Abstract
We improve the potency of antibody-drug conjugates (ADCs) containing the HER2-specific antibody pertuzumab by reducing their affinity for HER2 by >250-fold at acidic endosomal pH relative to near neutral pH. These engineered pertuzumab variants show increased lysosomal delivery and cytotoxicity towards tumor cells expressing intermediate HER2 levels. In HER2int xenograft tumor models in mice, the variants show higher therapeutic efficacy than the parent ADC and a clinically-approved HER2-specific ADC.
Antibody-drug conjugates (ADCs) combine the high specificity of antibodies with the potent cytotoxicity of drugs1,2. However, therapeutic efficacy of current ADCs requires relatively high levels of expression of the tumor target1,2. Although increased payload delivery could be achieved by delivering higher ADC doses, this approach can lead to unacceptable toxicities towards normal cells3,4. In addition, ADC treatment can result in a reduction of surface biomarker levels, further narrowing the therapeutic window5.
A possible pathway towards overcoming these limitations is to generate ADCs that deliver their payload more efficiently to target cells. The majority of current ADCs are designed to enter lysosomes following internalization into cells1,2. Typically, ADCs have high affinity for their target at endosomal, acidic pH, and payload delivery is associated with target entry into degradative lysosomal compartments1,2. Engineering the ADC to confer endosomal dissociation from its target is expected to enable payload entry into lysosomes and recycling of unbound target. A possible strategy towards achieving this is to generate engineered ‘acid-switched’ ADCs that bind target with substantially higher affinity at near neutral, extracellular pH relative to acidic endosomal pH, leading to endosomal dissociation of the ADC following internalization into cells.
The growth factor receptor, HER2, is a validated target for the treatment of HER2-overexpressing tumors, and several antibody-based therapies that include pertuzumab6 and the ADC trastuzumab-DM1 (T-DM1)7 are approved for clinical use. However, targeting HER2 with T-DM1 has met with disappointing results for HER2-positive tumors that do not overexpress this receptor2,8, supporting the need to develop ADCs with improved drug delivery properties. Here, we have generated acid-switched variants of the HER2-specific antibody pertuzumab6 for use as ADCs. Analysis of the interactions of trastuzumab9 and pertuzumab6 with recombinant HER2-ECD (HER2 extracellular domain-Fc fusion) demonstrated that although the affinity of trastuzumab for HER2 is similar across the pH range 6.0-7.4, that of pertuzumab is around 10-fold higher at pH 7.4 than pH 6.0 (Supplementary Fig. 1). Based on earlier studies using parent antibodies with some intrinsic pH-dependence as starting templates to produce antibodies with increased pH-dependent binding to their soluble targets10-12, we therefore chose pertuzumab for engineering to generate acid-switched variants.
First, complementarity-determining region (CDR) residues that either directly interact with, or are in proximity to, HER2 residues6 were mutated to histidine (Supplementary Fig. 2a). Second, CDR residues were randomly mutated to generate phage display libraries followed by panning against HER2-ECD to isolate acid-switched variants (Supplementary Fig. 2b). Using histidine replacement, two mutated variants of pertuzumab (Y55H, CDRL2; S103H, CDRH3) were combined to generate the acid-switched variant ‘YS’ (Y55H-VL/S103H-VH) (Supplementary Fig. 2c). An acid-switched variant with S55H and G57E (‘SG’) mutations in CDRH2 was also selected using phage display (Supplementary Figs. 2d,3). The binding of YS and SG (as human IgG1/κ) to HER2 was analyzed at different pH values (Supplementary Fig. 4). YS and SG have similar affinities for HER2 at pH 7.4, whereas the binding affinities of SG decrease more than those of YS as the pH is lowered (Supplementary Fig. 4). Both YS and SG showed favorable stability, including in human serum (Supplementary Fig. 5).
We conjugated WT pertuzumab, YS and SG through hinge cysteines to maleimidocaproyl-valine-citrulline-p-aminobenzoyloxycarbonyl-monomethyl auristatin E (MC-VC-PAB-MMAE). We chose this over random conjugation of lysines to the maytansinoid DM1 to facilitate the generation of homogeneously conjugated ADCs. MMAE is also the most commonly used cytotoxic drug for ADCs currently in the clinic3. The cysteines that form disulfide bonds in the hinge or hinge/Cκ domain (Cys220, Cys229 of heavy chain; Cys214 of light chain) were mutated to serines to enable conjugation to Cys226 with a drug-antibody ratio (DAR) of two. This conjugation level was used to reduce the hydrophobicity reported for ADCs with higher DARs13. Hydrophobic interaction and size exclusion column analyses indicated that the DARs range from 1.7-1.9 with no indication of aggregation (Supplementary Fig. 6a). The ADCs were also stable in human serum and retained similar binding properties for HER2 as their parent, unconjugated antibodies (Supplementary Fig. 6b,c).
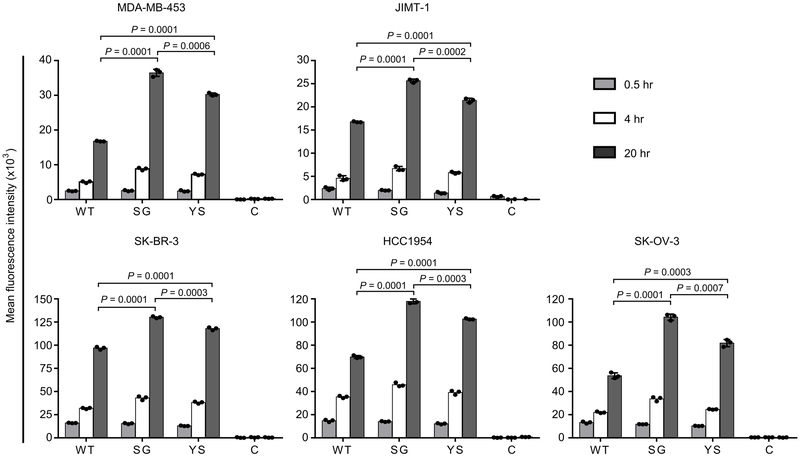
We next investigated the accumulation of SG-MMAE and YS-MMAE in tumor cell lines expressing HER2 at different levels14,15 (Supplementary Fig. 7). To quantitate the intracellular accumulation of the acid-switched pertuzumab-MMAE variants, cells were treated with Alexa 488-labeled ADCs followed by quenching of surface signal using an Alexa 488-specific antibody15,16 (Fig. 1). Higher intracellular levels of YS-MMAE and SG-MMAE were observed in all HER2-expressing cells following incubation for 4 and 20 hours compared with WT pertuzumab conjugated to MMAE (WT-MMAE), with greater levels of SG-MMAE relative to YS-MMAE (Fig. 1). In addition, both SG-MMAE and YS-MMAE accumulated more efficiently than WT-MMAE in lysosomes within HER2int MDA-MB-453 cells (Supplementary Fig. 8).
Figure 1.
Acid-switching results in increased accumulation of ADCs in HER2-expressing tumor cells. Cells were incubated with 10 nM Alexa 488-labeled MMAE-conjugated antibody (WT, SG, YS or control hen egg lysozyme-specific antibody, C) for the indicated times at 37 °C, washed, incubated with 5 μg/ml Alexa 488-specific antibody and analyzed by flow cytometry. Mean fluorescence intensities (mean values of independent triplicate cell samples) for Alexa 488 fluorescence are shown. Error bars indicate SD. Statistically significant differences are indicated by * (unpaired two-tailed t-test). Two independent experiments were carried out with similar results.
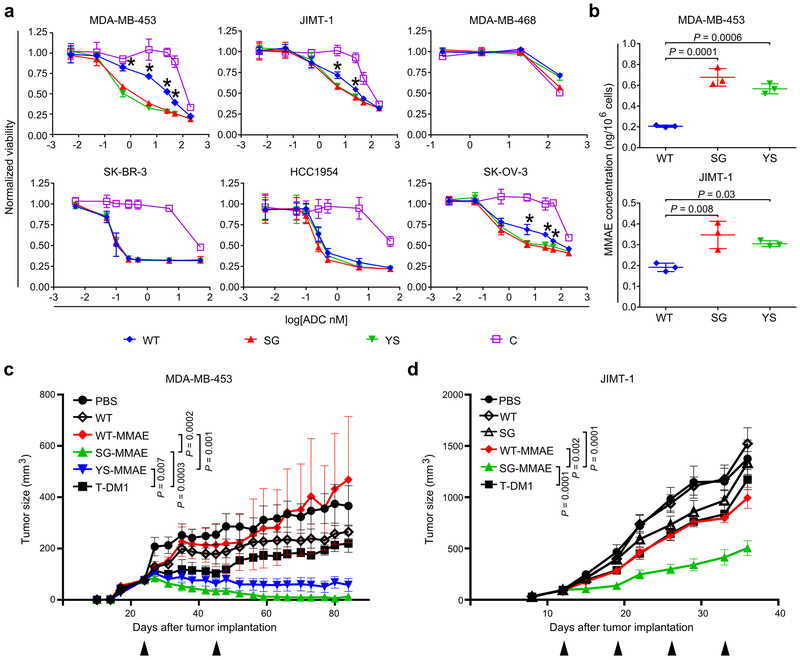
The acid-switched pertuzumab variants, or ALTAs (for ADCs with increased lysosomal trafficking activity), were more effective than WT-MMAE in reducing the viability of the HER2int cell lines, MDA-MB-453 and JIMT-1, and one HER2hi cell line, SK-OV-3 (Fig. 2a). The ALTAs showed similar cytotoxicity as WT-MMAE towards two other HER2hi cell lines, SK-BR-3 and HCC1954, and the HER2-negative cell line, MDA-MB-46814. It is possible that internalization of a relatively low proportion of surface HER2 molecules is sufficient for cytotoxicity towards HER2-overexpressing cells, leading to lower dependence on endosomal HER2:ADC dissociation and ADC reloading. Quantitative analyses using LC-MS/MS also demonstrated increased MMAE levels in MDA-MB-453 and JIMT-1 cells following treatment with SG-MMAE or YS-MMAE compared with WT-MMAE (Fig. 2b).
Figure 2.
ALTAs are more effective at reducing proliferation of HER2int-expressing cells and deliver increased levels of MMAE to target cells. (a) HER2-expressing cancer cells, or HER2-negative (MDA-MB-468) cells, were treated with MMAE-conjugated antibodies (WT, SG, YS or control hen egg lysozyme-specific antibody, C) for 4 days and cell viability determined. (b) MDA-MB-453 and JIMT-1 cancer cells were incubated with 10 nM MMAE-conjugated antibodies (WT, SG or YS) for 20 hours and cell-associated MMAE quantitated using LC-MS/MS. For (a) and (b), mean values of independent triplicate cell samples are shown and error bars indicate SD. Statistically significant differences between YS or SG and WT are indicated by * (one-way ANOVA with Tukey’s multiple comparison test; for panel (a), P values ranged from 0.001-0.0001 (MDA-MB-453), 0.003-0.008 (JIMT-1) and 0.001-0.05 (SK-OV-3)). (c) Female BALB/c SCID mice bearing MDA-MB-453 tumors were treated twice, with a 21 day interval (arrowheads), with 2 mg/kg ADC (WT-MMAE, SG-MMAE or YS-MMAE), T-DM1, unconjugated WT pertuzumab (WT) or vehicle (PBS) (n = 7 mice/group for YS-MMAE, T-DM1 or PBS; 8 mice/group for WT-MMAE, SG-MMAE or WT). (d) Female BALB/c SCID mice bearing JIMT-1 tumors were treated weekly (four times; arrowheads) with 2 mg/kg ADC (WT-MMAE or SG-MMAE), T-DM1, unconjugated WT or SG, or vehicle (PBS) (n = 6 mice/group for PBS; 7 mice/group for WT-MMAE, SG-MMAE, T-DM1, WT or SG). For (c) and (d), the mean tumor size for each treatment group is shown and error bars indicate SE. Statistically significant differences at the experimental endpoints are indicated by * (two-tailed Mann-Whitney U test and unpaired two-tailed t-test, respectively). For panels (a)-(d), at least two independent experiments were carried out with similar results.
Analyses in BALB/c SCID mice demonstrated that WT-MMAE, SG-MMAE and YS-MMAE have similar pharmacokinetic behavior (Supplementary Fig. 9). The therapeutic effects of the ALTAs were next compared with WT-MMAE and trastuzumab-DM1 (T-DM1) in mouse xenograft models expressing intermediate HER2 levels. We used T-DM1 instead of trastuzumab-MMAE as a comparator to benchmark the activity of the ALTAs with this clinically approved, HER2-specific ADC. A dose (2 mg/kg) that is suboptimal for T-DM114 was used to evaluate whether the ALTAs had greater anti-tumor effects. Treatment with SG-MMAE or YS-MMAE was more effective than WT-MMAE, T-DM1 or WT pertuzumab (unconjugated) in inhibiting the growth of MDA-MB-453 xenografts, with three mice in the SG-MMAE group (n = 8) having undetectable tumors at the experimental endpoint (Fig. 2c, Supplementary Fig. 10a). We also treated mice bearing JIMT-1 tumors, that are relatively resistant to T-DM114, with 2 mg/kg SG-MMAE, WT-MMAE, T-DM1 and, as controls, unconjugated WT pertuzumab or SG (Fig. 2d). The therapeutic effect of SG-MMAE was significantly higher than that of WT-MMAE, T-DM1, and unconjugated control antibodies (Fig. 2d, Supplementary Fig. 10b). In all therapy experiments, ADCs were well tolerated with no weight loss or indication of liver toxicity (Supplementary Fig. 10c,d).
We also carried out modeling studies to investigate the molecular basis of the reduced affinity at pH 5.8 of the lead acid-switched antibody, SG, for HER2 relative to WT pertuzumab (Supplementary Fig. 11). These analyses resulted in a model in which protonation of His55 in CDRH2, that replaces Ser55 in WT pertuzumab, at Nε of the imidazole ring predominates over that of Nδ1 at near neutral pH, whereas the protonated Nδ1 tautomer is favored at acidic pH (5.8). Protonation of Nδ1 leads to an electrostatic interaction with Glu57 in CDRH2 of SG in combination with interactions with Tyr252 of HER2. By contrast with the solvent exposure of Glu57 and Tyr252 in the SG:HER2 complex at pH 7.4, the change in protonation state of histidine at pH 5.8 results in reorientation of Glu57 and Tyr252, leading to an unfavorable desolvation penalty that is a primary contributor to the loss of affinity of SG at acidic pH.
Observations made during the use of ADCs in the clinic have indicated that dose-limiting toxicities for these therapeutics are usually ‘off-target’ rather than ‘on-target/off-tumor’3,4. Although the processes leading to off-target toxicities are currently not well understood, ADC entry into cells via non-specific pathways such as fluid-phase pinocytosis is a probable contributor. Further, endocytosis into FcγR-expressing cells may play a role3,4. Consequently, the generation of ADCs such as ALTAs that achieve therapeutic effects at lower doses is expected to lead to reduced undesirable toxicities. In this context, the activity of the IgG-recycling receptor, FcRn, in normal epithelium17,18, by contrast with reports that this receptor can be at low to undetectable levels in breast, prostate or lung cancer cells19,20, could also regulate the subcellular trafficking behavior of ALTAs to decrease both on- and off-target toxicities.
In sum, we have described ADCs comprising acid-switched pertuzumab variants that, based on improved intracellular trafficking, result in increased payload delivery to target cells. These ALTAs have higher therapeutic efficacy against HER2int tumor cells compared with ADCs containing the parent, WT antibody or T-DM1. This approach not only has the potential to treat tumors expressing a broad range of HER2 levels, but also promises to have general applicability in overcoming the current limitations of dose-limiting toxicities for other tumor targets.
ONLINE METHODS
Cell lines and reagents
The following cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA): SK-BR-3, SK-OV-3, MDA-MB-453, and MDA-MB-468. SK-BR-3 and SK-OV-3 cells were cultured in McCoy’s 5a medium, whereas MDA-MB-453 and MDA-MB-468 cells were cultured in RPMI1640 medium. The breast cancer cell line JIMT-1 was obtained from AddexBio and cultured in DMEM medium. The HCC1954 cell line was a generous gift from Drs. Adi Gazdar, John Minna, and Kenneth Huffman (Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center at Dallas) and was cultured in RPMI1640 medium. All media were supplemented with 1% penicillin/streptomycin, 1% GlutaMAX (Thermo Fisher Scientific, Waltham, MA, USA), 1% sodium pyruvate and 10% fetal calf serum. The cell lines were tested monthly for mycoplasma contamination and were authenticated annually at the University of Arizona Genetics Core through DNA fingerprint analysis.
Histidine scanning of pertuzumab
The sequences encoding the pertuzumab variable heavy (VH) and variable light (VL) chain domains6 were synthesized by GenScript (Piscataway, NJ, USA) and used to generate an expression vector to produce pertuzumab Fab fragments (human CH1, Cκ; C-terminal polyhistidine tag on Cκ domain) using standard methods of molecular biology. Residues in the complementarity-determining regions (CDRs) of the VH and VL domain genes of pertuzumab were selected for histidine scanning using the crystal structure of pertuzumab in complex with HER2 (Protein Data Bank accession code 1N8Z)6 in PyMOL (Schrödinger, New York, NY, USA) as a guide. The following residues, that are in proximity to or contact HER2, were targeted: Asp31, Tyr32, Asn54, Tyr60, Leu100, Gly101, Pro102, Ser103, Tyr105, Asp107 in the VH domain and Tyr55 in the VL domain. These residues were mutated to histidine using the PCR and standard methods, and the corresponding Fab fragments were expressed as periplasmically secreted proteins using E. coli as an expression host21. Fab fragments were purified using Ni2+-NTA-agarose21.
Generation of phage display libraries
To express pertuzumab as a single chain Fv fragment (scFv) using the phagemid vector, pHEN122, the expression vector for the pertuzumab Fab fragment was modified using standard methods of molecular biology to insert a linker peptide [(Gly4Ser)3Gly2Ser linker] between the pertuzumab VH and VL domain genes, followed by cloning into pHEN1 as a NotI-SfiI fragment. Libraries were generated with random mutations in CDR1, 2 and 3 of the VH domain (targeting Asp31, Tyr32 and Thr33; Asn75, Ser76, Gly77 and Gly78; Gly101, Ser103, Tyr105 and Tyr108) and CDR2 of the VL domain (targeting Ile48, Tyr49 and Ser50 or Ala51, Tyr53, and Tyr55). The following oligonucleotides were used in PCRs with standard methods to generate five libraries: Library 1: CDRH1Back, 5’ GCT TCT GGA TTC ACA TTC ACA NNB NNB NNB ATG GAT TGG GTG AGA CAG GCT 3’ and CDRH1For, 5’ TGT GAA TGT GAA TCC AGA AG C 3’; Library 2: CDRH2Back, 5’ TGG GTG GCT GAT GTG AAT CCT NNB NNB NNB NNB TCT ATC TAC AAT CAG AGA TTC 3’ and CDRH2For, 5’ AGG ATT CAC ATC AGC CAC CCA 3’; Library 3: CDRH3Back, 5’ TAC TAC TGT GCT AGA AAT CTG NNB CCT NNB TTC NNB TTC GAT NNB TGG GGA CAG GGA ACA CTG 3’ and CDRH3For, 5’ CAG ATT TCT AGC ACA GTA GTA 3’; Library 4: CDRL2-1Back, 5’ CCT AAG CTG CTG ATC TAC TCT NNB TCT NNB AGA NNB ACA GGA GTG CCT TCT AGA 3’ and CDRL2-1For, 5’ AGA GTA GAT CAG CAG CTT AGG 3’; Library 5: CDRL2-2Back, 5’ GGA AAG GCT CCT AAG CTG CTG NNB NNB NNB GCT TCT TAC AGA TAC ACA GGA 3’ and CDRL2-2For5, 5’ CAG CAG CTT AGG AGC CTT TCC 3’ (N = A, C, T or G; B = C, T, or G). The resulting gene libraries were transfected into E. coli TG1 (Lucigen, Middleton, WI, USA) to generate libraries of 2.6 ×106-1.7 × 108 clones. Each library was pooled and grown overnight at 30 °C in 4XTY broth supplemented with M13KO7 helper phage (New England Biolabs, Ipswich, MA, USA), 100 μg/ml ampicillin, and 50 μg/ml kanamycin. Phage were precipitated with a solution of PEG-8000 and NaCl at a concentration of 4% PEG-8000/0.5 M NaCl followed by centrifugation at 10,000 RCF. The precipitated phage were washed with 4% (w/v) PEG-8000/0.5 M NaCl prior to suspension in 1% (w/v) bovine serum albumin (BSA)/phosphate buffered saline (PBS). To select phage that display pertuzumab-derived scFvs that bind to HER2 with increased pH-dependence, recombinant phage were first pre-panned in Nunc 96-well flat-bottom immuno plates (Thermo Fisher Scientific) coated with 4% (w/v) milk powder/PBS (pH 7.4), and subsequently transferred to 96-well Nunc plates that had been coated with 2 μg/ml recombinant HER2-ECD (R&D Research, Minneapolis, MN, USA) and blocked with 4% (w/v) milk powder/PBS (pH 7.4). Following a 2 hour incubation, the plates were washed 5 times with PBS plus 0.1% Tween 20 (PBST) pH 7.4 followed by 5 washes with PBS pH 7.4. Bound phage were then eluted by the addition of 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 5.8 for 10 minutes. The eluted phage were amplified by re-infection of E. coli TG1, followed by two additional rounds of selection using 10 washes with PBST pH 7.4 and 10 washes with PBS pH 7.4. Eluted phage (pH 5.8) from the third round of panning were used to re-infect E. coli TG1, single clones expanded in 96 deep well plates and treated with helper phage using previously described methods22. Culture supernatants containing phage were used in ELISAs with plates coated with recombinant HER2-ECD, followed by washing with PBST pH 7.3 or pH 5.8. Bound phage were detected using anti-M13 antibody conjugated to horse radish peroxidase (HRP) (GE Healthcare, Chicago, IL, USA) at a 1:5,000 dilution in 4% milk powder in PBS, pH 7.3 or 5.8, followed by detection with 3’3’5,5’ tetramethylbenzidine substrate. For ELISAs using purified phage, phage suspensions were added to each well and binding at pH 5.8 and 7.4 was detected using the same protocol.
Recombinant antibodies
VH and VL domain genes encoding wild type or acid-switched pertuzumab variants (obtained by histidine scanning or phage display) were used to generate constructs for the expression of full length human IgG1, Cκ antibodies using standard methods of molecular biology and the vector, pcDNA3.4 (Life Technologies). The hinge disulfide bonds that link the Cκ domain to the hinge region and one hinge disulfide that links the two heavy chains to each another were removed by mutating the light chain cysteine (Cys214; EU numbering) and two heavy chain cysteines (Cys220, Cys229; EU numbering) to serine. Sequences of expression plasmids are available upon request.
For use as a control antibody, the VH and VL domains of the hen-egg lysozyme antibody, HuLys1023, were reformatted as heavy and light chain constructs to generate a full length human IgG1/κ antibody with one hinge disulfide (as above) using pcDNA3.4.
Recombinant antibodies were produced in Expi293F cells (Life Technology, Carlsbad, CA, USA) following transient transfection with the Expi293 expression system kit (Life Technology) as previously described24. Antibodies were purified from culture supernatants using protein G-Sepharose (GE Healthcare) and dialyzed against PBS. Purified antibodies were loaded onto a Hiload 16/600 Superdex 200 gel filtration column (GE Healthcare) to remove aggregates, followed by analyses of the ‘monomeric’ antibodies using a Yarra 3000 column (Phenomenex, Torrance, CA, USA).
Clinical grade trastuzumab and pertuzumab were acquired from the University of Texas Southwestern Medical Center Pharmacy.
Surface plasmon resonance analyses
Binding analyses were carried out using a Biacore 2000 or T200 (GE Healthcare). Flow cells of CM5 sensor chips were coupled using amine coupling chemistry with recombinant HER2-ECD (R&D Research) or coupling buffer only (10 mM sodium acetate pH 4.8) as a reference. Fab fragments or antibodies were injected over immobilized HER2-ECD (coupled to densities ranging from 426-956 relative units on flow cells of CM5 sensor chips) at a flow rate of 5 or 10 μl/min at 25 °C in PBS plus 0.01% (v/v) Tween 20 and 0.05% (w/v) sodium azide (pH 7.4, 7.0, 6.5, 6.0, 5.8). Flow cells were regenerated following each injection using 0.15 M NaCl/0.1 M glycine (pH 2.8) buffer. For full length antibodies, equilibrium dissociation constants were determined using previously described methods25 and custom written software (www.wardoberlab.com/software/sprtool) to yield apparent dissociation constants (due to bivalent binding of the antibodies to immobilized HER2). The KD of the YS mutant at pH 5.8 was too high to accurately determine (i.e. levels of binding close to saturation could not be reached), and was therefore estimated using the following equation: , where KD is the calculated equilibrium dissociation constant (nM), CK is the concentration of analyte flowed over the chip (nM), Rmax is the maximum analyte binding capacity (RU), and Req(Ck) is the equilibrium signal for concentration CK (RU).
Conjugation and labeling of antibodies
Antibodies (10 μM) in PBS were reduced by addition of 80 μM of tris(2-carboxyethyl)phosphine (TCEP) and 1 mM ethylenediaminetetraacetic acid (EDTA) for 2 hours at 37 °C. Reduced antibodies were cooled on ice and 40 μM maleimidocaproyl-valine-citrulline-p-aminobenzoylcarbonyl-monomethyl auristatin E (MMAE; Concortis, San Diego, CA, USA) was added. Following 4 hours incubation on ice, unconjugated MMAE was removed by extensive dialysis against PBS. Conjugated antibodies were analyzed using size exclusion chromatography (Yarra 3000) and hydrophobic interaction chromatography (HIC; TSKgel Butyl-NPR; Tosoh, Tokyo, Japan) to determine drug-to-antibody ratios (DARs). For HIC, solvent A was 10 mM potassium phosphate with 1.5 M ammonium sulfate at pH 7.0 and solvent B was 10 mM potassium phosphate at pH 7.0 with 20% (v/v) isopropanol. The gradient method used was 0-5 min (0% B), 5-15 min (0% to 100% B), and 15-20 min (100% B) at a flow rate of 0.5 ml/min.
ADCs were labeled with Alexa 488 Fluor (Thermo Fisher Scientific) or radiolabeled with 125I-Iodogen [Perkin Elmer or MP Biomedical (Solon, OH, USA)] as described previously26, except that a 1:1 molar ratio of Alexa 488:ADC was used during the labeling reaction.
Enzyme-linked immunosorbent assays
ADCs and antibodies were analyzed by ELISA for their binding to HER2-ECD (R&D Research). 96-well Nunc plates were coated with HER2-ECD at a concentration of 2 μg/ml overnight at 4 °C. ADCs and antibodies were diluted in 4% (w/v) milk powder/PBS (pH 5.8 or pH 7.0) and added to coated plates at concentrations ranging from 0.1 nM to 200 nM. Plate wells were washed 4 times with PBST (pH 5.8 or 7.0, as indicated). Antibodies and ADCs were detected with goat anti-human IgG (Fab-specific) conjugated with HRP (Sigma-Aldrich).
Serum stability analyses of antibodies and ADCs
Human serum (Sigma-Aldrich, St. Louis, MO, USA) was depleted of endogenous IgG by passage through a protein G-Sepharose column. Antibodies and ADCs were incubated at a concentration of 40 μg/ml in IgG-depleted serum for 0, 3, or 5 days at 37 °C followed by immunoprecipitation using agarose beads coupled to goat anti-human Fc-specific antibody (Sigma-Aldrich). Immunoprecipitated antibodies and ADCs were run on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (0.45 μm pore size; Genesee Scientific, San Diego, CA, USA). Immunoblotting was carried out using standard methods with goat anti-human IgG (H + L) antibodies conjugated with HRP (Jackson Immunoresearch, West Grove, PA, USA). HRP was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) followed by scanning with a C-DiGit blot scanner (LI-COR Biosciences, Lincoln, NE, USA).
ADC accumulation assay
Cancer cells were plated in 48-well plates and incubated overnight in a 37 °C incubator with 5% CO2, followed by treatment with 10 nM Alexa 488-labeled ADC in medium (pH 7.0) for 0.5, 4, and 20 hours. Treated cells were cooled on ice and surface fluorescence of Alexa 488-labeled ADC quenched with 5 μg/ml rabbit anti-Alexa 488 antibody (Thermo Fisher Scientific) for 30 minutes at 4 °C. The quenching efficiency was determined using separate cell samples which had been incubated with Alexa 488-labeled ADC on ice (to prevent internalization) followed by treatment with anti-Alexa 488 antibody or vehicle control. Samples were washed, harvested by trypsinization, resuspended in PBS, analyzed using a BD Accuri C6 flow cytometer (BD Biosciences, San Jose, CA, USA) and data processed with FlowJo (FlowJo, Ashland, OR, USA).
Fluorescence microscopy
MDA-MB-453 cells were plated in coverglass-bottomed Mattek dishes and allowed to adhere overnight in a 37 °C, 5% CO2 incubator. Cells were pre-treated with Alexa 647-labeled dextran (5 μM; pulsed for 2 hours and chased for 3 hours) followed by treatment with 10 nM Alexa 488-labeled WT-MMAE, SG-MMAE, or YS-MMAE for 4 or 20 hours. Samples were treated with 5 μg/ml rabbit anti-Alexa 488 antibody for 30 minutes on ice to quench surface Alexa 488 fluorescence, and subsequently fixed at room temperature with 3.4% (w/v) paraformaldehyde plus 0.025% (v/v) glutaraldehyde. Fixed cell samples were imaged with an Axio Observer Z1 inverted epifluorescent microscope (Zeiss, Oberkochen, Germany) equipped with a 63 X, 1.4 NA Plan-Apochromat objective (Zeiss), and a Zeiss 1.6 X internal optovar. A broadband LED lamp (X-cite 110LED, Excelitas Technologies, Waltham, MA, USA) was employed as the excitation source, and a digital CCD camera (Hamamatsu Orca-ER; Hamamatsu Photonics, Japan) as the detector. Standard fluorescent filter sets for Alexa 488 (FITC-3540C) and Alexa 647 (Cy5-4040C) were purchased from Semrock (Rochester, NY, USA). Images were acquired at room temperature using custom software written in Java and the C programming language. Acquired images were analyzed using the custom written software MIATool (www.wardoberlab.com/software/miatool)27. Acquired images for Alexa 488 were linearly adjusted with the same intensity adjustment settings. Independent linear adjustments were made for the Alexa 647 signal for each time point.
Cell viability assays
Cells were plated in 96-well plates at a density of 5,000 cells per well and treated with different concentrations of ADCs. Cells were treated for 4 days in a 37 °C, 5% CO2 incubator, followed by the use of CellTiter 96 AQueous One Solution Proliferation Assay kit (Promega, Madison, WI, USA) to determine cell viability using the manufacturer’s protocol.
Quantitation of MMAE using LC-MS/MS
Cancer cells were plated in 6 well plates and incubated overnight in a 37 °C, 5% CO2 incubator. Cells were treated with 10 nM ADC for 20 hours, followed by washing with PBS and harvesting by trypsinization. Trypsinized cells were counted and resuspended in 100% methanol containing 1.67 ng/ml deuterium-labeled MMAE (D8-MMAE; MedchemExpress, Monmouth Junction, NJ, USA) at a density of 8.3 × 105 cells/ml. Samples were lysed for 16 hours at −20 °C and then centrifuged at 21,100 RCF for 15 mins at 4 °C. The supernatants were transferred to 12 × 75 mm borosilicate glass tubes (Kimble Chase, Rockwood, TN, USA) and dried under a stream of nitrogen gas. The residue was resuspended in 20% methanol with 0.1% (v/v) formic acid (10% of original volume). To generate a standard curve, different concentrations of MMAE ranging from 0.017-3.33 ng/ml were mixed with 1.67 ng/ml D8-MMAE in lysates of untreated tumor cells and treated as above.
Targeted liquid chromatography mass spectrometry (LC-MS/MS) analysis was performed using a Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific) coupled to a binary pump HPLC (Ultimate 3000; Thermo Fisher Scientific). Chromatographic separation was achieved on a Hypersil Gold 5 μm, 50 mm × 3 mm column (Thermo Fisher Scientific) maintained at 30 °C using a solvent gradient method. Solvent A was H2O containing 0.1% (v/v) formic acid. Solvent B was acetonitrile containing 0.1% (v/v) formic acid. The gradient method used was 0-1 min (20% B to 60% B), 1-1.5 min (60% B to 95% B), 1.5-3 min (95% B), 3-4.1 min (95% B to 20% B), 4.1-5 min (20% B) with a flow rate of 0.5 ml/min. The sheath, auxiliary, and sweeping gasses were set at 50, 15, and 1 arbitrary units, respectively. The spray voltage was set to 3.5 kV and the vaporizer and capillary temperatures were maintained at 375 °C and 350 °C, respectively. Transitions of 718.8 m/z −134.1 m/z and 727.6 m/z – 134.1 m/z were used for the quantitation of MMAE and D8-MMAE, respectively. Sample acquisition and data analysis were performed using TraceFinder (Thermo Fisher Scientific).
Pharmacokinetic analyses
Pharmacokinetic experiments were carried out in 7-10-week old female BALB/c SCID mice using protocols approved by the Texas A&M University IACUC. Lugol solution (Sigma-Aldrich) was added to drinking water 48-72 hours prior to starting the experiment, and maintained throughout the experiment. Mice were injected intravenously with 15 μg 125I-labeled ADC in 200 μl PBS with 0.1% (w/v) BSA. Whole body counts were measured using an Atom Lab 100 dose calibrator (Biodex, Shirley, NY, USA) as described previously26 and β-phase half-lives determined by fitting exponentially decaying models to the data using gradient-based numerical optimization methods written in MATLAB. Mice were retroorbitally bled with 10 μl capillary tubes (Drummond Scientific, Broomall, PA, USA) and radioactive counts measured by gamma counting (Perkin Elmer, Waltham, MA, USA). Radioactivity counts were normalized to the values obtained immediately following injection.
Therapy experiments
Therapy experiments were carried out using protocols approved by the Texas A&M University IACUC. BALB/c SCID mice were purchased from Jackson Laboratories, bred and maintained in specific pathogen-free housing. 6-8-week old female mice were implanted with 3-5 × 106 MDA-MB-453 cells or 4-5 × 106 JIMT-1 cells suspended in 100 μl 50% RPMI1640 media and 50% Matrigel (Corning, Corning, NY, USA) in the mammary fat pad. When the tumor size reached a volume of 50-100 mm3, mice were randomized into groups and dosed intravenously with 2 mg/kg antibody, ADC, or vehicle twice with a three week interval (MDA-MB-453) or four times with one week intervals (JIMT-1). Tumor size was determined using the formula: , where the tumor length and tumor width were measured with digital calipers every 3-4 days for the duration of the experiment. When tumor volumes reached a size of ~2000 mm3, mice were euthanized. Alanine aminotransferase activity in serum samples from mice were analyzed using Alanine Aminotransferase Activity Assay Kits (Sigma-Aldrich) and the manufacturer’s protocol.
Modeling analyses
The X-ray crystallographic structure of the HER2-pertuzumab complex6 (PDB accession code: 1S78) was minimized using the molecular modeling software CHARMM v3628 in a CHARMM22 force field, and all missing residues and atoms were added. PROPKA v. 3.129 was used for empirical pKa prediction and protonation state determination for the minimized structure at pH 7.4 and 5.8, respectively. The resulting structure at either pH value was minimized for 5,000 steps with explicit solvent molecules and then subjected to a 10 ns molecular dynamics (MD) simulation using the computer program NAMD2 v. 2.1028. MD protocols followed similar details as described previously28. Snapshots were retained at the 0th ns (the beginning) and every ns from the 6th to the 10th ns to represent a conformational ensemble of the HER2-pertuzumab complex at either pH.
Structures of HER2 in complex with the SG mutant were modeled using iCFN30, an exact algorithm for multi-state protein design with substate ensembles, which was customized here for pH-dependence. First, the positive and negative substates were defined as conformational ensembles of the complex at pH 7.4 and 5.8, respectively. Substate energies were folding stabilities of the complex that include Coulomb electrostatics, van der Waals, calculated internal energies, and a nonpolar contribution to the hydration free energy based on solvent accessible surface area (SASA)30. A positive-substate stability cutoff was set at 10 kcal/mol and positive-versus-negative substate specificity was essentially not mandated with a cutoff of 1000 kcal/mol. Second, pertuzumab VH residues 55 and 57 were allowed to change to any amino acid, including histidines in δ−, ε−, and doubly protonated states. Residues within 5 Å from the mutated amino acids were allowed to be flexible. In addition, His245 of HER2 was allowed to be in any of the 3 possible protonation states and Asp50 of pertuzumab VH was allowed to be deprotonated or singly protonated at either oxygen atom on the side chain. Top conformations of each sequence-protonation combination in each substate (backbone conformation here) generated from iCFN for either pH were geometrically grouped into representatives and then screened by PROPKA for protonation states. Later, complex folding stabilities (G) and binding affinities (ΔG) of the top sequence-conformation ensembles retained for either pH were re-evaluated and re-ordered with a higher-resolution energy model where continuum electrostatics replaced Coulombic electrostatics30. Lastly, the representative conformation/protonation at either pH was chosen based on the best binding affinity (lowest ΔG) amongst those whose folding energies were within 1 kcal/mol of the most stable complex.
Each calculated binding energy relative to WT, ΔΔG, was further decomposed into contributions of van der Waals (vdW), continuum electrostatics, SASA-dependent nonpolar solvation interactions, and internal energy. vdW and electrostatics were found to be the most important contributors to pH-dependent binding. The calculated vdW contribution was largely due to the modeled clashes between the bulkier pertuzumab His55 and a HER2 loop at pH 5.8 whereas the clashes could be largely ameliorated by the flexible HER2 loop. Therefore, long-range electrostatic binding affinity, being less sensitive to conformational details, was further analyzed as an origin of the pH-dependent binding. Specifically, continuum electrostatics for binding were first decomposed into the contributions of the lost desolvation penalties and the gained solvent-screened intermolecular interactions. Desolvations were then decomposed into contributions of all residues and screened intermolecular electrostatic interactions into those of all residue pairs. To sum up screened intermolecular electrostatic interactions and avoid double counting for each residue, half of the pairwise interactions for each residue with all other residues were considered. Those residues with insignificant contributions (desolvation plus the half of the summed pairwise interactions) to electrostatic binding specificity ΔΔΔG (ΔΔΔG = ΔΔGpH 7.4 - ΔΔGpH 5.8) were not considered further. Since desolvation penalties dominated over screened electrostatic interactions in contributions to electrostatic binding specificity ΔΔΔG, the top 10 remaining residues with the highest desolvation contributions were reported for ΔΔGpH 7.4, ΔΔGpH 5.8, and ΔΔΔG, respectively.
Statistical analyses
Statistical significance was analyzed using two-tailed t-test, two-tailed Mann-Whitney U test or one-way ANOVA with Tukey’s multiple comparison test as indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that supports the findings in this study are available upon request from the corresponding authors.
Code availability
Software is available upon request at www.wardoberlab.com/software/sprtool and www.wardoberlab.com/software/miatool for SPR and microscopy data analyses, respectively. MATLAB-based software for fitting the pharmacokinetic data is available upon request from the corresponding authors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Gazdar, J. Minna, and K. Huffman (Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center at Dallas) for the gift of HCC1954 cells. We are grateful to L. Dangott, C. Klemashevich, and S. Shankar for help with the liquid chromatography mass spectrometry. R. Li and J. Heimann recloned the genes encoding the anti-hen egg lysozyme antibody (control) into the expression constructs used in this study. This work was supported in part by the Cancer Prevention and Research Institute of Texas (CPRIT; RP140141 to E.S.W.) and the National Institute of General Medical Sciences of the National Institutes of Health (R35GM124952 to Y.S.).
Footnotes
COMPETING FINANCIAL INTERESTS
J.C.K., W.S., R.J.O. and E.S.W. are co-inventors on a pending patent (PCT/US2018/013952) describing acid-switched ADCs that is owned by Texas A&M University.
REFERENCES
- 1.Sievers EL & Senter PD Annu. Rev. Med. 64, 15–29 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Lambert JM & Berkenblit A Annu. Rev. Med. 69, 191–207 (2018). [DOI] [PubMed] [Google Scholar]
- 3.de Goeij BE & Lambert JM Curr. Opin. Immunol. 40, 14–23 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hinrichs MJ & Dixit R AAPS J. 17, 1055–1064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G et al. Mol. Cancer Ther. 17, 1441–1453 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Franklin MC et al. Cancer Cell 5, 317–328 (2004). [DOI] [PubMed] [Google Scholar]
- 7.LoRusso PM, Weiss D, Guardino E, Girish S & Sliwkowski MX Clin. Cancer Res. 17, 6437–6447 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Burris HA III et al. J. Clin. Oncol. 29, 398–405 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Carter P et al. Proc. Natl. Acad. Sci. USA 89, 4285–4289 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igawa T et al. Nat. Biotechnol. 28, 1203–1207 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Devanaboyina SC et al. mAbs 5, 851–859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaparro-Riggers J et al. J. Biol. Chem. 287, 11090–11097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon RP et al. Nat. Biotechnol. 33, 733–735 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Li JY et al. Cancer Cell 29, 117–129 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Ram S, Kim D, Ober RJ & Ward ES mAbs 6, 1211–1219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Austin CD et al. Mol. Biol. Cell 15, 5268–5282 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward ES & Ober RJ Adv. Immunol. 103, 77–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyzik M, Rath T, Lencer WI, Baker K & Blumberg RS J. Immunol. 194, 4595–4603 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalloneau E et al. Oncotarget 7, 54415–54429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swiercz R et al. Oncotarget 8, 3528–3541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward ES J. Mol. Biol. 224, 885–890 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Hoogenboom HR et al. Nucleic Acids Res. 19, 4133–4137 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foote J & Winter G J. Mol. Biol. 224, 487–499 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Li R et al. Mol. Cancer Ther. 17, 169–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ober RJ & Ward ES Anal. Biochem. 306, 228–236 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Vaccaro C, Zhou J, Ober RJ & Ward ES Nat. Biotechnol. 23, 1283–1288 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Chao J, Ward ES & Ober RJ IEEE Trans. Inf. Technol. Biomed. 14, 1075–1087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toy W et al. Nat. Genet. 45, 1439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsson MH, Sondergaard CR, Rostkowski M & Jensen JH J. Chem. Theory Comput. 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Karimi M & Shen Y Bioinformatics 34, i811–i820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings in this study are available upon request from the corresponding authors.