Abstract
T follicular helper cells (Tfh), a subset of CD4+ T cells, provide requisite help to B cells in the germinal centers (GC) of lymphoid tissue. GC Tfh are identified by high expression of the chemokine receptor CXCR5 and the inhibitory molecule PD-1. Although more accessible, blood contains lower frequencies of CXCR5+ and PD-1+ cells that have been termed circulating Tfh (cTfh). However, it remains unclear whether GC Tfh exit lymphoid tissues and populate this cTfh pool. To examine exiting cells, we assessed the phenotype of Tfh present within the major conduit of efferent lymph from lymphoid tissues into blood, the human thoracic duct. Unlike what was found in blood, we consistently identified a CXCR5-bright PD-1–bright (CXCR5BrPD-1Br) Tfh population in thoracic duct lymph (TDL). These CXCR5BrPD-1Br TDL Tfh shared phenotypic and transcriptional similarities with GC Tfh. Moreover, components of the epigenetic profile of GC Tfh could be detected in CXCR5BrPD-1Br TDL Tfh and the transcriptional imprint of this epigenetic signature was enriched in an activated cTfh subset known to contain vaccine-responding cells. Together with data showing shared TCR sequences between the CXCR5BrPD-1Br TDL Tfh and cTfh, these studies identify a population in TDL as a circulatory intermediate connecting the biology of Tfh in blood to Tfh in lymphoid tissue.
Keywords: Immunology
Keywords: T cells
Introduction
T follicular helper cells (Tfh) are a subset of CD4+ T cells that provide B cell help within the follicles of secondary lymphoid tissues. Tfh promote B cell class switching and affinity maturation, and without Tfh help, germinal centers (GC) fail to form (1). The biology of Tfh is central to several fields of human health and disease (2). First, the ability of vaccines to elicit effective Tfh help is a major goal of rational vaccine design, given that antibodies are the primary correlates of protection for the majority of vaccines (3). Second, Tfh:B cell interactions are dysregulated in antibody-mediated autoimmune diseases (4), indicating that therapeutic opportunities may exist through modulation of Tfh. Finally, Tfh are a major cellular reservoir of replication-competent HIV, even in the setting of antiretroviral therapy (5–8). As a result, Tfh biology and measurement of Tfh responses to therapies and immunizations are of considerable interest in human immunology.
Tfh and B cell interactions occur within secondary lymphoid organs (SLO) (1). Considerable knowledge has been gained about human GC biology by studying human lymphoid tissues and human tonsillar tissue in particular. However, routine lymph node (LN) biopsies are impractical, and tonsillar tissue, although more easily accessible, may not accurately represent total body SLO biology (9, 10). Thus, the ability to interrogate Tfh biology in peripheral blood and understand how changes in blood link to events in SLO could provide valuable insights. Tfh in SLO are typically defined by coexpression of CXCR5, a chemokine receptor that allows cells to traffic into the B cell follicle, and the inhibitory receptor programmed death 1 (PD-1) (11). These CXCR5+PD-1+ SLO Tfh can then be divided into GC Tfh and non-GC Tfh (12, 13). This division reflects anatomical location, but can also be experimentally defined based on the amount of CXCR5 and PD-1 expressed, where GC Tfh have the highest expression of CXCR5 and PD-1 (refs. 11, 12; CXCR5-bright PD-1–bright [CXCR5BrPD-1Br]), an approach confirmed using histocytometry on human LNs (14).
Studies in the blood have focused on Tfh-like cells that also coexpress CXCR5 and PD-1. These circulating Tfh (cTfh), also called peripheral or pTfh, express lower amounts of CXCR5 and PD-1 than GC Tfh and are present in lower frequencies than are CXCR5+PD-1+ Tfh in SLO (12). In HIV, cTfh have been demonstrated to contain more virus than other circulating CD4+ T cell subsets (15). Because HIV can be enriched in SLO GC, these observations suggest a relationship between cTfh and SLO Tfh (7, 15). We and others have identified responses, following vaccination, in a population of cTfh that expresses ICOS (16–20). These ICOS+ cTfh increase after influenza vaccination and contain influenza-specific CD4+ T cells, with clones that dynamically reappeared after subsequent yearly influenza immunizations (17–21). Despite these studies identifying responses in the cTfh compartment, major questions remain about the ontologic relationship between cTfh in blood and GC Tfh in lymphoid tissues.
Questions about the origin of cTfh stem, at least in part, from an incomplete understanding of Tfh trafficking between SLO and the blood in humans. Tfh migration has been studied within LNs of mice in the setting of primary and secondary immune challenges (22, 23). Tfh from the GC can migrate into other GC within the same LNs (22), indicating that GC Tfh are not necessarily fixed residents of a single GC. Moreover, many mouse GC Tfh can migrate to the follicular mantle of the B cell follicle, and following secondary challenge, some of these follicular mantle Tfh exit into the subcapsular sinus of the LNs (23), suggesting a potential opportunity for Tfh with a history of participating in a GC reaction to exit the LNs altogether. However, the fate of these ex-GC Tfh is not known, and it is unclear whether such cells traffic only to other LNs or whether these ex-GC Tfh enter the systemic circulation. The answers to these questions could have important implications for the ability to monitor vaccine-induced immunity or track the effects of immune modulatory drugs in patients with autoimmunity. Studies in mice, and limited data in humans, do suggest that memory Tfh form after an initial reaction and that these memory Tfh can be found in peripheral blood (12, 17, 24–26). However, it is unclear whether these cTfh arise from LN Tfh and, if so, how closely related this pool might be to cells that participated in a GC reaction (12, 23).
In this study, we sought to address these questions and specifically interrogate whether the cTfh pool in blood contained cells that emigrated from LNs or, rather, represented a largely blood recirculating pool. Thus, we collected cells from the human thoracic duct, the anatomical structure that carries the terminal efferent lymph to the venous system (27), and examined Tfh in this compartment compared with blood and SLO. Our data demonstrate that the thoracic duct is enriched for Tfh compared with the blood and contains cells that are phenotypically and transcriptionally similar to GC Tfh. Although thoracic duct lymph (TDL) drains directly into the blood, cTfh with a CXCR5BrPD-1Br phenotype were rarely detected in the blood, but TCR clonotypes from this CXCR5BrPD-1Br TDL Tfh population could be found preferentially in the cTfh pool. To further address this issue, we examined Tfh from LNs, TDL, and blood to define a GC Tfh–specific epigenetic signature. We then identified components of this signature in TDL. Moreover, open chromatin regions (OCRs) common in GC Tfh and CXCR5BrPD-1Br TDL Tfh defined genes that were transcriptionally enriched in the activated subset of cTfh in the periphery known to contain vaccine-responding cells (17). Interrogation of cTfh may, therefore, serve as a periscope into Tfh biology within SLO.
Results
TDL carries Tfh-enriched lymphocytes to the peripheral blood.
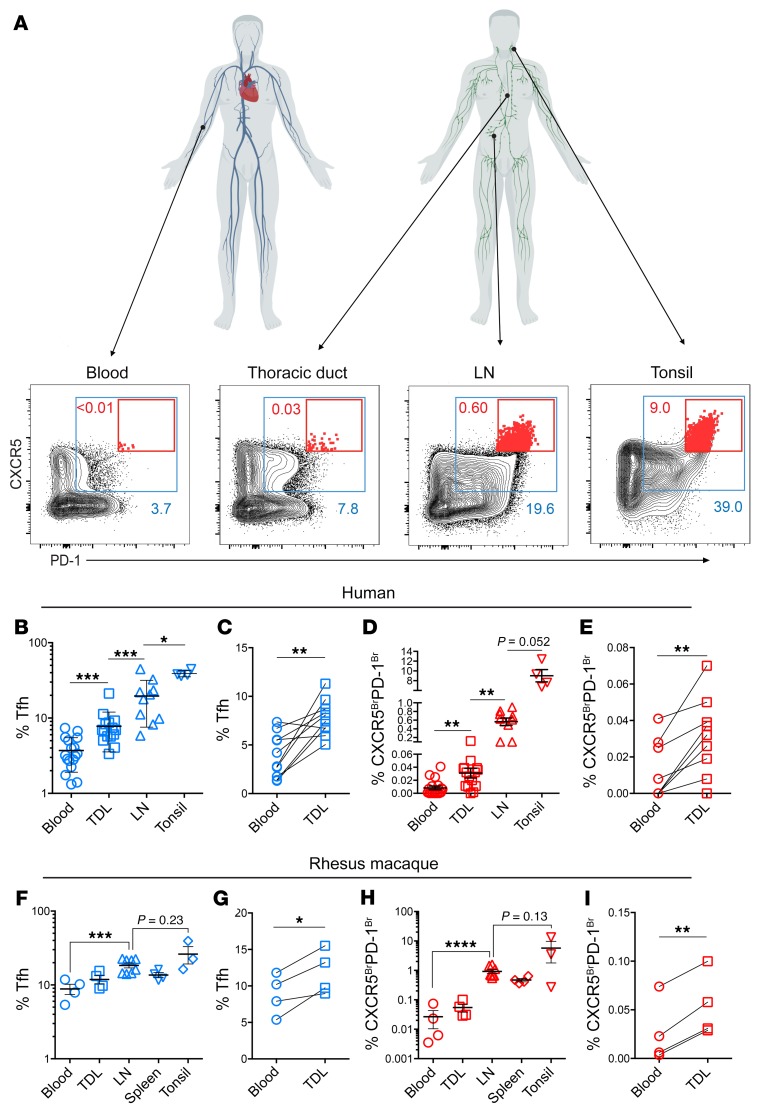
To better understand human Tfh trafficking, we first compared Tfh (CXCR5+PD-1+ CD4+ T cells) from multiple lymphoid tissues (gating strategy shown in Supplemental Figure 1A and Figure 1A). Approximately 40% of nonnaive CD4+ T cells in tonsils coexpressed the Tfh markers CXCR5 and PD-1 (Figure 1, A and B). The median frequency of Tfh in LN samples was approximately half the frequency of Tfh in tonsils, and blood had approximately 5-fold fewer Tfh than LNs (Figure 1, A and B). Given that the thoracic duct is a one-way conduit that empties lymph directly into the subclavian vein (28), we anticipated that CD4+ T cells in the blood would be similar to those found in TDL. Instead, we found that TDL CD4+ T cells were enriched for Tfh compared with those in peripheral blood (Figure 1, A–C), even when paired TDL and peripheral blood samples were directly compared (Figure 1C).
Figure 1. The thoracic duct carries Tfh-enriched lymphocytes to the blood, including a population of CXCR5BrPD-1Br Tfh.
(A) Nonnaive CD4+ T cells are gated as CXCR5+PD-1+ Tfh (blue boxes) and CXCR5BrPD-1Br Tfh (red boxes) with frequency (percentage) in matching color. Human figures created with BioRender. (B) Tfh frequencies in blood (n = 17), TDL (n = 15), LNs (n = 10), and tonsil (n = 4). (C) Tfh frequencies in TDL and blood from donors with paired samples (n = 10). (D) Frequency of CXCR5BrPD-1Br Tfh in the indicated tissues. (E) Paired analysis of CXCR5BrPD-1Br frequencies in TDL and blood. (F) RM Tfh frequency in the indicated tissues and (G) in paired PBMCs and TDL samples (n = 4, 2 LNs included from each animal, an iliac node and a mesenteric node). (H) CXCR5BrPD-1Br frequencies in the indicated tissues from RM (n = 4) with (I) paired frequencies. Error is reported as SD. Paired 2-tailed t tests were performed for data shown in C, E, G, and I. ANOVA with Holm-Šídák post-test was performed on log-transformed data shown in B, D, F, and H. In D, 0 values were replaced with 0.00001 for log transformation. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
A notable feature of Tfh in TDL was the frequency of high CXCR5-expressing cells and, in particular, in the consistent presence of CXCR5BrPD-1Br Tfh (Figure 1, D and E). Tonsils are commonly used to study GC Tfh biology in humans, in part because they are readily accessible. However, tonsils had a 10-fold higher frequency of GC Tfh than LNs (Figure 1, A and D). Peripheral blood samples rarely contain CXCR5BrPD-1Br Tfh (Figure 1, A and D) in patients undergoing lymphatic procedures as well as in healthy control subjects (Supplemental Figure 1B), and the frequency did not change when blood was sampled before or after the peripheral capillary beds (i.e., arterial versus venous blood; Supplemental Figure 1C). In contrast, the TDL contained a population of CXCR5BrPD-1Br Tfh, although the frequency of this population in the TDL was lower than in SLOs. Although this population was consistently present in TDL samples (Figure 1, A, D, and E), only half (4/9) of the PBMC samples contained any CXCR5BrPD-1Br Tfh (Figure 1, D and E).
To determine whether the presence of CXCR5BrPD-1Br Tfh was a generalizable observation in primates and to control for any possible perturbations due to the clinical indication for thoracic duct cannulation, we also collected tonsils, LNs, spleen, TDL, and blood from rhesus macaques (RM) (Supplemental Figure 1, D and E). As in humans, the frequency of Tfh in RM TDL was also higher than in PBMCs (Figure 1, F and G, and Supplemental Figure 1, D and E), although RM PBMCs contained a minor population of CXCR5BrPD-1Br cTfh that was more consistently identified than that in humans (Figure 1, H and I). Nevertheless, the frequencies of CXCR5BrPD-1Br Tfh in the TDL were considerably higher than in peripheral blood (Figure 1, H and I), in agreement with the findings in humans above. Together, these data demonstrate that efferent lymph contains higher frequencies of Tfh than peripheral blood and that, unlike that in blood, the TDL Tfh population includes a small but consistently detectable population of CXCR5BrPD-1Br Tfh.
Efferent lymphatic flow is required to sustain cTfh in blood.
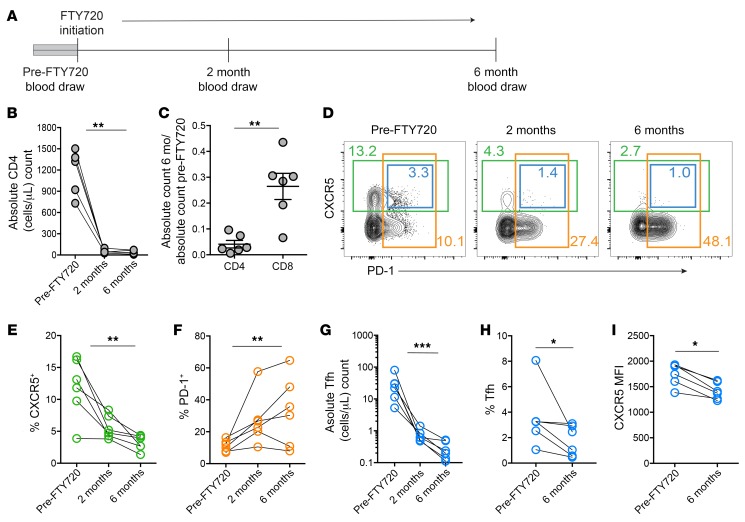
To determine whether or not Tfh from lymphatic flow were important to populate and sustain cTfh, we tested how blocking cellular egress into lymph would affect cTfh frequency. Thus, we longitudinally sampled patients receiving daily fingolimod (FTY720) for the treatment of multiple sclerosis (Figure 2A). FTY720 prevents cellular egress from lymphoid tissues by downregulating sphingosine-1-phosphate receptors (S1PRs), and treatment with FTY720 has been shown to decrease cTfh in mice (29). As expected, FTY720 treatment dramatically reduced the absolute number of circulating CD4+ T cells (Figure 2B) and affected CD4+ T cell numbers more than CD8+ T cells (Figure 2C). In mice, populations of memory T cells have been identified that are proposed to preferentially exist as mainly blood recirculating populations (30), with some evidence that similar populations might exist in humans (31). Such populations would be predicted to be affected less by FTY720, whereas populations dependent on lymphatic egress would be predicted to decrease in relative frequency. Thus, we next asked whether the remaining CD4+ T cells were changed in composition. Indeed, the proportion of CD4+ T cells that were CXCR5+ decreased over time, whereas there was an enrichment for CXCR5–CD4+ T cells and other PD-1+CD4+ T cells (Figure 2, D–F). In particular, cTfh decreased in absolute count as well as frequency (Figure 2, D, G, and H), and the cTfh remaining had decreased expression of CXCR5 per cell (Figure 2I), suggesting an enrichment for cells that were less efficient at migrating to chemokine cues in lymphoid tissues. In addition to preferential loss of cTfh, the frequency of CCR7+ and naive CD4+ T cells decreased in blood after FTY720 administration, as previously shown (Supplemental Figure 2, A–C, and ref. 32). Overall, these data support the idea that maintenance of human cTfh in peripheral blood is dependent on continuous efferent flow from the lymphatics.
Figure 2. Lymphatic egress is required to maintain cTfh populations in blood.
(A) Time line of blood draws in patients receiving FTY720. (B) Absolute numbers of human CD4+ T cells in the peripheral blood before and 2 and 6 months after FTY720 initiation in patients with multiple sclerosis (n = 6). (C) Fold change in absolute cell numbers at 6 months of therapy compared with pretherapy in CD4+ and CD8+ T cells. (D) Flow cytometric analysis of nonnaive CD4+ T cells over time, with CXCR5+ cells in green boxes, PD-1+ cells in orange boxes, and CXCR5+ PD-1+ cTfh in blue boxes. Frequency for each population noted in matching color. (E and F) Changes in the frequency of CXCR5+ and PD-1+ nonnaive CD4+ T cells over time. (G–I) Changes in the absolute count, frequency, and CXCR5 MFI of cTfh over time. Error is reported as SD. *P < 0.05; **P < 0.01; ***P < 0.001. Friedman’s test was used for data shown in B and E–G. A 2-tailed Student’s t test was performed for data shown in C, and paired t tests were performed for data shown in H and I.
Estimating residency time of various lymphocyte subsets in the blood.
We hypothesized that the absence of CXCR5BrPD-1Br Tfh cells in circulation might, in part, reflect short residence time in the blood. Thus, we developed a simple mathematical model of cell migration from the lymph to the blood (see Methods). Because we did not have measurements of the blood volume and lymph flow in the donors from Figure 1, we used typical estimates found for humans of 3 to 9 liters per day for lymph flow with a median of 5 liters per day (33) and 4.5 liters to 5.5 liters total blood volume for adults (34). In addition, we used estimates of T cell frequencies in TDL and blood, as listed in the Supplemental Methods. We therefore estimated that approximately 0.28 × 1010 to 1.7× 1010 CD4+ T cells enter the circulation daily from 2 lymphatic ducts, which is roughly consistent with previous estimates (35). Based on the frequencies listed in Supplemental Methods, approximately 1.25 × 105 to 1.55 × 105 Tfh and 0.64 × 105 to 1.55 × 105 CXCR5BrPD-1Br Tfh cells enter the circulation daily from the lymph. At steady state in blood, we estimate there are 3.6 × 109 to 8.8 × 109 CD4+ T cells, 0.64 × 108 to 1.55× 108 cTfh cells, and 0.64 × 105 to 1.55× 105 CXCR5BrPD-1Br cTfh cells at any given time. The discrepancy between relative frequencies of cells entering from lymph and those in circulation suggest differences in residency time. As described in the Supplemental Methods, we estimated the following residency time (T) of these cells in the blood: CD4+ T cells, T = 12.6 to 30.9 hours; cTfh, T = 5.0 to 12.2 hours; and CXCR5BrPD-1Br cTfh, T = 1.7 to 4.1 hours. Although the estimated residency times are dependent on the assumed rate of lymph flow and blood volume, the relative estimates between subsets suggest that CXCR5BrPD-1Br cTfh cells spend one-tenth of time that other CD4+ T cells spend in circulation. However, these estimates assume that the phenotype of CXCR5BrPD-1Br TDL Tfh remains constant upon blood entry.
TDL Tfh are phenotypically distinct from LN and blood Tfh.
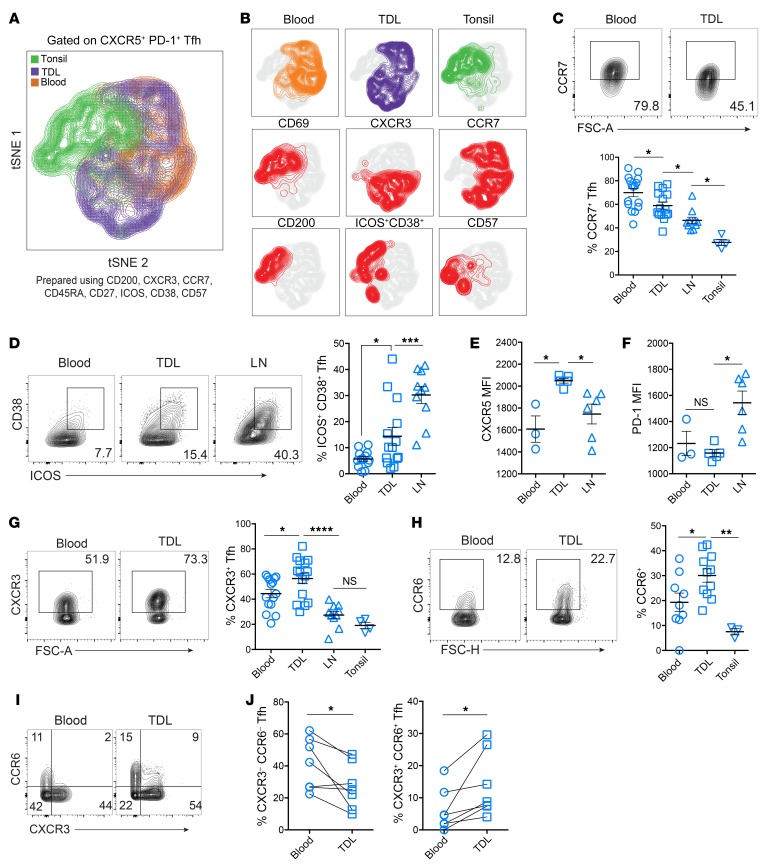
Tfh are heterogeneous in phenotype and function, but it is unclear whether components of the Tfh phenotype are fixed differentiation states or are, instead, dynamic and flexible responses to environmental stimuli. In lymphoid tissues, Tfh have been classified not only by the amount of CXCR5 and PD-1 expression (i.e., GC Tfh versus non-GC Tfh), but also by expression of CD200 (11) and CD57 (36, 37). In both lymphoid tissue and blood, Tfh have been divided into functional subsets based on surface markers, including ICOS, CXCR3, and CCR6 (38). Tfh that express higher ICOS have been shown to correlate with vaccine responses in blood (16–21, 39), and combinations of CXCR3 and CCR6 expression have been used to distinguish Tfh subsets that mirror CD4+ T cell effector subsets, termed Tfh1 (CXCR3+CCR6–), Tfh2 (CXCR3–CCR6–), and Tfh17 (CCR6+; refs. 38, 40). To interrogate the phenotypic diversity of Tfh from tonsils, TDL, and blood, we performed t-stochastic neighbor embedding (tSNE) of high-dimensional flow cytometric data. Clustering was performed using a set of surface markers defining Tfh subsets (Figure 3A). Overall, TDL Tfh had a phenotypic “landscape” that represented an intermediate profile between lymphoid tissue and blood cTfh, including intermediate expression of key proteins such as CCR7, ICOS, and CD38 (Figure 3, B–D). The increased pattern of activation for TDL Tfh, as illustrated by higher ICOS and CD38 expression, was also found in the B cell compartment, where TDL was more likely to contain activated CD27+CD38+ plasmablasts than paired blood (Supplemental Figure 3, A and B). Together, these data are consistent with the TDL containing transitional cell states between lymphoid tissue and blood.
Figure 3. TDL Tfh have an intermediate activation phenotype between LNs and blood, with increased expression of chemokine receptors.
(A) tSNE analysis of Tfh (all CXCR5+PD-1+) from tonsils (n = 4), TDL (n = 5), and blood (n = 5) using the indicated 8 surface markers. (B) Plots of individual marker expression within the tSNE space. (C) Frequency of CCR7+ Tfh in blood (n = 16), TDL (n = 15), LNs (n = 10), and tonsil (n = 4). (D) Frequency of ICOS+CD38+ Tfh in blood (n = 13), TDL (n = 14), and LNs (n = 10) (E and F) CXCR5 and PD-1 MFI of Tfh in blood (n = 3), TDL (n = 5), and LNs (n = 7). (G) Frequency of CXCR3 in blood (n = 15), TDL (n = 15), LNs (n = 10), and tonsil (n = 4). (H) CCR6 expression in paired blood and TDL (n = 7). (I) Expression of CXCR3 versus CCR6 by compartment in paired samples (n = 7), with (J) frequencies of Tfh expressing neither or both chemokine receptors. Error is reported as SD. Comparisons shown in C, D, G, and H were performed using ANOVA with the Holm-Šídák post-test. Comparisons shown in E and F were performed using Kruskal-Wallace with Dunn’s post-test. Paired 2-tailed t tests were performed for J. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Some aspects of the TDL Tfh phenotype were unexpected, however, and did not fit the pattern of a transitional or intermediate compartment. For example, TDL Tfh had significantly higher MFI of CXCR5 than either lymphoid or blood Tfh (Figure 3E). PD-1, on the other hand, was highest on LN Tfh (Figure 3F). Among the CXCR5+PD-1+ Tfh, CXCR3+ Tfh were more common in TDL Tfh than in blood or lymphoid Tfh (Figure 3G); however, when all CXCR5+ cells were analyzed without gating on PD-1, the lymphoid tissue and TDL contained similar CXCR3+ frequencies (Supplemental Figure 3, C and D). In addition to CXCR3, CCR6 expression in TDL Tfh was also higher than in blood (Figure 3H). When analyzed together, the increases in CXCR3 and CCR6 frequencies in TDL compared with blood reflected increases in TDL Tfh that expressed both chemokine receptors and decreases in CXCR3–CCR6– Tfh (Figure 3, I and J). Whether CXCR3+ cells in the TDL represent a transitory intermediate or a stable cell type (i.e., Tfh1) remains to be determined, but stimulation-induced IFN-γ expression was higher in blood than tonsil, consistent with higher CXCR3 in the blood cTfh (Supplemental Figure 3E). Our data indicate an increase in these chemokine receptors in lymph and might therefore suggest a role in exiting the LNs and/or populating the peripheral pool, consistent with the recently described role of PD-1 interactions in the GC suppressing CXCR3 expression to prevent GC Tfh egress (41). Indeed, another surface protein involved in both activation and residency programs, CD69, was downregulated in efferent lymphatic Tfh compared with those in lymphoid tissue (Supplemental Figure 3, F and G), suggesting that CD69 downregulation was important in Tfh egress from lymphoid tissues (42). Just as TDL Tfh were more likely to be CXCR3+ and CD69–, CXCR3 and CD69 defined 4 distinct populations in LN Tfh (Supplemental Figure 3G), with CXCR3+ Tfh more likely to lack CD69 (Supplemental Figure 3, G and H). Finally, we examined other markers of trafficking. As observed for CXCR5, CXCR3, and CCR6, TDL Tfh also had higher expression of CCR4 as well as the mucosal trafficking integrin α4β7 (Supplemental Figure 3I and ref. 43). Taken together, these data suggest that TDL Tfh represent a migrating population with shared and distinct phenotypic features compared with LNs and cTfh.
TDL CXCR5BrPD-1Br Tfh have a protein expression profile similar to that of GC Tfh.
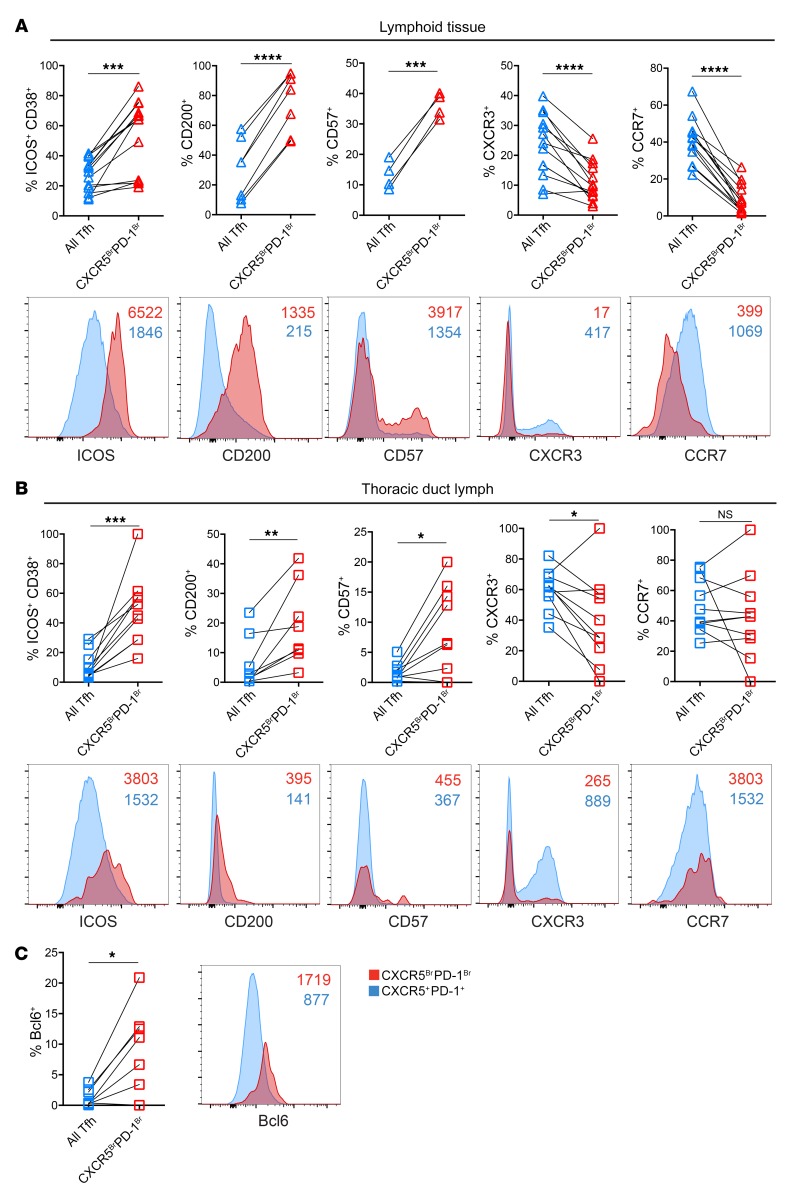
In lymphoid tissues, CXCR5BrPD-1Br Tfh are enriched for expression of several key molecules when compared with all CXCR5+ PD-1+ Tfh. We therefore next asked whether the CXCR5BrPD-1Br Tfh versus total Tfh in TDL also display the same phenotypic relationships as those observed in SLO. Indeed, higher expression of ICOS, CD200, and CD57 and lower expression of CXCR3 were observed for the CXCR5BrPD-1Br subset from LNs and TDL compared with all Tfh from the same locations (Figure 4, A and B). These observations suggest a relationship between these Tfh subpopulations in the TDL similar to what occurs in the SLO, though it should be noted that when CXCR5BrPD-1Br from each location were directly compared with each other, TDL CXCR5BrPD-1Br Tfh had lower expression of many of these proteins than GC Tfh. The one exception was the expression of CCR7, which was not different between CXCR5BrPD-1Br and all Tfh in TDL, perhaps because these cells are in the efferent lymphatics where there would not be expected to be any selective bias based on CCR7-mediated migration. Finally, Bcl6, the canonical transcription factor of GC Tfh, was higher both in frequency and MFI in TDL CXCR5BrPD-1Br Tfh compared with total Tfh in the lymph (Figure 4C). Together these data suggest that CXCR5BrPD-1Br Tfh in TDL maintain enrichment for proteins typical of GC Tfh.
Figure 4. TDL CXCR5BrPD-1Br Tfh have increased expression of proteins expressed by GC Tfh.
(A) Comparison of protein expression among all Tfh (including CXCR5BrPD-1Br Tfh) and GC Tfh from LNs and/or tonsils with representative plots of MFI comparisons for individual samples. Lymphoid ICOS+CD38+, CXCR3+, and CCR7+ analyses, n = 13; LN CD200+ analysis, n = 7; LN CD57+ analysis, n = 4. (B) Comparison of protein expression among all Tfh and CXCR5BrPD-1Br Tfh in TDL. TDL ICOS+CD38+, CXCR3+, and CCR7+ analyses, n = 10; CD200+ analysis, n = 8; CD57+ analysis, n = 9. (C) Bcl-6 frequency (n = 8) and MFI in TDL Tfh and TDL CXCR5BrPD-1Br Tfh. Colors reflect gates shown in Figure 1 (CXCR5+PD-1+ in blue = all Tfh). NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, paired, 2-tailed t tests.
TDL CXCR5BrPD-1Br Tfh are transcriptionally similar to GC Tfh.
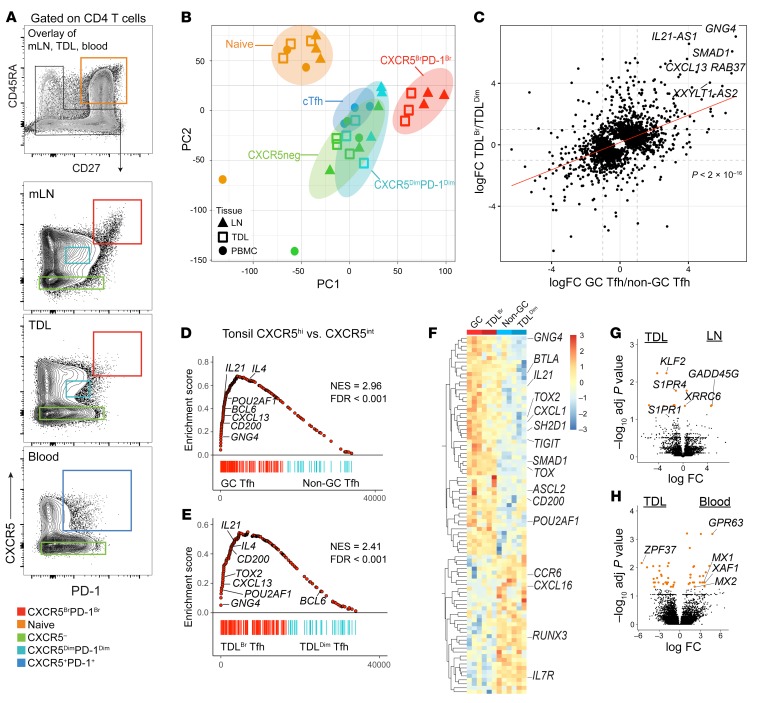
GC Tfh have a distinct transcriptional program compared with other CD4+ T cell subsets (12). We therefore investigated whether CXCR5BrPD-1Br TDL Tfh shared transcriptional similarities with GC Tfh. Prior transcriptional analyses of human GC Tfh have used GC Tfh from tonsils (12, 44). However, given that the thoracic duct collects lymph from the left upper quadrant and lower body (28), we instead compared TDL Tfh to mesenteric LN-derived Tfh. We isolated naive CD4+ T cells, CXCR5-negative nonnaive non-Tfh, and Tfh subsets from mesenteric LNs, TDL, and blood (Figure 5A) and performed RNA-Seq. To begin to understand the similarity in transcriptional programs among these Tfh populations, principle component analysis (PCA) was performed (Figure 5B). TDL CXCR5BrPD-1Br Tfh and LN GC Tfh clustered closely together, indicating a higher degree of transcriptional similarity compared with cTfh, CXCR5dimPD-1dim Tfh from TDL or LNs, or CXCR5– CD4+ T cells from all tissues (Figure 5B). We next examined specific transcriptional changes that were common to CXCR5BrPD-1Br TDL Tfh and LN GC Tfh compared with the CXCR5dimPD-1dim Tfh from each compartment (Figure 5C). Among the genes coordinately upregulated by the CXCR5BrPD-1Br TDL Tfh and LN GC Tfh were key GC Tfh transcripts including IL21-AS1 and CXCL13 (Figure 5C). To further interrogate the transcriptional programs of these Tfh populations, we used Gene Set Enrichment Analysis (GSEA) to compare these data sets to known GC Tfh signatures derived from tonsillar Tfh (NCBI’s Gene Expression Omnibus database [GEO] GSE50391). GSEA confirmed that both LN GC Tfh and TDL CXCR5BrPD-1Br Tfh were strongly enriched for the tonsillar GC Tfh signature (ref. 12 and Figure 5, D and E). Many genes in the leading edges driving the enrichment were common for GC Tfh and CXCR5BrPD-1Br TDL Tfh and included GC-Tfh–associated transcripts such as IL21, CXCL13, SH2D1A, and CD200 (Figure 5F). These analyses also demonstrated negative enrichment for Runx3, a transcription factor that can antagonize the follicular program in CD8+ T cells (45).
Figure 5. TDL CXCR5BrPD-1Br Tfh are transcriptionally similar to GC Tfh.
(A) Gating strategy for sort purification with an overlay of flow cytometry plots for blood (n = 3), TDL (n = 3), and mesenteric LNs (n = 3): naive (orange gate) and nonnaive (black gate). Nonnaive cells were sorted based on the gates shown for each tissue source: CXCR5BrPD-1Br (red), CXCR5DimPD-1Dim (teal), CXCR5– non-Tfh (green) and cTfh (blue). Plots for TDL and blood are shown in Figure 1A. mLN, mesenteric lymph node. (B) Principal component analysis of RNA-Seq data; colors represent populations from the matching gate color in A. (C) Correlation plot of the fold change from CXCR5BrPD-1Br Tfh to CXCR5DimPD-1Dim Tfh in TDL (TDLBr and TDLDim, respectively) and LNs (GC Tfh and non-GC Tfh, respectively). (D) GSEA of transcriptional signatures from tonsil GC Tfh versus tonsil non-GC Tfh (GSE50391) compared with differentially expressed genes in GC versus non-GC Tfh from LNs and (E) TDLBr Tfh versus TDLDim Tfh. Normalized enrichment scores (NES) and FDR marked on each GSEA. (F) Heatmap of leading edge genes from the LNs and TDL GSEAs in D and E were compared for overlap in both positively and negatively enriching genes. (G) Volcano plot of differentially expressed transcripts (adjusted P < 0.05 in orange) for LN Tfh (dim and bright) versus all TDL Tfh (dim and bright). (H) Volcano plot for differentially expressed transcripts (adjusted P < 0.05 in orange) for all TDL Tfh (dim and bright) versus blood cTfh.
Although CXCR5BrPD-1Br TDL Tfh and GC Tfh appeared to share a core transcriptional program, we also tested to determine whether anatomical location was associated with distinct transcriptional signatures. Comparing all TDL Tfh to all LN Tfh revealed compartment-related differences (Figure 5G). For example, Tfh in TDL had higher expression of genes linked to lymphatic egress, including S1PR1 and S1PR4 and Kruppel-like factor-2 (KLF2) (Figure 5G), consistent with reduced CD69 expression in CXCR5BrPD-1Br TDL Tfh (Supplemental Figure 3, F and G) and a difference in migratory biology. Upon entry into blood, cTfh display increased expression of the interferon stimulated genes (ISGs) MX1, MX2, and XIAF as compared with TDL Tfh (Figure 5H), similar to the increased ISGs previously observed in circulating mucosal-associated invariant T cells as compared with those in lymph (46). Together, these data suggested a common transcriptional program of Tfh biology between GC Tfh and CXCR5BrPD-1Br TDL Tfh with a set of differences between these Tfh populations that included evidence of altered trafficking and tissue residency programs. Overall, these data identified a core GC Tfh transcriptional program that could be identified in TDL.
Epigenetic accessibility defines stable GC Tfh program imprinting that can be traced into TDL.
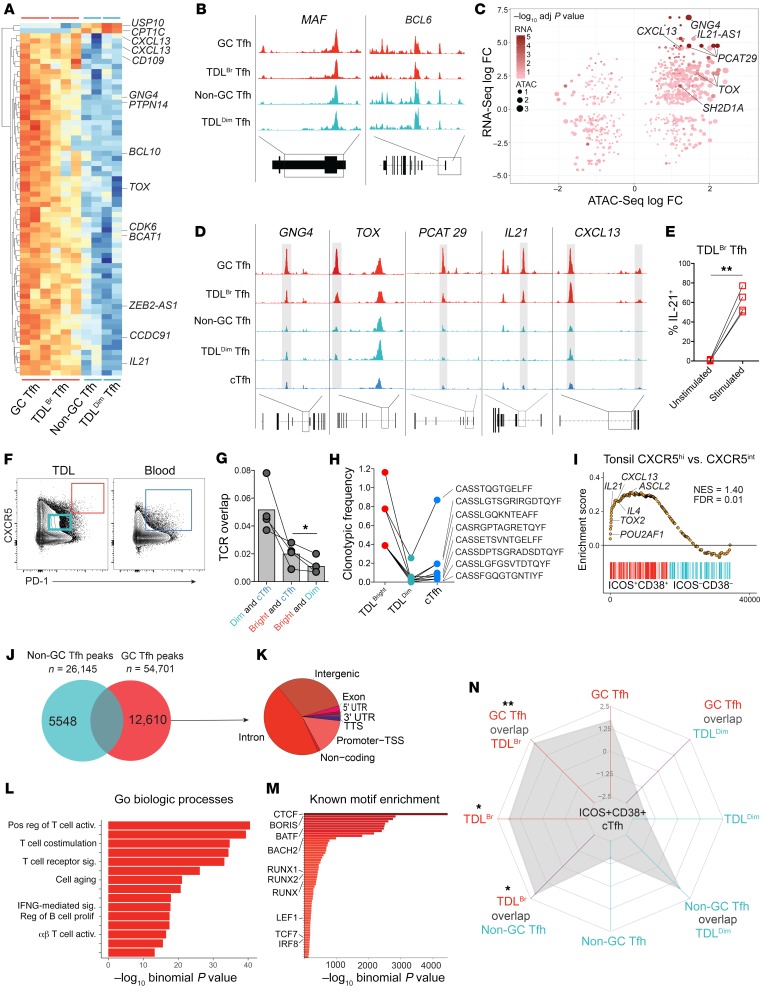
The protein and mRNA expression described above suggested a relationship between LN GC Tfh and CXCR5BrPD-1Br TDL Tfh. Whereas protein-based and transcriptional differences may be subject to transient changes based on environmental cues, epigenetic signatures may be more stable, allowing prior imprints of differentiation to be tracked, even when transcription and/or translation has changed (47). We therefore conducted assay of transposase accessible chromatin sequencing (ATAC-Seq) to interrogate the epigenetic relationship between GC Tfh and CXCR5BrPD-1Br TDL Tfh compared with non-GC Tfh and CXCR5DimPD-1Dim TDL Tfh (Supplemental Figure 4A). Shared chromatin accessibility regions were identified between GC Tfh and CXCR5BrPD-1Br TDL Tfh, including regions within or near CXCL13, IL21, and TOX (Figure 6A). In contrast, accessibility to genes encoding several core regulators of the Tfh lineage, including BCL6 and MAF, were similar across Tfh subsets and tissues (Figure 6B). By combining these data with the RNA-Seq data set used for Figure 5, we defined key genes in CXCR5BrPD-1Br Tfh displaying both epigenetic and transcriptional differences, where increased locus accessibility largely correlated with increased transcription and vice versa (Figure 6C). This combined RNA-Seq and ATAC-Seq analysis identified prominent transcriptional and epigenetic changes in CXCL13, TOX, IL21, PCAT29, and GNG4 in CXCR5BrPD-1Br GC Tfh that could also be observed in CXCR5BrPD-1Br TDL Tfh (Figure 6D), suggesting epigenetically regulated common transcriptional events between these anatomically separated Tfh. As would be predicted from these transcriptional and epigenetic data, CXCR5BrPD-1Br TDL Tfh readily expressed IL-21 protein upon in vitro stimulation (Figure 6E and Supplemental Figure 4B).
Figure 6. Epigenetic signature of GC Tfh can be traced into CXCR5BrPD-1Br TDL Tfh and is associated with transcriptional events in activated cTfh in blood.
(A) Heatmap of OCRs differentially accessible in CXCR5BrPD-1Br Tfh LNs and TDL compared with CXCR5DimPD-1Dim Tfh. (B) ATAC-Seq tracks of the BCL6 and MAF loci in CXCR5BrPD-1Br Tfh (red, merged from 3 samples for each population) and CXCR5DimPD-1Dim Tfh (teal, merged from 2 samples from each population). (C) Correlation plot of log fold changes in ATAC-Seq and RNA-Seq data sets. (D) ATAC-Seq tracks of loci defined in C, with cTfh in blue. Shaded bars are loci differentially accessible in CXCR5BrPD-1Br versus CXCR5DimPD-1Dim Tfh. (E) Percentage of CXCR5BrPD-1Br TDL Tfh producing IL-21 before and after stimulation with PMA/ionomycin. (F) Gating strategy for TCR sequencing sort (n = 4 paired samples). (G) TCR overlap between the Tfh populations as indicated in F. (H) Frequency of CDR3 sequences of overlapping clones between Tfh populations (TDL and blood) and gates (bright and dim) in a representative patient. (I) GSEA of transcriptional signatures from tonsil GC Tfh versus tonsil non-GC Tfh (GSE50391) compared with differentially expressed genes in ICOS+CD38+ versus ICOS–CD38– cTfh. Normalized enrichment scores, FDR, and selected leading edge genes noted on plot. (J) Unique peaks in GC Tfh versus non-GC Tfh. (K) Genomic distribution of GC Tfh unique peaks. (L) GREAT analysis to identify GO biologic processes within GC Tfh unique peaks. (M) Homer Motif prediction to identify known transcription factor–binding sites enriched in GC Tfh unique peaks. (N) Radar plot of GSEA enrichment for ICOS+CD38+ versus ICOS–CD38– cTfh signature using gene sets derived from unique peaks in GC Tfh, non-GC Tfh, CXCR5BrPD-1Br TDL Tfh, and CXCR5DimPD-1Dim Tfh as well as the overlap between the cell types as indicated. NES noted for each ring. *P < 0.05; **P < 0.01, paired 2-tailed t tests (E and G).
The data above support an ontological relationship between GC Tfh and CXCR5BrPD-1Br TDL Tfh, suggesting that at least some GC-experienced Tfh may emigrate from the SLO and enter the lymph. Because the TDL containing CXCR5BrPD-1Br TDL Tfh drains directly into the venous blood, these data also provoke the hypothesis that cells with a GC Tfh history might be found in circulation. However, given the low frequency of CXCR5BrPD-1Br Tfh in TDL and near absence of such cells in the blood, we questioned whether CXCR5BrPD-1Br Tfh epigenetic changes would be detectable in bulk cTfh populations. Indeed, the locus accessibility in key CXCR5BrPD-1Br Tfh genes in bulk cTfh was more similar to that of CXCR5DimPD-1Dim Tfh and non-GC Tfh (Figure 6D). These data could reflect a lack of GC Tfh progeny in the blood due to decreased residency times estimated above or may simply reflect the limit of detection of the epigenetic assays to discern a minor fraction of GC-derived cTfh in a much larger pool of non-GC–derived cTfh. To ask whether CXCR5BrPD-1Br TDL Tfh might enter the cTfh compartment using an alternative approach, we employed TCR sequencing to test whether TCR clonotypes found in CXCR5BrPD-1Br TDL Tfh cells could be found in blood. Thus, we sorted CXCR5BrPD-1Br Tfh and CXCR5DimPD-1Dim Tfh from TDL as well as cTfh from blood in paired samples from 4 donors (Figure 6F). Blood cTfh had the highest TCR overlap with CXCR5dimPD-1dim TDL Tfh (Figure 6G), identifying most cTfh as a pool of recirculating, resting Tfh cells that are not actively participating in an immune response. These data are consistent with the similarity of cTfh to CXCR5DimPD-1Dim TDL Tfh in the RNA-Seq (Figure 5B) and ATAC-Seq (Figure 6D) data. However, when TCR sequences from CXCR5BrPD-1Br TDL Tfh were examined, these sequences were more likely to be found in cTfh from paired blood than in the population of TDL CXCR5dimPD-1dim Tfh (Figure 6, G and H, and Supplemental Figure 4C). These data suggest that at least some CXCR5BrPD-1Br TDL Tfh enter the peripheral blood, even if a CXCR5BrPD-1Br phenotype population cannot be readily identified in circulation.
Our data suggested that most cTfh, like CXCR5dimPD-1dim TDL Tfh, represent a quiescent, recirculating pool. This interpretation is supported by our previous data showing that clonotypes of resting cTfh are highly stable in the blood over time (17). Nevertheless, TCR analysis suggested some CXCR5BrPD-1Br TDL Tfh-derived cTfh could be found in blood. Thus, to understand which cTfh populations in blood might enrich for CXCR5BrPD-1Br TDL Tfh descendants, we focused on the activated subset of cTfh defined by increased ICOS and CD38 expression. We and others have shown an increase in the frequency of ICOS+ cTfh following vaccination in multiple settings of human vaccination (16–21, 48, 49). For example, following influenza vaccination, we have shown that the ICOS+CD38+ cTfh subset contained the vaccine-specific cTfh population with TCR clonotypes that were repeatedly recalled into this cTfh subset upon subsequent yearly immunization (17). Because CXCR5BrPD-1Br TDL Tfh were enriched for expression of ICOS and CD38 (Figure 4B), we hypothesized that, as these cells entered the blood, they might populate the ICOS+CD38+ cTfh population, which represents approximately 5% of cTfh in blood (18). Moreover, we predicted that focusing on this cTfh subset might allow cells with a GC Tfh–like developmental imprint to be detected in circulation.
To begin to test this idea, we first examined the transcriptional signatures of the ICOS+CD38+ compared with the ICOS–CD38– cTfh subset using RNA-Seq. The transcriptional signature of tonsillar GC Tfh (ref. 12 and Figure 5, D and E) was enriched in the ICOS+CD38+ cTfh compared with the ICOS–CD38– cTfh, and genes at the leading edge of this enrichment included the Tfh cytokines IL21 and CXCL13 (Figure 6I). Indeed, when evaluated within the context of the broader Tfh data set, ICOS+CD38+ cTfh clustered more closely to CXCR5BrPD-1Br Tfh than did total cTfh or ICOS–CD38– cTfh (Supplemental Figure 4D). Further, the ICOS+CD38+ subset was more likely to express the lymphatic egress protein S1PR1 than were ICOS–CD38– cTfh, suggesting more recent lymphatic exit (Supplemental Figure 4E). Limited epigenetic analysis of the ICOS+CD38+ cTfh subset from one subject identified OCRs in both IL21 and CXCL13 (Supplemental Figure 4F). Moreover, consistent with our prior study (17), the ICOS+CD38+ subset was more likely to express IL-21 protein upon stimulation than the ICOS–CD38– cTfh (Supplemental Figure 4G). Finally, like GC Tfh, the ICOS+CD38+ cTfh subset was able to provide B cell help in vitro, although the ICOS–CD38– cTfh also provided help under these in vitro conditions (Supplemental Figure 4, H and I). Together these data reinforce the notion that ICOS+CD38+ cTfh contain a transcriptional and functional relationship to CXCR5BrPD-1Br Tfh.
To determine whether this ICOS+CD38+ cTfh transcriptional relationship to GC Tfh could be traced to a broader epigenetic imprint of the GC Tfh, we defined a unique OCR profile for GC Tfh and non-GC Tfh (Figure 6J). GC Tfh contained approximately 12,000 unique OCRs compared with non-GC Tfh (Figure 6J), and most of these OCRs were in introns or in intragenic regions (Figure 6K), as expected (50, 51). Using Genomic Regions Enrichment Annotation Tool (GREAT), the GC Tfh–specific OCRs were enriched for genomic regions associated with T cell activation, costimulation, and regulation of B cell proliferation (Figure 6L). Moreover, motif analysis (52) revealed that the GC Tfh–specific OCRs contained binding sites for the transcription factors LEF-1 and TCF1 as well as the RUNX family transcription factors (Figure 6M), consistent with Tfh biology in mice (53–55).
Using these data, we next determined the sets of OCR that defined the TDL Tfh subsets (Supplemental Figure 4J) and also sets of OCRs that were common to different pairs of Tfh (e.g., GC Tfh and CXCR5BrPD-1Br TDL Tfh, or non-GC Tfh and CXCR5dimPD-1dim TDL Tfh; Supplemental Figure 4K). Converting these Tfh subset–specific or shared OCR sets to gene lists, we then performed a multicell–type GSEA to investigate which cell types contained epigenetic changes with overlap in the ICOS+CD38+ cTfh signature (Figure 6N). This approach revealed several features. First, there was relatively little similarity between the transcriptional signature of ICOS+CD38+ cTfh and the epigenetically identified gene profiles of CXCR5dimPD-1dim TDL Tfh or non-GC LN Tfh or the features shared by GC Tfh and the CXCR5dimPD-1dim TDL Tfh (Figure 6N). There was, however, some enrichment with profiles common to non-GC Tfh and CXCR5dimPD-1dim TDL Tfh, although this overlap did not meet statistical significance. Second, the transcriptional signature of ICOS+CD38+ cTfh enriched most strongly for epigenetically identified genes shared between CXCR5BrPD-1Br TDL Tfh and non-GC Tfh and those shared between CXCR5BrPD-1Br TDL Tfh and GC Tfh. The genes driving these overlaps included CD38, KI67, POU2AF1, IL21-AS1, TOX2, IFNG, IL21, S1PR4, and SH2D1A. Thus, a subset of cTfh, in particular the ICOS+CD38+ subset, has transcriptional evidence of similarity to both transcriptional and epigenetic signatures found predominantly in GC Tfh and CXCR5BrPD-1Br TDL Tfh. Therefore, our data support the concept that some Tfh imprinted during GC responses in lymphoid tissue traffic through the lymph and are present in blood and that these cTfh with an imprint of GC biology are enriched in the ICOS+CD38+ subset. These data suggest that this subset of cTfh may provide a window into dynamic events that occur during the humoral response in SLO.
Discussion
A central challenge in the study of Tfh in humans is the localization of these cells to the largely inaccessible B cell follicles of secondary lymphoid tissues, rather than blood where most human immune responses are monitored. How the more accessible cTfh in the blood relate to those Tfh in the B cell follicles and GC of LNs, and the relationship between Tfh populations in lymphoid organs and those in the blood in humans, remains poorly understood. In particular, it is unclear whether Tfh that have participated in a GC reaction leave the LNs and enter the lymphatics and, if so, it remains unclear whether some of these cells eventually migrate into the blood. Here, we used human TDL to define the dynamic relationships between Tfh in SLO, lymph, and blood. Our study suggests that Tfh with phenotypic, TCR clonotype and transcriptional similarity to GC Tfh exit the LNs and enter circulation.
Our studies defined transcriptional and epigenetic signatures of Tfh subsets in LNs, TDL, and blood. Several transcriptional and epigenetic features were shared in these Tfh subpopulations. Key among shared features between different Tfh subsets were common epigenetic changes observed in LN GC Tfh and TDL CXCR5BrPD-1Br Tfh, including in the Tfh genes CXCL13 and IL21. Although insufficient material was available for extensive epigenetic analysis of the ICOS+CD38+ cTfh in blood, these cells had increased transcription of these and other genes that had epigenetic changes in GC Tfh and TDL CXCR5BrPD-1Br Tfh, suggesting an ontologic relationship between events in the GC and this cTfh subset, likely through the TDL CXCR5BrPD-1Br Tfh intermediate. CXCL13 is produced by both GC Tfh and GC B cells, and the CXCR5-CXCL13 axis regulates Tfh migration (56). Of note, CXCL13 expression in human Tfh is driven by Bcl-6 (57), and this transcription factor is increased in ICOS+CD38+ cTfh compared with quiescent ICOS-CD38-cTfh (17). Havenar-Daughton et al. found that plasma CXCL13 levels correlate with GC responses in lymphoid tissues and with antibody responses 4 weeks after immunization (48). Moreover, these authors showed that CXCL13 also directly correlated with increases in ICOS+ cTfh at 7 days after immunization (48), highlighting the importance of CXCL13 in Tfh biology. IL-21 is also a signature cytokine for GC Tfh function and is required for adequate B cell help (1). IL-21 had more epigenetic accessibility, was increased in expression in GC Tfh and CXCR5BrPD-1Br TDL Tfh, and was transcriptionally increased in ICOS+CD38+ cTfh. These data are consistent with our recent studies demonstrating that ICOS+CD38+ cTfh have greater ability to produce IL-21 protein than quiescent ICOS–CD38– cTfh upon stimulation (17). In addition to changes in IL21 itself, there were also prominent transcriptional and epigenetic alterations in IL21-AS1. IL21-AS1 encodes a long noncoding RNA (lncRNA) that overlaps with IL21 by 2KB. lncRNAs have been shown to modulate gene expression via multiple mechanisms and often function to regulate expression of neighboring genes (58). The function of IL21-AS1 in Tfh is currently unknown, but our data may suggest a distinct role in the GC-derived Tfh populations. Together, these data demonstrate that CXCR5BrPD-1Br Tfh in the TDL are poised to transcribe cytokines and chemokines involved in GC Tfh function and that this potential is also preserved in ICOS+CD38+ cTfh.
Despite similarities between GC Tfh and TDL CXCR5BrPD-1Br Tfh or ICOS+CD38+ cTfh, it remains challenging to directly determine ontologic relationships in the absence of true lineage-tracing experiments in humans. For example, although our data support the idea that CXCR5BrPD-1Br TDL Tfh reflect cells exiting the LN follicle after a GC response, it is also possible that these are cells that exit from the follicular mantle or the T-B border after a secondary response, as has been suggested in mice and humans, respectively (24, 53). However, other possibilities also exist. The CXCR5BrPD-1Br TDL Tfh could represent CD4+ T cells that were initially polarized toward Tfh, but could not complete their differentiation into GC Tfh due to a failure to productively engage B cells (59, 60). These cells could also represent the product of asymmetrical division, wherein one cell enters the follicle and/or GC and the other daughter cell enters lymphatic circulation or is diverted to create memory. Regardless of the precise developmental origin of the CXCR5BrPD-1Br TDL Tfh population, these cells clearly are more closely related transcriptionally to GC Tfh than are other Tfh populations, and they retain epigenetic accessibility in genes central to GC Tfh function. Moreover, the CXCR5BrPD-1Br TDL Tfh population can be clonally traced into the blood using TCR sequencing. Thus, although there is likely heterogeneity in the developmental origin of different subsets of Tfh, our data suggest at least some cTfh are derived from cells with a GC history. ICOS+CD38+ cTfh enrich for features found in both the transcriptional and epigenetic programs of GC Tfh and contain vaccine-responding cells, suggesting that this population — while still likely heterogenous — would enrich for cells that provide a dynamic window into SLO events.
Another unresolved question is the loss of the CXCR5BrPD-1Br phenotype in the transition from TDL to blood. One possibility was that the CXCR5BrPD-1Br TDL Tfh simply become numerically diluted as they enter the blood pool and are below detection. Our mathematical modeling suggests that paucity of CXCR5BrPD-1Br TDL Tfh in the blood might be explained by a short residency time of these cells in circulation (estimated to be one-tenth of the residency time of resting CD4+ T cells). Why CXCR5BrPD-1Br TDL Tfh have such a short blood-residence time is unclear, but could be related to their chemokine receptor expression pattern leading to rapid reextravasation into other lymphoid or nonlymphoid tissues. Indeed, given the increased expression of CXCR3, CXCR5, CCR6, CCR4, and α4β7 in TDL Tfh compared with Tfh in both blood and lymphoid tissue, TDL Tfh may have an increased capacity for tissue homing. Such a model would be compatible with a notion of spreading available T cell help to other anatomical sites. It is also possible, however, that CXCR5BrPD-1Br TDL Tfh die or change phenotype rapidly upon exit to the blood. It is interesting to note that, even though there are few CXCR5BrPD-1Br cTfh in the blood, we were still able to detect the transcriptional signature of these cells in the ICOS+CD38+ cTfh. These data might indicate different sensitivity of RNA-Seq and flow cytometry to detect small populations of T cells in the blood. Another possibility suggested by differences in residency time was that CXCR5BrPD-1Br Tfh preferentially exited the bloodstream in the peripheral capillary beds to facilitate body-wide distribution before entry into venous circulation. However, paired analysis of arterial and venous blood indicated a similar profile of cTfh CXCR5 and PD-1 expression, suggesting that these 2 “sides” of the circulatory loop could not explain the difference between TDL and blood. In addition, similarity between cTfh in arterial and venous blood argue against the possibility that a difference in oxygenation causes a change in CXCR5 and/or PD-1 expression.
We also considered that the CXCR5BrPD-1Br TDL Tfh were lost in pulmonary capillary beds after leaving the thoracic duct, but before entering the systemic circulation, given that TDL enters circulation just before venous return to the right side of the heart. Our ability to detect clones from CXCR5BrPD-1Br TDL Tfh in blood suggests that pulmonary exit is not a uniform mechanism of cell loss, and although possible, loss of CXCR5BrPD-1Br Tfh in the pulmonary beds would suggest that antigen-specific Tfh leaving lymphoid tissues would preferentially accumulate in the lungs, a scenario that would seem inefficient for maintaining systemic immunity. Nevertheless, changes due to passage through the lungs remains a possible contributor to differences between TDL and blood. Mathematical modeling of data from rats has suggested that lymphocytes isolated from lymph traffic to the lungs, but then exit, with a dwell time in respiratory vasculature of approximately 30 seconds (61). In contrast, modeling of data using activated thoracic duct lymphocytes from mice suggests that these activated cells could instead spend as much as 30 minutes in the lung (62). Whether these differences are due to different data sets modeled or species differences is not clear. However, differential epithelial-rolling times of T cell subsets has been described (63) and could play a role in the transition of CXCR5BrPD-1Br Tfh from TDL to blood in humans. A final possibility is that the highest expression of CXCR5 and PD-1 is only maintained in the presence of antigen stimulation (or other signals) present in GC and that the decrease in expression of these molecules correlates temporally with leaving the GC or lymphoid tissue. Such a model would be consistent with a durable epigenetic imprint, but downregulation of a phenotypic signature of GC biology. Future studies will be necessary to resolve this question.
In conclusion, our data support a model wherein CXCR5BrPD-1Br Tfh exit into efferent lymphatics, either as recent participants in a GC reaction or as sentinels of LN follicular biology. Animal models may be required to fully understand Tfh trafficking throughout these compartments. However, technologies that allow study of TCR and transcriptional and epigenetic profiles in individual cells would likely advance our knowledge of Tfh trafficking, especially if paired with an immunologic challenge, such as immunization. These efforts will then enhance our ability to use Tfh studies in blood as precise clinical monitors of LN events.
Methods
Human sample collection and processing.
TDL and paired blood were obtained from adult and pediatric patients with clinical indication for thoracic duct cannulation at the University of Pennsylvania and Children’s Hospital of Philadelphia, as indicated in Supplemental Table 1. Access to TDL was performed as previously described (64, 65). In short, under ultrasound guidance, a 25-gauge spinal needle was advanced into inguinal LNs bilaterally and under fluoroscopic guidance. An oil-based contrast agent (Lipiodol, Gurbert Group) was injected into the inguinal LNs. Once opacified, the cisterna chyli was accessed through a transabdominal approach using a 21-gauge Chiba needle (Cook Inc.). A V18 guide wire (Boston Scientific) was then advanced into the thoracic duct and manipulated cephalad. Over the wire, a 60 cm 2.3F Rapid Transit microcatheter (Cordis Corp.) was advanced further into the thoracic duct and TDL was then aspirated through this catheter. Paired blood was obtained by venipuncture and in some cases through an existing central venous catheter. TDL was stored in sodium heparin tubes and purified the same day using Ficoll-Hypaque density gradient centrifugation in order to remove contrast agents used during the fluid collection procedure. Normal donor PBMCs were obtained from healthy donors through the Human Immunology Core at the University of Pennsylvania. PBMCs were purified from whole blood or leukapheresis products by Ficoll-Hypaque density gradient centrifugation. LN mononuclear cells (LNMCs) were obtained from mesenteric LNs that were removed from patients undergoing abdominal surgery at Case Western Reserve Medical Center (Supplemental Table 1) and were deemed by the surgeon to be grossly normal. LNMCs were isolated by mechanical disruption followed by density gradient centrifugation. Tonsil samples were obtained from adult patients from the University of Pennsylvania (Supplemental Table 1) and processed as were the LNMCs. Fingolimod (FTY720) peripheral blood samples were obtained from patients with multiple sclerosis undergoing treatment at McGill University. PBMCs were collected before clinically indicated dosing of FTY720 as well as 2 and 6 months into treatment. Mononuclear cells were cryopreserved and stored at –140°C.
Nonhuman primate sample collection and processing.
RM samples from 4 monkeys of Indian origin were obtained from the University of Pennsylvania. TDL and veins were first cannulated under anesthesia, and tissues were subsequently removed at necropsy. Samples were processed for mononuclear cells as in human sample processing, above.
Flow cytometry and cell sorting.
Cryopreserved PBMCs, LNMCs, and TDL were thawed and rested overnight in RPMI-1640 media supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin/streptomycin and in the presence of DNAse and MgCl2. After the resting period, cells were washed with 1× PBS, followed by prestaining of chemokine receptors and adhesion molecules at 37°C, 5% CO2 for 10 minutes. An aqua amine–reactive dye (Invitrogen) to assess cell viability was added with an antibody cocktail diluted in FACS buffer (FACS buffer was replaced by media for sorting experiments) to stain for surface markers for an additional 20 minutes. The cells were washed with the staining buffer prior to analysis or progression to intracellular staining (ICS). For ICS, cells were then fixed and permeabilized using the Cytofix/Cytoperm Buffer Kit (BD) or the FoxP3 Transcription Factor Buffer Kit (eBioscience). An antibody cocktail was added to stain for intracellular markers for 1 hour at room temperature. Stained cells were fixed in PBS containing 1% paraformaldehyde (Sigma-Aldrich) and stored at 4°C. For intracellular cytokine staining, PMA/ionomycin stimulation was performed for 5 hours. Stimulated cells were first fixed with 1% paraformaldehyde to enhance detection of IL-21 before proceeding with surface and intracellular stains. B cell subset and S1PR1 stains were performed on freshly isolated mononuclear cells, with S1PR1 staining performed in charcoal stripped media. All fixed samples were analyzed within 3 days. Flow cytometry and frequency data shown in Figures 1, 3, and 4 were obtained from samples run across 9 separate experiments, each with either blood and TDL, or blood, TDL, and SLO mononuclear cells stained and analyzed simultaneously. Samples were analyzed on 3 cytometers: LSRII and Symphony A5 for phenotype as well as a FACSAria II (all BD) for phenotype and sorting. Samples run in common between experiments were used to standardize gating across dates and instruments and are only reflected once, using the average frequency between analysis dates. Paired analyses and MFI analyses include only those samples run simultaneously. Data were analyzed with FlowJo software (version 10.1.1 or higher, TreeStar). tSNE plots were generated using FlowJo with downsampling. Antibodies are noted in Supplemental Table 2.
RNA-Seq.
Cryopreserved samples were thawed, rested overnight, and stained. T cell populations of interest were sorted, at 250 cells each, directly into lysis buffer (Takara) and snap-frozen. RNA-Seq libraries were prepared using the Takara SMART-Seq v4 Ultra Low Input RNA Kit followed by the Nextera XT DNA Library Prep kit (Illumina, FC-131-1096) according to the manufacturers’ protocols. The libraries were pooled and sequenced on an Illumina NextSeq using 75 bp, paired-end sequencing on a 150 cycle high-output flow cell (FC-404-2002). FASTQ files from 3 sequencing runs were concatenated and aligned using STAR 2.5.2a and hg38 for a final unique paired mapped read depth between 8 million and 13.6 million reads per sample. The aligned files were normalized using PORT, and differential expression analyses were performed using Limma-Voom after filtering for genes with at least 50 cumulative reads across all samples. GSEA was performed against the gene signatures indicated in each figure (66, 67), and leading edge analyses were performed to identify the common cluster of genes between enriched gene sets for comparisons of interest. The normalized enrichment scores from GSEA for the gene signatures were plotted as a radarplot using R (3.5.1) package fmsb_0.6.3. Heatmaps were created using R (3.5.1) package pheatmap_1.0.12 and plots for PCA were created using ggplot2_3.1.0.
ATAC-Seq.
Mononuclear cells from mesenteric LNs, TDL, and PBMCs were thawed, rested overnight, and sorted in cell numbers of 5000 to 25,000 into complete RPMI with 50% FBS. Cells were then processed for ATAC-Seq as previously described (68). Briefly, nuclei from cell pellets (5,000–25,000 cells) were isolated using a lysis buffer and pelleted in low-bind 1.5 mL tubes (Eppendorf) before resuspending in 25 μl TD buffer with Tn transposase (Illumina). The transposition reaction was continued for 45 minutes at 37°C. The resulting DNA fragments were purified using the MinElute PCR Purification Kit (QIAGEN). Libraries were sequenced by the Children’s Hospital of Philadelphia partnership with the Beijing Genomics Institute on 2 lanes of a Hiseq 4000 paired-end sequencing (100 bp read length). The data processing pipeline is available on GitHub (https://github.com/wherrylab/jogiles_ATAC/blob/master/Giles_Wherry_ATAC_pipeline_hg19_UPennCluster.sh). Briefly, FASTQ files were aligned to hg19 using Bowtie2. Unmapped, unpaired, and mitochondrial reads were removed using samtools. Duplicates and blacklist regions were removed using PICARD and bedtools subtract, respectively. Peaks were called with Macs2 at an FDR of 0.01. A union peak list was generated with all CD4+ Tfh samples using bedtools merge. Normalization and differential peak analysis were performed using DESeq2. A unique peak list for each Tfh subset was determined using bedtools intersect if the OCR was present in all samples of the primary subset and not present in any samples of the comparator subset.
TCR sequencing.
Tfh subpopulations defined as indicated were sorted into 100% FBS for TCR sequencing. Cells were spun for FBS removal, and the cell pellet was then processed for DNA extraction using the QIAamp DNA Micro Kit (QIAGEN). DNA was processed by Adaptive Biotechnologies for TCR sequencing, and both overlap scores and recurring clones were determined using the Adaptive ImmunoSeq Analyzer 2.0 and as described previously (17).
B cell cocultures.
CD4+ T cell subsets from mesenteric LNs (Supplemental Table 1) and from the blood of healthy donors were sorted and plated with purified allogeneic naive peripheral blood B cells from a single donor (Naïve B Cell Isolation Kit; EasySep STEMCELL Technologies) at a 3:1 ratio and cocultured for 7 days in the presence or absence of SEB (0.1 μg/mL; Toxin Technology Inc.). For quantification of antibody production in vitro, Immunol 4HBX plates (Thermo Scientific) were coated with goat anti-human IgG+IgM (Jackson ImmunoResearch Laboratories). Bound Abs were detected by biotinylated anti-human IgG (Jackson ImmunoResearch Laboratories), alkaline-phosphatase–conjugated streptavidin (Jackson ImmunoResearch Laboratories), and phosphatase substrate (Sigma Aldrich), respectively.
Statistics, nonsequencing data.
Normality of frequency and MFI distribution were determined using the D’Agostino-Pearson omnibus normality test. Unpaired samples were compared using Mann-Whitney U test or Student’s t test. Paired samples were compared using Wilcoxon’s matched-pairs single-rank or paired t tests. All Student’s t tests were performed as 2-tailed tests. Samples obtained over time were compared with Friedman’s test. Tests with multiple comparisons were performed using ANOVA with Holm-Šídák post-test or Kruskal-Wallace with Dunn’s post-test. In Figure 1, B, D, F, and H, the variances were proportional to the means and the data therefore did not satisfy the equal variance assumption. To correct, multiple comparisons in Figure 1, B, D, F, and H, were performed on log-transformed data. The threshold for significance was set as P ≤ 0.05. Nonsequencing statistical analyses were performed in Prism, version 7.0 (Graphpad).
Study approval.
All human participant research was performed after informed consent in accordance with the institutional review boards at the following institutions: University of Pennsylvania, Children’s Hospital of Philadelphia; Case Western Reserve University; and McGill University. All macaque studies were performed under a protocol approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania. All procedures were performed in accordance with the Animal Welfare Act and other federal statutes and regulations relating to animals.
Data and materials availability.
Sequencing data reported in this manuscript have been deposited in the NCBI’s GEO (GSE130794).
Author contributions
LAV, MB, MRB, and EJW conceived of the overall design and designed experiments. MRB provided essential materials for research. LAV and MB — with assistance from RSH, IBB, HK, SN, IS, LKC, AM, TML, and KCO — designed and performed experiments. ABO, JPA, DHC, AN, MGI, and YD developed and executed patient recruitment protocols. LAV, SM, JRG, RSH, MEJ, and ADW contributed to processing and analysis of sequencing data. VVG contributed mathematical modeling. All authors critically reviewed the manuscript.
Supplementary Material
Acknowledgments
We thank members of the Wherry and Betts laboratories for discussions and critical review of the data and manuscript, T. Roberston of the J. Burkhardt laboratory at the Children’s Hospital of Philadelphia for guidance with S1PR1 staining, and all of the patients for participating in human subjects research. LAV was supported by the Pediatric Infectious Diseases Society Fellowship Award, the National Center for Advancing Translational Sciences of the NIH (award number KL2TR001879), and a Mentored Research Scholar Award from the Penn Center for AIDS Research (CFAR), an NIH-funded program (P30 AI 045008), and a Mentored Clinical Scientist Career Development Award from the National Institute of Allergy and Infectious Diseases (K08AI136660). MB was funded through the Swedish Research Council (Dnr 53720146829) and the Karolinska Institute. Collection of samples from patients undergoing FTY720 therapy was supported by Novartis Canada as part of the prospective, investigator-initiated (ABO, JPA) IGLOO study. Work in TML’s laboratory was supported in part by Merit Review Award I01 BX002644 from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Program and by RO1 AI23539 from the National Institute of Allergy and Infectious Diseases. This work was additionally supported by NIH grants GM118553 (to VVG) and R01 AI076066 and R01 118694 (to MRB) as well as AI105343, AI112521, AI082630, and AI117950 (to EJW). EJW is also supported by the Parker Institute for Cancer Immunotherapy, which supports the cancer immunology program at the University of Pennsylvania. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Version 1. 07/02/2019
Electronic publication
Version 2. 08/01/2019
Print issue publication
Footnotes
Conflict of interest: EJW has consulting agreements with and/or is on the scientific advisory board for Merck, Roche, Pieris Pharmaceuticals, Elstar Therapeutics, and Surface Oncology, is a founder of Surface Oncology and Arsenal Biosciences, and has a patent licensing agreement on the PD-1 pathway with Roche/Genentech (8,652,465).
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(8):3185–3200.https://doi.org/10.1172/JCI125628.
See the related Commentary at Searching for the origin of the enigmatic circulating T follicular helper cells.
Contributor Information
Marcus Buggert, Email: marcus.buggert@ki.se.
Sasikanth Manne, Email: sasim@pennmedicine.upenn.edu.
Ismail Sayin, Email: ixs115@case.edu.
Leticia Kuri-Cervantes, Email: kuri@pennmedicine.upenn.edu.
Hagop Kaprielian, Email: kaprielian.hagop@gmail.com.
Josephine R. Giles, Email: jogiles@pennmedicine.upenn.edu.
Son Nguyen, Email: sonn@pennmedicine.upenn.edu.
Alexander Muselman, Email: alex.muselman3@gmail.com.
Amit Bar-Or, Email: amitbar@pennmedicine.upenn.edu.
Ali Naji, Email: ali.naji@uphs.upenn.edu.
Vitaly V. Ganusov, Email: vitaly@utk.edu.
Terri M. Laufer, Email: tlaufer@pennmedicine.upenn.edu.
Yoav Dori, Email: doriy@email.chop.edu.
References
- 1.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 3.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueno H. T follicular helper cells in human autoimmunity. Curr Opin Immunol. 2016;43:24–31. doi: 10.1016/j.coi.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Perreau M, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210(1):143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pantaleo G, et al. Lymphoid organs function as major reservoirs for human immunodeficiency virus. Proc Natl Acad Sci U S A. 1991;88(21):9838–9842. doi: 10.1073/pnas.88.21.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boritz EA, et al. Multiple origins of virus persistence during natural control of HIV infection. Cell. 2016;166(4):1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohler SL, et al. Germinal center T follicular helper cells are highly permissive to HIV-1 and alter their phenotype during virus replication. J Immunol. 2016;196(6):2711–2722. doi: 10.4049/jimmunol.1502174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-Ephraim YE, et al. Cross-tissue transcriptomic analysis of human secondary lymphoid organ-residing ILC3s reveals a quiescent state in the absence of inflammation. Cell Rep. 2017;21(3):823–833. doi: 10.1016/j.celrep.2017.09.070. [DOI] [PubMed] [Google Scholar]
- 10.Alcántara-Hernández M, et al. High-dimensional phenotypic mapping of human dendritic cells reveals interindividual variation and tissue specialization. Immunity. 2017;47(6):1037–1050.e6. doi: 10.1016/j.immuni.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41(4):529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locci M, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma CS, et al. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87(8):590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 14.Sayin I, et al. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J Exp Med. 2018;215(6):1531–1542. doi: 10.1084/jem.20171940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pallikkuth S, et al. Peripheral T follicular helper cells are the major HIV reservoir within central memory CD4 T cells in peripheral blood from chronically HIV-infected individuals on combination antiretroviral therapy. J Virol. 2015;90(6):2718–2728. doi: 10.1128/JVI.02883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abudulai LN, et al. Production of IgG antibodies to pneumococcal polysaccharides is associated with expansion of ICOS+ circulating memory T follicular-helper cells which is impaired by HIV infection. PLoS ONE. 2017;12(5):e0176641. doi: 10.1371/journal.pone.0176641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herati RS, et al. Successive annual influenza vaccination induces a recurrent oligoclonotypic memory response in circulating T follicular helper cells. Sci Immunol. 2017;2(8):eaag2152. doi: 10.1126/sciimmunol.aag2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herati RS, et al. Circulating CXCR5+PD-1+ response predicts influenza vaccine antibody responses in young adults but not elderly adults. J Immunol. 2014;193(7):3528–3537. doi: 10.4049/jimmunol.1302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bentebibel SE, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5(176):176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentebibel SE, et al. ICOS(+)PD-1(+)CXCR3(+) T follicular helper cells contribute to the generation of high-avidity antibodies following influenza vaccination. Sci Rep. 2016;6:26494. doi: 10.1038/srep26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, et al. Metabolic phenotypes of response to vaccination in humans. Cell. 2017;169(5):862–877.e17. doi: 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulman Z, et al. T follicular helper cell dynamics in germinal centers. Science. 2013;341(6146):673–677. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suan D, et al. T follicular helper cells have distinct modes of migration and molecular signatures in naive and memory immune responses. Immunity. 2015;42(4):704–718. doi: 10.1016/j.immuni.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Hale JS, et al. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38(4):805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier N, et al. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J Immunol. 2011;186(10):5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 26.Choi YS, et al. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013;190(8):4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randolph GJ, Ivanov S, Zinselmeyer BH, Scallan JP. The lymphatic system: integral roles in immunity. Annu Rev Immunol. 2017;35:31–52. doi: 10.1146/annurev-immunol-041015-055354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen E, Itkin M. Thoracic duct embolization for chylous leaks. Semin Intervent Radiol. 2011;28(1):63–74. doi: 10.1055/s-0031-1273941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sage PT, Alvarez D, Godec J, von Andrian UH, Sharpe AH. Circulating T follicular regulatory and helper cells have memory-like properties. J Clin Invest. 2014;124(12):5191–5204. doi: 10.1172/JCI76861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerlach C, et al. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity. 2016;45(6):1270–1284. doi: 10.1016/j.immuni.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thome JJ, et al. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 2014;159(4):814–828. doi: 10.1016/j.cell.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehling M, et al. FTY720 therapy exerts differential effects on T cell subsets in multiple sclerosis. Neurology. 2008;71(16):1261–1267. doi: 10.1212/01.wnl.0000327609.57688.ea. [DOI] [PubMed] [Google Scholar]
- 33.Starzl TE, Weil R, Koep LJ, Iwaki Y, Terasaki PI, Schröter GP. Thoracic duct drainage before and after cadaveric kidney transplantation. Surg Gynecol Obstet. 1979;149(6):815–821. [PMC free article] [PubMed] [Google Scholar]
- 34. ICRP. Report of the Task Group on Reference Man. ICRP Publication 23. Oxford, Pergamon Press; 1975. [Google Scholar]
- 35.Pabst R. The spleen in lymphocyte migration. Immunol Today. 1988;9(2):43–45. doi: 10.1016/0167-5699(88)91258-3. [DOI] [PubMed] [Google Scholar]
- 36.Alshekaili J, et al. STAT3 regulates cytotoxicity of human CD57+ CD4+ T cells in blood and lymphoid follicles. Sci Rep. 2018;8(1):3529. doi: 10.1038/s41598-018-21389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193(12):1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cárdeno A, Magnusson MK, Quiding-Järbrink M, Lundgren A. Activated T follicular helper-like cells are released into blood after oral vaccination and correlate with vaccine specific mucosal B-cell memory. Sci Rep. 2018;8(1):2729. doi: 10.1038/s41598-018-20740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei Y, Feng J, Hou Z, Wang XM, Yu D. Flow cytometric analysis of circulating follicular helper T (Tfh) and follicular regulatory T (Tfr) populations in human blood. Methods Mol Biol. 2015;1291:199–207. doi: 10.1007/978-1-4939-2498-1_17. [DOI] [PubMed] [Google Scholar]
- 41.Shi J, Hou S, Fang Q, Liu X, Liu X, Qi H. PD-1 controls follicular T helper cell positioning and function. Immunity. 2018;49(2):264–274.e4. doi: 10.1016/j.immuni.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buggert M, et al. Identification and characterization of HIV-specific resident memory CD8. Sci Immunol. 2018;3(24):eaar4526. doi: 10.1126/sciimmunol.aar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazarovits AI, et al. Lymphocyte activation antigens. I. A monoclonal antibody, anti-Act I, defines a new late lymphocyte activation antigen. J Immunol. 1984;133(4):1857–1862. [PubMed] [Google Scholar]
- 44.Weinstein JS, et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014;124(25):3719–3729. doi: 10.1182/blood-2014-06-582700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shan Q, et al. The transcription factor Runx3 guards cytotoxic CD8. Nat Immunol. 2017;18(8):931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voillet V, et al. Human MAIT cells exit peripheral tissues and recirculate via lymph in steady state conditions. JCI Insight. 2018;3(7):e 98487. doi: 10.1172/jci.insight.98487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JL, Vahedi G. Do memory CD4 T cells keep their cell-type programming: plasticity versus fate commitment? epigenome: a dynamic vehicle for transmitting and recording cytokine signaling. Cold Spring Harb Perspect Biol. 2018;10(3):a028779. doi: 10.1101/cshperspect.a028779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Havenar-Daughton C, et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci U S A. 2016;113(10):2702–2707. doi: 10.1073/pnas.1520112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heit A, et al. Vaccination establishes clonal relatives of germinal center T cells in the blood of humans. J Exp Med. 2017;214(7):2139–2152. doi: 10.1084/jem.20161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354(6316):1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu K, et al. Individuality and variation of personal regulomes in primary human T cells. Cell Syst. 2015;1(1):51–61. doi: 10.1016/j.cels.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi YS, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16(9):980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu T, et al. TCF1 is required for the T follicular helper cell response to viral infection. Cell Rep. 2015;12(12):2099–2110. doi: 10.1016/j.celrep.2015.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol. 2015;16(9):991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 56.Ansel KM, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406(6793):309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 57.Kroenke MA, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188(8):3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mowel WK, Kotzin JJ, McCright SJ, Neal VD, Henao-Mejia J. Control of immune cell homeostasis and function by lncRNAs. Trends Immunol. 2018;39(1):55–69. doi: 10.1016/j.it.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barnett LG, et al. B cell antigen presentation in the initiation of follicular helper T cell and germinal center differentiation. J Immunol. 2014;192(8):3607–3617. doi: 10.4049/jimmunol.1301284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi H. T follicular helper cells in space-time. Nat Rev Immunol. 2016;16(10):612–625. doi: 10.1038/nri.2016.94. [DOI] [PubMed] [Google Scholar]
- 61.Ganusov VV, Auerbach J. Mathematical modeling reveals kinetics of lymphocyte recirculation in the whole organism. PLoS Comput Biol. 2014;10(5):e1003586. doi: 10.1371/journal.pcbi.1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganusov VV, Tomura M. Experimental and mathematical approaches to quantify recirculation kinetics of lymphocytes. bioRxiv. doi: 10.1101/268326. doi: 10.1101/268326. [published online ahead of print]. [DOI] [Google Scholar]
- 63.Abadier M, et al. Effector and regulatory T cells roll at high shear stress by inducible tether and sling formation. Cell Rep. 2017;21(13):3885–3899. doi: 10.1016/j.celrep.2017.11.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nadolski GJ, Itkin M. Feasibility of ultrasound-guided intranodal lymphangiogram for thoracic duct embolization. J Vasc Interv Radiol. 2012;23(5):613–616. doi: 10.1016/j.jvir.2012.01.078. [DOI] [PubMed] [Google Scholar]
- 65.Nadolski GJ, Itkin M. Thoracic duct embolization for nontraumatic chylous effusion: experience in 34 patients. Chest. 2013;143(1):158–163. doi: 10.1378/chest.12-0526. [DOI] [PubMed] [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 68.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.