Abstract
Cystic fibrosis (CF) is a multiorgan progressive genetic disease caused by loss of functional cystic fibrosis transmembrane conductance regulator (CFTR) channel. Previously, we identified a significant dysfunction in CF cells and model mice of the transcription factor nuclear factor E2–related factor-2 (Nrf2), a major regulator of redox balance and inflammatory signaling. Here we report that the approved F508del CFTR correctors VX809 and VX661 recover diminished Nrf2 function and colocalization with CFTR in CF human primary bronchial epithelia by proximity ligation assay, immunoprecipitation, and immunofluorescence, concordant with CFTR correction. F508del CFTR correctors induced Nrf2 nuclear translocation, Nrf2-dependent luciferase activity, and transcriptional activation of target genes. Rescue of Nrf2 function by VX809/VX661 was dependent on significant correction of F508del and was blocked by inhibition of corrected channel function, or high-level shRNA knockdown of CFTR or F508del CFTR. Mechanistically, F508del CFTR modulation restored Nrf2 phosphorylation and its interaction with the coactivator CREB-binding protein (CBP). Our findings demonstrate that sufficient modulation of F508del CFTR function corrects Nrf2 dysfunction in CF.
Keywords: Pulmonology
Keywords: Chloride channels, Genetic diseases
Introduction
Cystic fibrosis (CF) is an autosomal recessive disease characterized by progressive loss of lung function, pancreatic exocrine dysfunction, and gastrointestinal complications, which culminate in dramatically shortened life expectancy. CF patients have a loss of functional cystic fibrosis transmembrane conductance regulator (CFTR) channel, an important Cl–/HCO3– channel in epithelial cells (1). The most common mutation in CFTR is the F508del variant, which is a class II mutation, resulting in defective protein folding (2). F508del CFTR is retained in the endoplasmic reticulum (ER) and not transported to the cell membrane. Furthermore, F508del exhibits reduced gating and stability at the plasma membrane. CF airway epithelia exhibit many secondary defects, including abnormalities in pathways involved in inflammatory responses (3–5). However, no direct link between CFTR and regulators of inflammation, such as nuclear factor E2–related factor-2 (Nrf2) (4), has yet been reported.
Transforming CF care, several small-molecule compounds that target CFTR dysfunctions have been developed (6). VX809 (lumacaftor) is a “corrector” that is reported to bind to and partially correct the folding of F508del CFTR, increasing its delivery to the cell membrane (7). Alone, VX809 significantly improves sweat chloride levels (a measure of CFTR function in sweat glands), but does not elicit significant changes in nasal epithelial CFTR function or in lung function (8). In CF patients homozygous for the F508del mutation, combination therapy with VX809 and the CFTR potentiator VX770 (ivacaftor) significantly improves lung function, and reduces the number of events leading to hospitalization by 39%–61% (9). VX661 (tezacaftor, a corrector with efficacy similar to that of lumacaftor), when combined with VX770, reduced pulmonary exacerbations by 35% with an improved safety profile in a phase III clinical trial (10).
Secondary defects in CF are important contributors to the pathophysiology of the disease. Previously, we discovered the dysfunction of Nrf2 in CF airway epithelia (11, 12). Nrf2 is a transcription factor that regulates redox balance, and its dysfunction is associated with numerous disease states, including acute lung injury (13), emphysema (14), chronic obstructive pulmonary disease (COPD) (15, 16), pulmonary fibrosis (17), and asthma (18). The primary regulator of Nrf2 is Kelch-like erythroid cell–derived protein with cap’n’collar homology-associated protein 1 (Keap1), which sequesters Nrf2 in the cytoplasm (19, 20). Keap1 is a substrate adaptor of a cullin-3 E3 ligase complex that polyubiquitinates Nrf2 and thereby targets it to the 26S proteasome for degradation. As a consequence, Nrf2 levels and half-life are suppressed under basal conditions (21, 22). Reactive oxygen species (and xenobiotics) oxidize Keap1 cysteine residues and induce a conformational change that dissociates Keap1 and Nrf2 from the E3 ligase complex, resulting in Nrf2 stabilization (23–25). Stabilized Nrf2 then translocates into the nucleus and binds to antioxidant response elements (AREs) present in the promoters of its cognate target genes, stimulating the transcription of numerous antioxidant and detoxifying genes (26, 27).
Previously, we discovered that CF epithelia have reduced nuclear Nrf2 protein expression and activity, and corresponding increases in steady-state oxidants and enhanced inflammatory signaling (11). We found that inhibition of CFTR (with CFTRinh-172) decreases Nrf2 activity, and increases H2O2 levels in non-CF cells (11). Conversely, Nrf2 knockdown significantly decreases CFTR expression (28). We showed that the cAMP competitor Rp-cAMPS, which decreases phosphorylated CREB (p-CREB), stimulates Nrf2 activity (29). Mechanistically, we demonstrated that in CF cells increased p-CREB binding to CREB-binding protein (CBP) decreased its interaction with Nrf2 and diminished Nrf2 activation (29). Concomitantly, we found that an increased p-CREB–CBP in CF cells promoted the CBP-mediated activation of NF-κB, implicating Nrf2 dysfunction in inflammatory signaling in CF. In vivo, we found that activation of Nrf2 with the triterpenoid CDDO in CF mice significantly reduced inflammatory responses to Pseudomonas aeruginosa–derived LPS or flagellum (30).
To further investigate the relationship between CFTR and Nrf2, we evaluated whether VX809 or VX661 impacts Nrf2 dysregulation. We discovered that (a) VX809 and VX661 activate Nrf2; (b) Nrf2 and CFTR colocalize to within 40 nm and coimmunoprecipitate; (c) Nrf2-CFTR interaction is decreased in CF; (d) VX809/VX661 correction of Nrf2 activity and colocalization with CFTR are dependent on CFTR function at the cell membrane; and (e) mechanistically, CFTR correctors rescue Nrf2 interaction with its coactivator CBP. In addition to serving as tools to study F508del correction and Nrf2 biology, our studies of both VX809 and VX661 are clinically relevant as both drugs are prescribed regularly to CF patients with 2 copies of the F508del CFTR variant. Nrf2 activity and colocalization with CFTR were further confirmed in established murine models of CF. These findings represent the first evidence of a direct interface between CFTR and Nrf2. Given the association of Nrf2 with the resolution of inflammation (11, 12, 30–39), our studies suggest that sufficient correction of CFTR function has the potential to modulate inflammatory signaling in CF.
Results
VX809 increases Nrf2 nuclear translocation and activity.
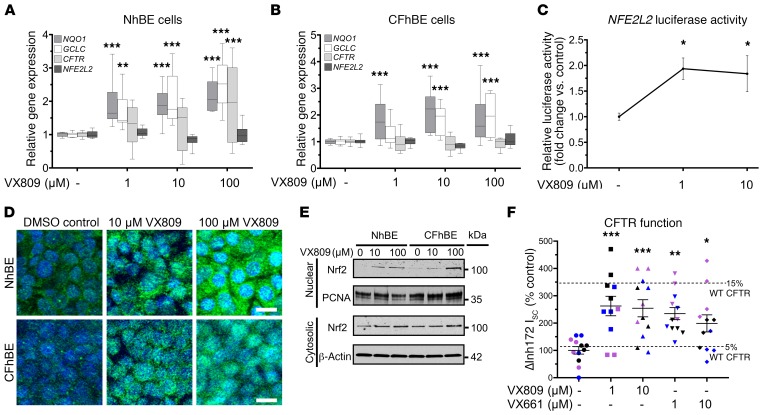
To determine the effects of CFTR modulation on Nrf2, primary non-CF and CF human bronchial epithelial (NhBE and CFhBE) cells were incubated with a range of doses of VX809 (1–100 μM) for 48 hours. The CFhBE cells used in this and subsequent experiments were homozygous for the F508del CFTR variant, the most common genotype in CF (2). VX809 induced the expression of Nrf2-regulated genes, including NQO1 [NAD(P)H quinone dehydrogenase 1] and GCLC (glutamate-cysteine ligase catalytic subunit) (Figure 1, A and B). Furthermore, VX809 increased Nrf2-dependent luciferase activity in CFhBE cells, indicating that the compound promotes Nrf2 activation (Figure 1C). We next investigated whether VX809 also regulates CFTR gene expression and found that after incubation with 1–100 μM VX809 for 48 hours, CFTR gene expression significantly increased in primary NhBE cells, but did not change in CFhBE cells (Figure 1, A and B). Consistent with increased Nrf2 function, we found that VX809 treatment enhanced the nuclear localization of Nrf2 by immunofluorescence (IF) (Figure 1D).
Figure 1. CFTR modulation dose-dependently increases Nrf2 nuclear localization and activity.
(A and B) Gene expression of CFTR, NFE2L2, and Nrf2-regulated genes GCLC and NQO1 in primary NhBE (A) and CFhBE (B) cells, after incubation with DMSO control or the indicated doses of VX809 for 48 hours, determined by real-time quantitative PCR (qPCR). Data presented as fold changes versus untreated cells; calculated from cycle threshold measurements and normalized to 18S rRNA for n = 3 independent experiments and 3 donors per cell type. Data are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. (C) VX809 induces Nrf2-mediated luciferase expression in CFhBE cells. Cells were transiently transfected with a plasmid containing the Nrf2-binding promoter ARE driving firefly and a transfection control Renilla luciferase plasmid, then treated with VX809 (1–10 μM) for 48 hours. Expression in cell lysates was measured by luminometer. Relative activity is firefly/Renilla luciferase activity normalized to total protein, and is expressed as fold change versus DMSO control. Mean ± SEM for n = 7 independent experiments. (D) Representative micrographs of basolateral view of immunofluorescence for Nrf2 (green) or DAPI nuclear staining (blue). Cells were treated with VX809 (10–100 μM) for 48 hours. Scale bars: 10 μm. (E) Western blotting for Nrf2 protein in nuclear and cytosolic fractions of NhBE and CFhBE cells after VX809 treatment for 48 hours; β-actin and proliferating cell nuclear antigen (PCNA) are loading controls. (F) Aggregate short-circuit current data for cells pretreated with VX809 or VX661 (symbols color-coded by donor). Mean ± SEM for 3–4 replicates per condition per donor, from at least 3 unique patient donors, normalized as percentage of donor DMSO control. Dashed lines represent percentage of NhBE control average. For A–C, *P < 0.05, **P < 0.01, ***P < 0.001 vs. DMSO control cells by 1-way ANOVA and Dunnett’s multiple-comparisons test.
VX809 treatment also increased Nrf2 cytosolic and nuclear protein levels (Figure 1, D and E), but not Nrf2 gene (NFE2L2) expression (Figure 1, A and B), in both CF and non-CF cells. In agreement with this, VX809 dose-dependently increased Nrf2 protein levels in the cytosolic and nuclear fractions of CFhBE cells up to 2-fold and almost 30-fold, respectively, by Western blot analysis (Figure 1E). Nrf2 fluorescence in the nucleus significantly increased after incubation with VX809 for 48 hours in both NhBE and CFhBE cells (Figure 1D), which agrees with the increase in nuclear Nrf2 observed by Western blotting (Figure 1E). An MTT cell viability assay confirmed that VX809 (10–100 μM) is not cytotoxic (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI96273DS1). Both the IF and Western blot data are consistent with the stimulation of Nrf2 activity following VX809 treatment. These data demonstrate that VX809, especially at higher doses, is able to correct Nrf2 dysfunction in CF by inducing translocation of Nrf2 to the nucleus and increasing Nrf2 transcriptional activity. While 100 μM VX809 induced more nuclear Nrf2 accumulation than lower doses (Figure 1, D and E), we used the lowest effective doses of 1–10 μM (Figure 1, A–C) in the remaining studies for physiological and clinical relevance.
Prolonged incubation with corrective modulators improves F508del CFTR function.
Ussing chamber studies were performed to confirm that incubation with CFTR corrector compounds improves stimulated F508del CFTR function in primary CFhBE cells. In the absence of corrector compounds, cells from five F508del CFTR homozygous donors exhibited stimulated (ΔcAMP+VX770) CFTR currents at 6.2% of those from four NhBE donors (Supplemental Figure 2, A and B). Preincubation with 1–10 μM VX809 for 48 hours increased F508del CFTR function 3- to 4-fold, and a similar trend of improvement was observed with VX661 preincubation (Supplemental Figure 2, A and B). In cells pretreated with 1 μM VX809, stimulated F508del CFTR function reached 17.3% of that of NhBE donors, similar to findings of previously published studies (40). In cells also pretreated with VX770, alone or in combination with corrector, very little stimulated F508del CFTR current was observed because of baseline CFTR activation in the presence of VX770. For these groups, however, a statistically significant increase in inhibited CFTR function (ΔInh172) was noted in cells pretreated with VX770 combined with 1–10 μM VX809, with a trend toward an increase in current in cells pretreated with VX770 and 1 μM VX661 (Supplemental Figure 2, C and D). Across all studies of stimulated CFTR function, no corrector-induced difference was consistently noted in other electrophysiological measures, including baseline resistance, current, amiloride-inhibited current, low-chloride-induced current, or ATP-stimulated currents (Supplemental Figure 3).
To mirror the nonstimulated conditions in our colocalization studies, we also examined the corrector-induced change in Inh172-inhibited, unstimulated CFTR current (i.e., in the absence of cAMP or VX770). Unstimulated F508del CFTR function in 3 CFhBE donors was 4.1% of that in NhBE donors in the absence of corrector compounds (Figure 1F and Supplemental Figure 4). Like in our stimulated studies, this was rescued to more than 10% of NhBE function following 48-hour preincubation with 1–10 μM VX809 or VX661 (Figure 1F and Supplemental Figure 4). These results demonstrate that both corrector drugs alone increase baseline F508del CFTR function without stimulation by cAMP activators or VX770. As with studies of stimulated CFTR function, there was no consistent corrector-induced differences in baseline resistance or baseline, amiloride-sensitive, low-chloride-induced, DIDS-sensitive, or ATP-stimulated currents (Supplemental Figure 5).
Prolonged incubation with VX809 or VX661 increases mature F508del CFTR.
Primary CFhBE vehicle control cells had a weak or undetectable mature band C, compared with a readily detectable band C in non-CF NhBE cells (Supplemental Figure 6, A and B). However, treatment for 48 hours with 1–10 μM VX809 or VX661 produced a detectable band C for F508del CFTR, indicating an increase in glycosylated mature protein with corrector drugs (Supplemental Figure 6, A and B). These data are consistent with correction of F508del CFTR trafficking, increased CFTR function at the plasma membrane (by Ussing chamber assay; Figure 1F), and increased Nrf2 activity following treatment with CFTR correctors (Figure 1, A–C).
Colocalization of Nrf2 and F508del CFTR is decreased in primary human CF airway epithelia.
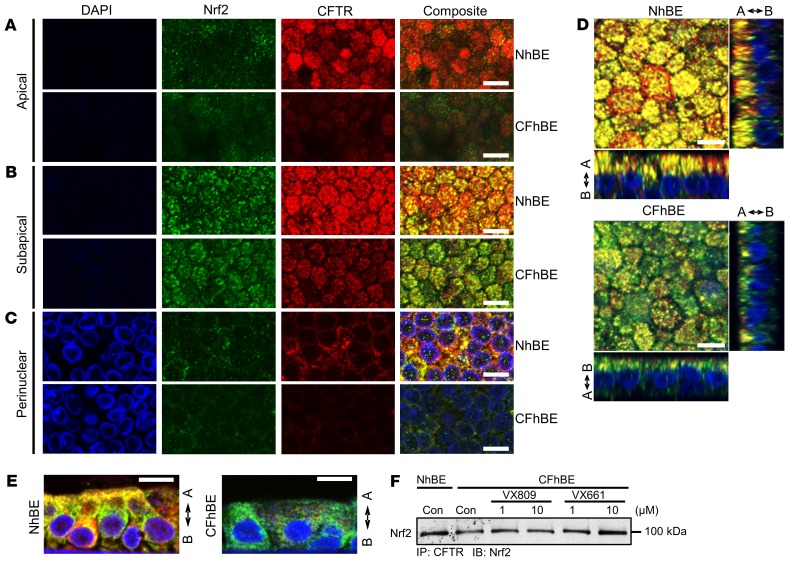
To explore the mechanism by which VX809 activates Nrf2, we examined the localization of Nrf2 with respect to F508del CFTR, since VX809 and VX661 change F508del CFTR localization. Dual IF with antibodies against Nrf2 (green) and CFTR (red) reveals that the 2 proteins colocalized (yellow) in primary NhBE cells (Figure 2, A–C). To focus on membrane and cytoplasmic staining, cells were minimally permeabilized. As expected, IF for F508del CFTR protein was significantly lower in primary CFhBE cells compared with NhBE cells, and was mainly limited to the perinuclear to subapical planes (Figure 2, A–C, and Supplemental Figure 7). Importantly, the colocalization of Nrf2 with CFTR was significantly diminished in CF cells compared with non-CF cells (Figure 2, A–E, and Supplemental Figure 7). Colocalization (yellow) in both the primary NhBE and CFhBE cells appeared highest in the perinuclear to subapical plane, although colocalization was observed from the basolateral plane to below the apical plane in the NhBE cells (Figure 2, A–E, and Supplemental Figure 8A).
Figure 2. Nrf2 and CFTR colocalize in NhBE cells, and this colocalization is diminished in CFhBE cells.
(A–C) Representative photographs of IF for Nrf2 (green), CFTR (red), or DAPI (blue) in the apical (A), subapical (B), and perinuclear (C) views of NhBE and CFhBE cells, at ×60 original magnification. Merged composite images are shown in the far right column, with colocalization of Nrf2 and CFTR in yellow. To avoid nuclear permeabilization, mild detergent was used. (D) Slices view of IF in NhBE and CFhBE cells stained as above. Arrows indicate apical (A) and basolateral (B) sides. Each experiment was performed at least 10 times. (E) Representative IF images of NhBE and CFhBE cells, XY side sectioned after paraffin embedding, with staining as above. (F) CFTR association with Nrf2 was also analyzed by immunoprecipitation. NhBE and CFhBE cells were treated with the indicated doses of DMSO control, VX809, or VX661 for 48 hours. Whole cell lysates were incubated with anti-CFTR antibody, immunoprecipitated, subjected to SDS-PAGE, transferred to PVDF membrane, and probed with anti-Nrf2 antibody. Data are representative of 3–4 CF and 3 non-CF donors, and 3–4 replicates per treatment per donor. Scale bars: 10 μm.
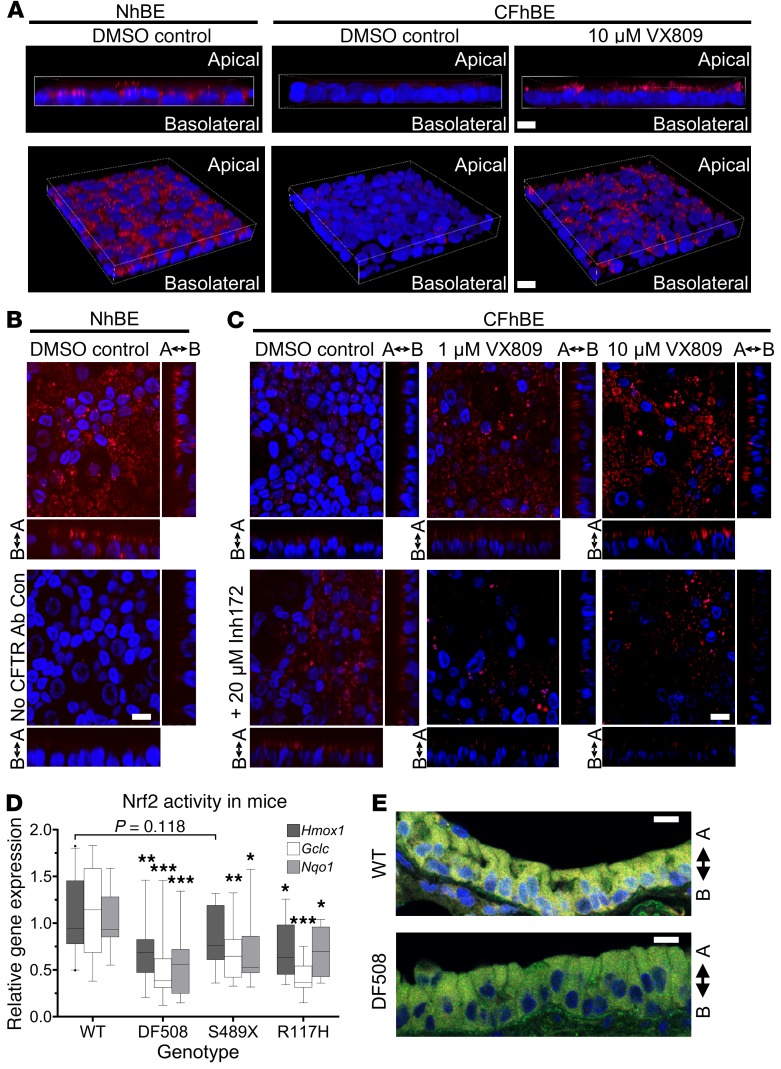
To directly test whether CFTR and Nrf2 interact, we conducted immunoprecipitations from whole cell lysates with an antibody against CFTR, followed by immunoblotting with an anti-Nrf2 antibody. Consistent with our IF studies, Nrf2 was pulled down with CFTR, and the association between CFTR and Nrf2 in CFhBE cells was decreased compared with that in NhBE cells, while 48 hours of treatment with 1 or 10 μM VX809 or VX661 increased this association (Figure 2F and Supplemental Figure 8, B and C). We also examined the interaction between CFTR and Nrf2 by proximity ligation assay (PLA), in which a red fluorescent signal indicated that the 2 proteins were less than 40 nm (~3 times the length of the average 100-kDa protein) apart. In WT NhBE cells, Nrf2 and CFTR colocalized to within 40 nm, as indicated by readily detectable red PLA signals (Figure 3, A and B). Conversely, CFhBE cells displayed a very weak PLA signal using antibodies for Nrf2 and CFTR, suggesting little interaction between these 2 proteins in the CF condition (Figure 3, A and C). The negative control (mouse IgG used instead of primary anti-CFTR antibody) did not produce a red fluorescent signal (Figure 3B). The PLA data corroborated our IF (Figure 2, A–C) and immunoprecipitation findings regarding CFTR and Nrf2 interactions (Figure 2F), and together, the results reveal a reduced association between F508del CFTR and Nrf2 in CF versus non-CF cells. Close Nrf2-CFTR association from the perinuclear compartment through the subapical membrane suggests that the interaction may occur early in the proteins’ life cycles and continues while they traffic to the membrane. In CFhBE cells, treatment with VX809 significantly increased PLA signal (Figure 3, A and C). Inhibition of CFTR function with 20 μM CFTRinh-172 blocked the VX809-stimulated Nrf2–F508del CFTR interaction, as assessed by diminished red fluorescent PLA signal, in comparison with cells treated with VX809 alone (Figure 3C).
Figure 3. Nrf2 dysfunction and diminished colocalization with F508del occur physiologically and can be rescued in primary CFhBE cells by CFTR modulation.
(A) Representative 3D micrographs of a proximity ligation assay (PLA) showing that Nrf2 interacts closely with CFTR in NhBE, but not CFhBE, cells (red dots: PLA signals), and that 10 μM VX809 treatment rescues Nrf2-CFTR interaction, visualized by increased red PLA signal. (B) PLA for NhBE cells incubated with antibodies for Nrf2 and either CFTR (top panel) or mouse IgG (bottom panel) as negative control. (C) CFhBE cells incubated with DMSO control or 1–10 μM VX809, and/or 20 μM CFTRinh-172 (Inh172), a CFTR inhibitor, for 48 hours. CFTR-Nrf2 interaction determined by PLA and fluorescent imaging. Arrows indicate apical and basolateral sides. Each experiment repeated in 3–4 CF and 3 non-CF donors with 3–5 replicates per treatment per donor. (D) Nrf2-activated gene expression is decreased in whole lungs from CF mouse models compared with WT mice, as determined by qPCR. Data for n = 5–10 mice per group are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. Outliers are indicated by points outside the box plots. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT mice by 1-way ANOVA and Dunnett’s multiple-comparisons test. (E) Nrf2-CFTR colocalization is decreased in the lung airway epithelial cells of DF508 CF mice versus WT mice. Representative micrographs of IF for Nrf2 (green), CFTR (red), and DAPI (blue), with Nrf2-CFTR colocalization in yellow, at ×60 original magnification, for at least n = 4 mice per group with 3–5 replicates per donor. Scale bars: 10 μm.
Nrf2 target gene expression and Nrf2-CFTR colocalization are decreased in CF mouse models.
To confirm the physiological relevance of crosstalk between Nrf2 and CFTR, we examined Nrf2 function and interaction with CFTR in CF mutant mouse lungs. In F508del CFTR (DF508) and knockout S489X-CFTR mice, expression of the Nrf2 target genes Hmox1, Nqo1, and Gclc was decreased to about 40%–75% of expression levels measured in WT mice (Figure 3D). Reduced Nrf2 function was also evident in mice with the less severe R117H-CFTR class IV variant, in which R117H-CFTR expression is about 10% of CFTR expression in WT mice (Craig Hodges, Case Western Reserve University, Cleveland, OH; personal communication). Furthermore, IF imaging for Nrf2 (green) and CFTR (red) demonstrated significant colocalization (yellow) in airway epithelial cells of WT mice versus reduced levels in DF508 mice (Figure 3E and Supplemental Figure 9). Further studies of the mechanism of activation of Nrf2 by VX809/VX661 were conducted in air-liquid interface cultures of primary human cells.
VX809 and VX661 rescue F508del CFTR and Nrf2 interaction in CF cells.
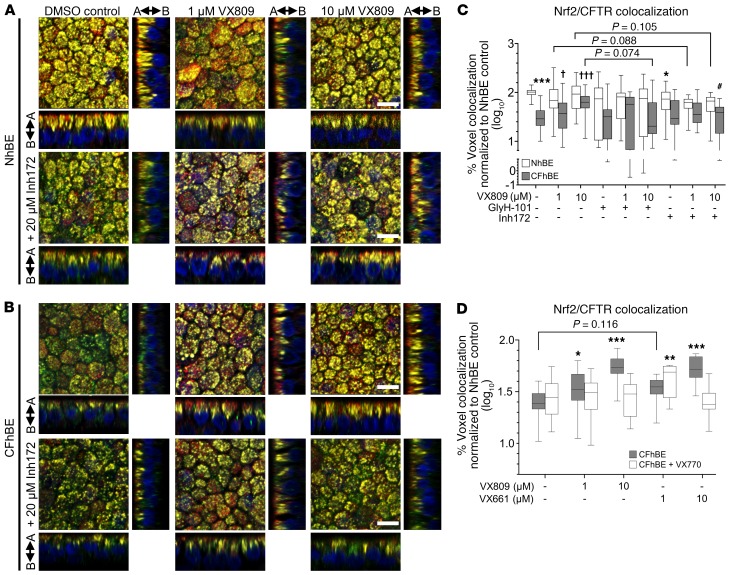
VX809 affects the cellular localization of F508del CFTR, and activates Nrf2, possibly by promoting the interaction of the transcription factor with CFTR (Figure 3). To test this hypothesis, we investigated whether VX809 modulates the association of Nrf2 and CFTR in primary CF cells using IF. Consistent with the PLA data, IF reveals F508del CFTR and Nrf2 colocalization (yellow) with 1 or 10 μM VX809 in CFhBE cells, to levels similar to those observed in control NhBE cells, following 72 hours of drug exposure (Figure 4, B and C, and Supplemental Figure 10). Quantitation revealed that CFhBE control cells had less than 40% of the Nrf2-CFTR colocalization observed in NhBE control cells (Figure 4C). VX809 (1–10 μM) treatment significantly and dose-dependently increased Nrf2–F508del CFTR colocalization in CFhBE cells, and at the 10-μM dose VX809 increased colocalization to approximately 67% of NhBE control levels (Figure 4C). VX809 treatment increased F508del CFTR levels (shown in red, Figure 4B). Incubation with 1–10 μM VX661 for 48 hours had a similar corrective effect on Nrf2-CFTR colocalization in CFhBE cells (Figure 4D). When the potentiator VX770 was added to either VX809 or VX661, colocalization was not significantly increased over that in CFhBE vehicle control and was similar to or lower than that seen with either corrector alone (Figure 4D), consistent with previous reports that VX770 reduces VX809 correction of F508del CFTR function in vitro (41). None of the treatments fully corrected Nrf2-CFTR colocalization.
Figure 4. VX809 increases colocalization of CFTR and Nrf2 in CFhBE cells, which is blocked by coincubation with inhibitors of CFTR.
(A and B) Representative photographs, at ×60 original magnification, of IF for Nrf2 (green), CFTR (red), and nuclei (DAPI, blue) in primary NhBE (A) and CFhBE (B) cells treated with indicated doses of VX809 and/or CFTRinh-172 (Inh172) for 72 hours. Colocalization of CFTR and Nrf2 is in yellow, and arrows indicate apical (A) and basolateral (B) sides. Cells permeabilized as in Figure 2. (C and D) Quantification of Nrf2-CFTR colocalization in NhBE and CFhBE cells after incubation with 1–10 μM VX809 and/or 20 μM CFTRinh-172 or GlyH-101 for 72 hours (11 independent experiments with 6 CF and 5 non-CF donors, with 3 technical replicates per treatment) (C), or 1–10 μM VX809 or VX661 and/or 1 μM VX770 for 48 hours (4 independent experiments with 4 CF and 3 non-CF donors, with 3 replicates per treatment per donor) (D). Data are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. For C, *P < 0.05, ***P < 0.001 vs. NhBE vehicle control; †P < 0.05, †††P < 0.001 CFhBE treatments vs. CFhBE vehicle control; #P < 0.05 vs. same donor cells with same dose of VX809 by mixed-effects ANOVA and Dunnett’s multiple-comparisons test. For D, *P < 0.05, **P < 0.01, ***P < 0.001 vs. CFhBE vehicle control by mixed-effects ANOVA and Dunnett’s multiple-comparisons test. Scale bars: 10 μm.
Prolonged inhibition of CFTR decreases Nrf2 and CFTR interaction.
To investigate the role of CFTR function in regulating Nrf2-CFTR interaction, we inhibited the channel following treatment with correctors. Colocalization of Nrf2 with CFTR was decreased in NhBE cells after incubation with CFTRinh-172, suggesting that functional CFTR modulates interaction between CFTR and Nrf2 in WT cells (Figure 4, A and C). Consistent with PLA studies shown in Figure 3C, treatment of CFhBE cells with 10 μM VX809 combined with 20 μM CFTRinh-172 trended (P = 0.07) towards reducing Nrf2-CFTR colocalization versus treatment with 10 μM VX809 alone (Figure 4, B and C), and incubation with 1–10 μM VX809 and 20 μM CFTRinh-172 did not significantly increase Nrf2-CFTR colocalization over that of the inhibitor alone. In order to confirm that the inhibitor effect is not due to competition with VX809, we used a second inhibitor of CFTR, GlyH-101. While the thiazolidinone CFTRinh-172 interacts with the intracellular Arg347 residue of CFTR, GlyH-101 is a glycine hydrazide analog that blocks the extracellular region of CFTR’s pore (37, 38). Treatment of CFhBE cells with 10 μM VX809 plus 20 μM GlyH-101 significantly reduced Nrf2-CFTR colocalization versus treatment with 10 μM VX809 alone (Figure 4C). VX809 failed to significantly increase Nrf2-CFTR colocalization in CFhBE cells treated with GlyH-101 over GlyH-101 alone or vehicle control. We also investigated whether VX809 altered Keap1 protein expression, the primary regulator of Nrf2 degradation and activation. We found no impact of VX809 treatment, suggesting that the effects of VX809 are not mediated by changes in total Keap1 levels (Supplemental Figure 6C). These data suggest that CFTR function, even during trafficking, is required for Nrf2 colocalization throughout the cell.
Acute manipulation of CFTR function modulates Nrf2-CFTR colocalization.
Based on the evidence above, we further investigated acute activation of CFTR and its role in Nrf2 interactions. In NhBE cells, acute treatment with CFTRinh-172 significantly reduced colocalization, while forskolin trending towards increasing it over that in vehicle control cells, and did so significantly in combination with VX809 (Supplemental Figure 11A). In conjunction with our other CFTR inhibitor studies, results with acute forskolin treatment support the notion that Nrf2-CFTR colocalization is CFTR function dependent. In CFhBE cells, acute treatment with CFTRinh-172 abrogated VX809-induced Nrf2-CFTR colocalization, similar to the effects of prolonged inhibition, further indicating a rapid sensitivity of colocalization to CFTR function (Supplemental Figure 11B). Acute treatment with amiloride, which inhibits ENaC, trended to increase Nrf2 colocalization with CFTR. In addition to corrector, both amiloride and DIDS, a non-CFTR chloride channel inhibitor, resulted in VX809-induced Nrf2-CFTR colocalization that significantly differed from that of CF vehicle control (Supplemental Figure 11B).
Knockdown of F508del expression in CFhBE cells blocks VX809 activation of Nrf2.
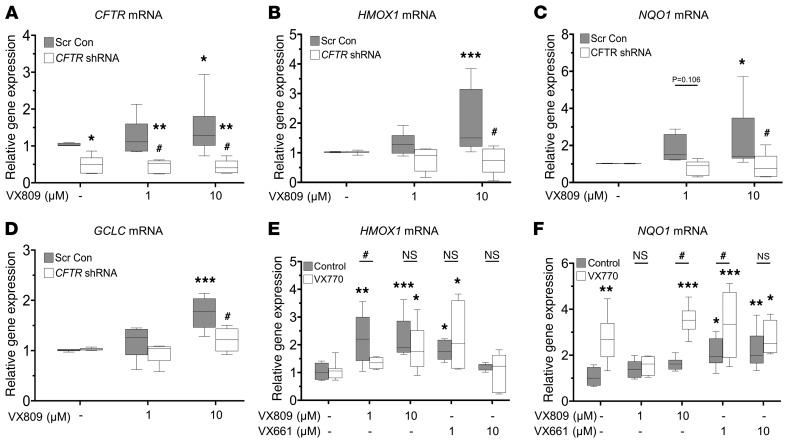
To further examine the CFTR dependence of VX809 activation of Nrf2, we used shRNA lentivirus to knock down F508del gene expression in CFhBE cells. When CFTR gene expression was reduced by more than 50% in comparison with scrambled control (Figure 5A), VX809-induced increase in HMOX1, NQO1, and GCLC mRNA levels (Figure 5, B–D) was significantly inhibited. Importantly, knockdown combined with VX809 did not change Nrf2 gene expression (Supplemental Figure 12), supporting the notion that CFTR-dependent VX809 activation of Nrf2 occurs at the protein level.
Figure 5. F508del knockdown in CFhBE cells blocks CFTR modulator–mediated activation of Nrf2.
(A–D) CFhBE cells were infected with CFTR shRNA or scrambled control (Scr Con) lentivirus for 4 days, then treated with vehicle (DMSO) control or 1–10 μM VX809 for 48 hours. Gene expression was determined for CFTR (A), HMOX1 (B), NQO1 (C), and GCLC (D), by real-time qPCR, with mRNA levels shown as fold changes versus Scr Con cells incubated with vehicle control (DMSO). Data for 4 independent experiments from 3 CF donors with 3 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 vs. Scr Con cells treated with DMSO control, or #P < 0.05 vs. Scr Con cells treated with the same dose of VX809, by mixed-effects ANOVA and Dunnett’s multiple-comparisons test. (E and F) Primary CFhBE cells were incubated with vehicle control (DMSO) or the indicated doses of VX809 or VX661, with or without 1 μM VX770, for 48 hours, and gene expression of HMOX1 (E) or NQO1 (F) was determined by qPCR as above. Data for 3–4 independent experiments from 3 CF donors with 3–4 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 vs. DMSO control cells by 1-way ANOVA (Control group) or 2-way ANOVA (VX770 group) and Dunnett’s multiple-comparisons test; #P < 0.05 vs. control cells treated with the same dose of VX809/VX661 by 1-way ANOVA.
Correction of F508del CFTR with VX661 stimulates Nrf2 transcriptional activity.
Similarly to VX809, treatment of primary CFhBE cells with the newer corrector drug VX661 for 48 hours also increased expression of the Nrf2 target genes HMOX1 and NQO1 (Figure 5, E and F). The combination of VX661 and the potentiator VX770 also stimulated expression of both HMOX1 and NQO1 significantly compared with DMSO control, but not over the same dose of VX661 alone except for NQO1 at the 1-μM dose (Figure 5, E and F). Potentiation by VX770 of VX809/VX661–corrected F508del CFTR was able to further increase Nrf2-activated gene expression over the effects of corrector alone, but failed to do so consistently. Nevertheless, these data further support the importance of CFTR functional correction rather than localization for Nrf2 activation.
Activation of Nrf2 by VX809/VX661 is dependent on CFTR function.
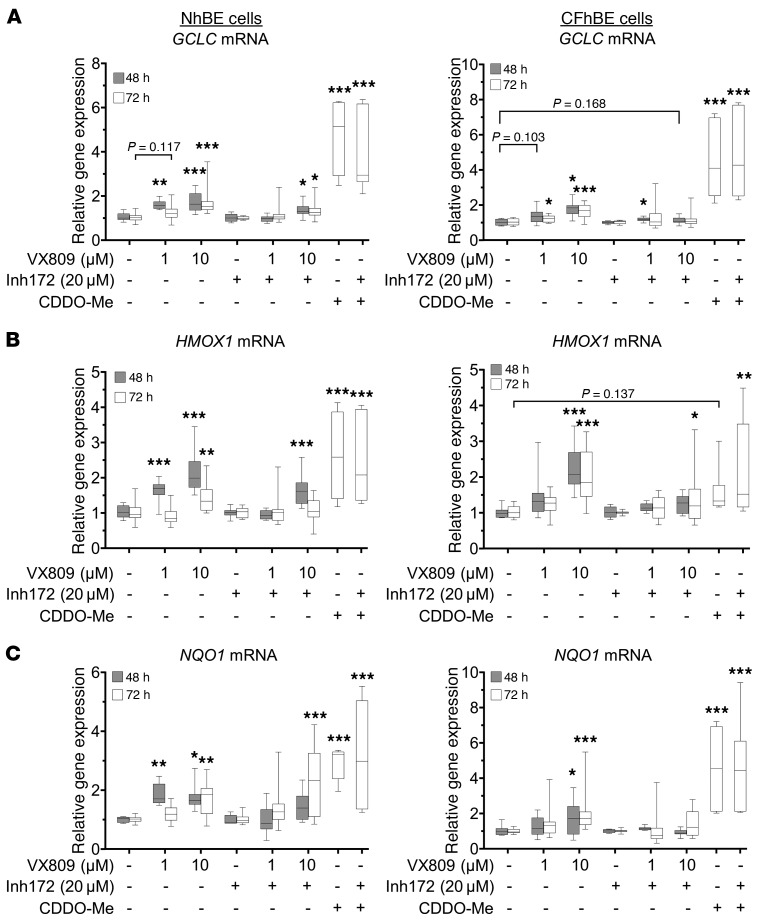
To examine whether CFTR function was necessary for Nrf2 transcriptional activation, we cotreated primary differentiated cultures with VX809/VX661 and CFTRinh-172. After 48–72 hours, 20 μM CFTRinh-172 blocked the VX809- or VX661-induced increase in gene expression of GCLC, HMOX1, and NQO1 in CFhBE cells, with a partial blockade in HMOX1 expression by VX809 at 48 hours (Figure 6 and Figure 7A). Taken together with shRNA knockdown experiments (Figure 5), inhibitor studies indicate that VX809 correction of Nrf2 is CFTR function dependent. Importantly, CDDO-Me, a triterpenoid activator of Nrf2 that is not dependent on CFTR function, served as positive control and increased GCLC, HMOX1, and NQO1 mRNA levels after 72 hours (Figure 6, A–C). Treatment with 20 μM CFTRinh-172 did not reverse the Nrf2 stimulatory effect of CDDO-Me (Figure 6, A–C), indicating that the inhibitory action of CFTRinh-172 is mediated through CFTR and not by direct action on Nrf2.
Figure 6. Functional inhibition of CFTR reverses VX809-induced expression of Nrf2 target genes.
NhBE and CFhBE cells were treated with DMSO control, 1–10 μM VX809, or 30 nM CDDO, a Nrf2 activator, with or without 20 μM CFTRinh-172 for 48 or 72 hours. Gene expression of Nrf2-activated genes, GCLC (A), HMOX1 (B), and NQO1 (C), was determined by real-time qPCR. Gene expression is expressed as fold changes versus control cells (DMSO control or CFTRinh-172 alone), and was calculated from cycle threshold and normalization to the control gene, 18S rRNA. For 48-hour experiments (3 independent experiments from 3 patient donors each for non-CF and CF cells) and 72-hour experiments (7 independent experiments from 4 CF and 4 non-CF donors with 3 replicates per treatment per donor) and for 72-hour CDDO experiments (3 independent experiments from 4 CF and 4 non-CF donors with 3 replicates per treatment per donor), data are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 by mixed-effects ANOVA with Dunnett’s multiple-comparisons test.
Figure 7. Inhibition of CFTR function blocks CFTR modulator–induced expression of Nrf2 target genes.
(A) Primary CFhBE cells were incubated with vehicle (DMSO) control or 1 μM VX661 and/or CFTRinh-172 (Inh172) for 48 hours. (B and C) Primary NhBE (B) or CFhBE (C) cells were treated with DMSO control or the indicated doses of VX661 and/or GlyH-101 for 72 hours. Gene expression of Nrf2-activated genes (HMOX1 and NQO1) was determined by real-time qPCR. Gene expression is expressed as fold changes versus control cells (DMSO control, Inh172, or GlyH-101 alone), and was calculated from cycle threshold and normalization to the control gene, 18S rRNA. Data for 3–6 independent experiments from 3 CF and 3 non-CF donors with 3–4 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 by mixed-effects ANOVA with Dunnett’s multiple-comparisons test.
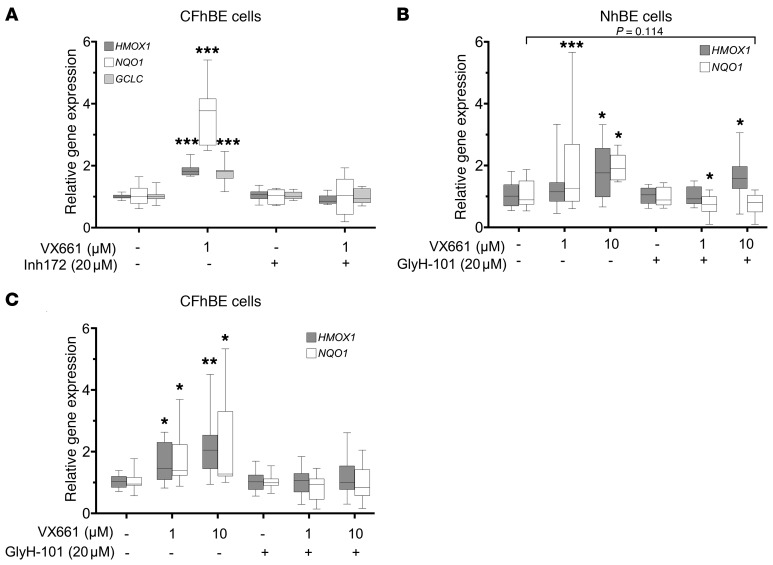
Coincubation with 20 μM GlyH-101 blocked the VX661-induced stimulation of Nrf2 target gene expression, including HMOX1 (at 1 μM VX661) and NQO1, in CFhBE cells after 72 hours (Figure 7, B and C), similar to the block of the increase in Nrf2-CFTR colocalization observed in imaging studies (Figure 4C). The similar inhibitory effects of CFTRinh-172 and GlyH-101 on corrector-induced Nrf2 activity and Nrf2-CFTR colocalization further support the notion that corrector effects are CFTR function dependent.
Conversely, activation of CFTR stimulated Nrf2 activity. Incubation of NhBE cells with forskolin for 6 hours stimulated HMOX1 or GCLC gene expression over 2- and 5-fold, respectively, indicating that activation of WT CFTR function increased Nrf2 transcriptional activity (Supplemental Figure 13A). In CFhBE cells, 6 hours of treatment with CFTRinh-172 blocked HMOX1 and GCLC gene expression stimulated by 48 hours of pre- or cotreatment with 10 μM VX809, while 6 hours of cotreatment with amiloride (ENaC inhibitor) and DIDS (non-CFTR chloride channel inhibitor) did not significantly inhibit VX809-induced Nrf2 activity (Supplemental Figure 13B).
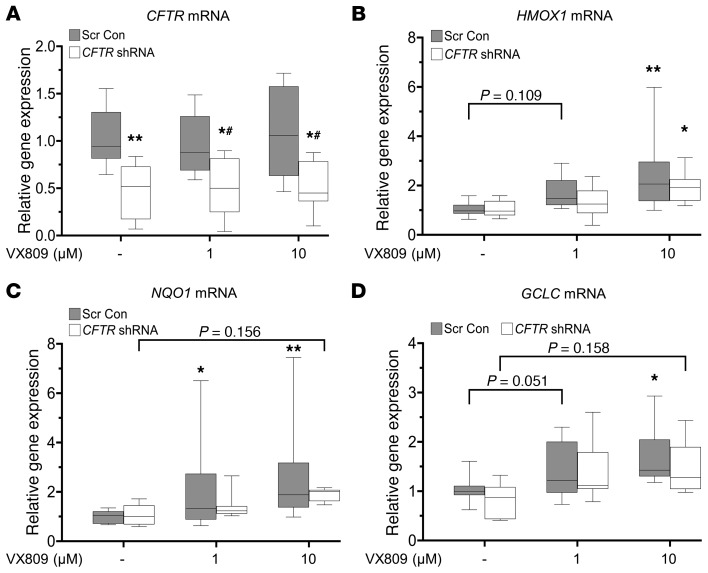
Partial CFTR knockdown in NhBE cells does not inhibit Nrf2 activity induced by VX809.
Infection of NhBE cells with CFTR shRNA lentivirus decreased CFTR gene expression by approximately 60% compared with scrambled control (Figure 8A). Partial knockdown experiments allow us to compare the effect of decreased CFTR function and Nrf2 CFTR colocalization, and are complementary to the studies using the inhibitors of CFTR function, CFTRinh-172, and GlyH-101. Partial knockdown of CFTR in NhBE cells did not decrease the basal gene expression of Nrf2 target genes (HMOX1, NQO1, and GCLC), but did block VX809-mediated activation of Nrf2, although high doses of corrector trended toward Nrf2 activation (Figure 8, B–D). These data support the importance of CFTR function over localization for corrector- mediated activation of Nrf2.
Figure 8. Partial knockdown of CFTR in NhBE cells decreases VX809-mediated activation of Nrf2.
Primary NhBE cells were infected with CFTR shRNA or scrambled control (Scr Con) lentivirus for 4 days, then treated with DMSO control or 1–10 μM VX809 for 48 hours. (A) Gene expression of CFTR was determined by real-time qPCR, with mRNA levels expressed as fold changes versus Scr Con with vehicle control (DMSO). (B–D) Gene expression of Nrf2 target genes (HMOX1, NQO1, or GCLC) was determined by qPCR, with mRNA levels shown as fold changes versus Scr Con or CFTR shRNA with vehicle control (DMSO), respectively. Data for 3 independent experiments on 3 non-CF donors with 4 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05 and **P < 0.01 vs. no drug control, while #P < 0.05 vs. same drug concentration scrambled shRNA control by mixed-effects ANOVA with Dunnett’s multiple-comparisons test.
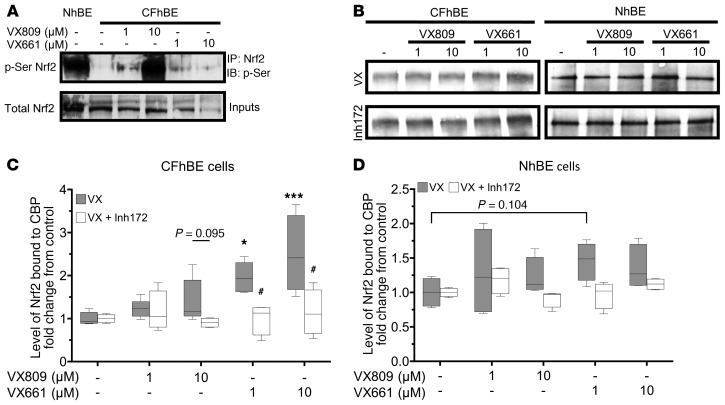
CFTR modulation stimulates Nrf2 phosphorylation and increased interaction with CBP.
To explore the mechanism by which correction of F508del CFTR function increases Nrf2 activity, we immunoprecipitated Nrf2, followed by immunoblotting with antibody against phosphoserine to assess serine phosphorylation of Nrf2 in CFhBE versus NhBE cells (Figure 9A). Nrf2 serine phosphorylation is required for its translocation to the nucleus (42, 43). Incubation of CFhBE cells with 1–10 μM VX809 or VX661 significantly increased Nrf2 phosphoserine levels (Figure 9A), consistent with increases in Nrf2 nuclear localization and activity observed in our other studies CFTR modulation studies (Figures 1 and 6–8). Furthermore, previous studies demonstrated that lack of CFTR function resulted in an increase in phospho-CREB, which increasingly bound CBP and decreased CBP-Nrf2 interaction, in primary CF epithelial cells and F508del mutant mice (12). CBP, a transcriptional coactivator, binds and maximally activates Nrf2 (44, 45). In previous studies, artificial restoration of Nrf2-CBP interaction corrected a large portion of the Nrf2 dysfunction observed in CF (12). We hypothesized that correction of F508del CFTR function with VX809/VX661 would also correct Nrf2-CBP interaction. To test this, we immunoprecipitated CBP, and probed for Nrf2 in CF and non-CF primary epithelia treated for 48 hours with VX809 or VX661. We found that, in a dose-dependent manner, VX661 significantly increased (while VX809 treatment trended towards increasing) CBP interaction with Nrf2 (Figure 9, B and C) 1.5- to 3-fold. This increase of Nrf2 interaction with CBP was CFTR function dependent, as coincubation with CFTRinh-172 abrogated any increase in Nrf2-CBP association over that observed with DMSO alone (Figure 9, B and C). No significant changes were observed in complementary studies with NhBE cells (Figure 9, B and D). These data are consistent with the increase in Nrf2 activation observed with F508del CFTR modulation and demonstrate a link between CFTR function and Nrf2 activation.
Figure 9. CFTR modulation increases Nrf2 phosphorylation and interaction with CBP.
NhBE and CFhBE cells were treated with the indicated doses of VX809 or VX661 for 48 hours. (A and B) Cell lysates were immunoprecipitated at 4°C with anti-Nrf2 antibody, then subjected to Western blot analysis with anti-phosphoserine antibody (A); or with anti-CBP antibody, then subjected to Western blot analysis with anti-Nrf2 antibody (B). (C and D) Average levels of Nrf2 immunoprecipitated with CBP from cell lysates from 4 CF (C) and 3 non-CF (D) donors following treatment with DMSO, VX809, VX661, or treatments plus CFTRinh-172, demonstrate that CFTR modulation increases Nrf2-CBP interaction in a CFTR function–dependent manner. Aliquots of the input lysates were analyzed by Western blot with antibody against total Nrf2 to confirm the presence of equal levels of Nrf2 in lysates subjected to immunoprecipitation (Supplemental Figure 14). Representative blots are shown for 4 independent experiments in 4 CF and 3 non-CF donors. Data are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, ***P < 0.001 vs. vehicle control; or #P < 0.05 vs. cells treated with the same dose of VX809/VX661 by mixed-effects ANOVA with Dunnett’s multiple-corrections test.
Discussion
The primary novel findings of this study are that the F508del CFTR correctors VX809 and VX661 reverse the dysregulation of Nrf2 activity in primary human CF epithelial cells, and that this rescue is CFTR function dependent. Previously, we found that dysfunction of Nrf2 significantly contributes to the dysregulation of inflammatory signaling in CF airway epithelia (11, 12, 30). This is consistent with a large body of evidence that demonstrates dysregulation of Nrf2 inflammatory signaling in the airway for a variety of diseases, including asthma and COPD (46–53). The crucial role of Nrf2 in regulating inflammation and the present findings that CFTR interacts with and regulates Nrf2 suggest a link between CFTR and inflammatory signaling. The implication of a direct relationship between CFTR and Nrf2 is that clinical modulation of F508del CFTR has the potential to also correct Nrf2 abnormalities and mitigate inflammatory signaling in CF patients. As inflammation continues to be a major cause of lung function decline in patients receiving F508del CFTR corrector therapy, the implications of this study are clinically relevant. Complete normalization of Nrf2 activity, which may not be necessary for significant modulation of inflammation in patients, was only achieved at high doses of corrector exposure, such as 100 μM VX809, which elevated Nrf2 nuclear localization to non-CF levels. However, 100 μM is approximately 7-fold higher than the maximum serum concentrations of correctors that can be attained in vivo in patients (achievable serum concentrations are 1–14 μM) (8). Nevertheless, at these clinically relevant doses of VX809 and VX661 (54), we found that while Nrf2 activity was not completely normalized, it was significantly stimulated, supporting the notion that F508del CFTR correction that can be achieved clinically may positively impact Nrf2 activation. The development of more effective modulators of CFTR function holds promise for complete correction of Nrf2, and associated regulation of inflammatory signaling (11, 12, 30).
Throughout our studies we found that the abilities of VX809 and VX661 to activate Nrf2 are CFTR dependent. To explore the mechanism by which VX809 and VX661 correct Nrf2 function in CF epithelial cells, we dissected the impact of the modulators’ effects on CFTR processing versus CFTR function. Others have shown that VX809 increases CFTR levels at the plasma membrane by binding directly to the first nucleotide binding domain (NBD1) and stabilizing its interaction with membrane-spanning domain-1 (MSD1) in F508del CFTR, allowing for better folding (55, 56). In examining whether changes in the processing of CFTR contributed to Nrf2 correction, we found that CFTR and Nrf2 colocalize in perinuclear compartments and the subapical membrane space (Figure 2) to within 40 nm (Figure 3, A–C) by PLA. Multiple IF studies (totaling n = 360 confocal Z-stacks from multiple CF and non-CF subjects, corrected for multiple comparisons) show that the colocalization of Nrf2 and CFTR is significantly decreased in CF compared with non-CF airway epithelial cells (Figures 2 and 4). Moreover, PLA (Figure 3) and CFTR immunoprecipitation experiments (Figure 2F) also demonstrate that the close association between Nrf2 and CFTR is diminished in CF versus non-CF cells. Our present studies do not distinguish between direct interaction and one that involves intermediary proteins. Nrf2 and/or Keap1 have not been shown to bind to any PDZ interactors of CFTR. The interaction is unlikely to occur at the C-terminal PDZ motif of CFTR, as our pull-down studies used the 24-1 anti-CFTR antibody, which recognizes CFTR’s C-terminus. We also found that Nrf2 interaction with CFTR is physiologically relevant in vivo, where it is significantly higher in the lung airway epithelia of WT mice versus the airways of F508del CFTR mice (Figure 3E and Supplemental Figure 9). The decreased levels of mutant CFTR in F508del homozygous cells (57) do not fully account for the significant decrease in the fraction of F508del CFTR and Nrf2 that colocalize in CFhBE compared with NhBE cells.
Our exploration of the functional consequence of VX809/VX661 treatment revealed that Nrf2 activation by CFTR modulators is CFTR function dependent (Figures 6–8). Even the Nrf2-CFTR interaction is modulated, at least in part, by CFTR function, and is reduced by CFTR inhibition in NhBE cells (Figures 3 and 4 and Supplemental Figures 11 and 13). Furthermore, approximately 50% knockdown of F508del CFTR expression by shRNA or blockade of corrected F508del CFTR function by CFTRinh-172 or GlyH-101 inhibits Nrf2 activity in CFhBE cells stimulated with VX809/VX661 (Figures 5–7). The use of 2 inhibitors of CFTR that bind at different sites confirmed that the loss of VX809/VX661 effect is not due to the blocking of modulator binding sites on CFTR. Conversely, activation of CFTR function increases both corrector-mediated Nrf2 activation and Nrf2-CFTR colocalization in NhBE cells with WT CFTR (Supplemental Figure 11A and Supplemental Figure 13A), and partial CFTR knockdown in NhBE cells does not completely block 10 μM corrector activation of Nrf2 (Figure 8). Taken together, our data of CFTR expression or functional inhibition indicate that modulator effects on Nrf2 activity are mediated through CFTR activity, and not necessarily by altering the interaction with CFTR or the levels of CFTR. This dependence on CFTR function is consistent with our previous studies that showed that the lack of CFTR function reduces Nrf2 activity, leading to increased steady-state intracellular hydrogen peroxide and inflammatory cytokine production (11, 12).
Mechanistically, our data indicate that one link that exists between CFTR function and Nrf2 in non-CF cells is restored in CF cells by CFTR modulation. Nrf2 interaction with its coactivator CBP in the nucleus is necessary for maximal transcriptional activity (44, 45, 58). In the context of CF, lack of CFTR function results in a feedback response that increases levels of cAMP and phosphorylated CREB (59, 60). Increases in p-CREB bind CBP and sequester it from interaction with Nrf2, reducing Nrf2 activity (58, 61). Previously, we reported that in CF cell lines, primary cells, and animals Nrf2 exhibits a reduced interaction with CBP compared with non-CF controls, and that artificial reversal of reduced Nrf2-CBP interaction corrects Nrf2 dysfunction (12). In the present studies, we examined the 3 major steps in Nrf2 activation: inhibition by Keap1 (Supplemental Figure 6C), serine phosphorylation (Figure 9A), and interaction with CBP (Figure 9, B–D). The data show that F508del CFTR modulation–mediated activation of Nrf2 was only coupled to increased serine phosphorylation and increased interaction with CBP in a CFTR function–dependent fashion. The implication of our studies is that VX809 and VX661 normalize CFTR colocalization with Nrf2, but rescue of Nrf2 activity in CF is due to modulation of CFTR function, which relieves feedback responses to CFTR dysfunction, allowing for sufficient levels of CBP to interact with Nrf2.
With present clinical doses and modulators, our studies suggest that it would be necessary to combine VX809 or VX661 and VX770 with a Nrf2-acitvating drug to attain sufficient Nrf2 rescue to significantly impact Nrf2-regulated inflammatory pathways in F508del homozygotes. The combination of VX809 and VX770 moderately improves lung function in CF patients homozygous for the F508del mutation, which is not observed with VX809 therapy alone (9). Addition of VX770 to VX809/VX661 did improve Nrf2-CFTR colocalization (Figure 4D) and Nrf2 activity (Figure 5, E and F), but rarely did so over either VX809 or VX661 alone. VX770 in combination with VX809/VX661 does not completely restore CFTR function in patients (62); however, new-generation modulators that more efficiently correct CFTR expression, localization, and function may fully correct Nrf2 dysfunction and downstream regulation of inflammatory signaling.
Our studies demonstrate a significant correction of a secondary defect directly linked to inflammation in CF primary cells following correction of F508del CFTR with VX809. Although partial restoration of Nrf2 is achieved at clinical concentrations of VX809 and VX661, full correction requires supratherapeutic clinical doses. Nevertheless, the important implication of our findings is that full clinical correction of CFTR dysfunction would correct Nrf2 regulation, and influence inflammatory signaling. In the absence of full mutant CFTR correction, additional treatment with Nrf2 activators may be necessary to significantly modulate Nrf2 function to influence inflammation in CF patients. VX809/VX661 activation of Nrf2 is dependent on the correction of CFTR function, but also involves the correction of Nrf2 colocalization with CFTR. Given the role of Nrf2 in deactivating inflammatory signaling (39, 47, 50), our data suggest a link between CFTR and inflammatory signaling in CF epithelial cells.
Methods
Cell culture.
Primary non-CF or CF human bronchial epithelial cells (NhBE or CFhBE cells, respectively) were purchased from ChanTest/Charles River Laboratories, received from the Pulmonary Medicine CF RDP Translational Core at Cincinnati Children’s Hospital Medical Center (CCHMC), or provided by Scott H. Randell (Marsico Lung Institute, Tissue Procurement and Cell Culture Core, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA; UNC IRB#03-1396), and prepared as described previously (63). Cells were cultured at air-liquid interface (ALI) on semipermeable filters, as described previously (29, 57), in media containing Ultroser G (Pall), following the Vertex Pharmaceuticals Inc. formula (64). The cells were differentiated, forming tight junctions and cilia. The non-CF cells from 6 donors had the following codes: DD007K, DD005K, DD021K, DD029J, DD032L, and DD053K. The CF cells from 12 donors had the following codes: KK002L, KK003K, KK003M, KK004i, KK006F, KK006G, KK011i, KK012B, KK013F, KK022M, KK024N, and KK027H. The primary cells used were all deidentified, and their use was approved under CCHMC IRB 2014-8600.
RNA isolation and quantitative real-time PCR.
Differentiated NhBE and CFhBE cells were treated with the indicated doses of VX809, VX661, VX770 (Selleck Chemicals), CDDO methyl ester (Cayman Chemical), CFTRinh-172, and/or GlyH-101 (Tocris) for 48 or 72 hours. Alternatively, differentiated NhBE or CFhBE cells were incubated on the basolateral side with DMSO control or VX809 (10 μM) for 48 hours, then coincubated acutely for 6 hours with apical forskolin (Tocris), amiloride (Sigma-Aldrich), DIDS (Sigma-Aldrich), or CFTRinh-172. Negative controls were treated with DMSO. Total RNA from cells was isolated using Aurum Total RNA Mini Kit (Bio-Rad), and reverse-transcribed in a 20-μL reaction containing random primers and iScript Reverse Transcriptase (Bio-Rad). Real-time PCR was performed with a StepOnePlus instrument using Power SYBR Green PCR Master Mix (Applied Biosystems). Primer pairs used for human samples were: HMOX1 (forward 5′-cttcttcaccttccccaaca-3′, reverse 5′-gctctggtccttggtgtcat-3′), GCLC (forward 5′-CCTCCAGTTCCTGCACATCT-3′, reverse 5′-GGGTAGGATGGTTTGGGTT-3′), NQO1 (forward 5′-caaatcctggaaggatggaa-3′, reverse 5′-ggttgtcagttgggatggac-3′), CFTR (forward 5′-TTGGATGACCTTCTGCCTCT-3′, reverse 5′-CTCCTGCCTTCAGATTCCAG-3′), NFE2L2 (NRF2) (forward 5′-GAGAGCCCAGTCTTCATTGC-3′, reverse 5′-TGCTCAATGTCCTGTTGCAT-3′), 18s rRNA (forward 5′-gtggagcgatttgtctggtt-3′, reverse 5′-cgctgagccagtcagtgtag-3′). Primer pairs used for mouse lung samples were: Hmox1 (forward 5′-GCCGAGAATGCTGAGTTCATG-3′, reverse 5′-TGGTACAAGGAAGCCATCACC-3′), Gclc (forward 5′-CTGCACATCTACCACGCAGT-3′, reverse 5′-TTCATGATCGAAGGACACCA-3′), Nqo1 (forward 5′-CGCCTGAGCCCAGATATTGT-3′, reverse 5′-GCACTCTCTCAAACCAGCCT-3′), 18s rRNA (forward 5′-GTAACCCGTTGAACCCCATT-3′, reverse 5′-CCATCCAATCGGTAGTAGCG-3′). All samples were run in duplicate or triplicate. The relative fold increase of specific RNA was calculated by the comparative cycle of threshold detection method, and values were normalized to 18S rRNA. Fold changes in gene expression were calculated using the 2–ΔΔCt method after normalization to 18S rRNA. Data were collected from 3–4 independent experiments.
Subcellular protein fractionation.
NhBE and CFhBE cells were treated with 0, 0.1, 1, 10, or 100 μM VX809 for 24 hours. Nuclear extracts were prepared as described previously (65). Briefly, NhBE and CFhBE cells were harvested in ice-cold PBS and centrifuged at 4000 g for 3 minutes at 4°C. The pellet was resuspended in buffer containing 10 mM HEPES (pH 7.0), 1.5 mM MgCl2, and 10 mM KCl on ice for 10 minutes, vortexed, and centrifuged at 4000 g, and the cytoplasmic protein in the supernatant was collected. Pellets were resuspended in buffer containing 20 mM HEPES (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 25% glycerol and incubated on ice for 20 minutes. Samples were centrifuged at 16,800 g, and the nuclear fraction in the supernatant was collected. Protein concentration was determined by Bradford assay (Bio-Rad). All buffers contained protease inhibitor cocktail (Roche) and 1 mM Na3VO4, 20 mM NaF, and 1 mM Na4P2O7. All extracts were stored at –80°C until analysis.
Western blot analysis.
The anti-CFTR antibody (mouse clone 570) was received from the CFTR antibody distribution program at the University of North Carolina, the anti-phosphoserine antibody was purchased from Invitrogen, and the anti-Nrf2 antibody was generated and validated previously (12, 66, 67). Differentiated NhBE and CFhBE cells grown on filters were treated with the indicated doses of VX809 or VX661 for 48 hours. Equal amounts of protein samples (20–30 μg) were separated on 7% SDS-PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked in 5% milk, incubated with primary antibodies overnight at 4°C, then washed and incubated with HRP-conjugated secondary antibodies for 1 hour. Products were exposed to the SuperSignal ECL detection system (Pierce) and photographed. β-Actin and proliferating cell nuclear antigen served as loading controls. Experiments were repeated at least 3 times.
Coimmunoprecipitation.
Differentiated NhBE and CFhBE cells grown on ALI were incubated with the indicated doses of VX809 or VX661 for 48 hours. Lysis of cells, and all precipitation procedures with the lysates, were conducted at 4°C. Protein concentration was measured using an aliquot of each lysate, and lysates were diluted at 4°C to equal concentrations with cold lysis buffer. Lysates (with equal concentrations of protein) were precleared with uncoated Dynabeads, then mixed with anti-CFTR antibody (mouse clone 24-1, R&D Systems), anti-Nrf2 antibody, or anti-CBP antibody (catalog sc-1211, Santa Cruz Biotechnology) conjugated to Dynabeads (5 μg antibody per 15 μL beads), and incubated overnight at 4°C. After at least 3 washes in cold buffer (25 mM Tris, pH 7.5, 150 mM NaCl, Roche Complete Protease Inhibitor, 1% Triton X-100), protein was eluted at 37°C in 50 μL SDS-PAGE sample loading buffer, subjected to gel electrophoresis on 7.5% gels, and analyzed by Western blot analysis for Nrf2 or CFTR as described above.
Immunofluorescence.
Differentiated NhBE and CFhBE cells grown on filters were treated with the indicated doses of VX809, VX661, VX770, and/or CFTRinh-172 for 48–72 hours. Alternatively, NhBE or CFhBE cells on ALI were treated on the basolateral side with DMSO control or VX809 (1 or 10 μM) for 48 hours, and then cotreated on the apical side for the last 2 hours with forskolin, amiloride, DIDS, or CFTRinh-172. Cells on filters were fixed in 4% paraformaldehyde (PFA) for 30 minutes. Cells were permeabilized with 0.1% Triton X-100 for 10 minutes, blocked with 2.5% horse serum (Vector Laboratories) for 1 hour, then incubated with anti-Nrf2 and/or anti-CFTR antibodies overnight at 4°C. For the Nrf2 nuclear localization experiments, cells were fixed in cold methanol for 15 minutes and permeabilized with 0.2% Triton X-100 for 20 minutes. Mouse lungs were fixed in 4% PFA, paraffin-embedded, and sectioned onto slides. Mouse sections were deparaffinized, rehydrated, and permeabilized with 0.1% Triton X-100 for 10 minutes, and antigen retrieval was done by microwave heating (1 minute full power, then 10 minutes 10% power) in sodium citrate buffer, followed by blocking. Samples were then incubated with species-matched Alexa Fluor–conjugated secondary antibodies (Alexa Fluor 594 goat anti-mouse IgG or Alexa Fluor 488 goat anti-rabbit IgG, Life Technologies), and nuclei were counterstained with DAPI. Filters were cut and mounted on slides. Specimens were visualized using a Nikon A1R Laser Scanning Confocal Microscope, and representative images were analyzed under the same conditions for each experiment, using NIS Elements Advanced Research software (Nikon) or Imaris software (Bitplane). Colocalization (shown in yellow) for Nrf2 and CFTR was quantified as percentage of colocalized voxels (voxels that were positive for both red and green channels above the threshold set for each channel) in the sample volume for each sample in Imaris software. All samples in each experiment were set to the same thresholds for each channel, and the thresholds set to include 40% of the data in each channel for the NhBE controls for each experiment. Data are normalized to NhBE control for each of 4–5 independent experiments.
Luciferase reporter assay.
Primary CFhBE cells were grown in 48-well plates until cells were 60%–80% confluent. Transient transfection assays were performed using Lipofectamine LTX reagent (Life Technologies) with 1 μg of a firefly luciferase plasmid (pNrf2-fluc) under the control of the Nrf2-dependent promoter for glutathione-S-transferase 1 (GST1) and/or 100 ng of Renilla luciferase plasmid (Promega) as an internal control for transfection efficiency. Six hours after transfection, cells were supplemented with complete media. One day after transfection, cells were incubated with the indicated doses of VX809 for 48 hours. Cells were lysed in Passive Lysis Buffer (Promega) and examined for firefly and/or Renilla luciferase activity using a SpectraMax L 1-channel luminometer (Molecular Devices). Protein concentration was determined by BCA protein assay. Firefly luciferase activity was normalized to protein levels or Renilla luciferase activity. Data are expressed as fold change versus vehicle control in 4 independent experiments.
CFTR electrophysiology assays.
Ion transport, including CFTR function, was quantified as short-circuit current (ISC) as previously described (68) with minor modifications. Mature primary NhBE and CFhBE cells grown on 0.33-cm2 semipermeable filters were mounted in Ussing chambers (Physiologic Instruments) and equilibrated in symmetric Ringer’s buffer. Cells were placed under voltage clamp conditions to measure baseline ISC and resistance, then transitioned to a 6-mM Cl– apical buffer (replacing sodium chloride with sodium gluconate) to create a basolateral-to-apical Cl– gradient. Culture quality was good with an average transepithelial resistance of 546.8 μΩ*cm2 (n = 127, SEM = 35.6); 7 cultures (5.5%) were excluded owing to transepithelial resistance measures less than 100 μΩ*cm2 with no predilection to any treatment condition. Apical compartments of all cells were then treated with amiloride (ENaC inhibitor, 100 μM). CFTR was stimulated in the apical and basolateral compartments with forskolin (10 μM) and IBMX (100 μM; Acros Organics) to increase cellular cAMP, and maximally potentiated with apical VX770 (1 μM). To confirm CFTR dependency of responses, CFTRinh-172 was added to the apical compartment (10 μM). Finally, calcium-activated Cl– conductance was assessed with apical ATP (100 μM; Sigma-Aldrich). For each step, the ISC tracing reached plateau before moving to the next treatment. Experiments were recorded using Acquire and Analyze 2.3 software (Physiologic Instruments), and the ISC change induced with each treatment was measured and transcribed into Prism software (GraphPad). To allow for comparisons between donors, all ISC values were normalized to the experimental DMSO control and are reported as percentage of internal control.
Baseline CFTR electrophysiology assays.
Unstimulated CFTR function was assessed using a modified version of the above stimulated protocol. Mature primary cells were mounted in Ussing chambers in a basolateral-to-apical Cl– gradient, and sodium transport was inhibited with amiloride. Alternate Cl– transporters were then blocked with 4,4′-diisothiocyano-2,2′-stilbenedisulfonic acid (DIDS; 100μM) in the apical compartment. CFTR function was then inhibited with CFTRinh-172 (10 μM) in the apical compartment; this CFTR inhibition was quantified as proxy for unstimulated CFTR function. For each step, the ISC tracing reached plateau before moving to the next treatment. Average transepithelial resistance was 703.3 μΩ*cm2 (n = 59, SEM = 45.9); 1 culture (1.7%) was excluded owing to transepithelial resistance measures less than 100 μΩ*cm2.
Proximity ligation assay.
NhBE and CFhBE cells differentiated on ALI on filters were treated with vehicle (DMSO) control or the indicated doses of VX809 and/or CFTRinh-172 for 48 hours. Cells were fixed with 10% formalin for 10 minutes and stored in 70% ethanol. Filters were cut out and placed in chamber slides. Cells were permeabilized with 0.1% Triton X-100 for 15 minutes at room temperature and then incubated in membrane blocking solution for 40 minutes, followed by overnight incubation with primary anti-Nrf2 antibody and anti-CFTR antibody (CF-3, Abcam) at 4°C. Then the proximity ligation assay (PLA) was carried out using the Duolink PLA in situ kit (Sigma-Aldrich) following the manufacturer’s instructions. The preparations were analyzed using a Nikon A1Rsi confocal microscope and NIS Elements Advanced Research or Imaris software. Experiments were repeated at least 3 times.
Knockdown of CFTR expression.
CFhBE or NhBE cells were plated in 48-well plates in media containing Ultroser G (Pall). Cells were infected with a panel of 5 shRNA lentiviral vectors, targeted against human CFTR (MISSION shRNA Lentiviral Transduction Particles SHCLNV-NM_000492, Sigma-Aldrich), in order to knock down CFTR gene expression. Scrambled control lentiviral vector (SH02, Sigma-Aldrich) served as negative control. After screening, plasmid TRCN0000082965 provided the best knockdown, as confirmed by quantitative PCR. Cells were infected with CFTR shRNA or scrambled control lentivirus for 4 days, and then incubated with the indicated doses of VX809 for 48 hours before harvesting for RNA. Each of 4 independent experiments contained 3–8 replicates.
Animals.
Lungs from CF mouse models with either the F508del Cftr mutation (Cftrtm1kth) (69), the R117H Cftr mutation (Cftrtm2Mrc) (70), or the S489X Cftr mutation (Cftrtm1Unc) (71) were received from the Case Western Reserve University Cystic Fibrosis Mouse Models Core (Cleveland, Ohio, USA). The mice were backcrossed for 10 generations on the C57BL/6J strain to make the mutations congenic. All animals were cared for according to a Case Western Reserve University–approved protocol and Institutional Animal Care and Use Committee guidelines. Animals were housed in standard polysulfone microisolator cages in ventilated units with corncob bedding. Mice were given ad libitum access to chow (Harlan Teklad 7960, Harlan Teklad Global Diets) and sterile water. All animals were maintained on a 12-hour light/12-hour dark schedule at a mean ambient temperature of 22°C.
Statistics.
Data were analyzed by 1- or 2-way ANOVA with Dunnett’s multiple-comparisons testing to measure significant differences. Values of P ≤ 0.05 are considered statistically significant. Data are presented by box plots ranging from the 25th to the 75th percentile with lines marking group medians and whiskers spanning the 5th to 95th percentiles. Data presented by scatter plot are shown including a mean line ± SEM. The number of subjects tested is denoted by the N number. The number of experiments of each condition is indicated in each figure legend, and each tested condition was replicated 2–4 times per experiment.
Study approval.
The studies with deidentified primary human bronchial epithelial cells were considered exempt (IRB 2014-8600) by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center (CCHMC). Studies with animals were approved by CCHMC’s Institutional Animal Care and Use Committee under IACUC protocol 2017-0095.
Author contributions
AGZ supervised the project. DCB, AGZ, and JB wrote the manuscript. AGZ, SL, DCB, and MES contributed to the conceptualization of the hypothesis and project studies presented in the paper. AGZ, DCB, MES, SL, and JB designed experiments and interpreted results. MES, DCB, SL, and JB performed and analyzed experimental data. SMP, HS, and JPC provided unique reagents and primary cells and aided with experimental design and interpretation of results.
Supplementary Material
Acknowledgments
We thank the CCHMC CF RDP Translational Studies Core for providing primary airway epithelial cells. Provision of primary cells to our sources was by Scott Randell at the University of North Carolina and was supported by grants from the Cystic Fibrosis Foundation (BOUCHE15R0) and NIH (DK065988). We also thank the CCHMC Confocal Imaging Core, directed by Matthew Kofron, for its help with confocal imaging techniques and analysis. We also thank the Case Western Reserve University Cystic Fibrosis Mouse Models Core for providing CF mouse tissues. Gail M. Macke provided assistance with histology. Rhonda Szczesniak at CCHMC aided with statistical analyses. Changsuk Moon and Anjaparavanda P. Naren at CCHMC provided technical advice on the proximity ligation assay. Technical edits to the manuscript and fact checking were provided by Katye M. Fichter of FichterScript LLC. This work was supported by the National Heart, Lung, and Blood Institute (1R01HL109362-01 to AGZ).
Version 1. 05/30/2019
In-Press Preview
Version 2. 07/22/2019
Electronic publication
Version 3. 08/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(8):3448–3463.https://doi.org/10.1172/JCI96273.
Contributor Information
Dana C. Borcherding, Email: dcborch@iastate.edu.
Matthew E. Siefert, Email: Matthew.Siefert@cchmc.org.
Songbai Lin, Email: songbai.lin@emory.edu.
John Brewington, Email: john.brewington@cchmc.org.
Hesham Sadek, Email: hesham.sadek@utsouthwestern.edu.
John P. Clancy, Email: jpclancy@cff.org.
Scott M. Plafker, Email: Scott-Plafker@omrf.org.
Assem G. Ziady, Email: assem.ziady@cchmc.org.
References
- 1.Riordan JR, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245(4922):1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Chen H, et al. [Inhibition of decomposing leaf litter of Cinnamomum camphora on growth of Capsicum annuum and the alleviation effect of nitrogen application] Ying Yong Sheng Tai Xue Bao. 2015;26(2):457–465. [PubMed] [Google Scholar]
- 3.Muhlebach MS, Stewart PW, Leigh MW, Noah TL. Quantitation of inflammatory responses to bacteria in young cystic fibrosis and control patients. Am J Respir Crit Care Med. 1999;160(1):186–191. doi: 10.1164/ajrccm.160.1.9808096. [DOI] [PubMed] [Google Scholar]
- 4.Cantin AM, Hartl D, Konstan MW, Chmiel JF. Inflammation in cystic fibrosis lung disease: pathogenesis and therapy. J Cyst Fibros. 2015;14(4):419–430. doi: 10.1016/j.jcf.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Wagener JS, Kahn TZ, Copenhaver SC, Accurso FJ. Early inflammation and the development of pulmonary disease in cystic fibrosis. Pediatr Pulmonol Suppl. 1997;16:267–268. doi: 10.1002/ppul.19502308138. [DOI] [PubMed] [Google Scholar]
- 6.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16(1):45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha C, et al. Capturing the direct binding of CFTR correctors to CFTR by using click chemistry. Chembiochem. 2015;16(14):2017–2022. doi: 10.1002/cbic.201500123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clancy JP, et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67(1):12–18. doi: 10.1136/thoraxjnl-2011-200393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wainwright CE, Elborn JS, Ramsey BW. Lumacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373(18):1783–1784. doi: 10.1056/NEJMc1510466. [DOI] [PubMed] [Google Scholar]
- 10.Taylor-Cousar JL, et al. Tezacaftor-ivacaftor in patients with cystic fibrosis homozygous for Phe508del. N Engl J Med. 2017;377(21):2013–2023. doi: 10.1056/NEJMoa1709846. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Kinter M, Shank S, Cotton C, Kelley TJ, Ziady AG. Dysfunction of Nrf-2 in CF epithelia leads to excess intracellular H2O2 and inflammatory cytokine production. PLoS One. 2008;3(10):e3367. doi: 10.1371/journal.pone.0003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziady AG, et al. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-κB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302(11):L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzec JM, et al. Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J. 2007;21(9):2237–2246. doi: 10.1096/fj.06-7759com. [DOI] [PubMed] [Google Scholar]
- 14.Sussan TE, et al. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc Natl Acad Sci U S A. 2009;106(1):250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui W, et al. Nrf2 attenuates inflammatory response in COPD/emphysema: Crosstalk with Wnt3a/β-catenin and AMPK pathways. J Cell Mol Med. 2018;22(7):3514–3525. doi: 10.1111/jcmm.13628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M. NRF2 targeting: a promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol Med. 2011;17(7):363–371. doi: 10.1016/j.molmed.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J. 2004;18(11):1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- 18.Brown RH, Reynolds C, Brooker A, Talalay P, Fahey JW. Sulforaphane improves the bronchoprotective response in asthmatics through Nrf2-mediated gene pathways. Respir Res. 2015;16:106. doi: 10.1186/s12931-015-0253-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakabayashi N, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35(3):238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 20.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8(4):379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24(16):7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 23.Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol. 2013;85(6):705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387(10-11):1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 25.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26(8):2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Stein TD, Johnson JA. Genetic dissection of systemic autoimmune disease in Nrf2-deficient mice. Physiol Genomics. 2004;18(3):261–272. doi: 10.1152/physiolgenomics.00209.2003. [DOI] [PubMed] [Google Scholar]
- 29.Ziady AG, et al. Interaction with CREB binding protein modulates the activities of Nrf2 and NF-κB in cystic fibrosis airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2012;302(11):L1221–L1231. doi: 10.1152/ajplung.00156.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols DP, Ziady AG, Shank SL, Eastman JF, Davis PB. The triterpenoid CDDO limits inflammation in preclinical models of cystic fibrosis lung disease. Am J Physiol Lung Cell Mol Physiol. 2009;297(5):L828–L836. doi: 10.1152/ajplung.00171.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuentes F, Paredes-Gonzalez X, Kong AN. Dietary glucosinolates sulforaphane, phenethyl isothiocyanate, indole-3-carbinol/3,3′-diindolylmethane: anti-oxidative stress/inflammation, Nrf2, epigenetics/epigenomics and in vivo cancer chemopreventive efficacy. Curr Pharmacol Rep. 2015;1(3):179–196. doi: 10.1007/s40495-015-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aubouy A, et al. Nrf2-driven CD36 and HO-1 gene expression in circulating monocytes correlates with favourable clinical outcome in pregnancy-associated malaria. Malar J. 2015;14:358. doi: 10.1186/s12936-015-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keleku-Lukwete N, et al. Amelioration of inflammation and tissue damage in sickle cell model mice by Nrf2 activation. Proc Natl Acad Sci U S A. 2015;112(39):12169–12174. doi: 10.1073/pnas.1509158112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho HY, Kleeberger SR. Association of Nrf2 with airway pathogenesis: lessons learned from genetic mouse models. Arch Toxicol. 2015;89(11):1931–1957. doi: 10.1007/s00204-015-1557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin S, Du R, Yin S, Liu X, Xu G, Cao W. Nrf2 is essential for the anti-inflammatory effect of carbon monoxide in LPS-induced inflammation. Inflamm Res. 2015;64(7):537–548. doi: 10.1007/s00011-015-0834-9. [DOI] [PubMed] [Google Scholar]
- 36.Gruber F, et al. Nrf2 deficiency causes lipid oxidation, inflammation, and matrix-protease expression in DHA-supplemented and UVA-irradiated skin fibroblasts. Free Radic Biol Med. 2015;88(pt B):439–451. doi: 10.1016/j.freeradbiomed.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Lin W, Wu RT, Wu T, Khor TO, Wang H, Kong AN. Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem Pharmacol. 2008;76(8):967–973. doi: 10.1016/j.bcp.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osburn WO, et al. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121(9):1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- 39.Cho HY, et al. Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol. 2002;26(2):175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 40.Van Goor F, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci U S A. 2011;108(46):18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cholon DM, et al. Potentiator ivacaftor abrogates pharmacological correction of ΔF508 CFTR in cystic fibrosis. Sci Transl Med. 2014;6(246):246ra96. doi: 10.1126/scitranslmed.3008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277(45):42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 43.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278(45):44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 44.Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol. 2009;29(10):2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6(10):857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 46.Rangasamy T, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114(9):1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iizuka T, et al. Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells. 2005;10(12):1113–1125. doi: 10.1111/j.1365-2443.2005.00905.x. [DOI] [PubMed] [Google Scholar]
- 48.Ishii Y, et al. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J Immunol. 2005;175(10):6968–6975. doi: 10.4049/jimmunol.175.10.6968. [DOI] [PubMed] [Google Scholar]
- 49.Mochizuki M, et al. Role of 15-deoxy delta(12,14) prostaglandin J2 and Nrf2 pathways in protection against acute lung injury. Am J Respir Crit Care Med. 2005;171(11):1260–1266. doi: 10.1164/rccm.200406-755OC. [DOI] [PubMed] [Google Scholar]
- 50.Rangasamy T, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med. 2005;202(1):47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thimmulappa RK, et al. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun. 2006;351(4):883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116(4):984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin W, et al. Influence of Nrf2 genotype on pulmonary NF-kappaB activity and inflammatory response after traumatic brain injury. Ann Clin Lab Sci. 2008;38(3):221–227. [PubMed] [Google Scholar]
- 54.Farinha CM, Sousa M, Canato S, Schmidt A, Uliyakina I, Amaral MD. Increased efficacy of VX-809 in different cellular systems results from an early stabilization effect of F508del-CFTR. Pharmacol Res Perspect. 2015;3(4):e00152. doi: 10.1002/prp2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren HY, et al. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol Biol Cell. 2013;24(19):3016–3024. doi: 10.1091/mbc.e13-05-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hudson RP, et al. Direct binding of the corrector VX-809 to human CFTR NBD1: evidence of an allosteric coupling between the binding site and the NBD1:CL4 interface. Mol Pharmacol. 2017;92(2):124–135. doi: 10.1124/mol.117.108373. [DOI] [PubMed] [Google Scholar]
- 57.Randell SH, Fulcher ML, O’Neal W, Olsen JC. Primary epithelial cell models for cystic fibrosis research. Methods Mol Biol. 2011;742:285–310. doi: 10.1007/978-1-61779-120-8_18. [DOI] [PubMed] [Google Scholar]
- 58.Kim SW, Lee HK, Shin JH, Lee JK. Up-down regulation of HO-1 and iNOS gene expressions by ethyl pyruvate via recruiting p300 to Nrf2 and depriving It from p65. Free Radic Biol Med. 2013;65:468–476. doi: 10.1016/j.freeradbiomed.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 59.Manson ME, Corey DA, Rymut SM, Kelley TJ. β-Arrestin-2 regulation of the cAMP response element binding protein. Biochemistry. 2011;50(27):6022–6029. doi: 10.1021/bi200015h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manson ME, Corey DA, White NM, Kelley TJ. cAMP-mediated regulation of cholesterol accumulation in cystic fibrosis and Niemann-Pick type C cells. Am J Physiol Lung Cell Mol Physiol. 2008;295(5):L809–L819. doi: 10.1152/ajplung.90402.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu GH, Qu J, Shen X. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta. 2008;1783(5):713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Veit G, et al. Some gating potentiators, including VX-770, diminish ΔF508-CFTR functional expression. Sci Transl Med. 2014;6(246):246ra97. doi: 10.1126/scitranslmed.3008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fulcher ML, Randell SH. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol Biol. 2013;945:109–121. doi: 10.1007/978-1-62703-125-7_8. [DOI] [PubMed] [Google Scholar]
- 64.Gentzsch M, et al. Pharmacological rescue of conditionally reprogrammed cystic fibrosis bronchial epithelial cells. Am J Respir Cell Mol Biol. 2017;56(5):568–574. doi: 10.1165/rcmb.2016-0276MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S, Saxena NK, Ding X, Stein LL, Anania FA. Leptin increases tissue inhibitor of metalloproteinase I (TIMP-1) gene expression by a specificity protein 1/signal transducer and activator of transcription 3 mechanism. Mol Endocrinol. 2006;20(12):3376–3388. doi: 10.1210/me.2006-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Mealey GB, Plafker KS, Berry WL, Janknecht R, Chan JY, Plafker SM. A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking. J Cell Sci. 2017;130(20):3467–3480. doi: 10.1242/jcs.203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plafker KS, Nguyen L, Barneche M, Mirza S, Crawford D, Plafker SM. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J Biol Chem. 2010;285(30):23064–23074. doi: 10.1074/jbc.M110.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun H, et al. Tgf-beta downregulation of distinct chloride channels in cystic fibrosis-affected epithelia. PLoS One. 2014;9(9):e106842. doi: 10.1371/journal.pone.0106842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeiher BG, et al. A mouse model for the delta F508 allele of cystic fibrosis. J Clin Invest. 1995;96(4):2051–2064. doi: 10.1172/JCI118253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Heeckeren AM, Schluchter MD, Drumm ML, Davis PB. Role of Cftr genotype in the response to chronic Pseudomonas aeruginosa lung infection in mice. Am J Physiol Lung Cell Mol Physiol. 2004;287(5):L944–L952. doi: 10.1152/ajplung.00387.2003. [DOI] [PubMed] [Google Scholar]
- 71.Snouwaert JN, et al. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257(5073):1083–1088. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.