 BET proteins, whereas ZL0580 selectively binds to BRD4 (Figure 5A); (b) even within BRD4, ZL0580 manifests high selectivity toward BD1 over BD2, whereas JQ1 binds to both BDs (Figure 5A) (33); and (c) docking analysis indicates that potential binding modes of ZL0580 and JQ1 to BRD4 BDs are notably different (Supplemental Figure 9). Therefore, we speculate that these binding differences may affect BRD4 protein conformation and its interactions with partnering proteins, leading to differential regulation of HIV transcription. Indeed, we show that ZL0580 and JQ1 treatments led to differential binding of BRD4 to CDK9 (Figure 6C) and to differential nucleosomal structures in HIV LTR (Figure 7), which is possibly due to interactions of BRD4 with different chromatin modifiers or remodelers following ZL0580 or JQ1 treatment. Together, our data demonstrate that BRD4 plays a predominant role and is specifically required in ZL0580-induced HIV suppression, although it remains to be determined whether BRD4 is directly modulated by ZL0580 or acts as an intermediate component of a pathway induced by ZL0580 that eventually leads to HIV suppression. Further delineating molecular interaction of ZL580 with BRD4 as compared with that of JQ1 can help define these mechanisms.
BRD4 has been described as playing an important role in HIV transcriptional regulation (29–32). It is known that during induced HIV transcription, BRD4 acts as a repressor and competes with Tat for cellular CDK9 to inhibit transcription elongation (30–32). A role of BRD4 in basal HIV transcription under resting or latent conditions when Tat levels are low remains controversial. Earlier studies using BRD4 overexpression and retroviral LTR-driven reporter genes suggested that BRD4 promotes basal HIV transcription (26). In our study, we observed that ablation of BRD4 in unstimulated J-Lat cells enhanced HIV activation (Figure 5, C and D), supporting the idea that BRD4 functions to repress basal HIV transcription. Our observation is consistent with several recent studies reporting that knockdown of BRD4 can reactivate latent HIV in resting Jurkat cells (29, 30). Mechanisms for BRD4 repression of basal HIV transcription are less clear. A recent report showed that BRD4 represses latent HIV by inducing a repressive HIV LTR structure (29). Based on our data and recent literature, we postulate that BRD4 generally plays a repressive role for both basal and induced HIV transcription. Further investigations are needed to better understand the molecular mechanisms underlying BRD4-mediated HIV repression and its associated epigenetic cofactors.
BRD4 interacts with an array of epigenetic and transcriptional regulators, including acetyl-histones (24, 25), transcriptional factors (27, 32), and chromatin remodeling proteins (29), to modulate gene expression. An established mechanism of BRD4 for suppressing HIV is sequestering cellular CDK9 from Tat and competitively inhibiting Tat transactivation (30–32). In line with this mechanism, we show that, in a PMA-induced HIV transcription model, ZL0580 does not appear to inhibit HIV transcription initiation (Figure 6A), but suppresses Tat-mediated transcription elongation (Figure 6). In addition to suppressing HIV in activated cells, our data also show that ZL0580 suppresses basal and latent HIV in resting cells (Figure 1G, Figure 2, E and F, and Figure 4), indicating that ZL0580 may induce epigenetic reprogramming. In fact, nucleosome structures of HIV LTR, which can markedly influence HIV transcription (45), can be modulated by BRD4 (29). It was recently reported that BRD4 represses latent HIV via a mechanism independent of Tat but engaging chromatin remodeling proteins (SWI/SNF) to induce the repressive nucleosome signature at HIV LTR. Of note, this repressive effect of BRD4 on HIV LTR can be relieved by JQ1 (29). Our study demonstrates that ZL0580 induces a more repressive chromatin structure at HIV LTR (Figure 7), suggesting an explanation for why ZL0580 also represses basal and latent HIV in resting cells. Nonetheless, mechanisms for how ZL0580 induces repressive HIV LTR remain unclear in the current study, but may involve engagement of histone modifiers and chromatin remodeling proteins (29, 45). This mechanism is of particular relevance to durably enforcing HIV latency in the block-and-lock approach and needs to be further explored.
Efficient HIV transcription elongation requires proper SEC assembly (47). Like CDK9, ELL2 is another important catalytic factor in SEC (48). In our study, we made an intriguing observation that ZL0580 reduces cellular levels of ELL2 (Figure 6, D–F). Together with the finding on ZL0580-induced reduction of CDK9 binding to Tat (Figure 6C), these data help explain why ZL0580 reduces RNAPII activation (Figure 6G). Our data also imply that ZL0580 inhibits ELL2 by reducing its protein stability (Figure 6, E and F), consistent with recent studies reporting that posttranslational ubiquitination and proteasome degradation (mediated by Siah1, an E3 ligase) is a major mechanism for cellular regulation of ELL2 (43, 44). In addition, we revealed that ZL0580 induces ELL2 inhibition via BRD4 (Figure 4D). These data together indicate that downregulation of ELL2 may represent another mechanism by which ZL0580 engages BRD4 to suppress HIV in addition to the established role of BRD4 in competing with Tat for cellular CDK9 as described earlier. Our data also raised several interesting issues that have not yet been addressed: (a) whether ELL2 deficiency plays a role in SEC assembly at the HIV promoter; (b) whether E3 ligases, for example Siah1 (43, 44), mediate ELL2 ubiquitination and destabilization after ZL0580 treatment; and (c) how BRD4 engages cellular mechanisms (e.g., E3 ligase) to destabilize ELL2 and thereby regulate HIV transcription. Addressing these gaps, especially the mechanistic connection between BRD4 and ELL2, in future research will not only improve our understanding of mechanisms of action (MoA) for ZL0580, but also provide insights into HIV proviral regulation and latency.
Another important finding of our study is that ZL0580 induces significant delay in viral rebound after ART removal ex vivo in aviremic PBMCs (Figure 4, A–C). Mechanisms for the prolonged effect of ZL0580 on viral rebound are not fully known, but may be attributed to the specific MoA of this compound, since it targets cellular protein to induce epigenetic alterations at HIV LTR, which typically induce a more durable effect on HIV compared with the conventional anti-HIV drugs. It is also possible that the intracellular half-life of ZL0580, which has not yet been determined in the present study, can be attributed to this prolonged effect. We speculate that the intracellular half-life of ZL0580 may not fully explain this, since we have observed that ZL0580 induces an average of more than 12 days of delay in viral rebound (Figure 4, A–C), which are typically longer than the half-lives of most small-molecule drugs. Detailed analyses of in vitro and in vivo pharmacokinetics properties of ZL0580 in future studies can help better define these mechanisms.
We noted that, despite significant delay, viral rebounds eventually occur in all the examined PBMCs after treatment cessation. This could be due to drug decay and loss of activity for ZL0580 after more than several days of drug removal. Another possible explanation is that, in this cell culture, T cells were stimulated and underwent continuous proliferation and turnover; following treatment removal, the newly generated T cells were not exposed to the drugs and might have contributed to viral release. This mechanism could be explored by comparing the epigenetic (e.g., LTR structure) and transcriptional profiles of T cells collected before and after viral rebound. Regardless of mechanisms, this observation indicates a potential limitation of ZL0580, since it cannot permanently or durably suppress the latent viruses. A similar finding was reported for the Tat inhibitor (dCA) in humanized mice (23). These data suggest that it may be challenging to “durably” silence HIV by a single “block” approach and that combination approaches may be needed. It is hoped that through comprehensive lead optimization and drug-development efforts (e.g., delivery, dosing, and combination use with other silencers), we can improve the durability and potency of this class of molecules in epigenetically suppressing HIV. Finally, for a block-and-lock HIV cure approach, while therapies targeting host proteins are considered promising and provide advantages (e.g., reduced drug resistance), off-target effects need to be carefully evaluated. To this end, a number of drug candidates targeting BET proteins have been tested in various disease models including HIV infection (35). The data obtained so far in our study support a safe profile of ZL0580 regarding cellular toxicity and off-target effects. Future pharmacokinetics and in vivo toxicity studies are warranted to better evaluate utility of this compound in HIV epigenetic suppression.
To summarize, we report identification of a small molecule that induces epigenetic suppression of HIV via BRD4. Our study provides a conceptual and translational basis for future development of this class of molecules as tools and/or potential therapeutic agents for HIV epigenetic silencing. Further studies are needed to advance the development of this class of molecules, including improved understanding of MoA, lead optimization, and efficacy testing in vivo in animal models of HIV/SIV infection.
Methods
Human PBMCs, cell lines, and HIV virus.
PBMCs of healthy donors and HIV-infected participants were obtained from the UTMB blood bank and the US Military HIV Research Program, respectively. Information about HIV-infected participants in RV21 and RV254 is presented in Supplemental Table 2 and Supplemental Table 3. J-Lat full-length cells (clone 10.6, catalog 9849) were obtained from the NIH AIDS Reagent Program and were maintained in complete RPMI medium (Gibco, Thermo Fisher Scientific). The HIV-1 virus used for in vitro infection was the R5 tropic US-1 strain (38).
Compound design and synthesis.
Compounds were designed and synthesized in the Chemical Biology Laboratory of UTMB. Detailed descriptions of compound design and synthesis are available in the Supplemental Methods.
Binding affinity analysis.
Binding affinities of ZL0580 and JQ1 to BDs of BRD4, BRD2, BRD3, and BRDT were determined using the TR-FRET assay as described (49). Details are available in the Supplemental Methods.
J-Lat cell treatments and HIV activation analysis.
J-Lat cells were used for various experiments in this study, including compound screening, measurement of latent HIV activation, and mechanistic analyses. In the latent HIV activation screening, cells were treated with individual compounds (10 μM) for 24 hours or with DMSO (NC), JQ1 (10 μM), or PMA (100 ng/mL) as controls. In HIV-suppression experiments, PMA-activated and resting J-Lat cells were used to examine the impact of ZL0580 on induced and basal HIV transcription. For activated J-Lat cells, cells were stimulated with PMA (100 ng/mL) and treated with ZL0580 (10 μM). Treatment with various ZL0580 concentrations (0, 1, 10, and 20 μM) was also conducted for dose-response analysis. For unstimulated J-Lat cells, cells were treated only with ZL0580 (10 μM) or not treated (NC). At different time points after treatments, cells were subjected to various analyses (cell viability, HIV expression, and mechanistic analysis), as indicated in individual figures. HIV expression in J-Lat cells was measured by flow cytometric analysis of GFP expression and/or qPCR quantification of HIV transcripts (Gag and 3′ LTR). In latent HIV reactivation experiments, J-Lat cells were treated with ZL0580 or not treated for 3 days, followed by reactivation with SAHA (2.5 μM) or prostratin (2.5 μM) for 24 hours.
In vitro HIV infection of human PBMCs.
PHA-activated and resting PBMCs were infected with HIV. To activate PBMCs, cells (0.4 × 106/well) were stimulated with PHA (10 μg/mL) for 2 days, followed by infection with pretitrated HIV (US-1 strain) (38) in the absence (0 μM) or presence of ZL0580 or ZL0454. Three days after HIV infection, cells were harvested and stained for viability (Aqua Blue LIVE/DEAD; Invitrogen), CD3 (APC-H7; 560176; BD Bioscience), CD4 (BV605; 30056; BioLegend), and CD8 (BV785; 301046; BioLegend). Following fixation and permeabilization, cells were stained for intracellular HIV p24 (KC57; Beckman Coulter). HIV infection of CD4+ T cells was measured by flow cytometry based on intracellular p24 expression. For HIV infection of resting PBMCs, cells were directly infected with pretitrated HIV (US-1) by spinoculation. Twenty-four hours after HIV inoculation, cells were washed and split into a 96-well plate (0.4 × 106/well) for ZL0580 treatment (0, 2, 4 μM). Six days after viral exposure, HIV infection in CD4+ T cells was measured by flow cytometry based on intracellular p24 expression. Cells were acquired using a BD LSR-Fortessa, and data were analyzed using FlowJo (Tree Star).
T cell phenotype and gene expression analysis.
Analysis of T cell phenotype and gene expression profile was conducted in both PHA-activated and resting PBMCs. Activated or resting cells were treated with ZL0580 or not treated, as indicated, followed by staining for viability (Aqua Blue; Invitrogen), CD3, CD4, CD8 (same antibodies as described above), CCR5 (Alexa Fluor 700; catalog 359116; BioLegend), CD25 (PE-Cy7; catalog 302608; BioLegend), CD38 (BV711; catalog 303528; BioLegend), and HLA-DR (V450; catalog 642276; BD Bioscience). T cell phenotypes were examined by flow cytometry. Gene expression was measured by qPCR using RNA extracted from the treated PBMCs. A list of genes and primer sequences is shown in Supplemental Table 4.
PCR quantification of cell-associated HIV DNA and RNA.
qPCR quantification of HIV DNA and RNA in cells was performed as previously reported with modifications (38, 50). Details of experimental procedures are available in the Supplemental Methods.
Treatments of PBMCs of aviremic HIV-infected participants (RV21).
Cryopreserved PBMCs (2 × 106) of each participant were stimulated with PHA (10 μg/mL) in the absence or presence of ZL0580 (8 μM). Two days after treatment, cells were lysed and RNA was extracted for qPCR quantification of HIV Gag RNA. Prior to cell harvesting and lysis, cell viability was examined using the TC20 Automated Cell Counter (Bio-Rad).
Treatments of PBMCs of ART-suppressed, aviremic HIV-infected participants (RV254).
PBMCs of aviremic RV254 participants (6 months after ART) were used in 2 experiments. First, PBMCs were stimulated with anti-CD3/CD8 (1 μg/mL) in culture medium containing IL-2 (100 U/mL) to activate latent HIV and to expand CD4+ T cells. Cells were equally split into 3 wells (1 × 106/well) in a 48-well culture plate and received mock treatment (NC), ART alone (Efavirenz 200 nM; Zidovudine 360 nM; Raltegravir 400 nM) (AIDS Reagents Program), or ART+ZL0580 (2.5 μM). Medium was replaced every 3 days, and HIV production in supernatants was measured by nested PCR. Treatments were stopped when ART fully suppressed HIV. HIV production in supernatants was continuously monitored after treatment cessation. Second, unstimulated PBMCs were treated with ZL0580 (2.0 μM) or not treated (NC) once every 3 days for a total of 3 times (days 0, 3, and 6). HIV production in supernatants was monitored by nested PCR. On day 18, cells were stimulated with PHA to reactivate latent HIV. HIV transcriptional activation in cells was measured by qPCR quantification of Gag RNA.
Ultrasensitive nested PCR for HIV quantification.
Two-step nested PCR for HIV quantification was established using a method similar to that described previously, with modifications (22). Briefly, HIV viral RNA in supernatants was extracted using the QIAamp Viral RNA Kit (QIAGEN). cDNA was synthesized from viral RNA and subjected to first-round PCR amplification (16 cycles) using a Gag-Out-F/R primer set (Supplemental Table 5). PCR products were serially diluted and subjected to second-round nested PCR (40 cycles) using a Gag-F/R primer set (Supplemental Table 5). After PCR amplification, HIV copies were quantified using the established standard curve. Detailed procedures are available in the Supplemental Methods.
CRISPR/Cas9 KO of BRD4 and BRD2.
BRD4 and BRD2 gene editing was carried out using CRISPR/Cas9, as we previously reported (51). Gene-specific guide RNA sequences targeting human BRD2 or BRD4 were designed using an online tool (http://crispr.mit.edu/) (Supplemental Table 6). The CRISPR/Cas9 strategy is illustrated in Supplemental Figure 10. Detailed procedures are available in the Supplemental Methods.
Exogenous BRD4 overexpression in BRD4-KO J-Lat cells.
Cells suspended in 200 μL electroporation buffer (Ingenio Solution, MIR50111; Mirus) (0.6 × 106 cells/200 μL) were transfected with 20 μg/mL plasmid (pcDNA5-Flag-BRD4-WT; Addgene catalog 90331, a gift from Kornelia Polyak) in a 200 μL cuvette (Bio-Rad) by nucleofection using the Bio-Rad Gene Pulser XceII Electroporation System (140V, 1000 microfarads). Following nucleofection, cells were cultured in RPMI medium. Cell viability and BRD4 overexpression were monitored daily. After confirmation of BRD4 expression, cells were used for compound treatments to evaluate the functional role of BRD4 in ZL0580-induced HIV suppression.
Western blotting.
Western blotting was conducted according to standard procedures, and experimental details are available in the Supplemental Methods. The following primary antibodies were used: Tat (catalog 160189, AIDS Reagent Program), NF-κB (catalog 33-9900, Thermo Fisher), phospho–NF-κB (catalog 13346, Cell Signaling Technology), BRD4 (catalog 730007, Thermo Fisher; 13440, Cell Signaling Technology), BRD2 (catalog PA5-15297, Thermo Fisher), BRD3 (catalog A302-368A-T, Bethyl Lab), BRDT (catalog PA5-40818; Thermo Fisher), anti-FLAG (catalog F7425, MilliporeSigma), CDK9 (catalog MA5-14912, Thermo Fisher), cyclin T1 (catalog PA5-24163, Thermo Fisher), cyclin T2 (catalog PA5-22200, Thermo Fisher), AFF4 (catalog PA5-46798, Thermo Fisher), ELL2 (catalog PA5-64949, Thermo Fisher), p-RNAPII-CTD (Ser2) (catalog MA5-23510, Thermo Fisher), and GAPDH (catalog 2118, Cell Signaling Technology).
Co-IP.
J-Lat cells were lysed in 1 mL NP-40 cell lysis buffer, followed by lysate clearance and supernatant collection. Total protein concentration was measured (Microplate BCA Protein Assay Kit), and equal amounts of proteins from different treatments were incubated with 2 μg anti-Tat (MA1-71509, Thermo Fisher), anti-BRD4 (730007, Thermo Fisher), or mouse control IgG (5415, Cell Signaling Technology) overnight at 4°C. Immune complexes were precipitated with 50 μL of protein G–conjugated magnetic beads (Thermo Fisher). Beads were washed 6 times and then subjected to SDS-PAGE electrophoresis. Following protein transfer, membranes were blotted for CDK9 (MA5-14912, Thermo Fisher).
ChIP and qPCR.
ChIP was performed using a ChIP-IT Express Kit (Active Motif) according to the manufacturer’s instructions. Detailed experimental procedures are available in the Supplemental Methods. Primer sequences for ChIP-qPCR are shown in Supplemental Table 5.
MNase nucleosomal mapping.
High-resolution MNase mapping of the HIV LTR was performed using a protocol described previously, with slight modifications (45). Detailed procedures are available in the Supplemental Methods. Primer sequences used in the MNase assay are shown in Supplemental Table 7.
Statistics.
Statistical analysis was performed using Prism 6.0 (GraphPad). Statistical comparison between the groups was performed using paired or nonpaired t test and 1-way ANOVA as appropriate for different data sets. Two-tailed P values were denoted, and P values of less than 0.05 were considered significant.
Study approval.
The project was reviewed and approved by the IRB of UTMB. The study involves the use of specimens of human participants with no code or link that could allow identification of subjects and was determined as nonhuman subject research. Informed consent was obtained from all subjects prior to their participation in the studies.
Author contributions
HH and QN conceived the study. QN, ZL, JZ, and HH designed the experiments. ZL, HC, and JZ synthesized the compounds. QN, ZL, EA, XF, BT, JZ, and HH performed experiments and analyzed the data. JE, BBG, JHK, NLM, MLR, and JA helped data analysis and/or provided samples. HH and QN prepared the manuscript with help from JE, BBG, JHK, NLM, MLR, JA, and JZ.
Supplementary Material
Acknowledgments
We thank human participants for providing cell samples, Istvan Boldogh for help with the ChIP assay, and Allan Brasier for help with the TR-FRET assay. The reagents J-Lat (clone 10.6, catalog 9849) and anti-Tat Ab (catalog 160189) were obtained through the AIDS Reagent Program. The study was funded by UTMB’s startup fund and grants from the NIH (R21AI110214) and the Robert Mapplethorpe Foundation (to HH) and was supported by NIH grant R01AI132674. During this study, QN was also affiliated with the Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, and was sponsored by the China Scholarship Council (no. 201603250037). ZL was supported by the Crohn’s & Colitis Foundation of America (grant 548813). The content of this publication does not necessarily reflect the views or policies of the NIH, the U.S. Army, or the Department of Defense, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The investigators have adhered to the policies for the protection of human subjects as prescribed in AR-70.
Version 1. 07/22/2019
Electronic publication
Version 2. 08/01/2019
Print issue publication
Footnotes
Conflict of interest: JA has received honoraria for participating in advisory meetings for ViiV Healthcare, Gilead Sciences, Merck, Roche, and AbbVie.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(8):3361–3373.https://doi.org/10.1172/JCI120633.
Contributor Information
Qingli Niu, Email: qiniu@UTMB.EDU.
Zhiqing Liu, Email: zhiqliu@UTMB.EDU.
Edrous Alamer, Email: edalamer@UTMB.EDU.
Xiuzhen Fan, Email: xifan@utmb.edu.
Haiying Chen, Email: haichen@UTMB.EDU.
Benjamin B. Gelman, Email: bgelman@utmb.edu.
Bing Tian, Email: bitian@utmb.edu.
Jerome H. Kim, Email: Jerome.Kim@ivi.int.
Jintanat Ananworanich, Email: jananworanich@hivresearch.org.
Jia Zhou, Email: jizhou@utmb.edu.
Haitao Hu, Email: haihu@utmb.edu.
References
- 1.Dahabieh MS, Battivelli E, Verdin E. Understanding HIV latency: the road to an HIV cure. Annu Rev Med. 2015;66:407–421. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siliciano RF, Greene WC. HIV latency. Cold Spring Harb Perspect Med. 2011;1(1):a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun TW, Moir S, Fauci AS. HIV reservoirs as obstacles and opportunities for an HIV cure. Nat Immunol. 2015;16(6):584–589. doi: 10.1038/ni.3152. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 7.Buzón MJ, et al. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med. 2010;16(4):460–465. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher CV, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111(6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzo-Redondo R, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530(7588):51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Besson GJ, McMahon D, Maldarelli F, Mellors JW. Short-course raltegravir intensification does not increase 2 long terminal repeat episomal HIV-1 DNA in patients on effective antiretroviral therapy. Clin Infect Dis. 2012;54(3):451–453. doi: 10.1093/cid/cir721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kearney MF, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10(3):e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deeks SG, Lewin SR, Havlir DV. The end of AIDS: HIV infection as a chronic disease. Lancet. 2013;382(9903):1525–1533. doi: 10.1016/S0140-6736(13)61809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyrer C, Pozniak A. HIV drug resistance - an emerging threat to epidemic control. N Engl J Med. 2017;377(17):1605–1607. doi: 10.1056/NEJMp1710608. [DOI] [PubMed] [Google Scholar]
- 14.Mbonye U, Karn J. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology. 2014;454-455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakre S, Chavez L, Shirakawa K, Verdin E. Epigenetic regulation of HIV latency. Curr Opin HIV AIDS. 2011;6(1):19–24. doi: 10.1097/COH.0b013e3283412384. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki K, et al. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1(2):66–78. [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagishi M, et al. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 2009;11(4):500–508. doi: 10.1016/j.micinf.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum Gene Ther. 2012;23(5):473–483. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner AM, De La Cruz J, Morris KV. Mobilization-competent lentiviral vector-mediated sustained transcriptional modulation of HIV-1 expression. Mol Ther. 2009;17(2):360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, et al. Promoter targeting shRNA suppresses HIV-1 infection in vivo through transcriptional gene silencing. Mol Ther Nucleic Acids. 2013;2:e137. doi: 10.1038/mtna.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mousseau G, et al. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe. 2012;12(1):97–108. doi: 10.1016/j.chom.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mousseau G, Kessing CF, Fromentin R, Trautmann L, Chomont N, Valente ST. The tat inhibitor didehydro-cortistatin A prevents HIV-1 reactivation from latency. MBio. 2015;6(4):e00465. doi: 10.1128/mBio.00465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessing CF, et al. In vivo suppression of HIV rebound by didehydro-cortistatin a, a “block-and-lock” strategy for HIV-1 treatment. Cell Rep. 2017;21(3):600–611. doi: 10.1016/j.celrep.2017.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 25.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149(1):214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 27.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29(5):1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Conrad RJ, et al. The short isoform of BRD4 promotes HIV-1 latency by engaging repressive SWI/SNF chromatin-remodeling complexes. Mol Cell. 2017;67(6):1001–1012.e6. doi: 10.1016/j.molcel.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep. 2012;2(4):807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41(1):277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci U S A. 2007;104(34):13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, et al. The KAT5-Acetyl-histone4-Brd4 axis silences HIV-1 transcription and promotes viral latency. PLoS Pathog. 2018;14(4):e1007012. doi: 10.1371/journal.ppat.1007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, et al. Drug discovery targeting bromodomain-containing protein 4. J Med Chem. 2017;60(11):4533–4558. doi: 10.1021/acs.jmedchem.6b01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spina CA, et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog. 2013;9(12):e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baell J, Walters MA. Chemistry: chemical con artists foil drug discovery. Nature. 2014;513(7519):481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 38.Liu F, et al. Sequential dysfunction and progressive depletion of candida albicans-specific CD4+ T cell response in HIV-1 infection. PLoS Pathog. 2016;12(6):e1005663. doi: 10.1371/journal.ppat.1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuetz A, et al. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog. 2014;10(12):e1004543. doi: 10.1371/journal.ppat.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm D, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12(3):452–462. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529(7586):413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282(18):13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 43.Yu D, Liu R, Yang G, Zhou Q. The PARP1-Siah1 axis controls HIV-1 transcription and expression of Siah1 substrates. Cell Rep. 2018;23(13):3741–3749. doi: 10.1016/j.celrep.2018.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu M, Hsu J, Chan C, Li Z, Zhou Q. The ubiquitin ligase Siah1 controls ELL2 stability and formation of super elongation complexes to modulate gene transcription. Mol Cell. 2012;46(3):325–334. doi: 10.1016/j.molcel.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 2011;9(11):e1001206. doi: 10.1371/journal.pbio.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Darcis G, Van Driessche B, Van Lint C. HIV latency: should we shock or lock? Trends Immunol. 2017;38(3):217–228. doi: 10.1016/j.it.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38(3):428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma CT, Sergienko EA. Time-resolved fluorescence assays. Methods Mol Biol. 2016;1439:131–142. doi: 10.1007/978-1-4939-3673-1_8. [DOI] [PubMed] [Google Scholar]
- 50.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20(4):425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu F, et al. Priming and activation of inflammasome by canarypox virus vector ALVAC via the cGAS/IFI16-STING-type I IFN pathway and AIM2 sensor. J Immunol. 2017;199(9):3293–3305. doi: 10.4049/jimmunol.1700698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.