Abstract
Clostridium [Clostridioides] difficile infection (CDI) is one of the leading causes of diarrhea associated with medical care worldwide, and up to 60% of patients with CDI can develop a recurrent infection (R-CDI). A multi-species microbiota biofilm model of C. difficile was designed to evaluate the differences in the production of biofilms, sporulation, susceptibility to drugs, expression of sporulating (sigH, spo0A), quorum sensing (agrD1, and luxS), and adhesion-associated (slpA and cwp84) pathway genes between selected C. difficile isolates from R-CDI and non-recurrent patients (NR-CDI). We obtained 102 C. difficile isolates from 254 patients with confirmed CDI (66 from NR-CDI and 36 from R-CDI). Most of the isolates were biofilm producers, and most of the strains were ribotype 027 (81.374%, 83/102). Most C. difficile isolates were producers of biofilm (100/102), and most were strongly adherent. Sporulation was higher in the R-CDI than in the NR-CDI isolates (p = 0.015). The isolates from R-CDI patients more frequently demonstrated reduced susceptibility to vancomycin than isolates of NR-CDI patients (27.78% [10/36] and 9.09% [6/66], respectively, p = 0.013). The minimum inhibitory concentrations for vancomycin and linezolid against biofilms (BMIC) were up to 100 times and 20 times higher, respectively, than the corresponding planktonic MICs. Expression of sigH, spo0A, cwp84, and agrD1 was higher in R-CDI than in NR-CDI isolates. Most of the C. difficile isolates were producers of biofilms with no correlation with the ribotype. Sporulation was greater in R-CDI than in NR-CDI isolates in the biofilm model of C. difficile. The R-CDI isolates more frequently demonstrated reduced susceptibility to vancomycin and linezolid than the NR-CDI isolates in both planktonic cells and biofilm isolates. A higher expression of sporulating pathway (sigH, spo0A), quorum sensing (agrD1), and adhesion-associated (cwp84) genes was found in R-CDI than in NR-CDI isolates. All of these factors can have effect on the recurrence of the infection.
Introduction
Clostridium [Clostridioides] difficile is one of the leading causes of healthcare-associated diarrhea worldwide. Since 2011, cases of C. difficile infection (CDI) have increased in the United States, with 453,000 infections and 29,000 deaths [1].
An estimated 20–35% of patients with CDI can develop a recurrent infection (R-CDI) within eight weeks of the first episode, with an incidence of 1,846–37,620 cases/year [2]. The development of R-CDI has been associated with the germination of spores of the strain that produced the initial infection in the colon or with the acquisition of a new strain [3–5].
The pathogenicity of chronic and recurrent infections has been associated with the production of biofilm in some bacterial species [6], and C. difficile has been shown to produce organized biofilm communities on abiotic surfaces in vitro [7–9]. Furthermore, C. difficile has shown to produce biofilms in the presence of other bacteria such as Finegoldia magna in vitro and participate in complex gut microbiota biofilms in vitro and during infection in vivo in a mouse model [10, 11]. Such biofilm formation could protect bacteria from cellular immune responses associated with toxin production and from antibiotics used for the treatment of CDI [10].
C. difficile spores have been found within the biofilms in a simulated chemostat gut model [11, 12], suggesting that the accumulation of spores within the biofilm of C. difficile could play a role in the development of R-CDI [13]. Lower spore germination rates have been reported in C. difficile biofilms than in vegetative cultures [14, 15], which may affect the persistence of infection.
The development of biofilms has been associated with the ability of bacteria to resist antimicrobial agents because they act as a physical barrier and decrease the effective concentration of antimicrobials [8, 10]. Indeed, the biofilms of C. difficile have shown 100 times greater resistance to metronidazole and 10 times greater resistance to vancomycin than cells cultured in liquid media [14].
Several factors have been described to have a key role in the formation of C. difficile biofilm, such as surface factors like S-layer protein, SlpA (encoded by slpA), the cell wall protein Cwp84 (encoded by cwp84), and the putative quorum sensing regulator LuxS (encoded by luxS) [8, 10, 16]. In addition, the master regulator of sporulation, Spo0A, has shown to determine the biofilm-producing phenotype [7, 14]. Recently, the sigma factor of sporulation, SigH, and the agr quorum sensing system, have been shown to regulate metabolism and virulence potential in C. difficile [17–20]. Nevertheless, their contributory role in R-CDI development remains unknown.
Therefore, the aim of this study was to evaluate the effect of biofilm production in a range of C. difficile ribotypes, their sporulation, antimicrobial susceptibilities, and expression of genes involved in sporulation and biofilm formation in isolates from recurrent and non-recurrent CDI patients.
Material and methods
Setting
Patients were recruited for this study who were treated at the following two hospitals in Mexico: The Civil Hospital of Guadalajara, "Fray Antonio Alcalde" a tertiary hospital with 1000 beds in Guadalajara; and the University Hospital "Dr. José Eleuterio González," a tertiary teaching hospital with 500 beds in Monterrey.
Ethics statement
The local ethics committee (Comité de Ética en Investigación del Antiguo Hospital Civil de Guadalajara “Fray Antonio Alcalde,” Jalisco, Mexico) approved this study with reference number 047/16. Informed consent was waived by the Ethics Committee because no intervention was involved and no patients identifying information was included.
Study population, CDI diagnosis, and classification of CDI
Patients with unexplained diarrhea (≥ 3 unformed stools, Bristol scale 5–7) within 24 h were included. For the diagnosis of CDI, fecal samples were collected, and C. difficile was investigated by real-time PCR (Cepheid Xpert C. difficile/Epi test, Cepheid, Sunnyvale CA). Patients were defined with CDI when patients were diarrheal and PCR was positive.
R-CDI was defined by the reappearance of diarrhea associated with CDI within eight weeks after the completion of antibiotic therapy or the resolution of the initial episode [21]. CDI was classified as NR-CDI when no new episode occurred within eight weeks. Data collected from patients with R-CDI and NR-CDI included epidemiological and clinical data, prior antibiotic therapy, and treatment for CDI. The study was reviewed and approved by the Local Ethics Committee (Approval: 047/16).
Culture of C. difficile and typing of isolates
Fecal specimens were cultured on C. difficile agar (Neogen Corporation, MI) with cefoxitin (16 mg/L) and incubated in an anaerobic chamber (10% CO2, 10% H2, and 80% N2) at 37°C for 48 h. Isolates were identified by polymerase chain reaction (PCR) with amplification of the triose phosphate isomerase (tpi) gene [22] and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). All isolates were stored at −80°C. The tcdA, tcdB, cdtA, and cdtB genes were amplified using a multiplex PCR method [22], and the ribotyping-PCR was performed as previously described [23].
Selected isolates were subjected to ribotyping by capillary electrophoresis at the C. difficile Ribotyping Network Reference Laboratory (CDRN) at Leeds Teaching Hospitals NHS Trust (Leeds, United Kingdom).
C. difficile biofilm model
The biofilm of C. difficile formation was conducted as reported previously with some modifications. [7, 24] Briefly, each isolate was cultured in brain heart infusion (BHI) broth supplemented with 0.5% yeast extract and 0.1% L-cysteine (BHIS) in 96-well microtiter plates and incubated in anaerobic conditions at 37°C for 7 days. The planktonic cells were removed, and absorbance of planktonic cultures was read at 590 nm using a microtiter plate spectrometer iMarK (Bio-Rad, Hercules, CA, USA). The biofilm was washed with sterile phosphate-buffered saline (PBS) (200 μl), fixed with 2% glutaraldehyde, and washed again with PBS. Then, 1% crystal violet was added and the biofilm was washed six more times with sterile water and de-stained with 30% acetic acid. The optical density was read at 590 nm. The adherence index (AI) was calculated, and the isolates were classified as strong adherents (AI > 1.20), moderate adherents (0.90 < AI <1.20), weak adherents (AI between 0.2 and 0.90) and non-adherents (AI < 0.2) [25]. The assays were performed in triplicate, and only the results with a variation coefficient greater than 20% were accepted.
Microbiota–C. difficile biofilm model
For this study, we designed a biofilm model containing C. difficile and species from the intestinal microbiota (Enterococcus faecalis ATCC 29212, E. faecium, Lactobacillus casei, Lactobacillus rahmnosus, and Lactobacillus acidophilus strains obtained from clinical specimens). Each strain was cultured in BHIS at 37°C in an anaerobic chamber for 24–48 h and subsequently diluted 1:10 in BHIS broth. A 200-μl mixture of microbiota and C. difficile (1:1:1:1:1:1 of each species) were added to each well of a microtiter plate, and the plates were incubated in an anaerobic chamber at 37°C for 7 days. The determination of the biofilm was carried out according to the biofilm model of C. difficile only.
Strains ATCC BAA-1805 (ribotype 027, strong adherent) and ATCC 9689 (ribotype 001, weak adherent) were used as controls in the biofilm assays.
Antimicrobial susceptibility testing
Antimicrobial susceptibilities were determined by the agar dilution method in selected isolates [26]. Antibiotics used to treat CDI infections, as well as those associated with the development of CDI, were included. The minimum inhibitory concentrations (MICs) were determined for metronidazole (ICN Biomedical, Costa Mesa, CA), vancomycin, linezolid, ciprofloxacin, moxifloxacin, erythromycin, clindamycin, rifampicin, and tetracycline (Sigma-Aldrich). The antimicrobial diluents used and the ranges tested were recommended by the Clinical and Laboratory Standards Institute (CLSI, 2019, M100-S28). An overnight culture in Schaedler broth (Neogen Corporation) of each isolate was inoculated using a multipoint inoculator (104 colony-forming units (CFU)/spot) on Wilkins-Chalgren agar (Neogen Corporation) [27]. The ATCC 700057 (ribotype 038) was used as a control strain.
Resistance breakpoints were defined according to the CLSI guidelines as follows: moxifloxacin and clindamycin ≥ 8 mg/L, tetracycline ≥ 16 mg/L, metronidazole ≥ 32 mg/L, (CLSI, 2019). The breakpoint for vancomycin was defined according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2019) as greater than 2 mg/L. For antimicrobial agents of which no standard breakpoints to C. difficile have yet to be defined, breakpoints were considered as follows: erythromycin ≥ 8 mg/L (CLSI, 2013), ciprofloxacin, ≥ 8 mg/L, [28] linezolid ≥ 16 mg/L [29], and rifampicin ≥ 32 mg/L [30].
Biofilm minimum inhibitory concentration
The BMIC was determined for vancomycin and linezolid. Briefly, the planktonic phase of a 7-day-old biofilm was removed, and the antibiotics were prepared in fresh BHIS. Each concentration (from 512 mg/L to 0.5 mg/L) was aliquoted into microtiter plates (one per concentration), and 200 μl per well were added. The biofilm was resuspended, and the plates were incubated at 37°C for 48 h in anaerobiosis.
The BMIC was defined as the lowest concentration of an antimicrobial that prevents growth. The ATCC 700057 C. difficile strain (ribotype 038) was used as a control.
Spore count in the biofilm
The 7-day-old biofilm was disrupted with a pipette and resuspended in 100 μl PBS. Serial ten-fold dilutions (10−1–10−7) were incubated at 65°C for 20 min to kill the vegetative cells. Both untreated and heat-treated suspensions were streaked on Clostridium difficile agar (Neogen Corporation, MI, USA) and incubated at 37°C for 48 h in anaerobic conditions. Total viable cells and spore counts were determined as CFU/biofilm.
Quantitative RT-PCR for spo0A, sigH, slpA, cwp84, agrD1, and luxS
Twenty selected strains (10 from R-CDI patients and 10 from NR-CDI patients) were analyzed. The biofilms incubated for 48 h were washed twice, then the pellet was resuspended in DEPC water (200 μl) and treated with lysozyme (10 mg/ml) (Bio-basic, Ontario, Canada) and proteinase K (Bio-basic). Total RNA was isolated using the Qiagen QIAamp DSP Viral RNA mini kit (QIAGEN, Hilden, Germany). The quantity and quality of the RNA were assessed using a NanoDrop spectrophotometer. The relative quantification of the expression of the RNA transcripts of the spo0A, sigH, slpA, cwp84, agrD1, and luxS genes normalized to 16S rRNA (rrs) was analyzed using the SuperScript III Platinum One-Step kit (Invitrogen, CA, USA).
Standard curves were generated using 5-fold dilutions of ATCC 9689 RNA for each gene to determine the efficiency of the reactions. RNA and diethyl pyrocarbonate (DEPC) water controls were also included. The real-time RT-PCRs were performed in two biological samples in duplicate; 200 ng RNA and 0.5 μl specific primers at 100 nM (listed on Table 1) in a 25-μL reaction volume were used. The Smart Cycler real-time PCR system (Cepheid) was used with cycling conditions as follows: 94°C for 8 min, then 45 cycles of 94°C for 30 s; 60°C for spo0A, sigH and cwp84; 54°C for slpA, agrD1 and 30 s for luxS and an extension cycle of 72°C for 25 s. Melting curves were determined to assure that only the expected PCR products had been generated. Relative expression ≥3 was classified as overexpression. Data were normalized and analyzed using the method described by Chang et al., and the ATCC 9689 was used as a calibrator [31].
Table 1. Primer pairs used to amplify the genes studied by real-time RT-PCR.
| Target gene | 5′primer | 3′primer | Source |
|---|---|---|---|
| rrs | GGGAGACTTGAGTGCAGGAG | GTGCCTCAGCGTCAGTTACA | [43] |
| spoA | CTCAAAGCGCAATAAATCTAGGAGC | TTGAGTCTCTTGAACTGGTCTAGG | [44] |
| sigH | GTTGGTAGCAAAAGAAAAAAGTTATGAG | GTACTCTAGTGCTATTTTATCCCCTTCAC | [45] |
| slpA | AATGATAAAGCATTTGTAGTTGGTG | TATTGGAGTAGCATCTCCATC | [43] |
| cwp84 | TGGGCAACTGGTGGAAAATA | TAGTTGCACCTTGTGCCTCA | [43] |
| luxS | GTGTACTTGATGGAGTAAAGGGAGA | TTCTACATCCCATTGGAGATAAGTC | [46] |
| agrD1 | TTTGCTAGCTCATTGGCACTT | GATTGCTGATTTCTTTGGGTACTT | Primer3 software |
Statistical analysis
Reduced susceptibility frequencies from planktonic and biofilm R-CDI cultures were compared with NR-CDI cultures using Pearson’s chi-square test and Fisher’s Exact test.
Differences in relative expression ratios mean of biofilm cultures from R-CDI and NR-CDI were analyzed using Student’s t-test. Non-parametric data were analyzed using the Mann-Whitney test and Spearman rank correlation test.
All statistical analysis were performed using the SPSS software package. A p value less than 0.05 was considered to be statistically significant.
Results
Study population
In total, 254 patients with CDI (35.29%, female and 64.70%, male; age range, 15–85) were confirmed by PCR, with 102 isolates of C. difficile recovered. Patients with R-CDI had more significant exposure to antibiotics before CDI (p = 0.037) than patients with NR-CDI. Fluoroquinolones and vancomycin were more frequently used in patients with NR-CDI than in those with R-CDI (p = 0.000 and 0.024, respectively). Cephalosporins were more frequently used in patients with R-CDI than in NR-CDI (p = 0.048) (Table 2).
Table 2. Clinical characteristics of patients with R-CDI and NR-CDI.
| R-CDI (n = 23) | NR-CDI (n = 31) | p value | |
|---|---|---|---|
| Hospitalization | |||
| Length of stay (mean days, range) | 29.55 (4–124) | 20.86 (4–59) | 0.181 |
| Intensive care unit, n (%) | 6 (26.09) | 10 (32.26) | 0.427 |
| Length of stay in ICU (mean days, range) | 14.77 (2–48) | 12.10 (2–48) | 0.460 |
| Prior antibiotics | |||
| Any antibiotic, n (%) | 30 (96.77) | 22 (95.65) | 0.675 |
| Length of exposure (mean days, range) | 21.48 (1–100) | 13.52 (1–52) | 0.132 |
| No. of antibiotics (mean) | 3.65 | 2.71 | 0.037* |
| Cephalosporins | 12 (54.54) | 8 (27.59) | 0.048* |
| Clindamycin | 18 (81.82) | 21 (72.41) | 0.329 |
| Macrolides | 21 (95.45) | 27 (91.10) | 0.605 |
| Fluoroquinolones | 10 (45.45) | 27 (91.10) | 0.000* |
| Vancomycin | 9 (40.91) | 21 (72.41) | 0.024* |
| Metronidazole | 19 (86.36) | 25 (86.21) | 0.657 |
| Carbapenems | 14 (63.64) | 17 (58.62) | 0.472 |
| CDI treatment | |||
| Vancomycin | 19 (86.36) | 24 (80.00) | 0.490 |
| Metronidazole | 13 (59.09) | 19 (63.33) | 0.415 |
| Metronidazole/vancomycin | 10 (45.45) | 16 (56.33) | 0.390 |
Data are no. (%) of patients, unless otherwise noted.
*Significant difference p value <0.05
Culture and ribotyping
Sixty-six isolates (64.70%) of patients with NR-CDI and 36 (35.29%) of patients with R-CDI were obtained. Most of the isolates were found to be ribotype 027 (83/102, 81.37%). The other isolates were found to be ribotypes 003 and 002 (3/102, 2.94% each); 001, 014, 078, 220, and 076 (2/102, 1.96% each one); and 353 (1/102, 0.98%) (Table 3). No significant differences in ribotype distribution between the R-CDI and NR-CDI groups was detected (p = 0.476).
Table 3. Ribotype distribution between R-CDI and NR-CDI strains.
| Genotype | PCR-Ribotype (n) | |
|---|---|---|
| R-CDI (n = 36) | tcdA+, tcdB+, tcdC Δ18+, cdtA+/cdtB+ | 027 (30) |
| tcdA+, tcdB+, tcdC Δ18−, cdtA-/cdtB- | 003 (1), 001 (1), 076 (1), NT (2) | |
| tcdA+, tcdB+, tcdC Δ18−, cdtA+/cdtB+ | 353 (1) | |
| NR-CDI (n = 66) | tcdA+, tcdB+, tcdC Δ18+, cdtA+/cdtB+ tcdA+, tcdB+, | 027 (53), |
| tcdA+, tcdB+, tcdC Δ18−, cdtA-/cdtB- | 002 (3), 014 (2) 003 (2), 220 (2), 076 (1), NT (1) | |
| tcdA+, tcdB+, tcdC Δ39+, cdtA+/cdtB+ | 078 (2) |
C. difficile biofilm sporulation
In the C. difficile biofilm model, the majority of C. difficile isolates were producers of biofilm (100/102), with 80.55% of R-CDI isolates and 90.90% of NR-CDI isolates being strongly adherent. No differences were detected in biofilm production among the isolates of R-CDI and NR-CDI (AIs geometric mean [GM], 53.87 and 54.06, respectively; p = 0.579).
Sporulation was higher in R-CDI than in NR-CDI isolates (5 log10 CFU/biofilm vs. 3.85 log10 CFU/biofilm; p = 0.015).
C. difficile–microbiota biofilm and sporulation
In the biofilm of C. difficile–microbiota, no difference was detected in biofilm production among the isolates of R-CDI and NR-CDI (AIs GM, 33.04 and 32.89, respectively; p = 0.677).
No significant difference was detected in sporulation between the R-CDI and NR-CDI isolates (6.21 log10 and 5.54 log10 CFU/biofilm, respectively; p = 0.565).
Minimum inhibitory concentrations
Drug susceptibility was evaluated on a selection of 65 isolates (26, R-CDI; 39, NR-CDI), and more than 70% of the isolates were resistant to ciprofloxacin (≥8 mg/L), moxifloxacin (≥8 mg/L), erythromycin (≥8 mg/L), clindamycin (≥8), and rifampin (≥32 mg/L). All isolates were susceptible to tetracycline (≤4 mg/L) and metronidazole (≤8 mg/L).
The isolates from R-CDI patients showed a greater reduced susceptibility to vancomycin (>2 mg/L) than the isolates from NR-CDI patients (27.78 and 9.09%, respectively). No other difference was observed between the R-CDI and NR-CDI isolates (Table 4).
Table 4. Antimicrobial susceptibility (mg/L) from R-CDI and NR-CDI strains.
| Antimicrobial agent | R-CDI | NR-CDI | p value | |
|---|---|---|---|---|
| Ciprofloxacin | GM | 111.43 | 61.71 | |
| Range | 8->128 | 1->128 | ||
| MIC90 | >128 | >128 | ||
| Resistant (%) | 96.15 | 89.74 | 0.342 | |
| Moxifloxacin | GM | 18.78 | 14.22 | |
| Range | 1–32 | 1–32 | ||
| MIC90 | 32.00 | 32.00 | ||
| Resistant (%) | 92.31 | 87.23 | 0.506 | |
| Erythromycin | GM | 190.21 | 87.70 | |
| Range | 1->128 | 1->128 | ||
| MIC90 | >128 | >128 | ||
| Resistant (%) | 81.81 | 95.23 | 0.152 | |
| Clindamycin | GM | 150.97 | 66.75 | |
| Range | 1->128 | 0.5->128 | ||
| MIC90 | >128 | >128 | ||
| Resistant (%) | 90.48 | 81.82 | 0.383 | |
| Vancomycin | GM | 2.16 | 1.76 | |
| Range | 1–4 | 0.25–4 | ||
| MIC90 | 4.00 | 2.00 | ||
| Resistant (%) | 27.78 | 9.09 | 0.013* | |
| Metronidazole | GM | 1.59 | 1.39 | |
| Range | 0.25–4 | 0.25–4 | ||
| MIC90 | 4.00 | 2.00 | ||
| Resistant (%) | 0.00 | 0.00 | NA | |
| Linezolid | GM | 3.92 | 4.00 | |
| Range | 0.5–32 | 0.03–32 | ||
| MIC90 | 16.00 | 16.00 | ||
| Resistant (%) | 38.89 | 19.70 | 0.036* | |
| Rifampin | GM | 13.55 | 16.02 | |
| Range | 0.001->128 | 0.002->128 | ||
| MIC90 | >128 | 128.00 | ||
| Resistant (%) | 70.59 | 79.41 | 0.484 | |
| Tetracycline | GM | 0.26 | 0.16 | |
| Range | 0.06–8 | 0.06–8 | ||
| MIC90 | 4.00 | 0.13 | ||
| Resistant (%) | 0.00 | 0.00 | NA | |
Breakpoints were as follows: moxifloxacin and clindamycin ≥8 mg/L, tetracycline ≥16 mg/L, metronidazole ≥32 mg/L according to CLSI (2019); vancomycin >2 mg/L according to EUCAST (2019), erythromycin ≥8 mg/L according to CLSI (2013); ciprofloxacin, ≥8 mg/L [28], linezolid ≥16 mg/L [29] and rifampicin ≥32 mg/L [30].
*Significant difference p value <0.05
Minimum inhibitory concentrations of the biofilm
In C. difficile biofilm isolates without microbiota, a reduced susceptibility to vancomycin was observed in 91.0% (101/102) of isolates and to linezolid in 89.21% (91/102) of isolates.
The BMICs were up to 100-fold higher for vancomycin and 20-fold higher for linezolid than the corresponding MICs. No differences between R-CDI and NR-CDI isolates were observed (Table 5).
Table 5. Distribution of MICs and BMICs between R-CDI and NR-CDI strains.
| Vancomycin | Linezolid | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Range | GM | Resistant (%) | p value | Range | GM | Resistant (%) | p value | ||
| R-CDI | MIC | 1.00–4.00 | 1.88 | 10/36 (27.78) | 0.000 | 0.50–32.0 | 3.92 | 3/36 (8.33) | 0.000** |
| BMIC | 2.00–256 | 109.08 | 35/36 (97.22) | 4.00–256.0 | 87.34 | 29/36 (80.55) | |||
| NR-CDI | MIC | 0.25–4.00 | 1.85 | 6/66 (9.09) | 0.000 | 0.03–32.0 | 4.02 | 13/66 (20.0) | 0.000** |
| BMIC | 2.00–256 | 108.35 | 66/66 (100) | 2.00–256.0 | 89.63 | 62/66 (95.38) | |||
Data are mg/L of an antimicrobial agent unless otherwise noted.
**Significant difference p value <0.01
Expression of spo0A, sigH, slpA, cwp84, agrD1, and luxS
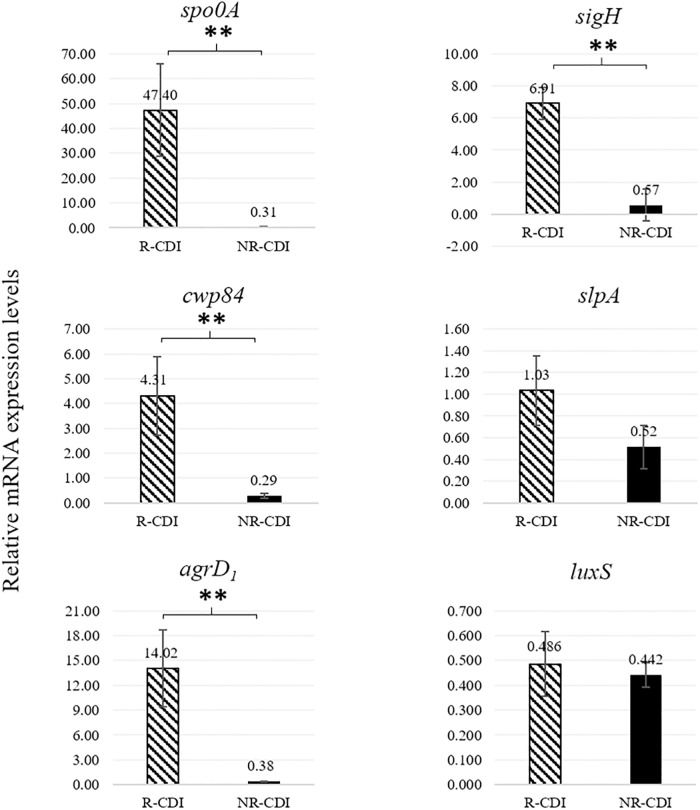
The relative expression of spo0A, sigH, cwp84, and agrD1 was higher in R-CDI than in NR-CDI isolates (Fig 1). Overexpression of spo0A (70%, 7/10), sigH (70%, 7/10), cwp84 (40%, 4/10), and agrD1 (70%, 7/10) was higher in R-CDI than in NR-CDI isolates. No significant difference was detected in the expression levels of slpA and luxS between R-CDI and NR-CDI isolates (Fig 1).
Fig 1. Expression levels of spo0A, sigH, cwp84, slpA, agrD1, and luxS transcripts between R-CDI and NR-CDI strains.
Relative mRNA transcripts expression means of spo0A (p = 0.003), sigH (p = 0.007), cwp84 (p = 0.001), slpA (p = 0.066), agrD1 (p = 0.001) and luxS (p = 0.400) from R-CDI and NR-CDI strains. **Significant difference p value <0.01.
Discussion
Although some bacterial species that cause recurrent or chronic infections have been studied for their ability to form biofilms in vivo and in vitro [6], C. difficile biofilms have not been widely studied. In the present study, we evaluated biofilm formation by C. difficile and detected that most isolates were biofilm producers (strong adherent), independent of the ribotype or whether the strains were isolated from R-CDI or NR-CDI patients. These results reflect those of other previous studies that found no correlation between the ribotype, the strain virulence, or relapse of infection [16].
The expression of quorum sensing regulators and adhesion-associated factors were determined from biofilm culture. agrD1 and cwp84 were overexpressed in R-CDI isolates, both of which were previously shown to regulate colonization, virulence, and relapses in in vivo models [8, 16, 18–20]. By contrast, luxS and slpA expression was similar in R-CDI and NR-CDI isolates. It would be valuable in the future, to analyze the transcription of toxins A and B involving isolates overexpressing agrD1 and cwp84 from R-CDI and NR-CDI isolates.
Differences in spore formation in biofilms aged 7–10 days compared to vegetative cultures have been reported previously, including higher viable counts, higher temperature tolerance, and pleiomorphic biofilm structures (thin, thick exosporium surrounding the spores in the biofilm) [32, 33]. Further studies to compare germination efficiency with co-germinants in this biofilm model need to be done. In the present study, we analyzed 7-day old biofilms and found that sporulation was greater in the R-CDI strains than in the NR-CDI strains. In addition, sporulation was associated with overexpression of the key regulators of the initial steps of the sporulation pathway, spo0A and sigH, suggesting their involvement in the overproduction of spores in the biofilm. According to our results, the production of spores can be associated with recurrent CDI isolates.
In the present study, a high proportion of resistance was detected against ciprofloxacin, moxifloxacin, erythromycin, clindamycin, and rifampin, and all isolates were susceptible to tetracycline and metronidazole. No differences were detected between NR-CDI and R-CDI strains for these antimicrobial susceptibilities.
Conversely, lower susceptibility to linezolid was observed in R-CDI strains than in NR-CDI strains, and this result is relevant because linezolid is considered to be a possible drug for treatment with CDI. Interestingly, our study population has no records of exposure to this antimicrobial agent in the 12 weeks before the diagnosis of CDI. The cfr gene has been associated with resistance to linezolid and has been detected in C. difficile with a MIC up to 16 mg/L [34]. Further studies are underway to clarify the molecular mechanisms associated with this drug resistance.
In our study, the isolates from R-CDI patients showed lower susceptibility to vancomycin (MIC > 2 mg/L) more frequently than isolates from NR-CDI patients (27.78 and 9.09%, respectively); and this result is important given the wide use of vancomycin for treatment of CDI.
High MICs have been reported for moxifloxacin, rifampicin, vancomycin, and clindamycin in ribotypes 001, 017, 027, 176, 078, and 014 [35, 36]. In the present study, high MICs were detected for the same antibiotics in addition to ciprofloxacin and erythromycin. Most of the isolates obtained were ribotype 027. No difference in ribotype was found between the R-CDI and NR-CDI groups. In our population, 027 strains are associated with higher mortality rates and greater probability of R-CDI [37].
C. difficile isolates from the present study had high exposure to clindamycin, and this exposure has been associated in prior studies with a high excretion of C. difficile spores [38]. Therefore, the high use of clindamycin in our clinical setting of Mexico may be associated with the high sporulation detected.
In a 3-day biofilm model of C. difficile, the susceptibility to vancomycin has been reported, with BMICs up to 100 times higher than the corresponding planktonic MICs [8, 39, 40]. In the present study, we confirmed a reduced susceptibility to vancomycin in most strains, with BMICs up to 100 times higher than MICs. Reduced susceptibility of C. difficile [27, 41] and BMIC values six times higher than MIC have been reported for metronidazole [14]. Despite our patients having been treated with metronidazole before the diagnosis of CDI, we did not find isolates with reduced susceptibility to this antimicrobial agent in either planktonic or biofilm MIC experiments.
Several risk factors have been described for the development of CDI [42]. In our study, the consumption of cephalosporins and a greater number of previous antibiotics were risk factors for R-CDI. In addition, patients with NR-CDI more frequently used fluoroquinolones (p = 0.000) and vancomycin (p = 0.024) than those with R-CDI.
The primary limitation of this study was that cultures were not performed on fresh samples. Instead, they were frozen and stored at −20°C for up to two years with at least two freeze-thaw cycles, which could explain the low recovery of C. difficile.
In conclusion, the sporulation and overexpression of sigH, spo0A, and agrD1 were greater in R-CDI than in NR-CDI isolates. The R-CDI isolates had more reduced susceptibility to vancomycin and linezolid than the NR-CDI isolates in both planktonic cells and biofilm isolates. These factors may affect the recurrence of the infection because a greater sporulation in the protected biofilm may facilitate less spore washout from the gut and a higher likelihood of C. difficile remaining after CDI therapy has ceased.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially supported by Mexico’s National Council for Science and Technology, CONACYT, grant 284042. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–34. 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFarland LV, Elmer GW, Surawicz CM. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol. 2002;97(7):1769–75. [DOI] [PubMed] [Google Scholar]
- 3.Mac Aogain M, Moloney G, Kilkenny S, Kelleher M, Kelleghan M, Boyle B, et al. Whole-genome sequencing improves discrimination of relapse from reinfection and identifies transmission events among patients with recurrent Clostridium difficile infections. J Hosp Infect. 2015;90(2):108–16. 10.1016/j.jhin.2015.01.021 [DOI] [PubMed] [Google Scholar]
- 4.Figueroa I, Johnson S, Sambol SP, Goldstein EJ, Citron DM, Gerding DN. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin Infect Dis. 2012;55 Suppl 2:S104–9. 10.1093/cid/cis357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gil F, Pizarro-Guajardo M, Alvarez R, Garavaglia M, Paredes-Sabja D. Clostridium difficile recurrent infection: possible implication of TA systems. Future Microbiol. 2015;10(10):1649–57. 10.2217/fmb.15.94 [DOI] [PubMed] [Google Scholar]
- 6.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013;(136):1–51. 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- 7.Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS One. 2012;7(12):e50527 10.1371/journal.pone.0050527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ethapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol. 2013;195(3):545–55. 10.1128/JB.01980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donelli G, Vuotto C, Cardines R, Mastrantonio P. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol Med Microbiol. 2012;65(2):318–25. 10.1111/j.1574-695X.2012.00962.x [DOI] [PubMed] [Google Scholar]
- 10.Dapa T, Unnikrishnan M. Biofilm formation by Clostridium difficile. Gut Microbes. 2013;4(5):397–402. 10.4161/gmic.25862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowther GS, Chilton CH, Todhunter SL, Nicholson S, Freeman J, Baines SD, et al. Development and validation of a chemostat gut model to study both planktonic and biofilm modes of growth of Clostridium difficile and human microbiota. PLoS One. 2014;9(2):e88396 10.1371/journal.pone.0088396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80(8):2704–11. 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paredes-Sabja D, Cofre-Araneda G, Brito-Silva C, Pizarro-Guajardo M, Sarker MR. Clostridium difficile spore-macrophage interactions: spore survival. PLoS One. 2012;7(8):e43635 10.1371/journal.pone.0043635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenyuk EG, Laning ML, Foley J, Johnston PF, Knight KL, Gerding DN, et al. Spore formation and toxin production in Clostridium difficile biofilms. PLoS One. 2014;9(1):e87757 10.1371/journal.pone.0087757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mora-Uribe P, Miranda-Cardenas C, Castro-Cordova P, Gil F, Calderon I, Fuentes JA, et al. Characterization of the Adherence of Clostridium difficile Spores: The Integrity of the Outermost Layer Affects Adherence Properties of Spores of the Epidemic Strain R20291 to Components of the Intestinal Mucosa. Front Cell Infect Microbiol. 2016;6:99 10.3389/fcimb.2016.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pantaleon V, Soavelomandroso AP, Bouttier S, Briandet R, Roxas B, Chu M, et al. The Clostridium difficile Protease Cwp84 Modulates both Biofilm Formation and Cell-Surface Properties. PLoS One. 2015;10(4):e0124971 10.1371/journal.pone.0124971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol. 2011;193(13):3186–96. 10.1128/JB.00272-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, et al. The agr locus regulates virulence and colonization genes in Clostridium difficile 027. J Bacteriol. 2013;195(16):3672–81. 10.1128/JB.00473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darkoh C, Odo C, DuPont HL. Accessory Gene Regulator-1 Locus Is Essential for Virulence and Pathogenesis of Clostridium difficile. MBio. 2016;7(4). 10.1128/mBio.01237-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darkoh C, DuPont HL, Norris SJ, Kaplan HB. Toxin synthesis by Clostridium difficile is regulated through quorum signaling. MBio. 2015;6(2):e02569 10.1128/mBio.02569-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013;108(4):478–98; quiz 99. 10.1038/ajg.2013.4 [DOI] [PubMed] [Google Scholar]
- 22.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42(12):5710–4. 10.1128/JCM.42.12.5710-5714.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett. 1999;175(2):261–6. http://www.ncbi.nlm.nih.gov/pubmed/10386377. 10.1111/j.1574-6968.1999.tb13629.x [DOI] [PubMed] [Google Scholar]
- 24.Hammond EN, Donkor ES, Brown CA. Biofilm formation of Clostridium difficile and susceptibility to Manuka honey. BMC Complement Altern Med. 2014;14:329 10.1186/1472-6882-14-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115(8):891–9. 10.1111/j.1600-0463.2007.apm_630.x [DOI] [PubMed] [Google Scholar]
- 26.Baines SD, O’Connor R, Freeman J, Fawley WN, Harmanus C, Mastrantonio P, et al. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother. 2008;62(5):1046–52. 10.1093/jac/dkn313 [DOI] [PubMed] [Google Scholar]
- 27.Freeman J, Stott J, Baines SD, Fawley WN, Wilcox MH. Surveillance for resistance to metronidazole and vancomycin in genotypically distinct and UK epidemic Clostridium difficile isolates in a large teaching hospital. J Antimicrob Chemother. 2005;56(5):988–9. 10.1093/jac/dki357 [DOI] [PubMed] [Google Scholar]
- 28.Buchler AC, Rampini SK, Stelling S, Ledergerber B, Peter S, Schweiger A, et al. Antibiotic susceptibility of Clostridium difficile is similar worldwide over two decades despite widespread use of broad-spectrum antibiotics: an analysis done at the University Hospital of Zurich. BMC Infect Dis. 2014;14:607 10.1186/s12879-014-0607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein EJ, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. In vitro activities of daptomycin, vancomycin, quinupristin- dalfopristin, linezolid, and five other antimicrobials against 307 gram-positive anaerobic and 31 Corynebacterium clinical isolates. Antimicrob Agents Chemother. 2003;47(1):337–41. http://www.ncbi.nlm.nih.gov/pubmed/12499210. 10.1128/AAC.47.1.337-341.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, et al. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother. 2008;52(8):2813–7. 10.1128/AAC.00342-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang LL, Chen HF, Chang CY, Lee TM, Wu WJ. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother. 2004;53(3):518–21. 10.1093/jac/dkh094 [DOI] [PubMed] [Google Scholar]
- 32.Pickering DS, Wilcox MH, Chilton CH. Biofilm-derived spores of Clostridioides (Clostridium) difficile exhibit increased thermotolerance compared to planktonic spores. Anaerobe. 2018;54:169–71. 10.1016/j.anaerobe.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 33.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol. 2009;191(17):5377–86. 10.1128/JB.00597-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marin M, Martin A, Alcala L, Cercenado E, Iglesias C, Reigadas E, et al. Clostridium difficile isolates with high linezolid MICs harbor the multiresistance gene cfr. Antimicrob Agents Chemother. 2015;59(1):586–9. 10.1128/AAC.04082-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger FK, Rasheed SS, Araj GF, Mahfouz R, Rimmani HH, Karaoui WR, et al. Molecular characterization, toxin detection and resistance testing of human clinical Clostridium difficile isolates from Lebanon. Int J Med Microbiol. 2018;308(3):358–63. 10.1016/j.ijmm.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 36.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21(3):248 e9–e16. 10.1016/j.cmi.2014.09.017 [DOI] [PubMed] [Google Scholar]
- 37.Camacho-Ortiz A, Lopez-Barrera D, Hernandez-Garcia R, Galvan-De Los Santos AM, Flores-Trevino SM, Llaca-Diaz JM, et al. First report of Clostridium difficile NAP1/027 in a Mexican hospital. PLoS One. 2015;10(4):e0122627 10.1371/journal.pone.0122627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77(9):3661–9. 10.1128/IAI.00558-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathur H, Rea MC, Cotter PD, Hill C, Ross RP. The efficacy of thuricin CD, tigecycline, vancomycin, teicoplanin, rifampicin and nitazoxanide, independently and in paired combinations against Clostridium difficile biofilms and planktonic cells. Gut Pathog. 2016;8:20 10.1186/s13099-016-0102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James GA, Chesnel L, Boegli L, deLancey Pulcini E, Fisher S, Stewart PS. Analysis of Clostridium difficile biofilms: imaging and antimicrobial treatment. J Antimicrob Chemother. 2018;73(1):102–8. 10.1093/jac/dkx353 [DOI] [PubMed] [Google Scholar]
- 41.Pelaez T, Alcala L, Alonso R, Martin-Lopez A, Garcia-Arias V, Marin M, et al. In vitro activity of ramoplanin against Clostridium difficile, including strains with reduced susceptibility to vancomycin or with resistance to metronidazole. Antimicrob Agents Chemother. 2005;49(3):1157–9. 10.1128/AAC.49.3.1157-1159.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abou Chakra CN, Pepin J, Sirard S, Valiquette L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One. 2014;9(6):e98400 10.1371/journal.pone.0098400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deneve C, Delomenie C, Barc MC, Collignon A, Janoir C. Antibiotics involved in Clostridium difficile-associated disease increase colonization factor gene expression. J Med Microbiol. 2008;57(Pt 6):732–8. 10.1099/jmm.0.47676-0 [DOI] [PubMed] [Google Scholar]
- 44.Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun. 2014;82(10):4276–91. 10.1128/IAI.02323-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson S, Burman LG, Akerlund T. Induction of toxins in Clostridium difficile is associated with dramatic changes of its metabolism. Microbiology. 2008;154(Pt 11):3430–6. 10.1099/mic.0.2008/019778-0 [DOI] [PubMed] [Google Scholar]
- 46.Yun B, Oh S, Griffiths MW. Lactobacillus acidophilus modulates the virulence of Clostridium difficile. J Dairy Sci. 2014;97(8):4745–58. 10.3168/jds.2014-7921 [DOI] [PubMed] [Google Scholar]