Summary
Monitor lizards are unique among ectothermic reptiles in that they have high aerobic capacity and distinctive cardiovascular physiology resembling that of endothermic mammals. Here, we sequence the genome of the Komodo dragon (Varanus komodoensis), the largest extant monitor lizard, and generate a high resolution de novo chromosome-assigned genome assembly for V. komodoensis using a hybrid approach of long-range sequencing and single molecule optical mapping. Comparing the genome of V. komodoensis with those of related species, we find evidence of positive selection in pathways related to energy metabolism, cardiovascular homeostasis, and hemostasis. We also show species-specific expansions of a chemoreceptor gene family related to pheromone and kairomone sensing in V. komodoensis and other lizard lineages. Together, these evolutionary signatures of adaptation reveal genetic underpinnings of the unique Komodo dragon sensory and cardiovascular systems, and suggest that selective pressure altered hemostasis genes to help Komodo dragons evade the anticoagulant effects of their own saliva. The Komodo dragon genome is an important resource for understanding the biology of monitor lizards and reptiles worldwide.
Introduction
The evolution of form and function in non-avian reptiles contains numerous examples of innovation and diversity. There are an estimated 10,000 reptile species worldwide, found on every continent except Antarctica, with a diversity of lifestyles and morphologies1 corresponding to a broad range of anatomic and physiological adaptations. Understanding how these adaptations evolved through changes to biochemical and cellular processes will reveal fundamental insights into areas ranging from anatomy and metabolism to behavior and ecology.
The varanid lizards (genus Varanus, or monitor lizards) are an unusual group within squamate reptiles (lizards and snakes). Varanids exhibit the largest range in size among reptiles, varying in mass by over five orders of magnitude (8 grams–100 kilograms)2. Varanids have a unique cardiopulmonary physiology and metabolism with numerous parallels to the mammalian cardiovascular system. For example, their cardiac anatomy is characterized by well-developed ventricular septa (“muscular ridge” and “bulbus lamellae”) resulting in a functionally divided heart3. This enables a dual pressure cardiovascular system characterized by high systemic and low pulmonary blood pressures3. Furthermore, varanid lizards can achieve and sustain higher aerobic metabolic rates and endurance capacity than similarly size non-varanid squamates, which enables intense, sustainable movements while hunting prey or in bouts of male-male combat. The largest of the varanid lizards, the Komodo dragon (Varanus komodoensis), can grow to 3 meters in length and run up to 20 kilometers per hour, allowing them to hunt large prey including deer and boar4. Komodo dragons have a higher metabolism than predicted by allometric scaling relationships for varanid lizards5, which may explain their capacity for daily movement to locate prey6. Their ability to locate injured or dead prey through scent tracking over several kilometers is enabled by a powerful olfactory system4, and their hunting is aided by serrate teeth, sharp claws, and saliva with anticoagulant and shock-inducing properties7,8. Further, Komodo dragons engage in aggressive intraspecific conflicts over mating, territory, and food, and wild individuals often bear scars from previous conflicts4.
To understand the genetic underpinnings of Komodo dragon physiology, we sequenced its genome and present a de novo assembly, generated with a hybrid approach of Illumina short-read sequencing with long-range sequencing using 10x Genomics, PacBio, and Oxford Nanopore sequencing, and single-molecule optical mapping using the Bionano platform. This suite of technologies allowed us to confidently assemble a high-quality reference genome for the Komodo dragon, which can serve as a template for other varanid lizards. We used this genome to understand the relationship of varanids to other reptiles using phylogenomics. We uncovered Komodo dragon-specific positive selection for genes encoding regulators of muscle metabolism, cardiovascular homeostasis, and hemostasis. Further, we discovered multiple lineage-specific expansions of a family of chemoreceptor genes in several squamates. Finally, we generated a high-resolution chromosomal map by assigning genomic scaffolds to chromosomes, enabling us to address questions about karyotype and sex chromosome evolution in squamates.
Results
De novo genome assembly
We sequenced the Komodo dragon genome principally from DNA isolated from peripheral blood of two male Komodo dragons housed at Zoo Atlanta: Slasher, offspring of the first Komodo dragons given to US President Reagan from President Suharto of Indonesia, and Rinca, an unrelated juvenile. A third individual from Gran Canaria was used for PacBio DNA sequencing. The Komodo dragon genome is distributed across 20 pairs of chromosomes, comprising eight pairs of large chromosomes and 12 pairs of microchromosomes9,10. De novo assembly was performed with a combination of 10x Genomics linked-read sequencing, Bionano optical mapping data, PacBio sequencing, and Oxford Nanopore MinIon sequencing (see Methods). The final assembly contained 1,411 scaffolds (>10 kb) with an N50 scaffold length of 24 Mb (longest scaffold: 138 Mb) (Table 1). The assembly is 1.51 Gb in size, ~32% smaller than the genome of the Chinese crocodile lizard (Shinisaurus crocodilurus)11, the closest relative of the Komodo dragon for which a sequenced genome is available, and ~15% smaller than the green anole (Anolis carolinensis)12, a model squamate lizard (Table S1). An assembly-free error corrected k-mer counting estimate of the Komodo dragon genome size13 is 1.69 Gb, while a flow cytometry-based estimate of the Komodo dragon genome size is 1.89 Gb14 (estimated 3.86 picograms of DNA per nucleus, with a conversion factor of 978 Mb per picogram15). Gaps comprise 0.97% of the assembly. We assessed the completeness of the Komodo dragon genome assembly by searching for 2,586 single-copy vertebrate genes using BUSCO16. The Komodo dragon genome has a similar distribution of single-copy (95.7%), duplicated (0.4%), fragmented (2%), and missing (1.9%) universal vertebrate genes as other reptile genomes (Table S3). The GC content of the Komodo dragon genome is 44.0%, similar to that of the S. crocodilurus genome (44.5%) but higher than the GC content of A. carolinensis (40.3%) (Table S1). Repetitive elements accounted for 32% of the genome, most of which were transposable elements (Table S2). As repetitive elements account for 49.6% of the S. crocodilurus genome11, most of the difference in size between the Komodo dragon genome and that of its closest sequenced relative can be attributed to repetitive element content.
Table 1.
Genome statistics of the Komodo dragon genome.
| Assembly size | 1.51 Gb (1,507,945,839bp) |
| Number of scaffolds | 1,411 |
| Minimum scaffold length | 10 Kb |
| Maximum scaffold length | 138 Mb |
| N50 scaffold length | 29 Mb (29,129,838) |
| Number of protein-coding genes | 18,457 |
| GC content | 44.04% |
Chromosome scaffold content
We isolated chromosome-specific DNA pools from a female Komodo dragon embryo of from Prague zoo stock through flow sorting10 and performed Illumina short-read sequencing on 15 DNA pools containing all Komodo dragon chromosomes (VKO1–20, VKOZ, VKOW) (Table S4). For each chromosome, we determined scaffold content and homology to A. carolinensis and chicken (Gallus gallus) chromosomes (Table 2 and Tables S5–6). For pools where chromosomes were mixed we determined partial scaffold content of single chromosomes. A total of 243 scaffolds containing 1.14 Gb (75% of total 1.51 Gb assembly) were assigned to 20 Komodo dragon chromosomes. As sex chromosomes share homologous pseudoautosomal regions, scaffolds enriched in both mixed 17/18/Z and 11/12/W chromosome pools most likely contained sex chromosome regions. As male varanid lizards are homogametic (ZZ) and the embryo used for flow sorting was female (ZW), scaffolds from the male-derived assembly enriched in these pools were assigned to the Z chromosome. Scaffold 79, which was assigned to the Z chromosome, contains an ortholog of amh (anti-Müllerian hormone) gene, which plays a crucial role in testis differentiation in vertebrates17. Scaffolds assigned to the Z chromosome were homologous to A. carolinensis chromosome 18, and mostly to G. gallus chromosome 28, in agreement with recent transcriptome analysis18.
Table 2.
Results of scaffold assignments to chromosomes of V. komodoensis.
| V. komodoensis chromosome | Gallus gallus homology | Anolis carolinensis homology | Total number of assigned scaffolds | Total length of assigned scaffolds (bp) |
|---|---|---|---|---|
| Chr1 | Chr1, 3, 5, 18, Z | Chr1, 2, 3 | 94 | 245,019,529 |
| Chr2 | Chr1, 3, 5, 7 | Chr1, 2, 6 | 14 | 156,023,568 |
| Chr3 | Chr1, 4 | Chr3, 5 | 11 | 115,571,927 |
| Chr4 | Chr1, 2, 5, 27 | Chr1, 4, 6 | 39 | 117,170,416 |
| Chr5 | Chr1 | Chr3 | 6 | 75,951,376 |
| Chr6, 7, 8 | Chr2, 6, 8, 9, 20 | Chr1, 2, 3, 4 | 25 | 200,178,831 |
| Chr9, 10 | Chr11, 22, 24 | Chr7, 8 | 8 | 69,008,218 |
| Chr11, 12 | Chr4, 10 | Chr10, 11 | 6 | 52,491,606 |
| Chr13 | Chr1, 5, 23 | Chr9 | 9 | 19,625,567 |
| Chr14 | Chr14 | Chr12 | 3 | 21,537,982 |
| Chr15 | Chr15 | ChrX | 4 | 14,821,201 |
| Chr16 | Chr17 | Chr16 | 2 | 13,367,238 |
| Chr17, 18 | Chr1, 19, 21 | Chr1, 9, 15, 17 | 10 | 17,262,365 |
| Chr19 | Chr1, 3, 25 | Chr14 | 6 | 11,765,548 |
| ChrZ | Chr1, 28 | Chr18 | 6 | 10,642,498 |
G. gallus homology: homology of scaffolds to G. gallus chromosomes; A. carolinensis homology: homology of scaffolds to A. carolinensis chromosomes; Total number of assigned scaffolds: total number of scaffolds assigned to each chromosome; Total length of assigned scaffolds (bp): size in base pairs of the sum of all scaffolds assigned to each chromosome.
Gene annotation
To annotate genes in the Komodo dragon genome, we performed RNA sequencing of heart tissue, and then used the MAKER pipeline with assembled RNA-seq transcripts, protein homology, and de novo predictions as evidence (see Methods). A total of 18,457 protein coding genes were annotated in the Komodo genome, 17,189 (93%) of which have at least one annotated Interpro functional domain (Table 1). Of these protein-coding genes, 63% were expressed (RPKM > 1) in the heart. Most (89%) of Komodo dragon protein-coding genes are orthologous to A. carolinensis genes. The median amino acid identity of single-copy orthologs between Komodo dragon and A. carolinensis is 68.9%, whereas it is 70.6% between one-to-one orthologs in Komodo dragon and S. crocodilurus (Figure S1).
Phylogenetic placement of Komodo Dragon
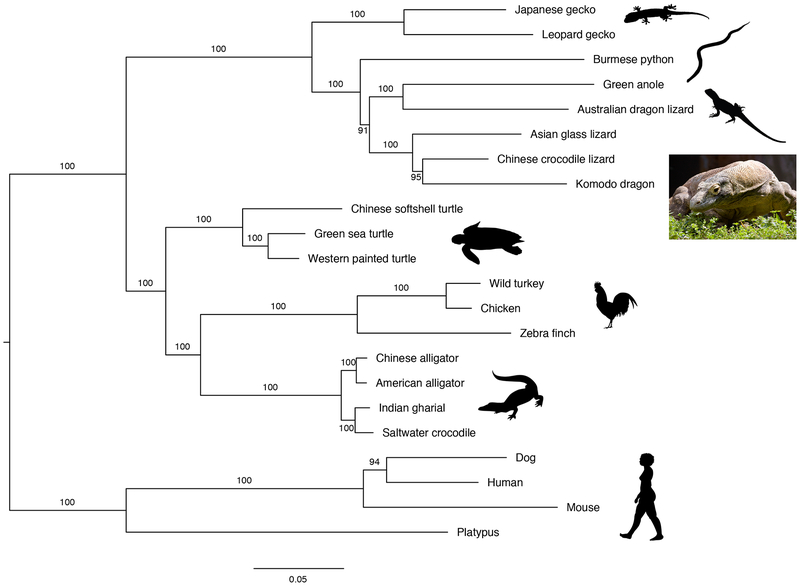
Recent analyses estimate that varanid lizards and their closest extant relative, the earless monitor lizard of the Lanthanotus genus, diverged 62 MYA, and that varanid lizards and the Shinisauridae family diverged 115 MYA19–38. We used 1,394 orthologous proteins from the Komodo dragon genome, 14 representative non-avian reptile species (seven squamates, three turtles, and four crocodilians), three avian species (chicken, wild turkey, and zebra finch), and four mammalian species (platypus, mouse, dog, and human) to estimate a species tree (Figure 1). Our analysis supports a sister relationship between anguimorphs (monitor lizards, anguids, Chinese crocodile lizards and relatives) and iguanians (dragon lizards, chameleons and iguanas), with snakes as sister to these two groups. This is in agreement with previously published analyses, including the most comprehensive marker gene based molecular phylogenetic analyses39–41, and in disagreement with a proposed sister relationship between anguimorphs and snakes or other topologies42,43.
Figure 1. Estimated species phylogeny of 15 non-avian reptile species, 3 avian species, and 4 mammals.
Maximum likelihood phylogeny constructed from 1,394 one-to-one orthologous proteins. Support values from 10,000 bootstrap replicates are shown. All silhouettes obtained from PhyloPic.org. Photograph of Slasher, a Komodo dragon sampled for DNA in this study. Photo courtesy of Adam K Thompson/Zoo Atlanta, with permission.
Expansion of vomeronasal genes across squamate reptiles
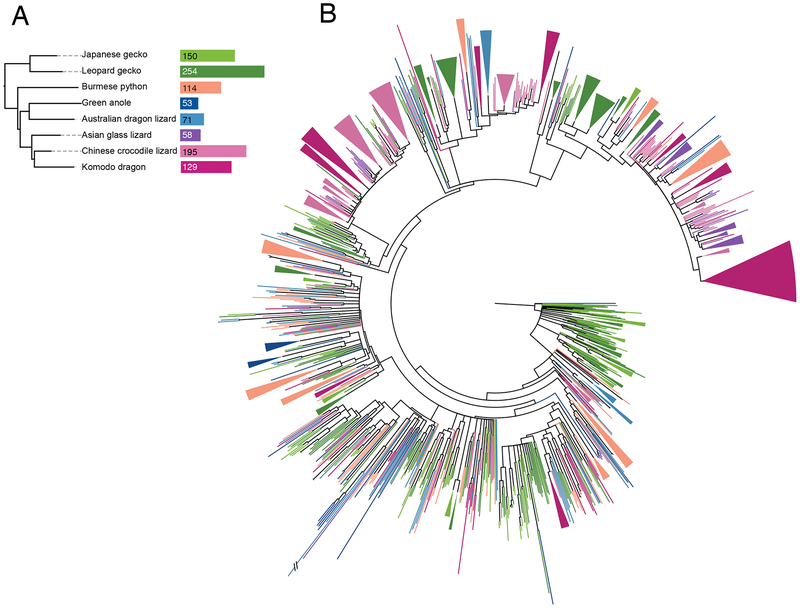
The vomeronasal organ is a chemosensory tissue shared across most amphibians, reptiles, and mammals that detects chemical cues including pheromones and kairomones. There are two classes of vomeronasal chemosensory receptors, both of which have undergone repeated gene family expansions and contractions across vertebrate evolution. The gene family encoding Type 2 receptors (V2Rs) has expanded in amphibians, snakes, and some mammalian lineages44,45. In contrast, crocodilian and turtle genomes contain very few V1R and V2R genes, and birds have entirely nonfunctional vomeronasal organs46,47. To clarify the relationship between vomeronasal organ function and the evolution of vomeronasal receptor gene families, we analyzed the coding sequences of 15 reptiles, including the Komodo dragon, for presence of V1R and V2R genes (Figure 2A). We found a large repertoire of V2Rs, comparable to that of snakes, in the Komodo dragon, other anguimorphan lizards, and geckos. We confirmed that there are few V1R genes across reptiles generally, and few to zero V2R genes in crocodilians and turtles (Table S7). The low number of V2R genes in A. carolinensis and the Australian dragon lizard (Pogona vitticeps) suggests that V2R genes are infrequently expanded in iguanians, though more iguanian genomes are needed to test this hypothesis.
Figure 2. Type 2 vomeronasal receptors have expanded in Komodo dragons and several other squamate reptiles.
(A) Type 2 vomeronasal gene counts in squamate reptiles. (B) Unrooted gene phylogeny of 1,024 vomeronasal Type 2 receptor transmembrane domains across squamate reptiles. The topology of the tree supports a gene expansion ancestral to squamates (i.e., clades containing representatives from all species) as well as multiple species-specific expansions through gene duplication events (i.e., clades containing multiple genes from one species). Branches with bootstrap support less than 60 are collapsed. Colors correspond to species in (A). Clades containing genes from a single species are collapsed.
We next constructed a phylogeny of all V2R gene sequences across squamates (Figure 2B) to understand the dynamic evolution of this gene family. The topology of this phylogeny supports the hypothesis that V2Rs expanded in the common ancestor of squamates, as there are clades of gene sequences containing members from all species45. In addition, there are many well-supported single species clades (i.e., Komodo dragon only or Burmese python only) dispersed across the gene tree, consistent with multiple duplications of V2R genes later in squamate evolution, including in the Komodo dragon and gecko lineages (Figure 2B).
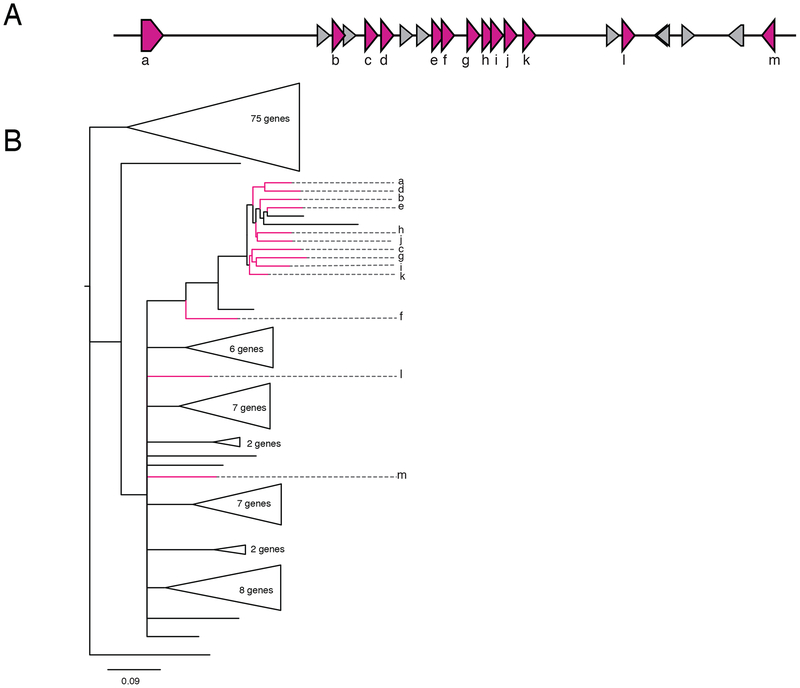
Because V2Rs expanded in rodents through tandem gene duplications that produced clusters of paralogs48, we examined clustering of V2R genes in our Komodo dragon assembly to determine if a similar mechanism was at play. Of 129 V2 genes, 77 are organized into 21 gene clusters ranging from 2 to 13 paralogs (Figure 3A, Table S8). A phylogeny of all Komodo dragon V2R genes (Figure 3B) showed that the genes in the largest 13-gene cluster group together in a gene tree of Komodo dragon V2R genes (Figure 3). Of the remaining 52 V2R genes, 35 are on scaffolds less than 100 Kb in size, so our estimate of V2R clustering is a lower bound due to fragmentation in the genome assembly (Table S8). These results support the hypothesis that expansions of V2R genes in multiple squamate reptile lineages arose through tandem gene duplication.
Figure 3. Gene clusters of Type 2 vomeronasal receptors evolved through gene duplication.
(A) Cluster of 13 vomeronasal Type 2 receptor genes in the Komodo dragon genome. Pink genes are V2R genes and gray genes are non-V2R genes. Gene labels correspond to labels in (B). (B) Unrooted phylogeny of 129 vomeronasal Type 2 receptor genes in Komodo dragon. As most of the genes in this gene cluster group together in a gene phylogeny of all Komodo dragon V2R genes, it is likely that this cluster evolved through gene duplication events. Branches with bootstrap support less than 80 are collapsed. Clades without genes in this V2R gene cluster are collapsed. Genes in the V2R cluster are colored pink and labeled as in (A).
Positive selection
To evaluate adaptive protein evolution in the Komodo dragon genome, we tested for positive selection across one-to-one orthologs in squamate reptiles using a branch-site model (Table S9). Our analysis revealed 201 genes with signatures of positive selection in Komodo dragons (Table S10). Of these, 188 had a one-to-one ortholog in humans; 93 mapped to pathways in the Reactome database, and 34 had an annotated functional interaction with at least one other positively selected gene (Figure S2)49,50. These 34 genes are enriched for 12 pathways (false discovery rate < 5%), including three related to mitochondrial function, four related to coagulation, and five related to immune function (Table S11).
Many of the genes under positive selection point towards important adaptations of the Komodo dragon’s mammalian-like cardiovascular and metabolic functions, which are unique among non-varanid ectothermic reptiles. These include mitochondrial function and cellular respiration, hemostasis and the coagulation cascade, and angiotensinogen (Table S11, Figure S2)51. Innate and adaptive immunity genes, which are frequently under positive selection in vertebrates, are well represented amongst positively selected genes52. Finally, 106 positively selected genes do not have an annotated function, and 25% of positively selected genes were not detectably expressed in the heart and likely represent adaptations in other aspects of Komodo dragon biology.
Positive selection of genes regulating mitochondrial function
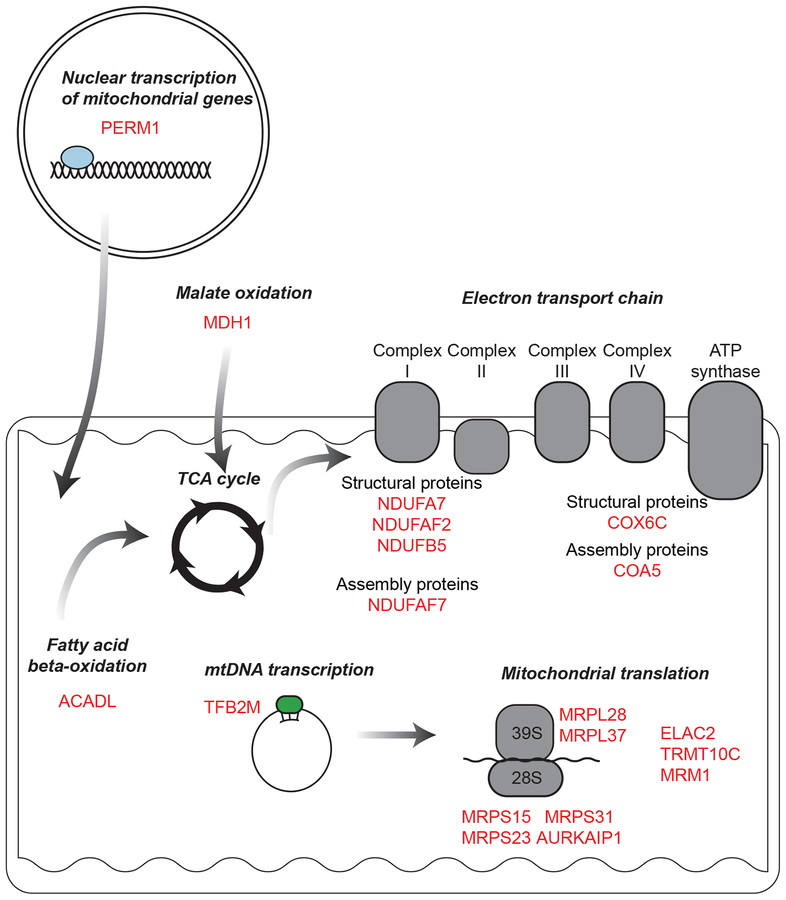
In the Komodo dragon genome we found evidence of positive selection of electron transport chain components including multiple subunits and assembly factors of the Type 1 NADH dehydrogenase— NDUFA7, NDUFAF7, NDUFAF2, NDUFB5— as well as components of the cytochrome c oxidase protein complexes, COX6C and COA5, (Figure 4, Figure S3, Table S10). We also found signatures of positive selection for other elements of mitochondrial function in the Komodo dragon lineage (Figure 4). For example, we detected positive selection for ACADL, which encodes a critical enzyme for mitochondrial fatty acid beta-oxidation, the major postnatal metabolic process in cardiac myocytes53. Further, two genes that promote mitochondrial biogenesis, TFB2M and PERM1, have undergone positive selection in the Komodo dragon. TFB2M regulates mtDNA transcription and dimethylates mitochondrial 12s rRNA54,55, while PERM1 regulates the expression of select PPARGC1A/B and ESRRA/B/G target genes with roles in glucose and lipid metabolism, energy transfer, contractile function, muscle mitochondrial biogenesis and oxidative capacity56. PERM1 also enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance when over-expressed in mice57. Finally, we observed positive selection of MDH1, encoding malate dehydrogenase, which together with mitochondrial MDH2 regulates the supply of NADPH and acetyl-CoA to the cytoplasm, thus modulating fatty acid synthesis58.
Figure 4. Positive selection of mitochondrial genes in the Komodo dragon.
Genes in the Komodo dragon genome under positive selection include components of the electron transport chain, regulators of mtDNA transcription, regulators of mitochondrial translation, and fatty acid beta-oxidation.
Multiple factors regulating mitochondrial translation have also undergone positive selection in the Komodo dragon (Figure 4). These include four components of the mitochondrial 28S small ribosomal subunit (MRPS15, MRPS23, MRPS31, and AURKAIP1) and two components of the mitochondrial 39S large ribosomal subunit (MRPL28 and MRPL37). We also found positive selection of ELAC2 and TRMT10C, which are required for maturation of mitochondrial tRNA, and MRM1, which encodes a mitochondrial rRNA methyltransferase59–61.
Overall, these instances of positive selection in genes encoding proteins important for mitochondrial function could underlie the remarkably high aerobic capacity in the Komodo dragon. Additional genome sequences are needed to determine whether these changes are specific to the Komodo dragon, shared across varanid lizards generally, or found in unsequenced reptiles.
Positive selection of angiotensinogen
We detected positive selection for angiotensinogen (AGT), which encodes the precursor of several peptide regulators of cardiovascular function, the most well-studied being angiotensin II (AngII) and angiotensin1–7 (A1–7). AngII has a vasoactive function in blood vessels and inotropic effects on the heart62. In mammals, the level of Angll increases in response to intense physical activity, contributing to arterial blood pressure and regional blood regulation63–66. Reptiles have a functional renin-angiotensin system (RAS) that is important for their cardiovascular response to aerobic activity67–69. The positive selection for AGT points to important adaptations in cardiovascular physiology and the renin-angiotensin system in the Komodo dragon.
Positive selection of hemostasis-related genes
We found evidence for positive selection across regulators of hemostasis, which reduces blood loss after injury. Four genes that regulate platelet activities, MRVI1, RASGRP1, LCP2, and CD63 have undergone positive selection in the Komodo dragon genome. MRVI1 is involved in inhibiting platelet aggregation70, RASGRP1 coordinates calcium dependent platelet responses71, LCP2 is involved in platelet activation72, and CD63 plays a role in controlling platelet spreading73. In addition, two coagulation factors, F10 (Factor X) and F13B (Coagulation factor XIII B chain) have undergone positive selection in the Komodo dragon genome. Activation of Factor X is the first step in initiating coagulation74, and Factor 13 is the final factor activated in the coagulation cascade75. Further, FGB, which encodes one of the three subunits of fibrinogen, the molecule converted to the clotting agent fibrin76, has undergone positive selection in the Komodo dragon genome.
Discussion
We have sequenced and assembled a high-quality genome of the Komodo dragon that will be a template for analysis of other varanid genomes, and for further investigation of genomic innovations in the varanid lineage. We were able to assign 75% of the genome to chromosomes, providing a significant contribution to comparative genomics of squamates and vertebrates generally. As the number of squamate whole genome sequences continues to grow, there will be opportunities to examine the evolution of non-coding DNA in these reptiles.
Varanid lizards have genotypic sex determination and share ZZ/ZW sex chromosomes with other anguimorphan lizards10,18. Here, we were able to assign genomic scaffolds to the Z chromosome of the Komodo dragon. All Z chromosome scaffolds were homologous to A. carolinensis chromosome 18 and mostly to chicken chromosome 28, in agreement with a recent transcriptome-based analysis18. Within Iguania, the sister group of anguimorphs20,40,77, there exist environmental sex determination systems without sex chromosomes as well as conserved XX/XY sex chromosomes homologous to anguimorphan autosomes78–82. Sex chromosomes in most snakes (pythons and all families of caenophidian snakes83) are homologous to chromosome 6 of A. carolinensis and thus to autosomes of the Komodo dragon, suggesting an independent origin of sex chromosomes in snakes and anguimorphs. However, the ancestral sex determination of snakes remains unresolved83,84. The regions of sex chromosomes shared by the common ancestor of varanids and several other lineages of anguimorphan lizards contain the amh (anti-Müllerian hormone) gene18, which plays a crucial role in vertebrate testis differentiation. Homologs of amh are strong candidates for sex-determining genes in several lineages of teleost fishes and in monotremes85–88, and should be considered candidate sex-determining genes in varanids and other anguimorphs.
Our comparative genomic analysis identified previously undescribed species-specific expansion of V2Rs across multiple squamates, including lizards and at least one snake. Komodo dragons, like other squamates, are known to possess a sophisticated lingual-vomeronasal systems for chemical sampling of their environment89. This sensory apparatus allows Komodo dragons to perceive environmental chemicals for social and ecological activities, including kin recognition, mate choice90,91, predator avoidance92,93, and hunting prey94,95. Komodo dragons are unusual as they adopt differential foraging tactics across ontogeny, with smaller juveniles preferring active foraging for small prey and large adult dragons targeting larger ungulate prey via ambush predation6. However, utilization of the vomeronasal system across ontogeny seems likely, given the exceptional capacity for Komodo dragons of all sizes to locate prey. Future work will be able to explore the role of V2R expansion in the behavior and ecology of Komodo dragons, including their ability to locate prey at long distances4.
We found evidence for positive selection in the Komodo dragon genome across genes involved in regulating mitochondrial biogenesis, cellular respiration, and cardiovascular homeostasis. Komodo dragons and other monitor lizards have a high aerobic capacity and exercise endurance, and our results reveal selective pressures on biochemical pathways that are likely to be the source of this high aerobic capacity. Reptile muscle mitochondria typically oxidize substrates at a much lower rate than mammalian mitochondria, partly based on substrate-type use96. The findings that Komodo dragons experienced selection in several genes encoding mitochondrial enzymes, including one involved in fatty acid metabolism, points towards a more mammalian-like mitochondrial function. Future work on additional varanid species, and other squamate outgroups, will test these hypotheses. Selective pressures acting on these mitochondrial genes in Komodo dragons is consistent with the increased expression of genes associated with oxidative capacity found in pythons after feeding97,98.
In addition, we found positive selection for angiotensinogen, which encodes two potent vasoactive and inotropic peptides with central roles in cardiovascular physiology. In mammals, AngII contributes to the mean arterial blood pressure and to the redistribution of cardiac output65,66. A compelling hypothesis is that these changes to angiotensinogen may be an important component in the ability of the Komodo dragon to rapidly increase blood pressure and cardiac output as required for hunting, extended periods of locomotion including inter-island swimming, and male-male combat during the breeding season. Direct measures of cardiac function have not been made in Komodo dragons, but in other varanid lizards, a large aerobic scope during exercise is associated with a large factorial increase in cardiac output99. Future physiological studies measuring the hemodynamic responses to exercises with respect to AngII expression can test this hypothesis. Giraffes, which have evolved high blood pressure to maintain cardiovascular homeostasis in their elongated bodies, have experienced positive selection on several blood pressure regulators, including the angiotensin-converting enzyme (ACE)100. It is possible that positive selection in animals with high blood pressures converges on angiotensin regulators. Overall, the evolution of these genes suggests a profoundly different cardiovascular and metabolic profile relative to other squamates, endowing the Komodo dragon with unique physiological properties.
We also found evidence for positive selection across genes that regulate blood clotting. Like other monitor lizards, the saliva of Komodo dragons contains anticoagulants, which is thought to aid in hunting7,8. During conflict with conspecifics over food, territories, or mates, Komodo dragons use their serrate teeth to inflict bite wounds, raising the possibility that these anticoagulants may enter their bloodstream. The extensive positive selection of genes encoding their coagulation system may reflect a selective pressure for Komodo dragons to evade the anticoagulant and hypotensive effects of the saliva of conspecifics. While all monitor lizards tested contain anticoagulants in their saliva, the precise mechanism by which they act varies8. It is possible that different species of monitor lizards evolved adaptations that reflect the diversity of their anticoagulants, or that co-evolution has occurred between monitor lizard coagulation systems and anti-coagulant saliva. Further, as Komodo dragons have high blood pressure, changes to their coagulation system may reflect increased protection from vascular damage.
Materials and Methods
DNA isolation and processing for Bionano optical mapping
Komodo dragon whole blood was obtained from one of two individuals housed at Zoo Atlanta (Rinca). Samples from the animals at Zoo Atlanta were collected with the approval of the Zoo’s Scientific Research Committee. High molecular weight genomic DNA was extracted for genome mapping. Blood was centrifuged at 2000g for 2 minutes, plasma was removed, and the sample was stored at 4°C. 2.5μl of blood was embedded in 100μl agarose gel plugs to give ~7μg DNA/plug, using the BioRad CHEF Mammalian Genomic DNA Plug Kit (Bio-Rad Laboratories, Hercules, CA, USA). Plugs were treated with proteinase K overnight at 50°C. The plugs were then washed, melted, and then solubilized with GELase (Epicentre, Madison, WI, USA). The purified DNA was subjected to four hours of drop-dialysis. DNA concentration was determined using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA), and the quality was assessed with pulsed-field gel electrophoresis.
The high molecular weight DNA was labeled according to commercial protocols using the IrysPrep Reagent Kit (Bionano Genomics, San Diego, CA, USA). Specifically, 300 ng of purified genomic DNA was nicked with 7 U nicking endonuclease Nb.BbvCI (NEB, Ipswich, MA, USA) at 37°C for two hours in NEB Buffer 2. The nicked DNA was labeled with a fluorescent-dUTP nucleotide analog using Taq polymerase (NEB) for one hour at 72°C. After labeling, the nicks were repaired with Taq ligase (NEB) in the presence of dNTPs. The backbone of fluorescently labeled DNA was stained with DNA stain (Bionano).
Bionano mapping and assembly
Using the Bionano Irys instrument, automated electrophoresis of the labeled DNA occurred in the nanochannel array of an IrysChip (Bionano Genomics), followed by automated imaging of the linearized DNA. The DNA backbone (outlined by YOYO-1 staining) and locations of fluorescent labels along each molecule were detected using the Irys instrument’s software. The length and set of label locations for each DNA molecule defines an individual single-molecule map. Raw Bionano single-molecule maps were de novo assembled into consensus maps using the Bionano IrysSolve assembly pipeline (version 5134) with default settings, with noise values calculated from the 10x Genomics Supernova assembly. The Bionano optical mapping data comprised 80X genome coverage, and the scaffold N50 of the assembly was 1.2 Mb.
DNA processing for 10x Genomics linked read sequencing
Blood samples from two individuals housed at Zoo Atlanta (Slasher and Rinca) were used. High molecular weight genomic DNA extraction, sample indexing, and generation of partition barcoded libraries were performed by 10x Genomics (Pleasanton, CA, USA) according to the Chromium Genome User Guide and as published previously101. 1.2 ng of genomic DNA was used as input to the Chromium system.
10x Genomics sequencing and assembly
The 10X Genomics barcoded library was sequenced on the Illumina HiSeq2500. 660M of the raw reads comprising 57X genome coverage were assembled using the company’s Supernova software (version 1.0) with default parameters. Output fasta files of the Supernova assemblies were generated in pseudohap format, which links phased and unphased regions of the assembly into ‘pseudo-haplotype’ scaffolds. This generated an initial assembly with a scaffold N50 length of 10.2 Mb and a contig N50 length of 95 Kb.
Oxford Nanopore sequencing
DNA isolated from Slasher was sequenced to 0.75X coverage on an Oxford Nanopore MinIon sequencer following manufacturer’s instruction. MinKNOW was used for basecalling and output to FASTQ files.
DNA processing for PacBio sequencing
Komodo dragon whole blood collected in EDTA from an individual housed at Reptilandia Zoo, Gran Canarias under institutional approval, stored at −20 C, was used to extract high molecular weight DNA for single molecule real time sequencing. Extraction was performed using gravity-flow, anion-exchange tips (Qiagen genomic-tip 100/G kit) to a final DNA concentration of 130 ng/μl assessed using a Qubit 2.0 Fluorometer. Size of extracted DNA was determined by a 16-hour pulse field gel electrophoresis, which resolved high molecular weight fragments from 15 kb to 85 kb. We constructed a 10 kb and a 20 kb insert library using 20 μg of genomic DNA. The library was then size selected using a Blue Pippin (Sage Science) and resulting double-stranded DNA fragments were capped by hairpin loops at both ends to form a SMRTbell template. Single molecule SMRTbell templates were then loaded in 150K Zero Mode Waveguides SMRT cells of a PacBio RS II sequencing system using paramagnetic beads (MagBeads, PacBio). Sequencing was performed using a total of 29 SMRT cells. We obtained a total of 2,061,804 subread filtered sequences for a total sequence length of 11,907,672,561 bp. Average, maximum and minimum sequence lengths were 5,775 bp, 48,338 bp and 35 bp, respectively. Median sequence length was 4,486 bp. N50 read length was 12,457 bp. In total, PacBio sequencing data represented 6.3X genome sequencing coverage.
Merging datasets into a single assembly
Sequencing and mapping data types were merged together as follows. First, Bionano assembled contigs and the 10x Genomics assembly were combined using Bionano’s hybrid assembly tool with the -B2 -N2 options. SSPACE-LongRead (cite https://doi.org/10.1186/1471-2105-15-211) was used in series with default parameters to scaffold the hybrid assembly using PacBio reads, Nanopore reads, and unincorporated 10x Genomics Supernova scaffolds/contigs, resulting in the final assembly.
Genome completeness assessment
The BUSCO pipeline version 3.0.2 was used to determine the completeness of the Komodo dragon genome, using the 2,586 gene vertebrata gene set and the Augustus retraining parameters (--long)16. For comparison, BUSCO was also run with the same parameters on all reptile genomes used for comparative analyses (Shinisaurus crocodilurus, Ophisaurus gracilis, Anolis carolinensis, Pogona vitticeps, Python molurus bivittatus, Eublepharis macularius, Gekko japonicus, Pelodiscus sinensis, Chelonia mydas, Chrysemys picta bellii, Alligator sinensis, Alligator mississippiensis, Gavialis gangeticus, and Crocodylus porosus) (Table S3).
We obtained an assembly-free estimate of the Komodo dragon genome size using an sequencing error corrected k-mer counting method implemented in the PreQC component of the SGA assembler13.
Assignment of scaffolds to chromosomes
Isolation of Komodo dragon chromosome-specific DNA pools was performed as previously described10. Briefly, fibroblast cultivation of a female V. komodoensis were obtained from tissue samples of an early embryo of a captive individual at the Prague National Zoo. Embryos are obtained under the laws of the Czech Republic and of the European Union. Chromosomes obtained by fibroblast cultivation were sorted using a Mo-Flo (Beckman Coulter) cell sorter. Fifteen chromosome pools were sorted in total. Chromosome-specific DNA pools were then amplified and labelled by degenerate oligonucleotide primed PCR (DOP-PCR) and assigned to their respective chromosomes by hybridization of labelled probes to metaphases. V. komodoensis chromosome pools obtained by flow sorting were named according to chromosomes (e.g. majority of DNA of VKO6/7 belong to chromosomes 6 and 7 of V. komodoensis). V. komodoensis pools for macrochromosomes are each specific for one single pair of chromosomes, except for VKO6/7 and VKO8/7, which contain one specific chromosome pair each (pair 6 and pair 8, respectively), plus a third pair which overlaps between the two of them (pair 7). For microchromosomes, pools VKO9/10, VKO17/18/19, VKO11/12/W and VKO17/18/Z contained more than one chromosome each, while the rest are specific for one single pair of microchromosomes. The W and Z chromosomes are contained in pools VKO11/12/W and VKO17/18/Z, respectively, together with two pairs of other microchromosomes each.
Chromosome-pool specific genetic material was amplified by GenomePlex® Whole Genome Amplification (WGA) Kit (Sigma) following manufacturer protocols. DNA from all 15 chromosome pools was used to prepare Illumina sequencing libraries, which were independently barcoded and sequenced 125 bp paired-end in a single Illumina Hiseq2500 lane.
Reads obtained from sequencing of flow-sorting-derived chromosome-specific DNA pools were processed with the dopseq pipeline (https://github.com/lca-imcb/dopseq)102,103. Illumina adapters and WGA primers were trimmed off by cutadapt v1.13104. Then, pairs of reads were aligned to the genome assembly of V. komodoensis using bwa mem105. Reads were filtered by MAPQ ≥ 20 and length ≥ 20 bp, and aligned reads were merged into positions using pybedtools 0.7.10106,107. Reference genome regions were assigned to specific chromosomes based on distance between positions. Finally, several statistics were calculated for each scaffold. Calculated parameters included: mean pairwise distance between positions on scaffold, mean number of reads per position on scaffold, number of positions on scaffold, position representation ratio (PRR) and p-value of PRR. PRR of each scaffold was used to evaluate enrichment of given scaffold on chromosomes. PRR was calculated as ratio of positions on scaffold to positions in genome divided by ratio of scaffold length to genome length. PRRs >1 correspond to enrichment, while PRRs <1 correspond to depletion. As the PRR value is distributed lognormally, we use its logarithmic form for our calculations. To filter out only statistically significant PRR values we used thresholds of logPRR >0 and its p-value <=0.01. Scaffolds with logPRR > 0 were considered enriched in the given sample. If one scaffold was enriched in several samples we chose highest PRR to assign scaffold as top sample.
We also compared the genome organization at the chromosome level among V. komodoensis, Anolis carolinensis and Gallus gallus. We determined homology of each V. komodoensis scaffold to scaffolds of A. carolinensis (AnoCar2.0) and G. gallus (galGal3) genome generating alignment between genomes with LAST108 and subsequently using chaining and netting technique109. Homology to A. carolinensis microchromosomes was determined using scaffold assignments from an Anolis chromosome-specific sequencing project102. For LAST we used default scoring matrix and parameters of 400 for gap existence cost, 30 for gap extension cost and 4500 for minimum alignment score. For axtChain we used same distance matrix and default parameters for other chain-net scripts.
RNA sequencing
RNA was extracted from heart tissue obtained from an adult male specimen that died of natural causes at the San Diego Zoo. This was approved by the Institutional Animal Care and Use Committee (IACUC) and Biomaterials Review Group of San Diego Zoo. Trizol reagent was used to extract RNA following manufacturer’s instructions. Two RNAseq libraries were produced using a NuGen RNAseq v2 and Ultralow v2 kits from 100 ng total RNA each, and sequenced on an Illumina Nextseq 500 with 150 bp paired-end strand-specific reads.
Genome annotation
RepeatMasker v4.0.7 was used to mask repetitive elements in the Komodo dragon genome using the Squamata repeat database from the RepBase-Dfam combined database as reference110. After masking repetitive elements, protein-coding genes were annotated using the MAKER version 3.01.02111 pipeline, combining protein homology information, assembled transcript evidence, and de novo gene predictions from SNAP and Augustus version 3.3.1112. Protein homology was determined by aligning proteins from 15 reptile species (Table S1) to the Komodo dragon genome using exonerate version 2.2.0113. RNA-seq data was aligned to the Komodo dragon genome with STAR version 2.6.0114 and assembled into 900,722 transcripts with Trinity version 2.4.0115. Protein domains were determined using InterProScan version 5.31.70116. Gene annotations from the MAKER pipeline were filtered based on the strength of evidence for each gene, the length of the predicted protein, and the presence of protein domains. Clusters of orthologous genes across 15 reptile species were determined with OrthoFinder v2.0.0117. A total of 284,107 proteins were clustered into 16,546 orthologous clusters. In total, 96.4% of Komodo dragon genes were grouped into orthologous clusters. For estimating a species phylogeny only, protein sequences from Mus musculus and Gallus gallus were added to the orthologous clusters with OrthoFinder. tRNAs were annotated using tRNAscan-SE version 1.3.1118, and other non-coding RNAs were annotated using the Rfam database119 and the Infernal software suite120.
Phylogenetic analysis
A total of 1,394 one-to-one orthologous proteins across 15 reptile species, three birds, and four mammals were used to estimate a species phylogeny. The 15 reptile species used were Varanus komodoensis, Shinisaurus crocodilurus, Ophisaurus gracilis, Anolis carolinensis, Pogona vitticeps, Python molurus bivittatus, Eublepharis macularius, Gekko japonicus, Pelodiscus sinensis, Chelonia mydas, Chrysemys picta bellii, Alligator sinensis, Alligator mississippiensis, Gavialis gangeticus, and Crocodylus porosus. The three birds used were Gallus gallus, Meleagris gallopavo, and Taeniopygia guttata, and the four mammals were Ornithorhynchus anatinus, Mus musculus, Canis familiaris, and Homo sapiens. Each set of orthologous proteins were aligned using PRANK v.170427121. Aligned proteins were concatenated into a supermatrix, and a species tree was estimated using IQ-TREE version 1.6.7.1122 with model selection across each partition123 and 10,000 ultra-fast bootstrap replicates124.
To identify genes in this dataset of 1,394 orthologs that are evolving in a clock-like manner and thus useful for phylogenomic analyses like divergence time estimates, we created individual trees for each of the genes in our analysis using the procedure described above but without bootstrapping. We then used SortaDate125 to calculate a number of informative metrics, including root-to-tip variance, tree length, and bipartition support relative to the species phylogeny. This dataset can be used to find candidate marker genes for phylogenomic analyses and is available in a Figshare repository at doi: 10.6084/m9.figshare.7967300.
Gene family evolution analysis
Gene family expansion and contraction analyses were performed with CAFE v4.2126 for the squamate reptile lineage, with a constant gene birth and gene death rate assumed across all branches.
Vomeronasal type 2 receptors were first identified in all species by containing the V2R domain InterPro domain (IPR004073)127. To ensure that no V2R genes were missed, all proteins were aligned against a set of representative V2R genes using BLASTp128 with an e-value cutoff of 1e-6 and a bitscore cutoff of 200 or greater. Any genes passing this threshold were added to the set of putative V2R genes. Transmembrane domains were identified in each putative V2R gene with TMHMM v2.0129 and discarded if they did not contain 7 transmembrane domains in the C-terminal region. Beginning at the start of the first transmembrane domain, proteins were aligned with MAFFT v7.310 (auto alignment strategy)130 and trimmed with trimAL (gappyout)131. A gene tree was constructed using IQ-TREE122–124 with the JTT+ model of evolution with empirical base frequencies and 10 FreeRate model parameters, and 10,000 bootstrap replicates. Genes were discarded if they failed the IQ-TREE composition test.
Positive selection analysis
We analyzed 4,047 genes that were universal and one-to-one across all squamate lineages tested (Varanus komodoensis, Shinisaurus crocodilurus, Ophisaurus gracilis, Anolis carolinensis, Pogona vitticeps, Python molurus bivittatus, Eublepharis macularius, and Gekko japonicus) to test for positive selection (Table S9). An additional 2,013 genes that were universal and one-to-one across a subset of squamate species (Varanus komodoensis, Anolis carolinensis, Python molurus bivittatus, and Gekko japonicus) were also analyzed (Table S9). We excluded multi-copy genes from all positive selection analyses to avoid confounding from incorrect paralogy inference. Proteins were aligned using PRANK121 and codon alignments were generated using PAL2NAL132.
Positive selection analyses were performed with the branch-site model aBSREL using the HYPHY framework133,134. For the 4,081 genes that were single-copy across all squamate lineages, the full species phylogeny of squamates was used. For the 2,040 genes that were universal and single-copy across a subset of species, a pruned tree containing only those taxa was used. We discarded genes with unreasonably high dN/dS values across a small proportion of sites, as those were false positives driven by low quality gene annotation in one or more taxa in the alignment. We used a cutoff of dN/dS of less than 50 across 5% or more of sites, and a p-value of less than 0.05 at the Komodo dragon node. Each gene was first tested for positive selection only on the Komodo dragon branch. Genes undergoing positive selection in the Komodo dragon lineage were then tested for positive selection at all nodes in the phylogeny. This resulted in 201 genes being under positive selection in the Komodo dragon lineage (Table S10).
Data availability
The assembled Komodo dragon genome is available in NCBI under the accession SJPD00000000. All DNA sequencing used to generate the assembly is available in the NCBI SRA database under accession PRJNA523222. Illumina sequencing data from chromosome pools is available in the NCBI SRA under accession PRJNA529483. RNA-seq data of heart tissue is available in the NCBI SRA under accession PRJNA527313. Original protein annotations, noncoding RNA annotations, all alignments for phylogenetic analyses and selection analyses, and newick files of phylogenetic trees are available in the following Figshare repositories: doi:10.6084/m9.figshare.7961135.v1, doi:10.6084/m9.figshare.7955891.v1, doi:10.6084/m9.figshare.7955879.v1, doi:10.6084/m9.figshare.7949483.v1, doi:10.6084/m9.figshare.7759496.v1, doi:10.6084/m9.figshare.7967300. The project folder for all Figshare data is available at https://figshare.com/projects/Data_for_Komodo_dragon_genome_paper/61271.
Supplementary Material
Acknowledgements
Special thanks from B.G.B. to John Romano for inspiration and historical information. We are grateful to staff at Zoo Atlanta for care of Slasher and Rinca and help obtaining samples, Jim Pether from Reptilandia zoo in Gran Canaria in the Canary Islands for additional samples, and R. Chadwick and N. Carli (Gladstone Genomics Core) for DNA and RNA-seq library preparation and MinION sequencing. We also thank Kristina Giorda, Rabeea Abbas, Deanna Church (10x Genomics) for 10x Genomics Chromium sequencing and Supernova assembly. This work was supported by institutional funding from the Gladstone Institutes to B.G.B and K.S.P.; an NHLBI grant to K.S.P and B.G.B (UM1 HL098179); the Younger Family gift to B.G.B.; an NHGRI grant to P.-Y.K. (R01 HG005946); NIH training grants (T32 AR007175) to A.C.Y.M and (T32 HL007731) to Y.M. M.A. and M.R. were supported by GACR 17–22141Y, M.R. was additionally supported by Charles University projects PRIMUS/SCI/46 and Research Centre (204069).
Footnotes
Competing Interests: The authors have no competing interests to declare
References
- 1.Chapman AD Numbers of Living Species in Australia and the World (Australian Biological Resources Study, 2009). [Google Scholar]
- 2.Collar DC, Schulte JA & Losos JB Evolution of extreme body size disparity in monitor lizards (Varanus). Evolution (N. Y). 65, 2664–2680 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Jensen B, Wang T, Christoffels VM & Moorman AFM Evolution and development of the building plan of the vertebrate heart. Biochim. Biophys. Acta - Mol. Cell Res 1833, 783–794 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Auffenberg W The Behavioral Ecology of the Komodo Monitor (University Presses of Florida, 1981). [Google Scholar]
- 5.Green B, King D, Braysher M & Saim A Thermoregulation, water turnover and energetics of free-living komodo dragons, Varanus komodoensis. Comp. Biochem. Physiol. Part A Physiol 99, 97–101 (1991). [Google Scholar]
- 6.Purwandana D et al. Ecological allometries and niche use dynamics across Komodo dragon ontogeny. Sci. Nat 103, 27 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fry BG et al. A central role for venom in predation by Varanus komodoensis (Komodo Dragon) and the extinct giant Varanus (Megalania) priscus. Proc. Natl. Acad. Sci. U. S. A 106, 8969–8974 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koludarov I et al. Enter the Dragon: The Dynamic and Multifunctional Evolution of Anguimorpha Lizard Venoms. Toxins 9, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson Pokorná M et al. First Description of the Karyotype and Sex Chromosomes in the Komodo Dragon (Varanus komodoensis). Cytogenet. Genome Res 148, 284–291 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Iannucci A et al. Isolating Chromosomes of the Komodo Dragon: New Tools for Comparative Mapping and Sequence Assembly. Cytogenet. Genome Res 157, 42–50 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Gao J et al. Sequencing, de novo assembling, and annotating the genome of the endangered Chinese crocodile lizard Shinisaurus crocodilurus. Gigascience 6, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alföldi J et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477, 587–591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson JT Exploring Genome Characteristics and Sequence Quality Without a Reference (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishan A et al. DNA index, genome size, and electronic nuclear volume of vertebrates from the Miami Metro Zoo. Cytom. Part A 65A, 26–34 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Doležel J, Bartoš J, Voglmayr H & Greilhuber J Letter to the editor. Cytom. Part A 51A, 127–128 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV & Zdobnov EM BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Rey R, Lukas-Croisier C, Lasala C & Bedecarrás P AMH/MIS: what we know already about the gene, the protein and its regulation. Mol. Cell. Endocrinol 211, 21–31 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Rovatsos M, Rehák I, Velenský P & Kratochvíl L Shared ancient sex chromosomes in varanids, beaded lizards and alligator lizards. Mol. Biol. Evol (2019). doi: 10.1093/molbev/msz024 [DOI] [PubMed] [Google Scholar]
- 19.Welton LJ, Travers SL, Siler CD & Brown RM Integrative taxonomy and phylogeny-based species delimitation of Philippine water monitor lizards (Varanus salvator Complex) with descriptions of two new cryptic species. Zootaxa 3881, 201 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Zheng Y & Wiens JJ Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol 94, 537–547 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Douglas ME, Douglas MR, Schuett GW, Beck DD & Sullivan BK Conservation phylogenetics of helodermatid lizards using multiple molecular markers and a supertree approach. Mol. Phylogenet. Evol 55, 153–167 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Castoe TA et al. Dynamic Nucleotide Mutation Gradients and Control Region Usage in Squamate Reptile Mitochondrial Genomes. Cytogenet. Genome Res 127, 112–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townsend TM et al. Phylogeny of iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species-tree approaches for an ancient, rapid radiation. Mol. Phylogenet. Evol 61, 363–380 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Alfaro ME et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl. Acad. Sci 106, 13410–13414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders KL & Lee MSY Molecular evidence for a rapid late-Miocene radiation of Australasian venomous snakes (Elapidae, Colubroidea). Mol. Phylogenet. Evol 46, 1165–1173 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Okajima Y & Kumazawa Y Mitogenomic perspectives into iguanid phylogeny and biogeography: Gondwanan vicariance for the origin of Madagascan oplurines. Gene 441, 28–35 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Kumazawa Y Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene 388, 19–26 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Hugall AF, Foster R & Lee MSY Calibration Choice, Rate Smoothing, and the Pattern of Tetrapod Diversification According to the Long Nuclear Gene RAG-1. Syst. Biol 56, 543–563 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Wiens JJ, Brandley MC & Reeder TW Why does a trait evolve multiple times within a clade? Repeated evolution of snakelike body form in squamate reptiles. Evolution 60, 123–41 (2006). [PubMed] [Google Scholar]
- 30.Pyron RA, Burbrink FT & Wiens JJ A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol 13, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y & Wiens JJ Do missing data influence the accuracy of divergence-time estimation with BEAST? Mol. Phylogenet. Evol 85, 41–49 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Suleski M & Hedges SB TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Hsiang AY et al. The origin of snakes: revealing the ecology, behavior, and evolutionary history of early snakes using genomics, phenomics, and the fossil record. BMC Evol. Biol 15, 87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tolley KA, Townsend TM & Vences M Large-scale phylogeny of chameleons suggests African origins and Eocene diversification. Proc. R. Soc. B Biol. Sci 280, 20130184–20130184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones ME et al. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara). BMC Evol. Biol 13, 208 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portik DM & Papenfuss TJ Monitors cross the Red Sea: The biogeographic history of Varanus yemenensis. Mol. Phylogenet. Evol 62, 561–565 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Pyron RA A Likelihood Method for Assessing Molecular Divergence Time Estimates and the Placement of Fossil Calibrations. Syst. Biol 59, 185–194 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Vidal N et al. Molecular evidence for an Asian origin of monitor lizards followed by Tertiary dispersals to Africa and Australasia. Biol. Lett 8, 853–855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong Z et al. Draft genome of the leopard gecko, Eublepharis macularius. Gigascience 5, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streicher JW & Wiens JJ Phylogenomic analyses of more than 4000 nuclear loci resolve the origin of snakes among lizard families. Biol. Lett 13, 20170393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiens JJ et al. Combining Phylogenomics and Fossils in Higher-Level Squamate Reptile Phylogeny: Molecular Data Change the Placement of Fossil Taxa. Syst. Biol 59, 674–688 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Fry BG et al. Early evolution of the venom system in lizards and snakes. Nature 439, 584–8 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Lee MSY Hidden support from unpromising data sets strongly unites snakes with anguimorph ‘lizards’. J. Evol. Biol 22, 1308–1316 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Silva L & Antunes A Vomeronasal Receptors in Vertebrates and the Evolution of Pheromone Detection. Annu. Rev. Anim. Biosci 5, 353–370 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Brykczynska U, Tzika AC, Rodriguez I & Milinkovitch MC Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol. Evol 5, 389–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green RE et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science (80-.). 346, 1254449–1254449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zippel HP The ecology of vertebrate olfaction. Behav. Processes 7, 198–199 (2002). [Google Scholar]
- 48.Yang H, Shi P, Zhang Y & Zhang J Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics 86, 306–315 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Fabregat A et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fabregat A et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics 18, 142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu G & Haw R Functional Interaction Network Construction and Analysis for Disease Discovery. in Methods in molecular biology (Clifton, N.J.) 1558, 235–253 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Shultz AJ & Sackton T Immune genes are hotspots of shared positive selection across birds and mammals. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riquelme CA et al. Fatty Acids Identified in the Burmese Python Promote Beneficial Cardiac Growth. Science (80-.). 334, 528 LP – 531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falkenberg M et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet 31, 289–294 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Cotney J, McKay SE & Shadel GS Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum. Mol. Genet 18, 2670–2682 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho Y, Hazen BC, Russell AP & Kralli A Peroxisome Proliferator-activated Receptor γ Coactivator 1 (PGC-1)- and Estrogen-related Receptor (ERR)-induced Regulator in Muscle 1 (PERM1) Is a Tissue-specific Regulator of Oxidative Capacity in Skeletal Muscle Cells. J. Biol. Chem 288, 25207–25218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho Y et al. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. FASEB J 30, 674–687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S et al. Regulation of Cellular Metabolism by Protein Lysine Acetylation. Science (80-.). 327, 1000–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brzezniak LK, Bijata M, Szczesny RJ & Stepien PP Involvement of human ELAC2 gene product in 3’ end processing of mitochondrial tRNAs. RNA Biol 8, 616–626 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Holzmann J et al. RNase P without RNA: Identification and Functional Reconstitution of the Human Mitochondrial tRNA Processing Enzyme. Cell 135, 462–474 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Lee K-W & Bogenhagen DF Assignment of 2′-O-Methyltransferases to Modification Sites on the Mammalian Mitochondrial Large Subunit 16 S Ribosomal RNA (rRNA). J. Biol. Chem 289, 24936–24942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cingolani HE et al. The Positive Inotropic Effect of Angiotensin II. Hypertension 47, 727–734 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Forrester SJ et al. Angiotensin II Signal Transduction: An Update on Mechanisms of Physiology and Pathophysiology. Physiol. Rev 98, 1627–1738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S & Iwao H Molecular and Cellular Mechanisms of Angiotensin II-Mediated Cardiovascular and Renal Diseases. Pharmacol. Rev 52, 11 LP – 34 (2000). [PubMed] [Google Scholar]
- 65.Symons JD & Stebbins CL Effects of angiotensin II receptor blockade during exercise: comparison of losartan and saralasin. J. Cardiovasc. Pharmacol 28, 223–31 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Stebbins CL & Symons JD Role of angiotensin II in hemodynamic responses to dynamic exercise in miniswine. J. Appl. Physiol 78, 185–90 (1995). [DOI] [PubMed] [Google Scholar]
- 67.WILSON JX The Renin-Angiotensin System in Nonmammalian Vertebrates. Endocr. Rev 5, 45–61 (1984). [DOI] [PubMed] [Google Scholar]
- 68.Fournier D, Luft FC, Bader M, Ganten D & Andrade-Navarro MA Emergence and evolution of the renin–angiotensin–aldosterone system. J. Mol. Med 90, 495–508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mueller CA, Eme J, Tate KB & Crossley DA Chronic captopril treatment reveals the role of ANG II in cardiovascular function of embryonic American alligators (Alligator mississippiensis). J. Comp. Physiol. B 188, 657–669 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Antl M et al. IRAG mediates NO/cGMP-dependent inhibition of platelet aggregation and thrombus formation. Blood 109, 552–559 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Puetz J & Boudreaux MK Evaluation of the gene encoding calcium and diacylglycerol regulated guanine nucleotide exchange factor I (CalDAG-GEFI) in human patients with congenital qualitative platelet disorders. Platelets 23, 401–403 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Bezman NA et al. Requirements of SLP76 tyrosines in ITAM and integrin receptor signaling and in platelet function in vivo. J. Exp. Med 205, 1775–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Israels S & McMillan-Ward E CD63 modulates spreading and tyrosine phosphorylation of platelets on immobilized fibrinogen. Thromb. Haemost 93, 311–318 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Cooper DN, Millar DS, Wacey A, Pemberton S & Tuddenham EG Inherited factor X deficiency: molecular genetics and pathophysiology. Thromb. Haemost 78, 161–172 (1997). [PubMed] [Google Scholar]
- 75.Takahashi N, Takahashi Y & Putnam FW Primary structure of blood coagulation factor XIIIa (fibrinoligase, transglutaminase) from human placenta. Proc. Natl. Acad. Sci 83, 8019 LP – 8023 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mosesson MW The roles of fibrinogen and fibrin in hemostasis and thrombosis. Semin. Hematol 29, 177–188 (1992). [PubMed] [Google Scholar]
- 77.Pyron R, Burbrink FT & Wiens JJ A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol 13, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pokorná M & Kratochvíl L Phylogeny of sex-determining mechanisms in squamate reptiles: Are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc 156, 168–183 (2009). [Google Scholar]
- 79.Rovatsos M, Pokorna M, Altmanova M & Kratochvil L Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett 10, 20131093–20131093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gamble T et al. Restriction Site-Associated DNA Sequencing (RAD-seq) Reveals an Extraordinary Number of Transitions among Gecko Sex-Determining Systems. Mol. Biol. Evol 32, 1296–1309 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Nielsen SV, Banks JL, Diaz RE, Trainor PA & Gamble T Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae). J. Evol. Biol 31, 484–490 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Rovatsos M, Altmanová M, Pokorná M & Kratochvíl L Conserved sex chromosomes across adaptively radiated anolis lizards. Evolution (N. Y). 68, 2079–2085 (2014). [DOI] [PubMed] [Google Scholar]
- 83.Gamble T et al. The Discovery of XY Sex Chromosomes in a Boa and Python. Curr. Biol 27, 2148–2153.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Emerson JJ Evolution: A Paradigm Shift in Snake Sex Chromosome Genetics. Curr. Biol 27, R800–R803 (2017). [DOI] [PubMed] [Google Scholar]
- 85.Hattori RS et al. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci 109, 2955 LP – 2959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cortez D et al. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Bej DK, Miyoshi K, Hattori RS, Strüssmann CA & Yamamoto Y A Duplicated, Truncated amh Gene Is Involved in Male Sex Determination in an Old World Silverside. G3 Genes|Genomes|Genetics 7, 2489–2495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ieda R et al. Identification of the sex-determining locus in grass puffer (Takifugu niphobles) provides evidence for sex-chromosome turnover in a subset of Takifugu species. PLoS One 13, e0190635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Halpern M Nasal chemical senses in reptiles: structure and function Pp 423–523 in Gans C, Crews D (eds) Biology of the Reptilia, Vol. 18, Brain. Horm. Behav Chicago/IL Univ. Chicago Press Google Sch; (1992). [Google Scholar]
- 90.Martin J & Lopez P Chemoreception, symmetry and mate choice in lizards. Proc. R. Soc. B Biol. Sci 267, 1265–1269 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baeckens S, Martín J, García-Roa R & van Damme R Sexual selection and the chemical signal design of lacertid lizards. Zool. J. Linn. Soc 183, 445–457 (2018). [Google Scholar]
- 92.van Damme R, Bauwens D, Thoen C, Vanderstighelen D & Verheyen RF Responses of Naive Lizards to Predator Chemical Cues. J. Herpetol 29, 38 (1995). [Google Scholar]
- 93.van Damme R & Castilla AM Chemosensory predator recognition in the lizard Podarcis hispanica: effects of predation pressure relaxation. J. Chem. Ecol 22, 13–22 (1996). [DOI] [PubMed] [Google Scholar]
- 94.Cooper WE Correlated evolution of prey chemical discrimination with foraging, lingual morphology and vomeronasal chemoreceptor abundance in lizards. Behav. Ecol. Sociobiol 41, 257–265 (1997). [Google Scholar]
- 95.Cooper W Tandem evolution of diet and chemosensory responses in snakes. Amphibia-Reptilia 29, 393–398 (2008). [Google Scholar]
- 96.Hulbert AJ & Else PL Evolution of mammalian endothermic metabolism: mitochondrial activity and cell composition. Am. J. Physiol. Integr. Comp. Physiol 256, R63–R69 (1989). [DOI] [PubMed] [Google Scholar]
- 97.Castoe TA et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl. Acad. Sci. U. S. A 110, 20645–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duan J et al. Transcriptome analysis of the response of Burmese python to digestion. Gigascience 6, 1–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gleeson TT, Mitchell GS & Bennett AF Cardiovascular responses to graded activity in the lizards Varanus and Iguana. Am. J. Physiol. Integr. Comp. Physiol 239, R174–R179 (1980). [DOI] [PubMed] [Google Scholar]
- 100.Agaba M et al. Giraffe genome sequence reveals clues to its unique morphology and physiology. Nat. Commun 7, 11519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weisenfeld NI, Kumar V, Shah P, Church DM & Jaffe DB Direct determination of diploid genome sequences. Genome Res 27, 757–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kichigin IG et al. Evolutionary dynamics of Anolis sex chromosomes revealed by sequencing of flow sorting-derived microchromosome-specific DNA. Mol. Genet. Genomics 291, 1955–1966 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Makunin AI et al. Contrasting origin of B chromosomes in two cervids (Siberian roe deer and grey brocket deer) unravelled by chromosome-specific DNA sequencing. BMC Genomics 17, 618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011). [Google Scholar]
- 105.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM (2013). [Google Scholar]
- 106.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Quinlan AR, Pedersen BS & Dale RK Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics 27, 3423–3424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kielbasa SM, Wan R, Sato K, Horton P & Frith M Adaptive seeds tame genomic sequence comparison. Genome Res (2011). doi: 10.1101/gr.113985.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kent WJ, Baertsch R, Hinrichs A, Miller W & Haussler D Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci 100, 11484–11489 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smit A, Hubley R & Green P Repeatmasker Open-4.0 (2013). Available at: http://www.repeatmasker.org. (Accessed: 10th January 2015)
- 111.Cantarel BL et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18, 188–96 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stanke M & Morgenstern B AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33, W465–7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Slater G & Birney E Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haas BJ et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8, 1494–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jones P et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Emms DM & Kelly S OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16, 157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lowe TM & Eddy SR tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25, 955–64 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Griffiths-Jones S, Bateman A, Marshall M, Khanna A & Eddy SR Rfam: an RNA family database. Nucleic Acids Res 31, 439–41 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nawrocki EP & Eddy SR Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Löytynoja A Phylogeny-aware alignment with PRANK. in Methods in molecular biology (Clifton, N.J.) 1079, 155–170 (2014). [DOI] [PubMed] [Google Scholar]
- 122.Nguyen LT, Schmidt HA, Von Haeseler A & Minh BQ IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A & Jermiin LS ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoang DT, Chernomor O, von Haeseler A, Minh BQ & Vinh LS UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith SA, Brown JW & Walker JF So many genes, so little time: A practical approach to divergence-time estimation in the genomic era. PLoS One 13, e0197433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Han MV, Thomas GWC, Lugo-Martinez J & Hahn MW Estimating Gene Gain and Loss Rates in the Presence of Error in Genome Assembly and Annotation Using CAFE 3. Mol. Biol. Evol 30, 1987–1997 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Mitchell AL et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res (2018). doi: 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 129.Krogh A, Larsson B, von Heijne G & Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11Edited by F. Cohen. J. Mol. Biol 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 130.Katoh K & Standley DM MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Capella-Gutiérrez S, Silla-Martínez JM & Gabaldón T trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Suyama M, Torrents D & Bork P PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Smith MD et al. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol 32, 1342–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pond SLK, Frost SDW & Muse SV HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled Komodo dragon genome is available in NCBI under the accession SJPD00000000. All DNA sequencing used to generate the assembly is available in the NCBI SRA database under accession PRJNA523222. Illumina sequencing data from chromosome pools is available in the NCBI SRA under accession PRJNA529483. RNA-seq data of heart tissue is available in the NCBI SRA under accession PRJNA527313. Original protein annotations, noncoding RNA annotations, all alignments for phylogenetic analyses and selection analyses, and newick files of phylogenetic trees are available in the following Figshare repositories: doi:10.6084/m9.figshare.7961135.v1, doi:10.6084/m9.figshare.7955891.v1, doi:10.6084/m9.figshare.7955879.v1, doi:10.6084/m9.figshare.7949483.v1, doi:10.6084/m9.figshare.7759496.v1, doi:10.6084/m9.figshare.7967300. The project folder for all Figshare data is available at https://figshare.com/projects/Data_for_Komodo_dragon_genome_paper/61271.