Abstract
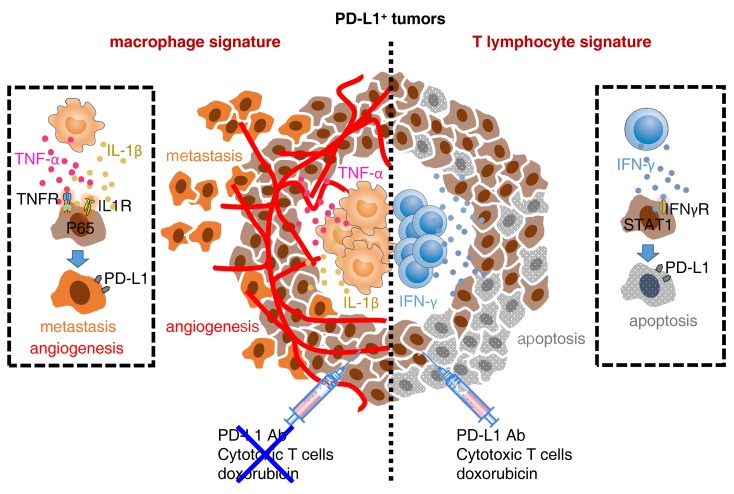
Programmed death-1 receptor ligand 1 (PD-L1) is a promising therapeutic target in aggressive cancers. However, immune landscapes and cancer hallmarks of human PD-L1+ tumors as well as their roles in determining therapeutic efficacies are unknown. Here, we showed, in detailed studies of gene data regarding 9769 patients of 32 types of human cancers, that PD-L1 could not exclusively represent the IFN-γ signature and potentially signified proinflammatory myeloid responses in a tumor. PD-L1 heterogeneity endowed by local immune landscapes controlled cancer hallmarks and clinical outcomes of patients. Mechanically, NF-κB signal elicited by macrophage inflammatory responses generated PD-L1+ cancer cells exhibiting capabilities to aggressively survive, support angiogenesis, and metastasize, whereas STAT1 signal triggered by activated T cells induced PD-L1+ cancer cells susceptive to apoptosis. Importantly, PD-L1+ cancer cells generated by macrophages established great resistance to conventional chemotherapy, cytotoxicity of tumor-specific effector T cells, and therapy of immune-checkpoint blockade. Therapeutic strategy combining immune-checkpoint blockade with macrophage depletion or NF-κB inhibition in vivo effectively and successfully elicited cancer regression. Our results provide insight into the functional features of PD-L1+ tumors and suggest that strategies to influence functional activities of inflammatory cells may benefit immune-checkpoint blockade therapy.
Keywords: Immunology, Inflammation
Keywords: Cancer immunotherapy
Introduction
Programmed death-1 receptor (PD-1) or its ligand-1 (PD-L1, B7-H1) is a promising therapeutic target in aggressive cancers (1, 2). PD-1 is mainly expressed by exhausted or suppressive lymphocytes and represents defects in adaptive immunity (3–5). Nevertheless, PD-L1 is chiefly detected in inflamed epithelial tissues or activated stromal cells; its expression is assumed to duplicate the IFN-γ signature (6, 7) and represents a predictive biomarker of benefit from PD-1/PD-L1 axis mAbs (8–12). However, in cancer patients with a PD-L1–positive signature, clinical response to PD-1/PD-L1 axis mAbs is still extremely low (13, 14), exposing the fact that PD-L1 expression may represent more intricate signatures in human cancers and cannot exclusively predict improved therapeutic efficacies. At present, immune landscapes and cancer hallmarks of human PD-L1+ tumors are in fact unclear. Furthermore, a related issue that must be addressed is whether immune landscapes of PD-L1+ tumors control the efficacies of cancer therapeutic strategies and, if so, how they exert that influence.
In this study, we have investigated the gene data in 37 cohorts of 9769 patients of 32 types of human cancers and isolated the signal released from the inflammatory infiltrates to characterize the immune signatures of PD-L1+ tumors. We identify a diversity of immune signatures of PD-L1+ tumors and show that balance of macrophage and T cell infiltration in PD-L1+ tumors controls patients’ clinical outcomes. Strikingly, we reveal that PD-L1+ cancer cells generated by macrophage-mediated inflammatory circumstance even acquire capabilities of supporting angiogenesis, metastasizing, and resisting killing by conventional chemotherapy and T cell cytotoxicity. In contrast, PD-L1+ cancer cells induced by activated T cells are debilitated and sensitive to therapy. Importantly, we demonstrate that a therapeutic strategy combining immune-checkpoint blockade with macrophage depletion or NF-κB inhibition effectively defeats tumors and even elicits complete regression in vivo.
Results
PD-L1 mirrors multiple immune signatures in human cancers.
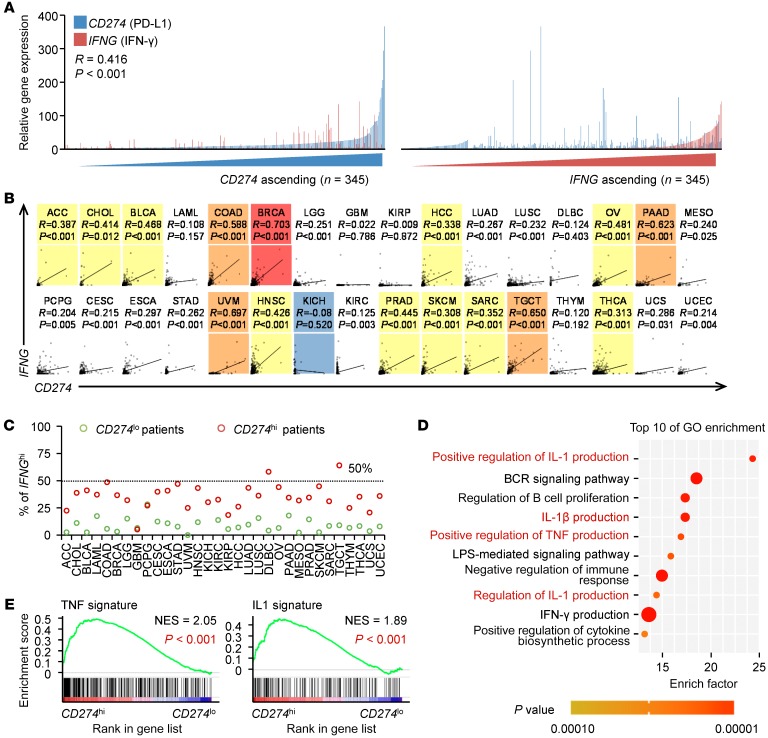
It is assumed that PD-L1 (CD274) mirrors the IFN-γ signature in cancers. However, we analyzed the relationship between PD-L1 and IFN-γ in human hepatocellular carcinoma (HCC) and observed that, in tumor tissues of 345 HCC patients (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI127726DS1), PD-L1 expression did not replicate the IFN-γ signature, although the tissue with a high IFN-γ signature frequently expressed marked PD-L1 (Figure 1A). We came to the same conclusion using the The Cancer Genome Atlas (TCGA) RNA-Seq data regarding 9138 patients with 32 types of cancers (15): in 27 types of human cancers analyzed, PD-L1 expression was either unrelated (P > 0.05 or R < 0.3) or weakly related (P ≤ 0.05, 0.3 ≤ R < 0.5) to IFN-γ expression (Figure 1B and Supplemental Table 2). We further divided patients with each type of cancers into 2 groups according to PD-L1 expression. Similarly, in those with the PD-L1hi signature, a very small fraction displayed the IFN-γhi signature (Figure 1C). These data prompted us to further investigate the immune signature of PD-L1hi tumors. We identified 53 genes correlated with PD-L1 expression in HCC tissues (n = 373, R ≥ 0.5, P ≤ 0.05) and annotated these genes using Gene Ontology (GO) (Figure 1D). Interestingly, among the top 10 enrichment GO terms, 4 pathways that related to proinflammatory TNF or the IL-1 signature were enriched intensively. Five pathways concerning innate activation, cytokine biosynthetic process, or immune regulation were also noted. Notably, although the pathway associated with the IFN-γ signature was also enriched, its priority was extremely low (Figure 1D). Using Gene Set Enrichment Analysis (GSEA) (Supplemental Table 3), we confirmed that genes indicating the IL-1 and TNF signatures were dominantly enriched in PD-L1hi HCC tumors (Figure 1E). Thus, these data suggest that PD-L1 also has great potential to predict proinflammatory innate response in human cancers.
Figure 1. PD-L1 reflects multiple immune signatures in human cancers.
(A) Expression of IFNG (IFN-γ) and CD274 (PD-L1) in 345 HCC tissues. Patients were ranked in ascending order of IFNG or CD274 expression. (B) Correlations between IFNG and CD274 in 9138 patients with 32 types of cancer samples from TCGA data set. P values and R values were calculated based on the analysis of Pearson’s correlation. (C) 9138 Patients with 32 types of cancer samples were divided into 2 groups according to the mean value of CD274 or IFNG expression within each tumor type. The ratios of CD274hi and CD274lo patients expressing higher levels of IFNG were shown. (D) Top 10 biological processes (GO terms) enriched in 53 genes strongly correlated with CD274 expression in HCC samples from TCGA data set (R > 0.5; P < 0.0001). (E) GSEA of TNF signature (left) and IL-1 signature (right) in CD274hi HCC samples versus CD274lo counterparts from TCGA data set. The enrichment scores and P values were calculated by GSEA with weighted enrichment statistics and ratio of classes for the metric as input parameters.
Immune landscapes of PD-L1hi tumors determine patients’ clinical outcomes.
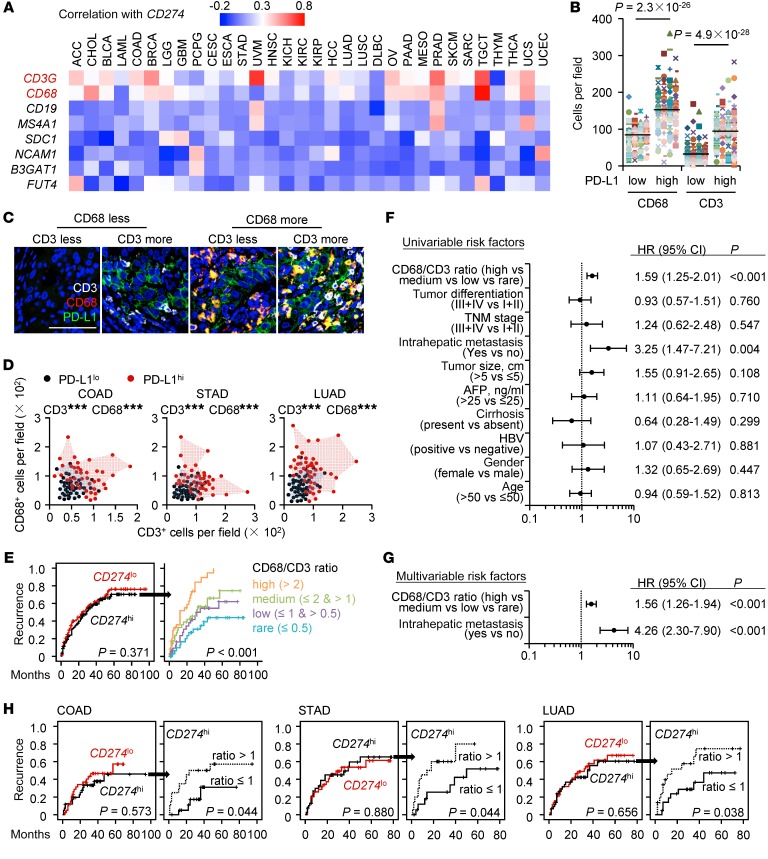
We next probed the composition of immune landscapes in PD-L1hi tumors. In 32 types of cancers analyzed, PD-L1 signatures, although not absolutely, did potentially reflect the infiltration of macrophages and T cells, but this was minimally correlated with the expression of lineage markers of B cells, NK cells, or neutrophils in most types of human cancers (Figure 2A). Using immunohistochemical and immunofluorescent staining, we confirmed that macrophages and T cells separately or together accumulated in PD-L1hi tumors, but not in PD-L1lo tumors (Figure 2, B–D). It should be emphasized that most macrophages in PD-L1hi tumors were also positive for PD-L1 (Figure 2C), sustaining the view that stromal cells contribute to PD-L1–associated suppression (16–19).
Figure 2. Immune landscapes of PD-L1hi cancers affect patients’ clinical outcomes.
(A) Correlations between CD274 and indicated genes were calculated in 9138 patients with 32 types of cancer samples from TCGA data set. (B) Correlations of macrophage and T cell densities with PD-L1 expression in HCC tissues (n = 276). Student’s t test. (C) Confocal microscopy analysis of PD-L1+ cells (green), CD68+ macrophages (red), and CD3+ T cells (white) in HCC tissue. Results represent 3 independent experiments (n = 8). Scale bar: 100 μm. (D) Densities of macrophages and T cells in PD-L1lo or PD-L1hi COAD (n = 82), STAD (n = 78), and LUAD (n = 89) tissues. (E) 276 Patients with HCC were divided into 2 groups according to the median value of CD274 expression in tumors: red lines, low expression (n = 138); black lines, high expression (n = 138). 138 CD274hi patients were further divided into 4 groups according to the ratio of macrophages to T cells in tumors: orange line, ratio value > 2, n = 39; green line, ratio value ≤ 2 and > 1, n = 30; purple line, ratio value ≤ 1 and > 0.5, n = 31; blue line, ratio value ≤ 0.5, n = 38. (F and G) Univariate (F) and multivariate (G) regression analyses of factors associated with recurrence. Cox proportional hazards regression model. (H) 82, 78, and 89 Patients with COAD, STAD, and LUAD, respectively, were analyzed for the prognosis value of CD274 expression plus macrophage/T cell ratio. Patients were divided into 2 groups according to the value of CD274 expression in tumors or ratio of macrophages to T cells in CD274hi tumors. Recurrence times were calculated by the Kaplan-Meier method and analyzed by the log-rank test. ***P < 0.001.
We afterward investigated the influence of immune landscapes on PD-L1–related clinical outcomes. 276 HCC patients who had received curative resection with follow-up data were divided into 2 groups according to the median value of PD-L1 expression in tumors. Unexpectedly, no correlation was found between PD-L1 and patients’ recurrence (Figure 2E). Moreover, 138 HCC patients in the same cohort with the PD-L1hi signature were further divided into 4 groups according to the ratio of macrophages to T cells in tumors. Although the intensities of PD-L1 expression among 4 groups were extremely similar (Supplemental Figure 1A), the clinical outcome of PD-L1hi patients inversely correlated with the ratio of macrophages to T cells (Figure 2E). Univariate (Figure 2F) and multivariate (Figure 2G) regression analyses demonstrated that the ratio of macrophages to T cells in PD-L1hi tumors was an independent predictor of HCC aggressiveness, with significant hazard ratios for predicting clinical outcome. We obtained parallel conclusions regarding validation cohorts of patients with lung adenocarcinoma (LUAD), colon adenocarcinoma (COAD), and stomach adenocarcinoma (STAD) (n = 89, 82, and 78, respectively) (Figure 2H, Supplemental Figure 1, B–D, and Supplemental Table 4). Thus, macrophages and T cells are present predominantly in tumor tissues with the PD-L1hi signature and their balances determine PD-L1–related clinical outcomes.
Distinct induction patterns of cancer cell PD-L1 by tumor macrophages and T cells.
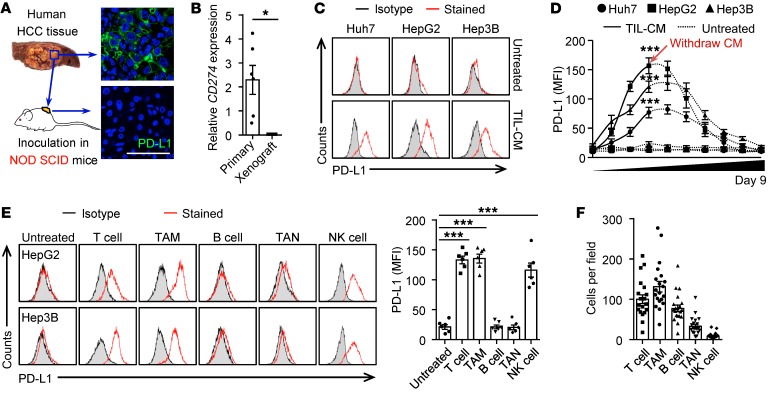
We now asked whether immune landscapes of PD-L1hi tumors mirrored the mechanisms regulating PD-L1 expression. Most tumor tissues from HCC patients were positive for cancer cell PD-L1, although intensities of that protein among patients were dissimilar (Figure 3, A and B). Inoculating with human hepatoma tissues (n = 6) in NOD SCID mice led to a complete loss of cancer cell PD-L1 (Figure 3, A and B). Correspondingly, exposing hepatoma cell line cells, including Hep3B, Huh7, and HepG2, to conditioned medium (CM) from a culture of tumor leukocytes resulted in rapid upregulation of PD-L1, reaching a maximum within 3 days and then gradually declining after removing the CM (Figure 3, C and D, and Supplemental Figure 2, A and B). More precisely, CM from HCC-derived macrophages (TAM-CM) or T cells (T cell–CM), but not that from B cells or neutrophils, individually upregulated cancer cell PD-L1 (Figure 3E and Supplemental Figure 2, A–C), which is consistent with the finding that PD-L1 expression is potent to replicate macrophage and T cell signature in tumors (Figure 2). Notably, although CM from tumor-infiltrating NK cells effectively induced cancer cell PD-L1 (Figure 3E and Supplemental Figure 2C), the NK cells were minimally detected in HCCs (Figure 3F).
Figure 3. Tumor macrophages and T cells effectively induce cancer cell PD-L1.
(A and B) PD-L1 expression in human HCC tissue before or after 25-day inoculation in NOD SCID mice (n = 6). Scale bar: 100 μm. (C–E) Human hepatoma cells were left untreated or were treated with CM from HCC-infiltrating leukocytes (TIL-CM) (C and D) or indicated immune cells isolated from HCC tumors (E). Expression of PD-L1 was determined by FACS on day 3 (C and E) or indicated time (D) (n = 5 for C and D; n = 6 for E). (F) Density of indicated cells in HCC tumor tissue (n = 20). Data represent mean ± SEM. Results are representative of 4 separate experiments. *P < 0.05; ***P < 0.001, Student’s t test (B and D) or 1-way ANOVA with Dunett’s post test (E).
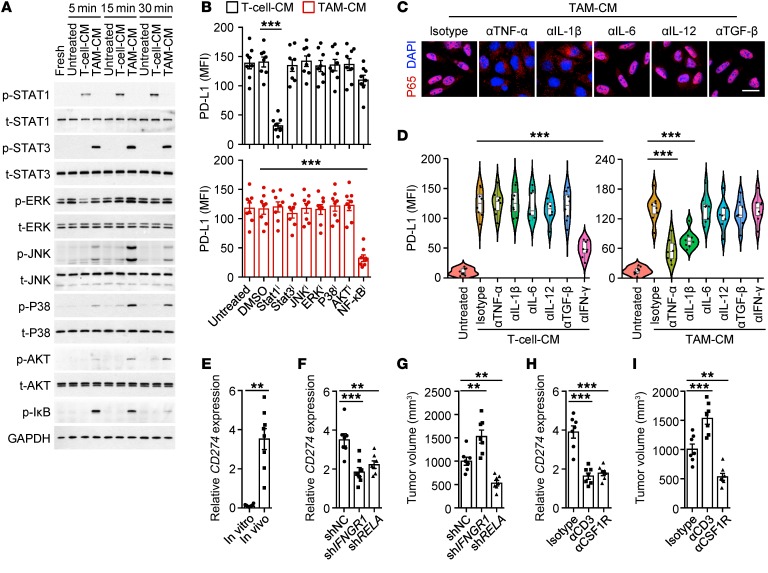
We further probed the signals involved in inducing cancer cell PD-L1 by immune landscapes. TAM-CM strongly elicited hepatoma STAT3 activation, while T cell–CM effectively triggered activation of STAT1, a downstream signal of IFN-γ (Figure 4A). Of note, TAM-CM selectively activated PI3K/AKT, MAPK, and NF-κB (Figure 4A), suggesting the possibility that the mechanisms employed by macrophages and T cells to trigger cancer cell PD-L1 are different. Consistently, blocking the activation of the NF-κB signal impaired TAM-CM–induced cancer cell PD-L1, whereas abolishing the activation of the STAT1 signal attenuated T cell–CM–mediated expression of that protein (Figure 4B and Supplemental Figure 2D). In contrast, suppressing MAPK, AKT, or STAT3 signals had no effect (Figure 4B). In human HCCs, macrophages showed inflammatory features with significant expression of TNFA, IL1B, IL12A, IL6, and TGFB (20). Accordingly, neutralizing the activity of either TNF-α or IL-1β, but not that of IL-12, IL-6, or TGF-β, reversed the TAM-CM–elicited NF-κB P65 nuclear translocation and suppressed subsequent PD-L1 upregulation in cancer cells (Figure 4, C and D, and Supplemental Table 5). Dissimilarly, IFN-γ was critical for T cell–CM in inducing hepatoma PD-L1 (Figure 4D).
Figure 4. Distinct induction patterns of cancer cell PD-L1 by tumor macrophages and T cells.
(A) HepG2 cells were left untreated or were incubated with T cell–CM or TAM-CM for indicated times. Activation of indicated pathways was analyzed by immunoblotting (n = 4). (B) Effects of signaling pathway inhibitor on HepG2 cell PD-L1 expression induced by T cell–CM or TAM-CM (n = 8). (C and D) Effects of cytokine neutralizing Ab on P65 nuclear translocation (C) or PD-L1 expression (D) in HepG2 cells induced by T cell–CM or TAM-CM (n = 4 for C and n = 8 for D). Scale bar: 20 μm. (E) Expression of CD274 on Hepa1-6 cells cultured in vitro or inoculated in liver of immune-competent mice (n = 8). (F and G) P65 knockdown (shRELA) or IFN-γ receptor knockdown (shIFNGR1) Hepa1-6 cells, as well as the control Hepa1–6 cells, were inoculated in liver of C57BL/6 mice as described (Supplemental Figure 2E). CD274 expression in tumor tissues (F) and tumor volume (G) were analyzed. (H and I) Mice bearing Hepa1-6 hepatoma were injected with isotype Ab, anti-CD3 Ab, or anti-CSF1R Ab as described (Supplemental Figure 2F). CD274 expression in tumor tissues (H) and tumor volume (I) were analyzed. Data represent mean ± SEM. Results are representative of at least 3 separate experiments (n = 7). **P < 0.01; ***P < 0.001, 1-way ANOVA with Bonferroni’s post test (B and D), Dunett’s post test (F–I), or Student’s t test (E).
We established autologous mouse models to investigate the regulation of cancer cell PD-L1 in vivo. Hepa1-6 hepatoma from liver of immune-competent mice considerably expressed PD-L1, although it was minimally expressed when cultured in vitro (Figure 4E). In support, knockdown of either IFNGR1 or P65 in Hepa1-6 hepatoma effectively impaired PD-L1 expression in vivo (Figure 4F and Supplemental Figure 2E). Notably, knockdown of P65 effectively suppressed hepatoma growth in mice, but knockdown of IFNGR1 promoted that process in mice (Figure 4G). The same conclusion was obtained when we depleted macrophages or T cells in mice bearing Hepa1-6 hepatoma or B16-F10 melanoma (Figure 4, H and I, and Supplemental Figure 2, F–K): macrophages and T cells individually triggered PD-L1+ tumors, but macrophages simultaneously accelerated the growth of PD-L1+ tumors, whereas T cells delayed the growth of PD-L1+ tumors (Figure 4, H and I, and Supplemental Figure 2, J and K).
Macrophages and T cells induce PD-L1+ cancer cells with distinct hallmarks.
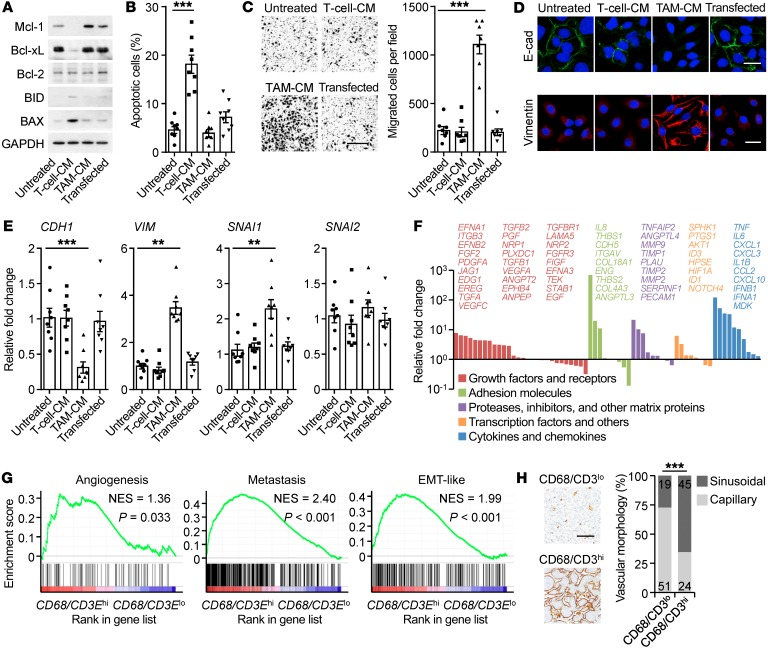
The results described above suggested that PD-L1+ cancer cells generated in different ways might exhibit distinct features during disease progression. We next determined and compared the functional features of PD-L1+ cancer cells triggered by tumor macrophages and T cells. We transduced hepatoma HepG2 cells with pBABE-Puro retroviral vector encoding human PD-L1 (Supplemental Figure 3A) and simultaneously generated PD-L1+ HepG2 cells through exposure to TAM-CM and T cell–CM (Figure 3E). T cell–CM–generated PD-L1+ hepatoma cells undergoing serum starvation displayed proapoptotic status with reduced Mcl-1 and Bcl-xl and increased Bax and Bid (Figure 5A). In contrast, TAM-CM–triggered PD-L1+ cells expressed higher prosurvival Mcl-1 and Bcl-xl (Figure 5A), and these cells resisted serum starvation–elicited apoptosis (Figure 5B). Measuring the metastatic potential revealed that TAM-CM–triggered PD-L1+ hepatoma cells displayed a 5-fold increase in motility (Figure 5C). Consistently, TAM-CM–triggered PD-L1+ hepatoma cells selectively expressed increased vimentin and SNAI1 and reduced E-cadherin, suggesting a process of epithelial-mesenchymal transition (EMT) (Figure 5, D and E). Using SuperArray Real-Time PCR, we identified marked upregulation of several sets of proangiogenic genes in PD-L1+ hepatoma cells induced by TAM-CM, but not by T cell–CM (Figure 5F and Supplemental Figure 3B). These data together reveal that macrophages not only generate PD-L1+ cancer cells, but also endow the cells with capabilities to aggressively survive, migrate, and support angiogenesis. Using GSEA (Supplemental Table 3), we confirmed that genes indicating EMT, metastasis, and angiogenesis were selectively enriched in PD-L1hi HCC, COAD, LUAD, and STAD tumors with a higher ratio of macrophages to T cells (Figure 5G and Supplemental Figure 3C). By analyzing the morphologic features of microvessels, we observed that PD-L1hi HCC patients with a high ratio of macrophages to T cells tended to develop a sinusoidal vasculature in tumor tissues (Figure 5H).
Figure 5. Macrophages and T cells induce PD-L1+ cancer cells with distinct cancer hallmarks.
(A–E) PD-L1+ HepG2 cells were generated by transducing with pBABE-Puro retroviral vector encoding human CD274 or incubating with T cell–CM or TAM-CM. Proteins of survival genes in serum-starvation cells (A), apoptosis of serum-starvation cells (B), migration of cells (C), proteins of EMT genes in cells (D), and expression of EMT markers (E) after 24 hours were determined. Data are representative of 4 separate experiments (n = 4 for A and D, n = 8 for B and E, and n = 7 for C). Scale bars: 500 μm (C); 20 μm (D). (F) Fold changes of protumorigenic gene mRNA levels in TAM-CM–generated PD-L1+ HepG2 cells compared with untreated HepG2 cells were analyzed by SuperArray Real-Time PCR. (G) GSEA of angiogenesis signature, metastasis signature, and EMT-like signature in CD68/CD3Ehi HCC samples versus CD68/CD3Elo counterparts within CD274hi HCC patients from TCGA data set. (H) Different levels of angiogenesis progression in CD68/CD3hi HCC samples versus CD68/CD3lo counterparts within CD274hi HCC patients (n = 139). Scale bar: 100 μm. Data represent mean ± SEM. **P < 0.01; ***P < 0.001, 1-way ANOVA with Dunett’s post test (B, C, and E) or χ2 test (H).
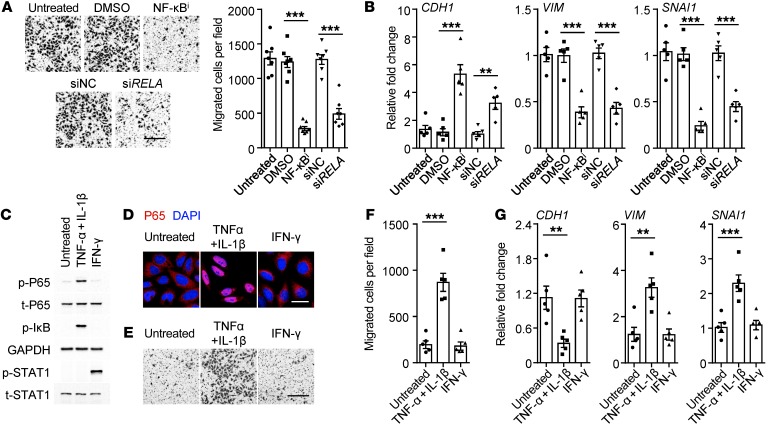
Considering that the NF-κB signal was required for macrophage-triggered PD-L1+ cancer cells (Figure 4B), we determined whether such a mechanism also contributes to aggressive features of a cancer. As expected, either suppressing the activation of NF-κB by inhibitor or knocking down the P65 NF-κB subunit significantly impaired the migration and EMT of hepatoma cells induced by TAM-CM (Figure 6, A and B). Analogously, induction of NF-κB activation in hepatoma cells by exposure to TNF-α and IL-1β (Figure 6, C and D), both of which contributed in a marked way to TAM-CM–upregulated cancer cell PD-L1 (Figure 4, C and D), not only generated PD-L1+ cancer cells (Supplemental Figure 4), but also endowed the cells with aggressive features (Figure 6, E–G). In contrast, IFN-γ could not trigger NF-κB activation and thus it marginally affected aggressive cancer features (Figure 6, C–G), though it induced PD-L1+ hepatoma cells (Supplemental Figure 4).
Figure 6. Macrophages generate aggressive PD-L1+ cancer cells via NF-κB signaling.
(A and B) Incubating with an inhibitor against NF-κB and knockdown of P65 NF-κB subunit (siRELA) attenuated migration (A) and EMT marker expressions of TAM-CM–generated PD-L1+ HepG2 cells (B) (n = 7 for A and n = 5 for B). (C–G) HepG2 cells were left untreated or were incubated with TNF-α plus IL-1β or IFN-γ. Activation of indicated pathways (C), P65 nuclear translocation (D), migration of cells (E and F), and EMT marker expressions (G) were analyzed (n = 5). Scale bars: 500 μm (A and E); 20 μm (D). Data represent mean ± SEM. Results are representative of 4 separate experiments. **P < 0.01; ***P < 0.001, 1-way ANOVA with Bonferroni’s post test (A and B) or Dunett’s post test (F and G).
PD-L1+ tumors generated differently respond to therapeutic strategies distinctly.
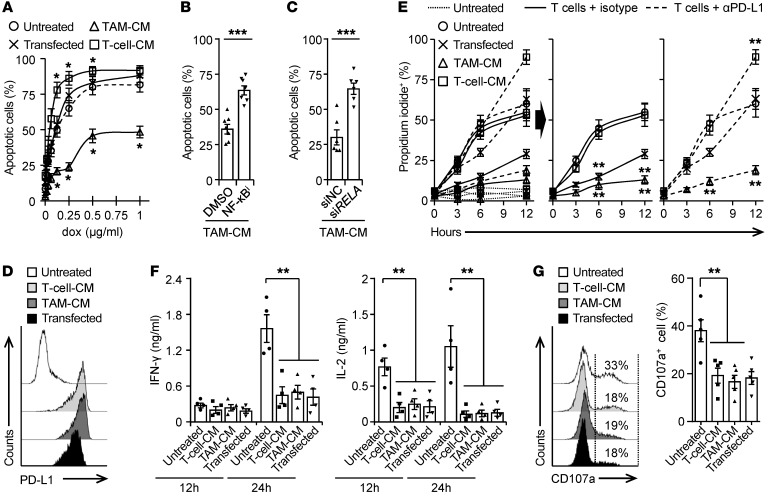
After establishing the regulation, immune landscapes, and functional and clinical relevance of PD-L1+ cancer cells, we hypothesized that PD-L1+ cancer cells generated differently would respond to therapeutic strategies differently. Doxorubicin, an anthracycline commonly used in cancer therapy, at a concentration of 0.25 μg/mL could result in marked apoptosis of untreated or PD-L1–transduced HepG2 cells (Figure 7A). However, at the same concentration, doxorubicin did not trigger the apoptosis of TAM-CM–induced PD-L1+ hepatoma cells, but it elicited a rapid and radical apoptosis of T cell–CM–induced PD-L1+ hepatoma cells (Figure 7A), suggesting that PD-L1+ hepatoma cells generated by macrophages establish resistance to conventional chemotherapy. Supporting our hypothesis that the NF-κB signal contributed to aggressive features of PD-L1+ cancer cells, either suppressing the activation of NF-κB or knocking down the P65 NF-κB subunit in hepatoma cells effectively abolished the TAM-CM–mediated resistance to chemotherapy (Figure 7, B and C).
Figure 7. PD-L1+ tumors generated differently respond to therapeutic strategies distinctly.
(A) PD-L1+ HepG2 cells were generated by transducing with pBABE-Puro retroviral vector encoding human CD274 or incubating with tumor T cell–CM or TAM-CM. Survival of cells after 48-hour exposure to doxorubicin was determined (n = 4). (B and C) Incubating with an inhibitor against NF-κB (B) and knockdown of P65 NF-κB subunit (siRELA, C) in HepG2 cells attenuated TAM-CM–mediated resistance to doxorubicin (0.25 μg/mL; n = 7 for B and n = 6 for C). (D–G) PD-L1+ hepatoma Hepa1-6 cells were generated by transducing with pBABE-Puro retroviral vector encoding mouse CD274 or by exposure to CM from Hepa1-6 hepatoma–derived macrophages or T cells (D). Thereafter, these cells were left untreated or cultured with Hepa1-6 hepatoma–derived CD8+ T cells in the presence of control IgG or an Ab against PD-L1. Survival of Hepa1-6 cells at indicated times (E), 24-hour production of IFN-γ and IL-2 by T cells (F), and 24-hour expression or CD107a on T cells (G) were determined. Data represent mean ± SEM. Results are representative of at least 4 separate experiments. *P < 0.05; **P < 0.01; ***P < 0.001, 1-way ANOVA with Dunett’s post test (A, F, and G), Student’s t test (B and C), or 2-way ANOVA with Bonferroni’s post test (E).
We also established an autologous ex vivo model to investigate the resistance of PD-L1+ hepatoma cells to tumor-specific T cell cytotoxicity. PD-L1+ hepatoma Hepa1-6 cells were generated by transducing with pBABE-Puro retroviral vector encoding mouse PD-L1 or by exposure to CM from Hepa1-6 hepatoma–derived macrophages or T cells (Figure 7D). Thereafter, we cultured these cells with CD8+ T cells directly isolated from mouse Hepa1-6 hepatoma. The untreated Hepa1-6 cells were effectively killed during a narrow time window, 3 to 6 hours after their exposure to CD8+ T cells (Figure 7E). The Hepa1-6 cells transducing PD-L1 could partially resist T cell cytotoxicity, but this process was abolished by blocking the PD-L1 signal (Figure 7E). Dissimilarly, in the same occasion, TAM-CM–generated PD-L1+ hepatoma cells successfully avoided tumor-specific T cell cytotoxicity, while T cell–CM–generated PD-L1+ hepatoma cells were killed at levels comparable to those seen in untreated Hepa1-6 cells (Figure 7E). Moreover, measuring the functional status of tumor-specific CD8+ T cells showed that those cells exposed to PD-L1+ hepatoma cells generated in different ways produced comparable IFN-γ and IL-2 and expressed similar levels of surface degranulation marker CD107a (Figure 7, F and G). Thus, TAM-CM–generated PD-L1+ hepatoma cells displayed the ability to resist the cytotoxicity of T cells, but not to additionally suppress the function of CD8+ T cells.
Suppressing macrophage-elicited NF-κB signal augments immune-checkpoint blockade efficacy.
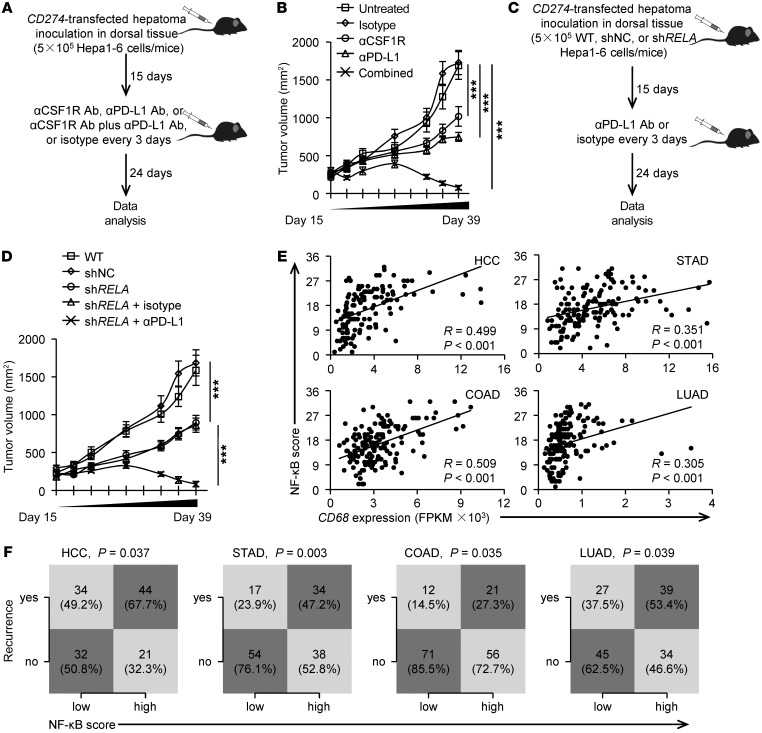
To determine whether macrophage-elicited NF-κB activation suppresses PD-L1–related cancer immunotherapeutic efficacy in vivo, we initially inoculated PD-L1–transduced Hepa1-6 hepatoma into the dorsal tissue of C57BL/6 mice for 15 days. Thereafter, we used a specific Ab to either deplete macrophages or shield the PD-L1 signal in those mice (Figure 8A). Depleting macrophages or injecting Abs to shield the PD-L1 signal in these mice partially affected the progression of hepatoma (Figure 8B). Supporting our findings in vitro that macrophages elicited resistance of hepatoma cells to therapeutic strategies (Figure 7), combined usage of anti–PD-L1 and anti-CSF1R Abs in hepatoma-bearing mice reduced tumor volumes at each measurement time point from day 18 (n = 8 mice per group; *P < 0.05 compared with control Ab), eventually eliminating the hepatoma in vivo (Figure 8B). Analogously, we obtained the same conclusion when using B16-F10 melanoma as a model system (Supplemental Figure 5)
Figure 8. Suppressing macrophage-elicited NF-κB activation augments immunotherapeutic efficacy of a PD-L1 Ab.
(A and B) Mice bearing Hepa1-6 hepatoma in dorsal tissues for 15 days were left untreated or were treated with isotype, αCSF1R Ab, αPD-L1 Ab, or αCSF1R Ab plus αPD-L1 Ab as described (A). Tumor sizes over the indicated time were analyzed (B, n = 8). (C and D) WT (untreated or shNC) or P65 knockdown (shRELA) Hepa1-6 cells were inoculated in dorsal tissues of C57BL/6 mice. Thereafter, mice bearing P65 knockdown (shRELA) Hepa1-6 hepatoma were untreated or treated with isotype or αPD-L1 Ab (C). Tumor sizes over the indicated time were analyzed (D, n = 8). (E) Correlation between CD68 expression and the scoring of the NF-κB pathway were calculated in HCC, STAD, COAD, and LUAD patients from the TCGA data set. (F) Statistical analysis was conducted based on the scoring of the NF-κB pathway and recurrence rate in HCC, STAD, COAD, and LUAD patients from the TCGA data set. P values and R values were calculated based on the analysis of Pearson’s correlation. Data represent mean ± SEM. Results are representative of 3 separate experiments. ***P < 0.001, 1-way ANOVA with Bonferroni’s post test (B and D) or χ2 test (F).
Considering that the NF-κB signal regulated aggressive features of PD-L1+ cancer cells (Figure 6), we subsequently knocked down the P65 gene in Hepa1-6 hepatoma and found that this treatment partially impaired the growth of hepatoma in vivo (Figure 8, C and D). In such a model, injecting anti–PD-L1 Abs led to complete regression of hepatoma (Figure 8D). Of note, we also found that the NF-κB signal signature score positively correlated with both macrophage marker CD68 expression in tumor tissues and recurrence in patients with PD-L1hi HCC, COAD, LUAD, and STAD tumors (Figure 8, E and F). Taken together, our data show that changing macrophage infiltration or suppressing the NF-κB signal in tumors augments the therapeutic efficacy of an anti–PD-L1 Ab.
Discussion
Immune landscapes shape progression of human cancers (21–24). In this work, we have shown that the multiplicity of immune signatures of PD-L1+ tumors controls clinical outcomes and cancer hallmarks as well as therapeutic efficacy in patients.
In the tumor microenvironment, PD-L1 can be upregulated in both tumor and host cells (16–18). To date, the prognostic significance of PD-L1 expression in human tumor progression is still unclear. It is noteworthy that PD-L1 upregulated on either cancer cells or host cells can lead to the exhaustion of tumor-specific T cells (16, 19, 25). In parallel, PD-L1 expression in tumor tissues is assumed to duplicate tumor-reactive T cell infiltration and represent a predictive biomarker of benefit from the PD-L1/PD-1 blockade (6, 7). However, our current study reveals that, although T cells selectively accumulate in PD-L1hi tumors, PD-L1 expression in tumor tissue cannot exclusively represent the IFN-γ signature or T cell infiltration. We demonstrate that PD-L1 expression in tumor tissue had no impact on patients’ recurrence. Interestingly, in patients with PD-L1hi tumors, an increased ratio of macrophages to T cells in tumor tissue predicts early recurrence. It is plausible that it is not PD-L1 per se, but rather the environments that induce PD-L1, that determine tumor recurrence (26). This notion is supported by our observation that infiltrated macrophages can generate PD-L1+ cancer cells exhibiting aggressive cancer hallmarks.
PD-L1 can be regularly induced or maintained by T cell–derived mediators, of which IFN-γ is the most potent (27–29). In this work, we demonstrate that macrophages can operate via IFN-γ–independent pathways to trigger PD-L1 on cancer cells, and this conclusion is supported by the results of 3 sets of experiments. First, TAM-CM individually upregulated cancer cell PD-L1, and this process was abrogated by neutralizing TNF-α or IL-1β, but not by IFN-γ. Second, TAM-CM induced PD-L1 on cancer cells by activation of NF-κB, whereas T cell–CM triggered PD-L1 on cancer cells by phosphorylating STAT1, a downstream signal of IFN-γ. Third, in mice with hepatoma, macrophages and T cells could individually trigger PD-L1+ hepatoma, but macrophages simultaneously accelerated the growth of PD-L1+ hepatoma, while T cells delayed the growth of PD-L1+ hepatoma. Moreover, we previously showed that autocrine antiinflammatory IL-10 was required for PD-L1 induction in tumor macrophages (16). Because the IL-10 receptor is unexpressed by most cancer cells, antiinflammatory IL-10 is not responsible for PD-L1 induction in cancer cells. Consistent with our findings, other investigators have identified proinflammatory TNF-α as a major factor triggering cancer cell immunosuppression against T cell surveillance via stabilization of PD-L1 (30, 31).
Both NF-κB and STAT signals are tightly associated with inflammatory responses in human cancers (32, 33), but the functions and regulation of these signals in human cancers remain unclear. The present study provided evidence that cytokine-mediated activation of either NF-κB or STAT1 can individually regulate the expression of PD-L1 in both human and mouse cancer models. Importantly, NF-κB signal–induced PD-L1+ cancer cells exhibit capabilities of aggressive proliferation, support angiogenesis, and metastasize, whereas STAT1 signal–triggered PD-L1+ cancer cells display a proapoptotic phenotype. In fact, although not directly related to human cancers, activation of NF-κB signaling has been shown to participate in regulating the multiple processes of cell proliferation, survival, migration, and invasion (34–36). Contrary to functions of NF-κB signaling, STAT1 signaling often possesses tumor suppressor functions (37, 38), constitutively active STAT1 can effectively induce apoptosis and inhibit cell growth (39, 40), and STAT1 is frequently downregulated in various human cancers (41, 42). Therefore, a better understanding of the signaling network of PD-L1 regulation in human tumor environments would be helpful for developing rational designs of anticancer therapies that can amplify the antitumorigenic function of immune-checkpoint blockade (43–46).
In 32 types of cancers analyzed, we only detected positive associations between PD-L1 and macrophages or T cell lineage markers in 21, showing that, in the other 11 types of cancers analyzed, major factors contributing to tumor PD-L1 induction were unrelated to the immune landscapes. In fact, besides being induced by immune landscapes, tumor PD-L1 might be regulated by many other biological processes, including chromosomal rearrangements, copy number alterations, oncogenic pathway dysregulation, and epigenetic modulation (47–50). In addition, although our current work focuses on the prognostic value and the immune landscapes of PD-L1+ cancer cells, PD-L1+ host cells, particularly macrophages and dendritic cells, also play very important roles in promoting cancer progression (16–18). Of note, activated APCs, but not suppressive APCs, are more potent in expressing PD-L1 (16, 51). Thus, blocking PD-L1 on activated APCs may not only abrogate coinhibitory functions of the PD-L1/PD-1 axis, but may also restore costimulatory properties of APCs to induce tumor-specific T cell responses. Studying the source, regulation, and function of PD-L1+ cells may help us better understand their roles in tumor pathogenesis.
Our results give important insights into the immune signature, induction, and functional status of PD-L1+ cancer cells in human cancers. Despite recent success in demonstrating the importance of macrophages and the PD-1/PD-L1 axis during tumor progression and therapy (1, 2, 52, 53), little is known about the regulating roles of macrophages in the clinic in PD-1/PD-L1 blockade. In our study, we have demonstrated that macrophages not only generate PD-L1+ cancer cells, but also endow those cells with capabilities to aggressively survive, support angiogenesis, and metastasize. In addition to its biological importance, our work may be relevant in clinical management of cancer patients. Our data raise an important clinical question: is immune-checkpoint blockade suitable for cancer patients with a high amount of macrophage infiltration? Alternatively, we suggest that cancer patients can be treated with immune-checkpoint blockade in combination with strategies targeting the “context” of macrophages and NF-κB signaling. In this study, depleting macrophages does directly impair NF-κB signaling–mediated cancer growth. It should be emphasized that depleting macrophages only abolishes their protumorigenic functions, but modulating the “context” of macrophages might restore their antitumorigenic properties. In support of this conclusion, we have recently observed that depleting B cells in hepatoma-bearing mice repurposed the macrophage polarization away from protumorigenic properties and toward antitumorigenic pathways and subsequently elicited antitumorigenic T cell responses (23, 24, 54). Analogously, NF-κB inhibitors in therapy may directly attenuate the growth of a tumor, but such a treatment also abrogates the activities of antitumorigenic effector T cells or APCs (55–57). Thus, studying the mechanisms that can specifically modulate functional activities of inflammatory stroma cells or cancer cells would be helpful for developing a novel strategy for anticancer therapy (58–60).
Methods
Patients and specimens.
Tissue samples were obtained from 631 patients with pathologically confirmed hepatocellular, pulmonary, colorectal, and gastric carcinomas at the Cancer Center of Sun Yat-sen University (Supplemental Tables 1 and 4). None of the patients had received anticancer therapy before the sampling, and those with concurrent autoimmune disease, HIV, or syphilis were excluded. Tumor tissues and paired blood samples (taken on day of surgery) from 37 HCC patients who underwent surgical resection between December 2015 and December 2018 were used for isolating tissue-infiltrating leukocytes (cohort 2; Supplemental Table 1). Other samples with complete follow-up data were enrolled for analysis of recurrence (Supplemental Tables 1 and 4). Clinical stages were classified according to the guidelines of the Union for International Cancer Control (61).
Immunohistochemistry and immunofluorescence.
Paraffin-embedded human HCC samples were cut in 5 μm sections, which were processed for immunohistochemistry. The sections were subsequently incubated with Abs against human CD3 (UMAB54, ZSBio), CD68 (KP1, Dako), CD79a (EP82, ZSBio), CD15 (MMA+BY87, ZSBio), CD57 (NK-1, ZSBio), or CD34 (10C9, ZSBio), then stained in an Envision System (DakoCytomation).
For immunofluorescence analysis of patient samples, frozen sections were stained with mouse anti-human PD-L1 (MIH1, eBioscience) together with rabbit anti-human CD68 and rat anti-human CD3, followed by Alexa Fluor 488–conjugated anti-mouse IgG plus Alexa Fluor 555–conjugated anti-rat IgG and Alexa Fluor 633–conjugated anti-rabbit IgG; or mouse anti-human PD-L1 followed by Alexa Fluor 488–conjugated anti-mouse IgG (Molecular Probes). Positive cells were detected by confocal microscopy (LSM 510, AxioObserver, Carl Zeiss).
For immunofluorescence analysis of cultured cells, cells growing on a cover slide were fixed, permeabilized, and incubated with mouse anti-human E-cadherin (36/E-Cadherin, BD Biosciences), mouse anti-human Vimentin (RV202, BD Biosciences), or rabbit anti-human P65 (D14E12, Cell Signaling Technology), followed by incubation with Alex Fluor 488–conjugated anti-rabbit IgG. Nuclei were counterstained with DAPI.
Evaluation of immunohistochemical variables.
The procedure for evaluation of immunohistochemical variables was established as previous described (16). Analysis was performed by 2 independent observers who were blinded to the clinical outcome. At a low-power field (×100), the tissue sections were screened, and the 5 most representative fields were selected using a Leica DM IRB inverted research microscope (Leica Microsystems). Thereafter, respective areas were measured at ×400 magnification (0.146 mm2 per field). The numbers of nucleated stromal cells in the tumor regions were then counted manually and expressed as cells per field. Positively stained cells that were smaller than the size of circulating T cells (10 μm) were excluded from counting. The average of counts by 2 investigators was applied in the following analysis to minimize interobserver variability.
Isolation of tumor leukocytes and preparation of CM from tumor leukocytes.
Fresh HCC biopsy specimens were cut into small pieces and digested in RPMI 1640 supplemented with 0.05% collagenase IV (Sigma-Aldrich), 0.002% DNase I (Roche), and 20% FBS at 37°C for 30 minutes. Dissociated cells were filtered through a 150 μm mesh and separated by density gradient centrifugation. Thereafter, the leukocytes were harvested and the tumor-infiltrating neutrophils, monocytes, T cells, NK cells, and B cells were sorted by FACS according to CD15, CD14, CD3, CD56, and CD19 expression and based on forward-scatter and side-scatter profiles (Supplemental Figure 2B). The Abs used are listed in Supplemental Table 6.
For preparation of CM from tumor leukocytes, 106 sorted CD45+ leukocytes were resuspended in 1 mL of CM and cultured. After 24 hours, the supernatants were harvested, centrifuged, and stored at −80°C. All specimens were from individuals without concurrent autoimmune disease, HBV, HCV, HIV, or syphilis. The digested tumor or liver cells were washed in medium containing polymyxin B (20 μg/mL; Sigma-Aldrich) to exclude endotoxin contamination.
Flow cytometry (FACS).
Huh7, HepG2, or Hep3B cells or mouse T cells from ex vivo culture were stained with fluorochrome-conjugated Abs according to the manufacturers’ instructions. For apoptosis assay, an annexin V apoptosis detection kit was used according to the manufacturer’s instructions (Biovision, 1001). Data were measured by flow cytometer (Gallios, Beckman Coulter) and analyzed using FlowJo software, version 10.0. The Abs used are listed in Supplemental Table 6.
ELISA.
Concentrations of IFN-γ (eBioscience, 88-7314) and IL-2 (eBioscience, 88-7024) in the supernatants from mouse T cell and hepatoma cell coculture systems were detected using ELISA kits according to the manufacturers’ instructions.
Immunoblotting.
Proteins were extracted as previously described (20). The Abs used are listed in Supplemental Table 6.
Real-time PCR.
TRIzol reagent (Invitrogen) was used to isolate total RNA of cancer cells from an ex vivo culture system. Aliquots (2 μg) of the RNA were reverse transcribed using MMLV Reverse Transcriptase (Promega). The specific primers used to amplify the genes are listed in Supplemental Table 7. PCR was performed in triplicate using SYBR Green Real-Time PCR MasterMix (TOYOBO) in a Roche LightCycler 480 System. All results are presented in arbitrary units relative to 18S rRNA expression.
Tumor cell lines.
Human hepatoma Huh7, Hep3B, and HepG2 cell lines were obtained in August 2015 from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The mouse hepatoma cell lines Hepa1-6 and B16-F10 were obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences in January 2018 and February 2019, respectively. Experiments were carried out within 6 months after purchase. All cells were tested for mycoplasma contamination using the single-step PCR method and maintained in CM composed of DMEM supplemented with 10% FBS.
Regulation of PD-L1 expression in cancer cells.
Huh7, HepG2, or Hep3B cells were untreated or stimulated with CM of tumor-infiltrating leukocytes or T cells, macrophages, B cells, neutrophils, or NK cells or with recombinant TNF-α (20 ng/mL) plus IL-1β (10 ng/mL) or IFN-γ (50 ng/mL) (R&D Systems) for indicated times. In some experiments, before exposure to TAM-CM or T cell–CM, cells were pretreated with neutralizing mAbs against TNF-α (10 μg/mL), IL-1β (10 μg/mL), IL-6 (40 μg/mL), IL-12 (10 μg/mL), TGF-β (10 μg/mL), or IFN-γ (5 μg/mL) (all from R&D Systems). Other cells were pretreated with a specific inhibitor for STAT1 (Fludarabine, 10 μg/mL, Enzo), STAT3 (AG490, 100 μM), JNK (SP 600125, 100 μM), ERK (U0126, 25 μM), P38 (SB 203580, 50 μM), AKT (AIP2, 100 μM), or NF-κB (BAY 11-7082, 20 μM) signal (Sigma-Aldrich). Thereafter, cells were subject to FACS analysis to detect surface PD-L1 protein. The Abs used are listed in Supplemental Table 6.
Construction of viral vectors and PD-L1 stable cell lines.
The candidate sequences for mouse shRELA (5′-GCGAATCCAGACCAACAATAA-3′) and shIFNGR1 (5′-GCCTGTACCGACGAATGTTCT-3′) and a scrambled sequence for shNC (5′-GACCAAACTCGACAATCAGAA-3′) were cloned into pSIF-H1-CopGFP-shRNA lentiviral vectors (System Biosciences). Thereafter, the lentiviral vectors were transfected into HEK293T cells together with their helper virus vectors pFIV-34N and pVSV-G (System Biosciences) using calcium phosphate. After 2 days, the viral particles were harvested and enriched through ultracentrifugation.
The CD274 gene was amplified by PCR from human and mouse cDNA and confirmed by DNA sequencing. The gene fragment was inserted into retrovirus vector pBABE-puro and cotransfected into the package cell HEK293T with helper virus vector pBABE-ampho in the context of Lipofectamine (Invitrogen). The supernatant of these HEK293T cells was used to infect original human HepG2 cells, mouse Hepa1-6 cells, and B16-F10 cells. The cell lines stably expressing PD-L1 and the mock transfectant were selected with puromycin (1 μg/mL, Sigma-Aldrich).
RNA interference.
Human hepatoma HepG2 cells were left untreated or transfected with 50 nM of a negative control siRNA (sense: 5′-UUGUACUACACAAAAGUACUG-3′; antisense: 5′-GUACUUUUGUGUAGUACAGUU-3′) or a P65-specific siRNA (sense: 5′-UGGAGUACCCUGAGGCUAUdTdT-3′; antisense: 5′-AUAGCCUCAGGGUACUCCAUC-3′) using Lipofectamine-RNAi MAX (Invitrogen, 13778150). After 36 hours of transfection, cells were exposed to CM of macrophages isolated from HCC tumors. All siRNA duplexes were purchased from GenePharma.
Cell migration assay.
The migration assay was performed in a 24-well Boyden chamber with an 8 μm polycarbonate membrane (Corning, 3422). HepG2 cells were left untreated, transduced with pBABE-Puro retroviral vector encoding human PD-L1, or incubated with TAM-CM or T cell–CM for 24 hours. In some cases, cells were pretreated with a specific inhibitor for NF-κB (BAY 11-7082, 20 μM) or transfected with P65-specific siRNA before exposure to TAM-CM or T cell–CM. Thereafter, cells (3 × 104) in 100 μl of serum-free DMEM were added to the upper compartment of the chamber, and the lower compartment was filled with 600 μl of DMEM containing 10% FBS (Gibco, Thermo Fisher Scientific). After 10 hours of incubation, the cells remaining on the upper surface of the membrane were removed. The migrated tumor cells on the lower surface of the membrane were rinsed with PBS and subsequently stained with crystal violet after fixation and then counted under a light microscope (Nikon, Ni-U).
Animal experiments.
WT female C57BL/6 mice (6 to 8 weeks old) were purchased from the Guangdong Medical Laboratory Animal Center (Guangzhou, China). NOD SCID mice were purchased from the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). All mice were maintained under specific pathogen–free conditions in the animal facilities of the Cancer Center of Sun Yat-sen University. All mice were randomly grouped. Mouse Hepa1-6 hepatoma was established as shown in Figure 8, A and C, Supplemental Figure 2, E, F, and H, and Supplemental Figure 5A. For the xenograft tumor model, primary tumor tissues from 6 HCC patients were implanted subcutaneously into the flanks of NOD SCID mice with Matrigel (R&D Systems). Three weeks after implantation, tumors were collected and subjected to immunofluorescence analysis to detect PD-L1 expression. The Abs used are listed in Supplemental Table 6.
Statistics.
Results are expressed as mean ± SEM. All statistical tests were performed as 2 sided. For data normally distributed, we applied Student’s t test, and the nonparametric exact Wilcoxon’s signed-rank test was used to compare data not normally distributed. For multiple comparisons (including multiple 2-group comparisons shown in the same panel), 1-way or 2-way ANOVA (for parametric data) followed by Bonferroni’s correction (when only 2 groups were compared), Dunnett’s test (when all groups were compared with 1 control group), Tukey’s multiple comparison test (when all groups were compared with each other), or Kruskal-Wallis test (for nonparametric data) followed by Dunn’s multiple comparison test was applied. Cumulative survival time was estimated by the Kaplan-Meier method, and the log-rank test was applied to compare the groups. P < 0.05 was considered statistically significant. No animal data were excluded.
Study approval.
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committee of Sun Yat-sen University. For experiments using human samples, all samples were anonymously coded in accordance with local ethical guidelines (as stipulated by the Declaration of Helsinki). Written informed consent was obtained from patients, and the protocol was approved by the Ethical Review Board of Sun Yat-sen University.
Author contributions
YW, QZ, XML, and DMK designed, performed, and analyzed experiments. WML, DPC, MM, CXH, and ZYL performed experiments and analyzed the data. BL, LZ, and ZG provided mice, clinical resources, and technical support. DMK supported and supervised the research. YW and DMK wrote the manuscript, and all authors contributed to editing the manuscript.
Supplementary Material
Acknowledgments
The study was supported by project grants from the National Key Research and Development Plan of China (2016YFA0502600 and 2018ZX10302205); the National Natural Science Foundation of China (31830025, 91642102, and 81422036); the Natural Science Foundation of Guangdong Province, China (2018B030308010); and the China Postdoctoral Science Foundation (2019M653191 and 2019M653169).
Version 1. 05/21/2019
In-Press Preview
Version 2. 07/15/2019
Electronic publication
Version 3. 08/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(8):3347–3360.https://doi.org/10.1172/JCI127726.
Contributor Information
Yuan Wei, Email: 278641920@qq.com.
Qiyi Zhao, Email: qiyizh@163.com.
Zhiliang Gao, Email: 40903562@qq.com.
Xiang-Ming Lao, Email: laoxming@mail.sysu.edu.cn.
Wei-Ming Lin, Email: 7911439@qq.com.
Dong-Ping Chen, Email: chdongp@mail2.sysu.edu.cn.
Ming Mu, Email: 935435805@qq.com.
Chun-Xiang Huang, Email: h_phileo@163.com.
Zheng-Yu Liu, Email: lzhyyoooo@sina.com.
Bo Li, Email: libo47@mail.sysu.edu.cn.
Limin Zheng, Email: 765025987@qq.com.
Dong-Ming Kuang, Email: kdming@mail.sysu.edu.cn.
References
- 1.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 2.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keir ME, Francisco LM, Sharpe AH. PD-1 and its ligands in T-cell immunity. Curr Opin Immunol. 2007;19(3):309–314. doi: 10.1016/j.coi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Xiao X, et al. PD-1hi identifies a novel regulatory B-cell population in human hepatoma that promotes disease progression. Cancer Discov. 2016;6(5):546–559. doi: 10.1158/2159-8290.CD-15-1408. [DOI] [PubMed] [Google Scholar]
- 5.Devulder J, Tsicopoulos A, Duez C. Neuroendocrine control of natural killer cell function through the checkpoint molecule PD-1. Cell Mol Immunol. 2019;16(2):109–111. doi: 10.1038/s41423-018-0180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilke CM, Wei S, Wang L, Kryczek I, Kao J, Zou W. Dual biological effects of the cytokines interleukin-10 and interferon-γ. Cancer Immunol Immunother. 2011;60(11):1529–1541. doi: 10.1007/s00262-011-1104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao Y, et al. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143(4):931–943. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 8.Topalian SL, et al. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst RS, et al. Predictive correlates of response to the anti–PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garon EB, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 11.Reck M, et al. Pembrolizumab versus chemotherapy for PD-L1–positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 12.Oliva M, et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann Oncol. 2019;30(1):57–67. doi: 10.1093/annonc/mdy507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robert C, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 14.Ribas A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA. 2016;315(15):1600–1609. doi: 10.1001/jama.2016.4059. [DOI] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang DM, et al. Activated monocytes in peritumoral stroma of hepatocellular carcinoma foster immune privilege and disease progression through PD-L1. J Exp Med. 2009;206(6):1327–1337. doi: 10.1084/jem.20082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin H, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128(2):805–815. doi: 10.1172/JCI96113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H, et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest. 2018;128(2):580–588. doi: 10.1172/JCI96061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, et al. Distinct contribution of PD-L1 suppression by spatial expression of PD-L1 on tumor and non-tumor cells. Cell Mol Immunol. 2019;16(4):392–400. doi: 10.1038/s41423-018-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuang DM, et al. B7-H1-expressing antigen-presenting cells mediate polarization of protumorigenic Th22 subsets. J Clin Invest. 2014;124(10):4657–4667. doi: 10.1172/JCI74381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XF, et al. Increased autophagy sustains the survival and pro-tumourigenic effects of neutrophils in human hepatocellular carcinoma. J Hepatol. 2015;62(1):131–139. doi: 10.1016/j.jhep.2014.08.023. [DOI] [PubMed] [Google Scholar]
- 23.Liu RX, et al. Chemokine (C-X-C motif) receptor 3-positive B cells link interleukin-17 inflammation to protumorigenic macrophage polarization in human hepatocellular carcinoma. Hepatology. 2015;62(6):1779–1790. doi: 10.1002/hep.28020. [DOI] [PubMed] [Google Scholar]
- 24.Chen MM, et al. Polarization of tissue-resident TFH-like cells in human hepatoma bridges innate monocyte inflammation and M2b macrophage polarization. Cancer Discov. 2016;6(10):1182–1195. doi: 10.1158/2159-8290.CD-16-0329. [DOI] [PubMed] [Google Scholar]
- 25.Juneja VR, et al. PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J Exp Med. 2017;214(4):895–904. doi: 10.1084/jem.20160801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, Qiao J, Fu YX. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370(1):85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loke P, Allison JP. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc Natl Acad Sci U S A. 2003;100(9):5336–5341. doi: 10.1073/pnas.0931259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SJ, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580(3):755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 29.Taube JM, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim SO, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30(6):925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asgarova A, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology. 2018;7(5):e1423170. doi: 10.1080/2162402X.2017.1423170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooks T, et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013;23(5):634–646. doi: 10.1016/j.ccr.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Shea JJ, Holland SM, Staudt LM. JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med. 2013;368(2):161–170. doi: 10.1056/NEJMra1202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruland J. Return to homeostasis: downregulation of NF-κB responses. Nat Immunol. 2011;12(8):709–714. doi: 10.1038/ni.2055. [DOI] [PubMed] [Google Scholar]
- 35.Bhat KPL, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Aravindan N, Xu J, Natarajan M. Inter- and intra-cellular mechanism of NF-kB-dependent survival advantage and clonal expansion of radio-resistant cancer cells. Cell Signal. 2017;31:105–111. doi: 10.1016/j.cellsig.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Koromilas AE, Sexl V. The tumor suppressor function of STAT1 in breast cancer. JAKSTAT. 2013;2(2):e23353. doi: 10.4161/jkst.23353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24(130):19–29. [PubMed] [Google Scholar]
- 39.Kim HS, Lee MS. STAT1 as a key modulator of cell death. Cell Signal. 2007;19(3):454–465. doi: 10.1016/j.cellsig.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Sironi JJ, Ouchi T. STAT1-induced apoptosis is mediated by caspases 2, 3, and 7. J Biol Chem. 2004;279(6):4066–4074. doi: 10.1074/jbc.M307774200. [DOI] [PubMed] [Google Scholar]
- 41.Chan SR, et al. STAT1-deficient mice spontaneously develop estrogen receptor α-positive luminal mammary carcinomas. Breast Cancer Res. 2012;14(1):R16. doi: 10.1186/bcr3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol Immunother. 2011;60(4):525–535. doi: 10.1007/s00262-010-0961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang H, et al. Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell. 2016;29(3):285–296. doi: 10.1016/j.ccell.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maj T, et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1–blockade resistance in tumor. Nat Immunol. 2017;18(12):1332–1341. doi: 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu G, et al. Low-dose decitabine enhances the effect of PD-1 blockade in colorectal cancer with microsatellite stability by re-modulating the tumor microenvironment. Cell Mol Immunol. 2019;16(4):401–409. doi: 10.1038/s41423-018-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Song W, Shao C, Shi Y, Han W. Emerging predictors of the response to the blockade of immune checkpoints in cancer therapy. Cell Mol Immunol. 2019;16(1):28–39. doi: 10.1038/s41423-018-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George J, et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res. 2017;23(5):1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaver TM, et al. Diverse, biologically relevant, and targetable gene rearrangements in triple-negative breast cancer and other malignancies. Cancer Res. 2016;76(16):4850–4860. doi: 10.1158/0008-5472.CAN-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapuy B, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869–881. doi: 10.1182/blood-2015-10-673236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng S, Hu Q, Zhang H, Yang F, Peng C, Huang C. HDAC3 inhibition upregulates PD-L1 expression in B-cell lymphomas and augments the efficacy of anti–PD-L1 therapy. Mol Cancer Ther. 2019;18(5):900–908. doi: 10.1158/1535-7163.MCT-18-1068. [DOI] [PubMed] [Google Scholar]
- 51.Curiel TJ, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9(5):562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- 52.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim YB, Ahn YH, Jung JH, Lee YJ, Lee JH, Kang JL. Programming of macrophages by UV-irradiated apoptotic cancer cells inhibits cancer progression and lung metastasis. Cell Mol Immunol. 2019:s41423-019-0209-1 : doi: 10.1038/s41423-019-0209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei Y, et al. Plasma cell polarization to the immunoglobulin G phenotype in hepatocellular carcinomas involves epigenetic alterations and promotes hepatoma progression in mice. Gastroenterology. 2019;156(6):1890–1904.e16. doi: 10.1053/j.gastro.2019.01.250. [DOI] [PubMed] [Google Scholar]
- 55.Buggins AG, et al. Microenvironment produced by acute myeloid leukemia cells prevents T cell activation and proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways. J Immunol. 2001;167(10):6021–6030. doi: 10.4049/jimmunol.167.10.6021. [DOI] [PubMed] [Google Scholar]
- 56.Hou Y, et al. Non-canonical NF-κB antagonizes STING sensor-mediated DNA sensing in radiotherapy. Immunity. 2018;49(3):490–503.e4. doi: 10.1016/j.immuni.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knudson KM, Pritzl CJ, Saxena V, Altman A, Daniels MA, Teixeiro E. NFκB-Pim-1-Eomesodermin axis is critical for maintaining CD8 T-cell memory quality. Proc Natl Acad Sci U S A. 2017;114(9):E1659–E1667. doi: 10.1073/pnas.1608448114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, et al. Effector T cells abrogate stroma-mediated chemoresistance in ovarian cancer. Cell. 2016;165(5):1092–1105. doi: 10.1016/j.cell.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, et al. Targeting the tumor microenvironment with interferon-β bridges innate and adaptive immune responses. Cancer Cell. 2014;25(1):37–48. doi: 10.1016/j.ccr.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sobin LH, Gospodarowicz, MK, Wittekind C. TNM Classification of Malignant Tumors, International Union Against Cancer. 7th ed. Oxford, UK: Wiley-Blackwell; 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.