Abstract
Paralytic Shellfish Poisoning (PSP) caused by the dinoflagellate Alexandrium catenella is a well-known global syndrome that negatively impacts human health and fishery economies. Understanding the population dynamics and ecology of this species is thus important for identifying determinants of blooms and associated PSP toxicity. Given reports of extensive genetic heterogeneity in the toxicity and physiology of Alexandrium species, knowledge of genetic population structure in harmful algal species such as A. catenella can also facilitate the understanding of toxic bloom development and ecological adaptation. In this study we employed microsatellite markers to analyze multiple A. catenella strains isolated from several sub-regions in the Gulf of Maine (GoM) during summer blooms, to gain insights into the sources and dynamics of this economically important phytoplankton species. At least three genetically distinct clusters of A. catenella were identified in the GoM. Each cluster contained representatives from different sub-regions, highlighting the extent of connectivity and dispersal throughout the region. This shared diversity could result from cyst beds created by previous coastal blooms, thereby preserving the overall diversity of the regional A. catenella population. Rapid spatiotemporal genetic differentiation of A. catenella populations was observed in local blooms, likely driven by natural selection through environmental conditions such as silicate and nitrate/nitrite concentrations, emphasizing the role of short-term water mass intrusions and biotic processes in determining the diversity and dynamics of marine phytoplankton populations. Given the wide-spread intraspecific diversity of A. catenella in GoM and potentially elsewhere, harmful algal blooms will likely persist in many regions despite global warming and changing environmental conditions in the future. Selection of different genetic lineages through variable hydrological conditions might impact toxin production and profiles of future blooms, challenging HAB control and prediction of PSP risk in the future.
Keywords: Alexandrium, harmful algal blooms, population structure, dispersal, environmental selection
1. Introduction
Paralytic Shellfish Poisoning (PSP) caused by the dinoflagellate Alexandrium catenella is a global issue that negatively impacts human health and fisheries (Hallegraeff, 1993; Van Dolah, 2007). The Gulf of Maine (GoM) is a continental shelf sea located in the northwest Atlantic where a large areas of shellfish resources are recurrently contaminated by PSP (Anderson et al., 2014a). As a consequence, the local commercial and recreational shellfishing industries regularly suffer from seafood safety issues, resulting in a loss of millions of dollars in some years. According to long-term monitoring data, A. catenella blooms in the western GoM usually start in late April, peak in June or mid-July with cell densities reaching or exceeding ten thousand cells per liter, and decline in late July (Li et al., 2014; Martin et al., 2014a). The life cycle of A. catenella includes an annual alternation between haploid vegetative cells that germinate in response to favorable conditions in early spring, and a resting stage (cyst) that is presumably formed in response to adverse environmental conditions in the field (Anderson et al., 1983; Anderson & Lindquist, 1985; Brosnahan et al., 2015). Previous physiological research on A. catenella in GoM revealed that strains from northern areas had higher toxin content and different toxin composition than strains from the south (Anderson et al., 1994; Maranda et al., 1985). Based on these differences in toxicity and other distinctions in bioluminescence and morphology, a northern and a southern cluster of A. catenella were proposed (Anderson et al. 1994). However, the dynamics of the population structure of A. catenella in GoM are still unclear, limiting our ecological understanding of the development and persistence of harmful algal blooms (HABs) in the region.
In recent years, many population genetics studies have observed spatiotemporal structure in a large variety of marine phytoplankton species (Alpermann et al., 2010; Casabianca et al., 2012; Godhe et al., 2016; Richlen et al., 2012), which have greatly challenged the idea that phytoplankton populations are panmictic. These observed spatial and/or temporal genetic differentiation of phytoplankton populations was attributed to three factors: (1) oceanographic connectivity: physical barriers such as circulation and benthic topography can determine dispersal pathways of planktonic cells, therefore determining the extent of gene flow between phytoplankton populations (Casabianca et al. 2012; Godhe et al. 2013; Sjöqvist et al. 2015); (2) Natural selection: environmental conditions can select for individuals with high fitness, resulting in adaptation of a population to the local environmental conditions and providing a competitive advantage of the native population over migrants (Blanquart et al., 2013; Sjöqvist et al., 2015); and (3) isolation by distance (IBD): gene flow and sexual recombination occur more frequently among geographically-close individuals, so distant populations may diverge due to limited gene flow (Hamilton, 2011).
Most previous studies about population genetics of marine HABs focused on spatial structure of phytoplankton populations across large spatial scales, and connectivity explains many instances of spatial differentiation of marine micro-algal populations (e.g. Nagai et al., 2009; Casabianca et al., 2012; Sjöqvist et al., 2015; Richlen et al., 2012). In addition to spatial structure, temporal differentiation has also been reported in several phytoplankton groups. In diatom populations, temporal genetic structure was linked to silicate depletion (Godhe et al. 2016; Rynearson et al., 2006), possibly indicating local adaptation. In dinoflagellate populations, temporal genetic differentiation of Gambierdiscus caribaeus in the Greater Caribbean Region was likely affected by salinity and benthic coverage of the habitat (Sassenhagen et al., 2018). Temporal population structure has also been examined in the genus Alexandrium. Dia et al. (2014) monitored the temporal population differentiation of the dinoflagellate Alexandrium minutum in two estuaries in France and showed that interannual genetic differentiation was greater than intra-bloom differentiation. Temporal differentiation in blooms of the dinoflagellate A. catenella in salt ponds near GoM was observed to occur on short (< 1 month) time scales, potentially driven by different timing of excystment, parasitism and grazing (Richlen et al., 2012). During the Alexandrium bloom in GoM in the year 2005, temporal succession of two genetically distinct sub-populations was observed on similar time scales (Erdner et al., 2011).
In this study, we investigated the genetic dynamics of Alexandrium catenella blooms in the GoM by sampling over three summer months in 2007, from several sub-regions, providing good spatial coverage of the blooms. The main objectives of this investigation were to: (1) examine temporal changes in genetic composition during local A. catenella blooms in the GoM; (2) determine the regional genetic structure and sources of A. catenella populations across the GoM; and (3) test associations between environmental conditions and genetic differentiation. Based on previous research we hypothesized that barriers to panmixia (e.g. hydrography) may weaken gene flow between northern and southern regions of the GoM and facilitate the formation of spatially-differentiated A. catenella sub-populations. Natural selection of adapted genotypes may cause short-term temporal genetic differentiation during local blooms in the GoM. By using microsatellite markers on A. catenella strains we sought to reveal the sources and dynamics of this economically important phytoplankton species in the GoM, providing insights for future bloom sustainability and toxicity under global warming and changing environmental conditions.
2. Method
2.1. Study sites
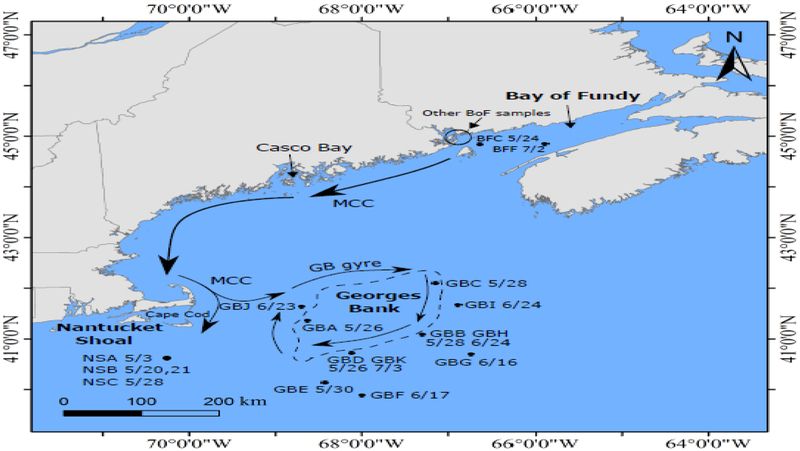
Bloom sampling focused on three sub-regions within the GoM area (Figure 1): Georges Bank (GB), Bay of Fundy (BoF) and Nantucket Shoal (NS). GB is characterized by a central bank, which is 60 meters deep and influenced by a strong clockwise gyre that retains water as long as 90 days (Brink et al., 2003). The BoF is characterized by extensive local cyst beds, a retentive hydrography (e.g. gyre), and regular nutrient inflow from tidal water masses (Anderson et al., 2014b; Martin et al., 2014a). NS is a submerged, shallow, sand and gravel ridge that acts as a topographic barrier to deep flow between the GoM and the New England continental shelf (Limeburner & Beardsley, 1982). The Main Coastal Current (MCC; Pettigrew et al., 2005) and the Gulf of Maine Coastal Plume (Keafer et al., 2005) are the dominant currents in the GoM, and they flow from the BoF in the Northeast to southern parts of the Gulf (Figure 1).
Figure 1.
The location of samples in GoM. Sampling sites are indicated with black dots/circle. GB’s location is shown in dashed outline, arrow lines indicate the Main Coastal Current (MCC) and GB gyre.
2.2. Sampling
In May 2007, an A. catenella bloom covered most parts of GB with cell concentrations up to 12,000 cells L−1, and then declined in June with advection to NS (McGillicuddy et al., 2014). In contrast, the inshore bloom along the BoF did not develop until June, after the GB bloom had traveled to NS (Martin et al., 2014b; McGillicuddy et al., 2014).
The A. catenella blooms in the surface water (0–1 m depth) were sampled at several stations in GB, BoF and NS during cruises EN435 and 437 in 2007 (Figure 1). Sampling dates and number of strains per sample were shown in Table 1. Eleven samples at GB (GBA-GBK), eight samples in BoF (BFA-BFH), and three samples in NS (NSA-NSC) were collected during the bloom season. GB samples were taken around the shallow central bank from late May to early July. Strains from GBB&GBH and GBD&GBK were obtained from the exactly same locations (Figure 1). Among BoF samples, BFC and BFF were taken during cruises, while other samples were taken from field work by Jennifer Martin. BFA, BFD and BFH were sampled from Passamaquoddy Bay, while BFG was collected near the Wolves Islands, and BFB and BFE were pooled samples from Passamaquoddy Bay and the Wolves Islands. Single A. catenella cells were isolated by micropipette and cultured in 96-well plates as in Erdner et al., 2011. A total of 761 clonal strains were established for this study, representing 18–40 clonal strains from each sample.
Table 1:
Sampling dates and number of strains per sample used in this study.
| Sample | Date | # of individuals |
|---|---|---|
| GBA | 5/26 | 32 |
| GBB | 5/28 | 31 |
| GBC | 5/28 | 33 |
| GBD | 5/26 | 36 |
| GBE | 5/30 | 24 |
| GBF | 6/17 | 30 |
| GBG | 6/16 | 40 |
| GBH | 6/24 | 30 |
| GBI | 6/24 | 30 |
| GBJ | 6/23 | 24 |
| GBK | 7/3 | 19 |
| NSA | 5/3 | 29 |
| NSB | 5/20–21 | 39 |
| NSC | 5/28 | 35 |
| BFA | 5/7 | 28 |
| BFB | 5/14 | 18 |
| BFC | 5/24 | 31 |
| BFD | 6/12 | 38 |
| BFE | 6/26 | 31 |
| BFF | 7/2 | 31 |
| BFG | 7/10 | 20 |
| BFH | 7/24 | 38 |
2.3. DNA extraction and microsatellite genotyping
In order to extract DNA, each clonal culture was harvested when the cell density exceeded 100 cells per well. DNA was extracted from 200 μL of each culture using the Generation Capture Column Kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. From the thirteen microsatellite loci found by Nagai et al. (2004), the four loci Atama 15, Atama 23, Atama 27 and Atama 39 were selected for this study based on amplification success and the observed number of alleles. These same microsatellite markers have previously been employed to examine the genetic diversity and differentiation of A. catenella populations sampled in 2005 to 2007 from the GoM and Nauset Estuary on Cape Cod, MA, USA (Erdner et al., 2011; Richlen et al., 2012). PCR reactions contained ~5 ng of template DNA, 0.2mmol L−1 of each dNTP, 0.5 μmol L−1 of each designed primer pair, with one primer labeled with 6FAM, NED, PET, or VIC, 1x PCR buffer (10mmol L−1 Tris-HCl, pH 8.3, 500 mmol L−1 KCl, 15 mmol L−1 MgCl2, 0.01% w/v gelatin), and 0.25 U of Ampli Taq Gold (Applied Biosystems, Foster City, CA), in 10 μL total volume. The PCR cycling conditions were as follows: 10 min at 94 °C, 10 cycles of 30 sec at 94°C, 30 sec at 60°C and 1 min at 72°C, and then 28 cycles of 30 sec at 94°C, 30 sec at the primer-specific annealing temperature (Nagai et al., 2004), and 1 min at 72°C, and a final elongation for 5 min at 72°C. In order to determine if amplification was successful, PCR products were run on a 1% TAE agarose gel. For allele size analysis, PCR products were diluted 3–5x with nuclease-free water, and 1 μL of diluted product was mixed with 0.25 μL 500 LIZ Size Standard and 8.75 μL Hi-Di Formamide, and then analyzed using an ABI 3730xi DNA Analyzer (Applied Biosystems, Foster City, CA). Allele sizes were determined using the program FPMiner (2005; Bioinfor-Soft LLC, Beaverton, OR). Microsatellite data is available from the Mendeley Data repository, doi: 10.17632/nhwkm3nczj.1.
2.4. Microsatellite Data Analysis
2.4.1. Genetic diversity
Allele diversity (Nei 1973) was calculated based on all individuals as well as only unique genotypes in each genetic sub-cluster using POPGENE v. 1.32 (Yeh et al., 1997). Clonal diversity was defined as the ratio of the number of unique four-locus genotypes (G) to the total number of genotypes analyzed (N) with a threshold of one base to distinguish clones in Arlequin v. 3.5.1.2 (Excoffier et al., 2005). This parameter was also calculated after adjusting the sample size of each genetic sub-cluster to N=59 by using the R function “sample”. Multilocus linkage disequilibrium was assessed by the Monte Carlo method in LIAN v. 3.5 with 10,000 resamplings (Haubold & Hudson, 2000), while linkage disequilibrium between pairs of loci was determined in GenePop v. 4.5 with Markov Chain parameters of 1000 dememorisation steps and 100 batches with 1000 iterations per batch (Rousset et al., 2008).
2.4.2. Inference of population structure
To identify the genetic clusters in the GoM dataset, PCAGEN was applied to run a principal components analysis (PCA) based on allele frequencies, with 10000 randomisations used to determine the significance of the inertia of each axis (Goudet, 1999).
Genetic composition and structuring were examined using Bayesian cluster analysis implemented in STRUCTURE 2.3.2 (Pritchard et al., 2000). The number of potential clusters (K) was first assessed using 10 runs performed at every K value from 1 to 10, each with a burn-in period of 100,000 steps and 200,000 Markov Chain Monte Carlo repetitions. Simulations were conducted using the admixture model with the locprior option, assuming correlated allele frequencies among populations. The calculation of the most likely value of K was performed in Structure Harvester Web v0.6.94 (Earl, 2012) by the method of ad hoc statistic Δ K (Evanno et al., 2005).
The partitioning of genetic variance within and among genetic clusters was assessed using the analysis of molecular variance (AMOVA) implemented in the software GenoDive 2.0b27 (Meirmans & Van Tienderen, 2004).
Genetic clustering of A. catenella populations in the GoM was further examined by performing pairwise comparisons. Wright’s FST (Wright, 1949) was used to identify fixation of genetic differences, calculated in Arlequin v. 3.5.1.2 with 10,000 permutations to test for significance. The Fisher’s exact test (Raymond, 1995) was used to assess differences in allele frequencies between clusters, performed in GenePop v. 4.5 using Markov Chain parameters of 1000 dememorisation steps and 100 batches with 1000 iterations per batch (Rousset et al., 2008). All p-values were Bonferroni corrected to account for multiple comparisons.
2.5. Environmental effects
Environmental data were obtained from http://grampus.umeoce.maine.edu/gomtox/gomtox.htm. We included nitrate and nitrite concentrations (μM), temperature (°C), salinity, water density anomaly measured as δT (kg m−3), ammonium (μM), chlorophyll a (μg L−1), phaeopigments (μg L−1), silicate (μM) and phosphate (μM) from surface water of a subset of the same sampling stations as used for isolation of algal strains (GBA, GBB, GBC, GBD, GBE, GBG, GBH, GBJ, GBK and BFC) (Table 2). Environmental data were not available from the other stations, and therefore could not be included in this analysis.
Table 2:
Environmental data of samples from GoM (obtained from the University of Maine GOMTOX Data Archives http://grampus.umeoce.maine.edu/gomtox/gomtox.htm).
| Nitrate+Nitrite (μM) |
Silicate (μM) |
Phosphate (μM) |
Ammonium (μM) |
Temp (°C) |
Salinity (psu) |
Sigmat (kg m−3) |
Chlorophyll a (μg L−1) |
Phaeopigments (μg L−1) |
|
|---|---|---|---|---|---|---|---|---|---|
| GBA | 0.10 | 0.40 | 0.09 | 0.20 | 8.62 | 32.98 | 25.59 | 3.76 | 0.05 |
| GBB | 0.09 | 0.90 | 0.10 | 0.21 | 9.62 | 32.97 | 25.43 | 2.16 | 0.32 |
| GBC | 1.47 | 1.22 | 0.38 | 0.59 | 8.17 | 33.14 | 25.79 | 4.61 | 0.60 |
| GBD | 0.15 | 10.53 | 0.20 | 1.35 | 9.29 | 33.06 | 25.55 | 5.05 | 0 |
| GBE | 0.09 | 6.83 | 0.09 | 0.40 | 10.62 | 32.80 | 25.12 | 1.82 | 0.13 |
| GBG | 0.09 | 1.13 | 0.11 | 0.72 | 6.99 | 31.95 | 25.02 | 0.80 | 0.67 |
| GBH | 0.62 | 0.95 | 0.10 | 0.61 | 11.57 | 32.81 | 24.97 | 2.11 | 0 |
| GBJ | 0.11 | 1.40 | 1.22 | 0.28 | 11.73 | 32.38 | 24.61 | 0.77 | 0.03 |
| GBK | 0.09 | 0.03 | 0.03 | 0.04 | 14.04 | 32.90 | 24.56 | 1.48 | 0.26 |
| BFC | 6.63 | 10.17 | 0.41 | 0.16 | 6.29 | 29.58 | 23.24 | 1.77 | 0 |
To understand the impact of environmental conditions on the population structure of A. catenella, the relationship between genetic structure and environmental variables was tested using a distance based redundancy analysis (dbRDA) (Legendre & Andersson, 1999). Distances between samples based on the genetic data were calculated with a principal coordinate analysis (PCoA) using the R package adegenet 1.0.1 (Jombart, 2008). The relationship between the coordinates of samples from the PCoA and the environmental variables was determined by dbRDA using the R package vegan 2.4–1 (Oksanen et al., 2016).
3. Results
3.1. Genetic differentiation and population dynamics
In this study, 761 isolates were genotyped using four different microsatellite loci. Strains that had any null alleles were removed from the data set, resulting in a final set of 667 individuals. All four loci were polymorphic in all samples. There were 158 unique genotypes in the entire data set resulting in an overall clonal diversity of 0.23. Multilocus linkage disequilibrium was not detected (IAS =0.0069, p=0.179), nor was linkage disequilibrium between pairs of loci.
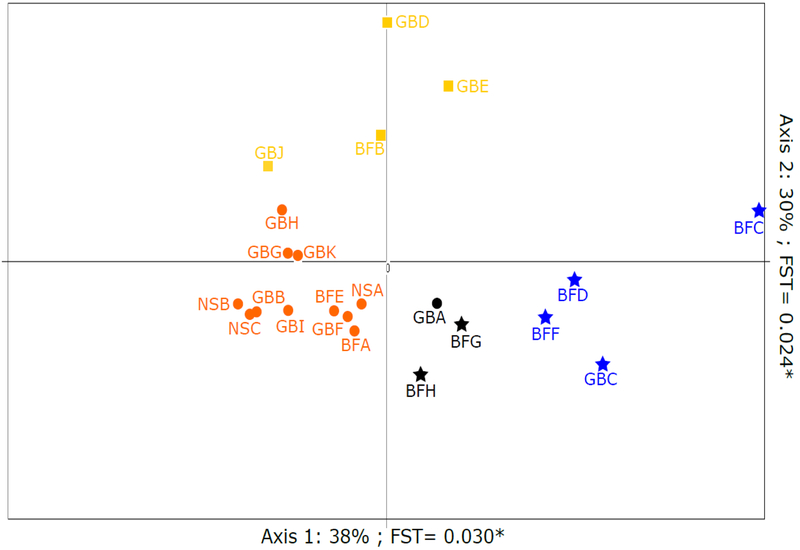
The A. catenella populations displayed more rapid and complicated differentiation over the course of the 2007 blooms compared with the temporal succession from early to late sub-populations as observed in the 2005 bloom across the GoM (Erdner et al., 2011). To identify genetic structure of A. catenella populations in the GoM, PCA implemented in the program PCAGEN was performed (Figure 2). The first two axes of the PCA were both significant and explained 38% and 30% of the total variation in FST, respectively. Three clusters of samples were identified: (1) GBB, GBF, GBG, GBH, GBI, GBK, BFA, BFE, NSA, NSB and NSC (orange samples in Figure 2); (2) GBC, BFC, BFD and BFF (blue samples in Figure 2); (3) BFB, GBJ, GBD and GBE (yellow samples in Figure 2). However, GBA, BFG and BFH (black samples in Figure 2) were in a transitional location between two of the clusters. In order to further examine the genetic differentiation among these three samples, the clustering pattern was investigated using an AMOVA. When GBA was added to Cluster (1) and BFH&BFG were added to Cluster (2), differentiation among clusters was significant (Rct=0.022, p=0.008), but not significant within each cluster (Rsc=0.009, p=0.122). Other grouping patterns were also tested but were less supported by AMOVA based on distribution of within-cluster variation and among-cluster variation (data not shown). The PCA and AMOVA analyses indicated differentiation of the GoM dataset into three genetic clusters based on their genetic similarity, and they were re-named by the location of the majority of samples: (1) Southern Cluster: GBA, GBB, GBF, GBG, GBH, GBI, GBK, BFA, BFE, NSA, NSB and NSC; (2) Northern Cluster: GBC, BFC, BFD, BFF, BFG and BFH; (3) Mixed Cluster: GBD, GBE, GBJ and BFB. These genetic clusters were not entirely grouped according to geography, but instead comprised samples from different sub-regions in the GoM.
Figure 2:
Principal Component Analysis (PCA) of a covariance matrix from the genetic data of GoM. Both axes are significant: Axis 1: percent inertia=38%, FST =0.030, p=0.004; Axis 2: percent inertia=30%, FST =0.024, p=0.000. Shapes correspond to the genetic clusters suggested by the AMOVA analyses (Circles=Southern Cluster; Stars=Northern Cluster; Squares=Mixed Cluster).
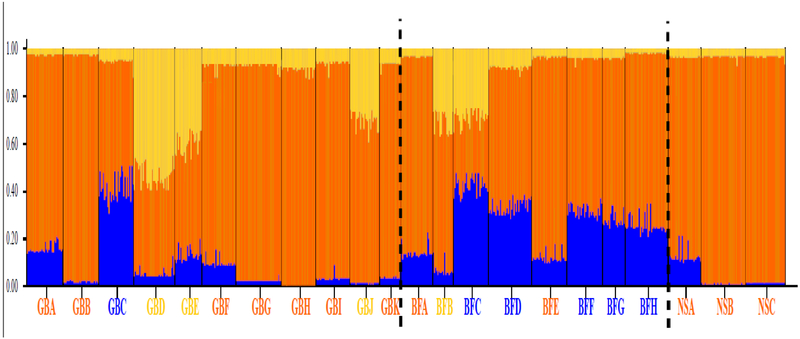
Bayesian analyses implemented in the program STRUCTURE were performed to examine the genetic composition and the relationships of samples (Figure 3). A K value of three was identified as the most likely number of “ancestral” populations in the GoM dataset using the Evanno method. The result of the STRUCTURE bar plot from “K=3” was similar to the “three-cluster” pattern described above, i.e. samples with similar proportional ancestry membership corresponded to the groupings identified by PCA and AMOVA. Subtle population structure was apparent within clusters. BFC’s unique genetic composition in STRUCTURE analysis was reflected in its divergence in the PCA plot. The transitional locations of BFG and BFH in PCA analysis were also shown in STRUCTURE with mixed ancestral populations.
Figure 3:
Population structure of GoM determined by Bayesian cluster analysis. Results of STRUCTURE analysis are presented for K=3. Dashed lines separate samples from different sub-regions in GoM. Colors of sample names correspond to the genetic clusters suggested by the PCA & AMOVA analyses (Orange=Southern Cluster; Blue=Northern Cluster; Yellow=Mixed Cluster).
Pairwise FST values were calculated between the Northern, Southern and Mixed Clusters. All pairwise FST values between the genetic clusters were significant and ranged from 0.051 to 0.088 (Table 3). Additionally, the degree of differentiation based on Fisher’s exact test was highly significant between these clusters. Allele frequencies from each genetic cluster were compared in detail. All loci in Southern and Northern Clusters had private alleles, whereas none were observed the Mixed Cluster. For example, of the 21 alleles detected at locus Atama15 in GoM, 8 alleles were found only in Southern Cluster while another 4 were found only in Northern Cluster. In order to understand the sources of the observed genetic differentiation, FST values based on each locus were calculated (data not shown). The divergence between Southern and Northern Cluster was driven by locus Atama27, as the differentiation was not significant based on other microsatellites loci. Specifically, allele size 162 (87%) was most common in Southern Cluster while allele sizes 162 (56%) and 160 (38%) dominated in Northern Cluster. All loci except Atama 39 were the driving force to differentiate Northern and Mixed Cluster, while only Atama 15 and Atama23 contributed significantly to the genetic differentiation between Southern and Mixed Cluster.
Table 3:
Pairwise comparisons of genotypic differentiation between clusters in GoM. Lower diagonal: FST values. Upper diagonal: p-values of the Fisher’s exact test. Bold characters denote significance at α=0.05 after Bonferroni correction.
| Southern cluster | Northern cluster | Mixed cluster | |
|---|---|---|---|
| Southern cluster | -- | Highly sign. | Highly sign. |
| Northern cluster | 0.051 | -- | Highly sign. |
| Mixed cluster | 0.061 | 0.088 | -- |
Within sub-regions of the GoM, significant changes in genetic composition were observed during local blooms. GB samples contained representatives from all of the three genetic clusters defined above: (1) GB-Southern: GBA, GBB, GBF, GBG, GBH, GBI and GBK; (2) GB-Mixed: GBD, GBE and GBJ; and (3) GB-Northern: GBC. The pairwise FST analysis was repeated for the sub-clusters in GB to clarify local population structure. The results confirmed the significant differentiation of this local bloom into the three distinct sub-clusters, with FST values ranging from 0.058 to 0.134 (data not shown). To gain insights into temporal structure of the GB bloom, early (5/26–5/30: GBA, GBB, GBC, GBD and GBE) and late samples (6/16–7/3: GBF, GBG, GBH, GBI, GBJ and GBK) were investigated separately. During the early bloom period, all three genetically distinct sub-clusters described above were represented: (1) GB-Southern: GBA and GBB; (2) GB-Northern: GBC; (3) GB-Mixed: GBD and GBE. Pairwise FST comparisons among sub-clusters that included only samples from the early bloom confirmed patchy distribution of spatially differentiated clusters over the bank during this period, with FST values ranging from 0.047 to 0.137. Twenty days later, during the late bloom period, a more homogenous distribution of A. catenella was observed, as all samples were identified as GB-Southern except GBJ (GB-Mixed).
BoF samples also contained all three genetic clusters based on the results of PCA and AMOVA analyses of the entire data set: (1) BF-Southern: BFA and BFE; (2) BF-Mixed: BFB; and (3) BF-Northern: BFC, BFD, BFF, BFG and BFH. The pairwise FST analysis was repeated for the BoF dataset, and indicated significant differentiation of these three sub-clusters, with FST values ranging from 0.029 to 0.058.
The genetic diversity of different sub-clusters varied, however (Table 4). The Southern sub-clusters from each sub-region exhibited the lowest Ht: GB (Ht=0.49) and BoF (Ht=0.47). However, the lowest values of Ht (unique) in the respective sub-regions were found in GB-Northern and BoF-Mixed. When strain numbers were equalized to 59, clonal diversity was higher in GB-Mixed (0.63), and distinctly lower in GB-Southern (0.46) among all sub-regions.
Table 4:
Number of strains per sub-clusters (N), number of different clonal lineages (G), overall clonal diversity (G/N), allele diversity calculated with all strains (Ht) and excluding replicated genotypes (Ht unique).
| N | G | G/N | G (N=59) | G/N (N=59) | Ht | Ht (unique) | |
|---|---|---|---|---|---|---|---|
| GB-Southern | 212 | 72 | 0.34 | 27 | 0.46 | 0.49 | 0.63 |
| GB-Mixed | 86 | 45 | 0.52 | 37 | 0.63 | 0.55 | 0.64 |
| GB-Northern | 31 | 19 | 0.61 | 19 | N/A | 0.52 | 0.5 |
| BoF-Southern | 59 | 35 | 0.59 | 35 | 0.59 | 0.47 | 0.59 |
| BoF-Mixed | 18 | 13 | 0.72 | 13 | N/A | 0.50 | 0.53 |
| BoF-Northern | 158 | 62 | 0.39 | 30 | 0.51 | 0.51 | 0.63 |
| NS-Southern | 103 | 43 | 0.42 | 33 | 0.56 | 0.45 | 0.59 |
3.2. Environmental Correlation
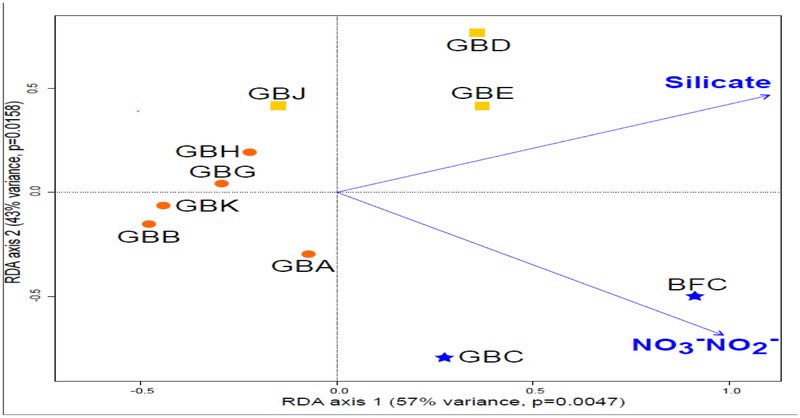
Nine environmental factors were collected from ten samples in the GoM. Some samples stood out due to relatively high nutrient concentrations: GBC with 1.47 μM of nitrate and nitrite; GBD with relatively high silicate concentrations (10.53 μM); and BFC with 6.63 μM of silicate and 10.17 μM of nitrate and nitrite (Table 2).
The impact of variable environmental conditions on the population genetic structure of A. catenella populations was assessed using a distance-based redundancy analysis (dbRDA). Among nine environmental variables, only silicate (P=0.0068), and nitrate and nitrite (P=0.0007) concentrations emerged as factors that were significantly associated with the genetic structure (Figure 4). Silicate concentration distinguished GBD and GBE (Mixed Cluster) from the other samples, while nitrate and nitrite concentration primarily drove the divergence of GBC and BFC (Northern Cluster), although the silicate concentration in BFC was also comparatively high.
Figure 4:
Environmental association analysis based on distance-based Redundancy Analysis (dbRDA). RDA plot for significant environmental factors (silicate, P=0.0068; nitrate+nitrite, P=0.0007). Colors correspond to the clusters suggested by the STRUCTURE analyses (Orange=Southern Cluster; Blue=Northern Cluster; Yellow=Mixed Cluster).
4. Discussion
This study investigated regional population genetic structure of A. catenella blooms distributed throughout the GoM. Spatiotemporal genetic differentiation of A. catenella populations was analyzed to describe the dispersal of A. catenella cells in the GoM as well as the influence of environmental conditions on their population structure. At least three genetically distinct clusters of A. catenella were identified in the GoM based on PCA analysis, STRUCTURE plots, fixation indices of genetic differentiation, and AMOVA. Significant short-term temporal and spatial differentiation was observed within local blooms, and linked to environmental conditions. Across the region as whole, representatives of the three genetic clusters were detected at all locations, indicating significant gene flow that could arise from hydrographic connectivity and the presence of resting (cyst) stages.
Circulation around GB is characterized by a strong gyre that is expected to homogenize A. catenella bloom populations, however, small-scale spatial differentiation into several sub-clusters was observed throughout the bloom on this bank. The early bloom period was characterized by three spatial sub-clusters, while the late bloom period was mostly characterized by one homogeneous population. This spatiotemporal variation in population structure at GB may be influenced by many factors, among which nitrate, nitrite and silicate concentrations may play a key role (Figure 4). During most of the bloom season in GB, abundant and stable supplies of ammonium and phosphate as well as low supply of nitrite and nitrate were found all over the bank (McGillicuddy et al. 2014; Townsend et al. 2014), which coincided with the presence of GB-Southern Cluster. Hence, the A. catenella cells from GB-Southern Cluster were likely utilizing ammonium instead of nitrite or nitrate as primary nitrogen source. However, GB-Northern Cluster, located at the Northeast Peak of GB, coincided with significantly higher nitrite and nitrate concentrations, which may have been derived from an inflow of nitrate and nitrite-rich deeper water masses, i.e. Warm Slope Water, onto the Bank along the Northern Flank and Northeast Peak (Hu et al., 2008). Since laboratory experiments have shown differential preference of A. tamarense strains for different nitrogen sources (Leong et al., 2004), high nitrite and nitrate concentrations may select for distinct Alexandrium strains, facilitating the prevalence of the selected alleles. Occasional increases in silicate concentration on GB may also indirectly select GB-Mixed genotypes, some of which were present at the beginning of the A. catenella bloom in early summer, co-existing and likely competing with the annual spring diatom bloom (Townsend et al., 2014). This distribution pattern of silicate may result from regeneration of silicate from diatom frustules produced in spring bloom (Gettings et al., 2014). Short-term variations in nutrient concentrations may thus have rapidly selected for distinct genotypes, which were present before in low concentrations, and disrupted the otherwise homogenous population structure at GB. Many previous studies have described the effect of natural selection on the change of genetic structure within short periods (Rynearson et al., 2006; Godhe et al., 2016; Sassenhagen et al., 2018). For example, changes in environmental conditions such as salinity could rapidly select favorable genotypes and therefore drive genetic differentiation of Gambierdiscus caribaeus in the Greater Caribbean Region (Sassenhagen et al., 2018). Succession of genetically distinct populations of the diatom Skeletonema marinoi was observed even within fifteen days after a decline in silicate concentrations in the Baltic Sea (Godhe et al., 2016).
NS samples were not genetically different from the dominant cluster on GB (GB-Southern), thus strong algal dispersal and gene flow strengthened by the Gulf of Maine Coastal Plume (Keafer et al., 2005) or other currents might occur between these two sub-regions. Stable genetic composition of the A. catenella population in NS during the sampling period may reflect overall stable local environmental conditions. However, since no information about the NS cyst bed and transport through sub-subface layers are available, the sources of NS populations still need to be explored.
The BoF is considered to retain phytoplankton cells through the Minas Basin cyclonic gyre (Aretxabaleta et al., 2009; Townsend et al., 2014) and an extensive cyst seedbed (Anderson et al., 2014b; Martin et al., 2014a), potentially restricting gene flow yet preserving high genetic diversity of algal species (Sundqvist et al., 2018). Our results showed significant temporal genetic differentiation in the BoF among samples collected at roughly the same location. This pattern may result from induction of sexual reproduction by unfavorable conditions. In Alexandrium, environmentally induced encystment could strengthen the effects of natural selection on dominant genotypes by removing lineages that are less successful under the prevailing conditions. At the same time, some genotypes will be favored and accelerate vegetative growth. This characteristic would facilitate the domination of the population by a few successful lineages, effectively reducing gene flow between sub-clusters (Erdner et al., 2011). The turnover rates of genetically distinct sub-clusters were as short as seven days in the BoF. Such rapid changes in population structure may facilitate the persistence of phytoplankton populations in variable marine environments (Rynearson & Armbrust, 2005). Temporal differentiation of phytoplankton populations has been observed in several different microalgal species from many locations around the world (Godhe et al. 2016; Richlen et al. 2012; Rynearson & Armbrust, 2005). In particular, rapid (õne week) temporal differentiation in populations of the dinoflagellate A. catenella was also observed in Cape Cod MA (USA) salt pond systems, which may have been facilitated by early induction of sexuality and different timing of germination that preserved the genetic diversity of local populations (Richlen et al., 2012).
Environmental data were unfortunately not available from most samples from BoF for correlation analysis with the observed population structure. The only sample with available environmental data (BFC) was characterized by much higher nitrate and nitrite conditions – as was GBC – which might have selected for comparatively similar genotypes in both locations. Thus, variable nutrient concentrations might have induced the observed rapid temporal genetic differentiation of A. catenella populations in the BoF throughout the sampling period, which could have been facilitated by local cyst beds with high genotype diversity. In order to test this hypothesis, further research should be directed to investigating the genetic diversity of the resting cysts in the BoF.
Although GB and the BoF are located at the southern and northeastern regions of the GoM respectively, at least 400 km apart from each other, several samples from both sites were very similar to each other genetically and belonged to the same clusters, which may indicate algal dispersal across the region. Studies of the population genetic structure of the dinoflagellate Cochlodinium polykrikoides across the Sea of Japan found evidence for strong dispersal and gene flow over a distance of more than 600 km, highlighting the important role of Tsushima Warm Current on the connectivity of algal populations (Nagai et al., 2009). The BoF is located upstream of the MCC, the prevailing current that flows from Northeast to southern parts of the Gulf, and the bloom in BoF is supported by an extensive local cyst bed and a retentive gyre at the mouth of the Bay. The gyre is leaky, however (Aretxabaleta et al., 2009), so there is potential for dispersal from this northern area to affect the initiation and genetic structure of distant GB blooms, for example through the transport of vegetative cells from BoF to GB during the bloom.
However, the bloom on GB was extensive in May 2007 when there was near absence of A. catenella in the coastal waters of the GoM including BoF (McGillicuddy et al., 2014). Furthermore, during the bloom season of that year no evidence of A. catenella cells was found in surface waters between Casco Bay and Cape Cod, which is the main circulation pathway connecting BoF and GB-NS (McGillicuddy et al., 2014). Additionally, Li et al. (2014) studied coastal ocean connectivity in the GoM using surface numerical particle tracking and found that the circulation significantly retained particles within the BoF in 2007. As the gyre-like circulation in the BoF may serve as a barrier reducing surface connectivity between BoF and south-western parts of the GoM in 2007, surface cells from the BoF may not be the source of cells blooming on GB. However, the transport of cells through sub-surface layers between the BoF and GB cannot be excluded due to a lack of information about this process.
Other potential explanations for observed patterns of genetic connectivity could be the formation of cyst beds on GB or inshore areas including BoF and mid-coast Maine (Anderson et al., 2014b) that contained a mix of resting stages from different genetic clusters. During summer 2005, the GoM experienced the largest A. catenella bloom in the preceding three decades, spanning over 700 km of coastline. In that year, temporal succession of different inshore A. catenella sub-populations was observed during the bloom (Erdner et al., 2011). These differentiated blooms could have resulted in the formation of cysts with high genotype diversity, allowing rapid changes in population structure through natural selection of germinating cells. Although the bloom in 2006 was smaller than in other years, numerical simulations showed a significant number of particles traveling from BoF to the western GoM that year (Li et al., 2014), which may have further enhanced genetic diversity in the downstream cyst beds. Similarly, low to moderate algal dispersal via the MCC across the GoM over many years might have contributed to mixed cysts beds throughout the region.
Previous studies have found significant numbers of A. catenella cysts in benthic nepheloid layers (BNLs) throughout the GoM, representing a deep interconnection between BoF, MCC and south-central regions of the gulf (Pilskaln et al., 2014). Resuspended cysts may therefore serve as bloom inoculum on GB. Although cyst concentrations in the sediments of GB were roughly two orders of magnitude smaller than in the coastal GoM (Anderson et al., 2014b; McGillicuddy et al., 2014), local germination might still contribute at least partially to the bloom on GB. Furthermore, even low cyst concentrations might be sufficient for inoculation of a bloom due to rapid exponential growth of microalgae under favorable environmental conditions. McGillicuddy et al. (2014) also hypothesized that cysts could be transported to the bank from the adjacent deep basins by tidal pumping. Our results suggest significant dispersal and gene flow throughout the GoM, resulting in the absence of endemic populations. Meanwhile, the results described herein also highlight the need for a comprehensive cyst bed investigation including cyst abundance, genetic diversity and transport pathway across the GoM.
5. Conclusions
At least three genetically distinct clusters of A. catenella were identified in the GoM. Each cluster contains representatives from different sub-regions, highlighting the extent of connectivity and dispersal throughout the GoM. This shared diversity could result from cyst beds containing a mix of resting stages from different genetic clusters. The cyst beds would preserve the overall diversity of the regional A. catenella population and allow selection of physiologically and phenotypically different genotypes based on local conditions. Rapid temporal and spatial genetic differentiation of A. catenella populations, as observed in local blooms, was likely associated with natural selection through environmental conditions such as silicate and nitrate/nitrite concentrations, emphasizing an important role of the changing nutrient environment in determining the diversity and dynamics of marine phytoplankton populations. This observed short-term genetic differentiation may be indicative of patchiness of different water masses and biotic processes during the development of the bloom. Given the wide-spread intraspecific diversity of A. catenella in the GoM and potentially elsewhere, blooms of this species will likely persist in many regions despite global warming and changing environmental conditions in the future. Furthermore, selection of different genetic lineages through variable environmental conditions might impact toxin production and profiles of future blooms, challenging HAB control and prediction of PSP risk with changeable toxicity effect and persistent bloom dynamics.
Highlights.
Each of the three genetically distinct clusters of Alexandrium catenella identified in the Gulf of Maine contained representatives from different sub-regions, highlighting the extent of connectivity and dispersal throughout the region.
This shared diversity could result from cyst beds containing a mix of resting stages from different genetic clusters with selection of different genotypes based on local conditions.
Rapid spatiotemporal genetic differentiation of A. catenella populations was observed in sub-regional blooms, likely driven by natural selection through nutrient concentrations
6. Acknowledgement
We thank David Townsend for collection and provision of environmental data of sampling stations, and Bruce Keafer, Kerry Norton and Dave Kulis for their hard work in obtaining the samples used in this study. We also gratefully acknowledge the work of captains and crews of the R/V Endeavor. This study was supported by National Science Foundation [grant numbers OCE-0430724, OCE-0850421, OCE-0911031, OCE-1314642, OCE1840381] and National Institutes of Health [grant numbers 1P50-ES012742–01, 1P50-ES021923–01, and 1P01-ES028938–01] through the Woods Hole Center for Oceans and Human Health. Funding was also provided by NOAA Grant [NA06NOS4780245, NA11NOS4780061 and NA15NOS4780181 ]. This is contribution ECO938 from the US ECOHAB program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alpermann TJ, Tillmann U, Beszteri B, Cembella AD, & John U (2010). PHENOTYPIC VARIATION AND GENOTYPIC DIVERSITY IN A PLANKTONIC POPULATION OF THE TOXIGENIC MARINE DINOFLAGELLATE ALEXANDRIUM TAMARENSE (DINOPHYCEAE)1. Journal of Phycology, 46(1), 18–32. [Google Scholar]
- Anderson DM, Chisholm SW, & Watras CJ (1983). Importance of life cycle events in the population dynamics of Gonyaulax tamarensis. Marine Biology, 76(2), 179–189. [Google Scholar]
- Anderson DM, Couture DA, Kleindinst JL, Keafer BA, McGillicuddy DJ Jr., Martin JL, … Solow AR (2014a). Understanding interannual, decadal level variability in paralytic shellfish poisoning toxicity in the Gulf of Maine: The HAB Index. Deep Sea Research Part II: Topical Studies in Oceanography, 103, 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Keafer BA, Kleindinst JL, McGillicuddy DJ, Martin JL, Norton K, … Butman B (2014b). Alexandrium fundyense cysts in the Gulf of Maine: Long-term time series of abundance and distribution, and linkages to past and future blooms. Deep Sea Research Part II: Topical Studies in Oceanography, 103, 6–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Kulis DM, Doucette GJ, Gallagher JC, & Balech E (1994). Biogeography of toxic dinoflagellates in the genus Alexandrium from the northeastern United States and Canada. Marine Biology, 120(3), 467–478. [Google Scholar]
- Anderson DM, & Lindquist NL (1985). Time-course measurements of phosphorus depletion and cyst formation in the dinoflagellate Gonyaulax tamarensis Lebour. Journal of Experimental Marine Biology and Ecology, 86(1), 1–13. [Google Scholar]
- Aretxabaleta AL, McGillicuddy DJ, Smith KW, Manning JP, & Lynch DR (2009). Model simulations of the bay of fundy gyre: 2. Hindcasts for 2005–2007 reveal interannual variability in retentiveness. Journal of Geophysical Research: Oceans, 114(9), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, & Gandon S (2013). A practical guide to measuring local adaptation. Ecology Letters, 16(9), 1195–1205. [DOI] [PubMed] [Google Scholar]
- Brink KH, Limeburner R, & Beardsley RC (2003). Properties of flow and pressure over Georges Bank as observed with near-surface drifters. Journal of Geophysical Research, 108(C11), 8001. [Google Scholar]
- Brosnahan ML, Velo-Suárez L, Ralston DK, Fox SE, Sehein TR, Shalapyonok A, … Anderson DM (2015). Rapid growth and concerted sexual transitions by a bloom of the harmful dinoflagellate Alexandrium fundyense (Dinophyceae). Limnology and Oceanography, 60(6), 2059–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casabianca S, Penna A, Pecchioli E, Jordi A, Basterretxea G, & Vernesi C (2012). Population genetic structure and connectivity of the harmful dinoflagellate Alexandrium minutum in the Mediterranean Sea. Proceedings of the Royal Society B: Biological Sciences, 279(1726), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dia A, Guillou L, Mauger S, Bigeard E, Marie D, Valero M, & Destombe C (2014). Spatiotemporal changes in the genetic diversity of harmful algal blooms caused by the toxic dinoflagellate Alexandrium minutum. Molecular Ecology, 23(3), 549–560. [DOI] [PubMed] [Google Scholar]
- Earl DA (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. [Google Scholar]
- Erdner DL, Richlen M, McCauley LAR, & Anderson DM (2011). Diversity and Dynamics of a Widespread Bloom of the Toxic Dinoflagellate Alexandrium fundyense. PLoS ONE, 6(7), e22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EVANNO G, REGNAUT S, & GOUDET J (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology, 14(8), 2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, & Schneider S (2005). Arlequin ( version 3 . 0 ): An integrated software package for population genetics data analysis, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Gettings RM, Townsend DW, Thomas MA, & Karp-Boss L (2014). Dynamics of late spring and summer phytoplankton communities on Georges Bank, with emphasis on diatoms, Alexandrium spp., and other dinoflagellates. Deep-Sea Research Part II: Topical Studies in Oceanography, 103, 120–138. [Google Scholar]
- Godhe A, Egardt J, Kleinhans D, Sundqvist L, Hordoir R, & Jonsson PR (2013). Seascape analysis reveals regional gene flow patterns among populations of a marine planktonic diatom. Proceedings of the Royal Society B: Biological Sciences, 280(1773), 20131599–20131599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godhe A, Sjöqvist C, Sildever S, Sefbom J, Harðardóttir S, Bertos-Fortis M, … Kremp A (2016). Physical barriers and environmental gradients cause spatial and temporal genetic differentiation of an extensive algal bloom. Journal of Biogeography, 43(6), 1130–1142. [Google Scholar]
- Goudet J (1999). PCA-GEN for Windows, V. 1.2 Institute of Ecology, Univ. of Lausanne. [Google Scholar]
- Hallegraeff GM (1993). A review of harmful algal blooms and their apparent global increase*. Phycologia, 32(2), 79–99. [Google Scholar]
- Hamilton M (2011). Population genetics. John Wiley & Sons. [Google Scholar]
- Haubold B, & Hudson RR (2000). multilocus data, 16(9), 847–848. [DOI] [PubMed] [Google Scholar]
- Hu S, Townsend DW, Chen C, Cowles G, Beardsley RC, Ji R, & Houghton RW (2008). Tidal pumping and nutrient fluxes on Georges Bank: A process-oriented modeling study. Journal of Marine Systems, 74(1–2), 528–544. [Google Scholar]
- Jombart T (2008). Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24(11), 1403–1405. [DOI] [PubMed] [Google Scholar]
- Keafer BA, Churchill JH, McGillicuddy DJ, & Anderson DM (2005). Bloom development and transport of toxic Alexandrium fundyense populations within a coastal plume in the Gulf of Maine. Deep-Sea Research Part II: Topical Studies in Oceanography, 52(19–21 SPEC. ISS.), 2674–2697. [Google Scholar]
- Legendre P, & Andersson MJ (1999). Distance-based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecological Monographs, 69(1), 1–24. [Google Scholar]
- Leong SCY, Murata A, Nagashima Y, & Taguchi S (2004). Variability in toxicity of the dinoflagellate Alexandrium tamarense in response to different nitrogen sources and concentrations. Toxicon, 43(4), 407–415. [DOI] [PubMed] [Google Scholar]
- Li Y, He R, & Manning JP (2014). Coastal connectivity in the Gulf of Maine in spring and summer of 2004–2009. Deep-Sea Research Part II: Topical Studies in Oceanography, 103, 199–209. [Google Scholar]
- Limeburner R, & Beardsley RC (1982). The Seasonal hydrography and circulation over Nantucket Shoals. J. Mar. Res, 40, 371–406. [Google Scholar]
- Maranda L, Anderson DM, & Shimizu Y (1985). Toxicity of Gonyaulax tamarensis clones from eastern North American waters. Toxic Dinoflagellates, 2, 349–350. [Google Scholar]
- Martin JL, Legresley MM, & Gidney ME (2014a). Phytoplankton monitoring in the Western Isles region of the Bay of Fundy during 2007–2013. Can. Tech. Rep. Fish. Aquat. Sci.3105. [Google Scholar]
- Martin JL, LeGresley MM, & Hanke AR (2014b). Thirty years - Alexandrium fundyense cyst, bloom dynamics and shellfish toxicity in the Bay of Fundy, eastern Canada. Deep-Sea Research Part II: Topical Studies in Oceanography, 103, 27–39. [Google Scholar]
- McGillicuddy DJ, Townsend DW, Keafer BA, Thomas MA, & Anderson DM (2014). Georges Bank: A leaky incubator of Alexandrium fundyense blooms. Deep Sea Research Part II: Topical Studies in Oceanography, 103, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIRMANS PG, & VAN TIENDEREN PH (2004). genotype and genodive: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology Notes, 4(4), 792–794. [Google Scholar]
- Nagai S, Lian C, Hamaguchi M, Matsuyama Y, Itakura S, & Hogetsu T (2004). Development of microsatellite markers in the toxic dinoflagellate Alexandrium tamarense (Dinophyceae). Molecular Ecology Notes, 4(1), 83–85. [Google Scholar]
- Nagai S, Nishitani G, Sakamoto S, Sugaya T, Lee CK, Kim CH, … Yamaguchi M (2009). Genetic structuring and transfer of marine dinoflagellate Cochlodinium polykrikoides in Japanese and Korean coastal waters revealed by microsatellites. Molecular Ecology, 18(11), 2337–2352. [DOI] [PubMed] [Google Scholar]
- Nei M (1973). Analysis of Gene Diversity in Subdivided Populations. Proceedings of the National Academy of Sciences, 70(12), 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, … & Stevens MHH (2016). Package “Vegan”—community ecology package. R-Package Version 24–6. [Google Scholar]
- Pettigrew NR, Churchill JH, Janzen CD, Mangum LJ, Signell RP, Thomas AC, … Xue H (2005). The kinematic and hydrographic structure of the Gulf of Maine Coastal Current. Deep Sea Research Part II: Topical Studies in Oceanography, 52(19–21), 2369–2391. [Google Scholar]
- Pilskaln CH, Hayashi K, Keafer BA, Anderson DM, & McGillicuddy DJ (2014). Benthic nepheloid layers in the Gulf of Maine and Alexandrium cyst inventories. Deep-Sea Research Part II: Topical Studies in Oceanography, 103, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, & Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M (1995). J. Hered 86:248–249. J. Hered, 86, 248–249. [Google Scholar]
- Richlen ML, Erdner DL, McCauley LAR, Libera K, & Anderson DM (2012). Extensive genetic diversity and rapid population differentiation during blooms of Alexandrium fundyense (Dinophyceae) in an isolated salt pond on Cape Cod, MA, USA. Ecology and Evolution, 2(10), 2588–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F, De S, Montpellier U, & Bataillon PE (2008). GENEPOP ’ 007 : a complete re-implementation of the GENEPOP software for Windows and Linux, 103–106. [DOI] [PubMed] [Google Scholar]
- Rynearson TA, & Armbrust EV (2005). Maintenance of clonal diversity during a spring bloom of the centric diatom Ditylum brightwellii. Molecular Ecology, 14(6), 1631–1640. [DOI] [PubMed] [Google Scholar]
- Rynearson T. a., Newton J. a., & Armbrust EV (2006). Spring bloom development, genetic variation, and population succession in the planktonic diatom Ditylum brightwellii. Limnology and Oceanography, 51(3), 1249–1261. [Google Scholar]
- Sassenhagen I, Gao Y, Lozano-Duque Y, Parsons ML, Smith TB, & Erdner DL (2018). Comparison of Spatial and Temporal Genetic Differentiation in a Harmful Dinoflagellate Species Emphasizes Impact of Local Processes. Frontiers in Marine Science, 5(October), 1–13.29552559 [Google Scholar]
- Sjöqvist C, Godhe A, Jonsson PR, Sundqvist L, & Kremp A (2015). Local adaptation and oceanographic connectivity patterns explain genetic differentiation of a marine diatom across the North Sea-Baltic Sea salinity gradient. Molecular Ecology, 24(11), 2871–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist L, Godhe A, Jonsson PR, & Sefbom J (2018). The anchoring effect—long-term dormancy and genetic population structure. The ISME Journal, 12(12), 2929–2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend DW, McGillicuddy DJ, Thomas MA, & Rebuck ND (2014). Nutrients and water masses in the Gulf of Maine–Georges Bank region: Variability and importance to blooms of the toxic dinoflagellate Alexandrium fundyense. Deep Sea Research Part II: Topical Studies in Oceanography, 103, 238–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dolah FM (2007). Marine Algal Toxins : Origins , Health Effects , and Their Increased Occurrence Frances M . Van Dolah. Environmental Health, 108, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S (1949). The genetical structure of populations. Annals of Human Genetics. Annals of Human Genetics, 15(1), 323–354. [DOI] [PubMed] [Google Scholar]
- Yeh FC, Yang RC, Boyle TB, Ye ZH, & Mao JX (1997). POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada, 10, 295–301. [Google Scholar]