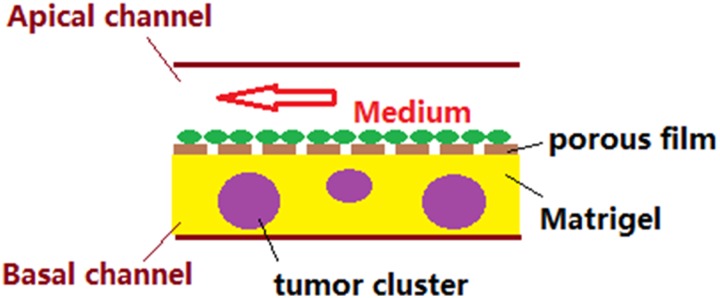
Abstract
Two-dimensional (2D) cell culture is not ideal for traditional drug screening, because 2D culture does not accurately mimic the physiological microenvironment of tumor cells. Thus, a drug-screening system which more closely mimics the microenvironment of in vivo tumors is necessary. Here, we present a biomimicking bilayer microfluidic device that can facilitate antitumor drug screening. The microfluidic device consists of two polydimethylsiloxane (PDMS) pieces with channels which are separated by a semipermeable membrane to allow water, oxygen, and nutrition supply, while preventing cell migration. The channels embedded on the two PDMS pieces overlap each other over a long distance to ensure a larger exchange area to mimic the blood vessel-tumor model. High concentrations of endothelial cells (EC) are first seeded onto the membrane through the apical channel, and after a two-day culture, a confluent EC monolayer forms. Tumor spheroid-laden Matrigel is then seeded into the basal channel. After the Matrigel is cured, the device is ready for drug testing. Paclitaxel is used as the model drug for testing. Confocal microscopy and ImageJ are used to assess the efficacy of different concentrations of paclitaxel, and optical coherence tomography (OCT) is employed to determine the tumor volumetric change after the drug treatment. The results indicate that the proposed bilayer microfluidic device in combination with confocal and OCT optical characterization provide an efficient platform for antitumor drug testing.
INTRODUCTION
Traditional tumor drug screenings utilize 2D cell monolayers. Although this model is easy-to-use, suitable for automation, and is useful for High Throughput Screening (HTS) systems,1–3 using 2D tumor cell monolayers for drug-screening purposes is not a realistic recreation of tumors in an in vivo environment. This is because 2D cell culture does not mimic the essential cell-cell or cell-extracellular matrix (ECM) interactions present in vivo, which presents a crucial limitation for accurately estimating the efficacy of a tumor drug in the human body. There is overwhelming evidence that in vitro three-dimensional (3D) tumor cell cultures more accurately reflect the complex in vivo microenvironment than simple 2D cell monolayers, with respect to gene expression profiles, signaling pathway activity, and drug sensitivity.4–7 For example, some tumor cell lines developed dense multicellular spheroids in 3D-culture and showed greater resistance to paclitaxel and doxorubicin as compared to the 2D-cultured cells.8–14 Therefore, utilization of 3D tumor models increases the accuracy of tumor drug-screening results.
Most of the 3D cellular models are focused on 3D spheroids, due to the ease of formation and enhanced manipulation.15–17 Of the 3D spheroid models, tumor spheroid models are the most successful and widely applied research tools for cancer studies and drug screening.18–21 Studies have been performed to make the morphology and physiology of the tumor spheroid similar to those in vivo, i.e., the network of cell-cell interactions, the 3D structure, and the presence of a natural ECM, nutrients, metabolites, and oxygen gradients.22 More evidence has been found that the tissue structure determines the growth rate of a tumor as well as its response to anticancer drugs.23 Despite a variety of advantages offered by 3D tumor spheroids for drug-screening applications as described above, they are infrequently utilized for drug screening. Limitations include a low number of effective spheroid cultivation methods, difficult quantitative determination of cellular responses in the 3D arrangement, and little drug transportation information. Microfluidic devices, which emerged recently, are believed to be a substantial solution to these limitations.24 Microfluidic devices can be fabricated to have multiple regions to facilitate increased spheroid formation and can allow nutrition or cell gradients to dictate the size of the spheroids produced. Additionally, drugs can be introduced in a gradient manner, which dramatically increases the device’s efficiency.25 The implementation of microfluidic systems into 3D tumor cell culture can compensate for the absence of vasculature, support nutrition supply, and allow for fluid dynamic studies.
Although plenty of work has been focused on microfluidic-based 3D tumor spheroid drug screening,26,27 there is no pilot study which pays attention to the transportation of the drug through blood vessels and its effects on drug efficacy. Our group has a series of well-defined studies on blood vessels in vitro which allow for mimicked drug or nanodrug transportation in the blood vessel environment, based on bilayer microfluidic devices.28–32 These studies show that the endothelial cell (EC) monolayer is an obstacle for drug transportation, and it is possible to manipulate the permeability of the endothelial cell monolayer by adding specific chemicals such as thrombin.
Thus, in this work, a bilayer microfluidic device for in vitro blood vessel mimicking and tumor drug screening was proposed. The bilayer microfluidic device consists of two polydimethylsiloxane (PDMS) pieces with embedded channels, and the two pieces are separated by a semipermeable membrane which allows for water, oxygen, and a nutrition supply to pass through, while preventing cell migration. The two channels on the two PDMS pieces have long sections of channels which overlap to ensure a larger exchange area to mimic the blood vessel-tumor model. High concentrations of EC were first seeded onto the membrane through the apical channel, and after a two-day culture which ensures the formation of a confluent EC monolayer, tumor spheroid-laden Matrigel was seeded into the basal channel. After the Matrigel was cured in an incubator for 30 min, the devices were ready for drug testing. Confocal imaging and ImageJ were used to assess the efficacy of different drug concentrations and combinations of drug therapies. Optical coherence tomography (OCT) was employed to determine the tumor volume shrinkage after drug treatment.41,45,46
MATERIALS AND METHODS
Materials
The media used for the HCT116 cells was Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technology), supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotic and antimycotic (ThermoFisher). Fluorescence dyes [Calcein AM and propidium iodide (PI)] were purchased from ThermoFisher. Dispase II was purchased from EMD Millipore. Paclitaxel was purchased from LC labs. PDMS is Sylgard 184 from Dow Corning. All chemicals that are not mentioned are directly used without purifying.
Drug
Paclitaxel is a microtubule-active drug that inhibits mitotic progression and arrests cells in mitosis.33,34 Following the mitosis arrest, cells may either exit from mitosis or undergo apoptosis. Evidence was found that low concentrations of paclitaxel induce cell-type-dependent p53, p21, and G1/G2 arrest. However, for HCT116 and other cell lines, paclitaxel cannot readily induce p53, and, therefore, these cells can undergo exclusively mitotic and postmitotic arrest after paclitaxel treatment.35
Design of microfluidic devices
To ensure sufficient nutrition and oxygen supply and optimized metabolite clearance, the microfluidic devices were designed as shown in Fig. 1. Both apical and basal pieces were made with PDMS, and the embedded channels were formed from a preobtained silicon wafer by photolithography. The two pieces were separated by a semipermeable membrane, which usually has dense 200–800 nm pores, allowing for water, nutrient, oxygen, and metabolite diffusion, while preventing cell transportation. Four outlets (diameter = 2 mm) on the apical piece were matched to the four circle areas for the apical and basal channels. The two channels share the same size (500 μm width and 200 μm height), and they have more than 80% overlapping area, as shown in Fig. 1. Since the two PDMS pieces are separated by the semipermeable polyurethane membrane, which has a surface that cannot be activated by plasma treating, an additional thin layer of PDMS was applied onto the whole membrane, excluding the channel area to ensure strong bonding.
FIG. 1.
Diagram of the microfluidic device using PDMS molds with basal and apical channels. The basal channel is the straight channel on the bottom layer of the device, where cells like human colon cancer cells (HCT116) can be grown. The next layer is the semipermeable membrane, and the top layer is the apical channel, which is the Z-shaped channel. As media is flowed through the apical channel, nutrients permeate the membrane and reach the cells in the basal channel. Waste can be transported back through the membrane to the apical channel and removed from the apical channel outlet. (a) Layer diagram, (b) top view, and (c) photo of the microfluidic device.
Fabrication of the microfluidic devices
PDMS (Sylgard 184) was obtained by mixing a base agent and curing agent at a ratio of 10:1 (v:v), respectively. PDMS was poured onto the prepared molds, and bubbles were removed via vacuum for 30 min. Molds were cured in a 60 °C oven for 30 min. PDMS with imprinted channels were peeled carefully from the mold and cut into rectangular pieces. Basal channels were on the top and bottom of the straight channel, while the apical channels formed the Z-shape. A piece with only the basal channels was paired with one piece with only apical channels; these were then aligned to have symmetrical outlets and to match well overall. The centers of the basal outlets were matched on the apical piece, and the apical piece was then removed. A biopsy punch was used to cut four holes for the four outlets on the apical piece; this includes two holes for the apical channel and two for the basal channel. In order to ensure smooth and efficient cutting, the channels were punched powerfully and quickly. Subsequently, the basal piece and a glass slide were cleaned using acetone and after drying, the surfaces were activated by an oxygen plasma handgun treatment for at least 30 s. The basal piece and glass slide were then attached and bubbles were removed. The channel side of the basal piece was then cleaned with acetone and plasma treated again, and the smooth side of a semipermeable membrane was attached to the channel side of the basal piece. A thin layer of uncured PDMS was then painted onto the film without touching the channel area. Being careful that PDMS was not applied to the channel was important because the PDMS should not spread to and permeate the channel. The thin layer of uncured PDMS was served as a glue to adhere the apical and basal PDMS pieces as well as the semipermeable membrane. The PDMS-coated pieces were then cured on a hot plate for 15 min. The film covering the two outlets on the basal pieces was then cut and removed to expose the outlets. The apical piece of the channel was then cleaned with acetone, and the PDMS-coated surface and apical piece were then plasma treated. Since the attachment process is irreversible, the apical piece was attached onto the film by matching the channels, holes, and outlets slowly and carefully. The piece was then cured in a 60 °C oven for 30 min. The devices were autoclaved at 100 °C for 20 min for sterilization purposes and dried for 5 min.
Endothelial layer seeding (seed EC first)
Bovine aortic endothelial cells (BAEC, we use EC for short) were used as the endothelium model. Since ECs need some cell adhesion motifs to help them attach and grow, both apical and basal channels were incubated in an incubator with 0.2% porcine gelatin solution overnight (or for at least 3 h). ECs from a confluent Petri dish were harvested to obtain a 1 × 107 cells/ml concentration. A droplet of EC suspension was applied to an outlet of the apical channel and was sucked through the channel by a negative pressure using a disposable pipette. After 3 h of incubation, which allowed the ECs to attach to the bottom (the semipermeable membrane) of the apical channel, EC media was introduced to sustain the nutrition supply. A confluent monolayer of EC formed in two days. The devices were placed in a large cell culture dish with drops of sterilized water placed around it to make the environment more humidified in order to prevent the devices from drying.
Tumor spheroid formation, harvest, filtering, and seeding
The tumor spheroids were formed and filtered to uniform sizes as described in our previous work.36 In brief, a low concentration of HCT116 cells was seeded into 6-cm diameter Petri dishes, and after 5–7 days of growth, dispase was used to detach the cell sheets, which were then orbitally-shaken in dispase-doped media in the same Petri dishes for at least 3 days. A set of custom filters were used for size uniformization. In this work, the HCT116 tumor spheroids were from 100 to 120 μm sieves, which means the spheroids which passed through the 100 μm sieve but did not pass through the 120 μm sieve were used in the study. The average size of the spheroids was 151 ± 24 μm. To create the tumor spheroid seeding mixture, the tumor spheroid suspension was diluted with Matrigel at a volume ratio of 1:1. The tumor spheroids were then seeded in the microfluidic device by placing the obtained mixture into an outlet of the basal channel and sucking on the opposite outlet of the channel with a disposable bulb pipette. This allowed for the cell suspension to flow throughout the whole basal channel. The device was placed in the 37 °C incubator for 30 min to cure the Matrigel. To apply the media, the disposable bulb pipette method was used to administer media to the apical channel. Since two cell types were involved, the EC media was used. HCT116 can be sustained in EC media, with no remarkable negative effects observed in using EC media to culture HCT116. After both cell seeding, the longitudinal cross-sectional diagram of the bilayer blood vessel mimicking microfluidic device is shown in Fig. 2.
FIG. 2.
Longitudinal cross-sectional diagram of the bilayer blood vessel mimicking microfluidic device.
Drug testing
Paclitaxel was used as a model drug for the HCT116 spheroid treatment. The drug displays a remarkable efficacy on HCT11633,34 but also on endothelial cells.37,38 Thus, the endothelial layer in the device not only mimicked the blood vessel and its permeability but could also serve as the indicator for the toxicity of drug therapy on healthy cells. The drug was dissolved in EC media and diluted to different concentrations [0M (control), 1 nM, 3 nM, 10 nM, 30 nM, 100 nM, 300 nM, and 1 μM], and the drug was applied using a syringe pump for a series of time frames (a few hours to a few days). To demonstrate the advantages of our platform, a comparison of the spheroids in a well plate, an EC-free device, and an EC device was made. To achieve the best drug efficacy—greater tumor cell death and less EC death—a comparison of viability between EC and tumor cells was also made.
Characterization
There are two main principles to assess the efficacy of a drug or therapy for tumor treatment. One is to measure the population of dead and live cells in a 3D tumor spheroid, which can be achieved via live/dead staining and confocal microscopy. The second way is to observe the shrinkage ratio (or volume change) of the 3D tumor spheroid, which can be achieved using Optical Coherence Tomography (OCT).
Live/dead staining
The control and drug-treated 3D tumors were stained with a Calcein AM and propidium iodide (PI) (live/dead) staining kit. To ensure a better staining effect, double the recommended concentrations of the dyes and longer staining time (2 h) were used. After staining, phosphate-buffered saline flow was introduced for at least 1 h to clear any unbound dye.
Confocal fluorescence microscopy was used to quantify the efficiency of the drugs by comparing the dead and live cells in a certain plane of the spheroids. Since the spheroid size is usually larger than 200 μm, only the 4× objective lens can cover this large area, though this lens lacks good z-resolution. Thus, the stacks obtained in the middle of spheroids are usually blurred and mixed with the signal from different layers, which were difficult to enumerate. In contrast, the confocal images taken from the surface of spheroids were more vivid. By comparing the different z-stacking images, we found that the live and dead cell ratio on the spheroid surface could roughly represent the overall live-dead cell ratio of the whole spheroid. The surface viability of the spheroids cannot comprehensively represent that of the whole 3D spheroid. However, in this study, we were focused on presenting a drug-screening device other than how to deal with the spheroid imaging, so we chose the surface viability to represent the overall viability of a tumor spheroid. Recently, a lot of studies were focused on how to image thick samples in a 3D manner.39,40 In the following studies, we can incorporate those techniques to improve the accuracy of the tumor cell viability.
Since the cells were highly compact in confocal images, ImageJ (FIJI) was used to process the images and enumerate the dead and live cells, as shown in Fig. 3. In brief, an image that could represent the surface of the spheroid was selected and transferred to a 16-bit gray scale image. The threshold was then tuned to ensure that most of the out-of-focus cells were removed. Usually, there would be some leftover cells that are packed together, and these could be distinguished by a plug-in algorithm called Watershed, which can build dams to separate packed cells. Followed by Watershed, the Analyze Particles function was run to enumerate the total in-focus cells. The pixel cut-off was selected as larger than 50 pixels, which correlates to half the size of a cell with 10 μm diameter. Both live and dead cells can be counted in this way.
FIG. 3.
Diagram of how to use FIJI to enumerate cells in highly packed conditions. (a) → (b): transfer color images to 16-bit gray scale images. (b) → (c): tune threshold to remove most of the out-of-focus cells and keep most of the in-focus cells. (c) → (d): run watershed in FIJI to distinguish the highly packed cells. Run Analyze Particles function to count the cells in image (d).
Based on the enumeration of dead and live cells, the relative viability and drug efficacy were defined as
where Drug Efficacy = 1 − Relative viability.
OCT for determination of 3D tumor shrinkage
OCT is an optical technique based on low-coherence interferometry, which is capable of obtaining 3D images. This technique does not require any staining and uses near-infrared light which allows for deeper light penetration into tissue samples during scanning in a nondestructive manner. It can also provide information about the volume of scanned objects. Because the surroundings of the tumor spheroids are transparent in this 3D tumor model, it is ideal to use OCT to monitor the size or volume changes of the 3D tumor spheroids.
The volumetric quantification of 3D tumor spheroids was performed by a voxel-based method, which was described in detail in Refs. 41 and 46. First, a 3D average filter was applied to remove the speckles within the tumor spheroids in the OCT images. Then, the tumor spheroid was segmented using a canny edge detection filter, frame by frame. Connective voxels were grouped, and the mean distance of grouped connective voxels to the manually chosen spheroid centroid was calculated. The group with the minimum mean distance was identified as the spheroid region. The voxels within this region were summed and then multiplied by the actual volume of an individual voxel (volume/voxel), yielding the total volume of the spheroid.
The volume change of the spheroids at different drug concentrations was defined as
RESULTS AND DISCUSSION
Dead and live staining
The viability and drug efficacy of both cell types can be assessed using dead and live staining and confocal microscopy. It is relatively easier to address such parameters for EC, since EC were in a 2D manner. But for HCT116 spheroids, which are in a 3D conformation, and the size of which are usually larger than 200 μm, a 4× objective lens had to be used for imaging, leading to a low resolution in the z direction. It is, therefore, difficult to obtain an overall viability using confocal 3D scanning. However, since cells on the spheroid surface can be clearly imaged, as shown in Fig. 4(a), the viability of the spheroid surface was used to represent that of the whole 3D spheroid. As shown in Fig. 4, a freshly prepared HCT116 spheroid of approximately a 150 μm diameter was stained with dead/live staining and showed a strong robustness on the surface close to the objective of the microscope, with only one red spot representing a dead cell. The spheroid was obtained using a set of 105 μm and 114 μm filters, and this spheroid size was chosen for all of the following studies. This intermediate size allowed for the spheroids to be accessibly seeded into the devices as well as never exceeding the imaging limit of our microscope as the spheroids grew in size. During Petri dish culture, the necrotic core will appear when the size of tumor spheroids is larger than about 400 μm,36 which the spheroids during the coculture usually would not exceed, so we did not consider the effect of necrosis.
FIG. 4.
Confocal and bright-field images of a HCT116 spheroid in the bilayer device. (a) Calcein AM stained, (b) PI stained, (c) stack of (a) and (b), and (d) bright-field image. The four images share the same scale bar in (a).
OCT scanning and volume information
As mentioned in the section on OCT for Determination of 3D Tumor Shrinkage, OCT is one of the best ways to obtain 3D information of biological samples in the range of submillimeters to 10 mm. As OCT utilizes the refractive index of transparent objects to reproduce 3D images, all the OCT scanning was performed through the glass bottom of the devices to avoid interference from the translucent semipermeable membrane. As shown in Fig. 5, (a) shows an image of a cross section perpendicular to the channel; (b) shows a vertical cross section, while (c) is a 3D rendered OCT image using the Amira software.
FIG. 5.
OCT scanning images of HCT116 spheroids in the device. (a) Cross section of a HCT116 spheroid perpendicular to the channels, (b) cross section of the spheroid parallel to the membrane, and (c) a 3D rendered OCT image of the spheroid. (a) and (b) share the same scale bar.
Coculture of EC and HCT spheroids in the device
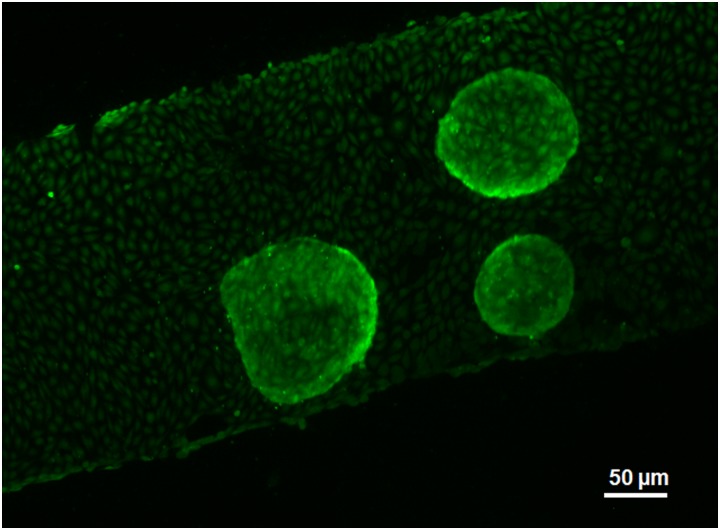
As a core part of the study, coculture of EC and HCT116 spheroids in the microfluidic device is rather important. Because of their nature, EC hardly adhere to Petri dishes as well as the polycarbonate membranes. EC always need a gelatin coating to promote the cell-substrate adhesion, and a confluent monolayer of EC is desired. Therefore, EC are always seeded at a high concentration prior to HCT116 spheroids. After 2–3 days of culture, EC can reach full confluency. Then, the spheroids may be seeded with the Matrigel. Figure 6 displays a device with HCT116 spheroids freshly seeded and stained with CellTracker Green. The EC layer on the membrane shows ample confluency, and the three spheroids are robust. The image was taken from the top of the device, in order to view the EC layer clearly. As the spheroids were seeded, media with different concentrations of drug, or drug-free media, were infused at a low flow rate for 3 days. The behavior of both EC and spheroids is discussed below.
FIG. 6.
Coculture of EC and HCT spheroids in the device. This is a fluorescence image in which both cell types were stained by CellTracker Green. The image was taken from the apical side of the device.
EC behavior under drugs
Paclitaxel of a high concentration not only has negative effects on the HCT116 cells but also affects EC growth. There is ample evidence that shows even low concentrations of paclitaxel can induce senescence of EC and change their phenotype. As shown in Fig. 7, maintaining a confluent EC monolayer in the device with drug-free media infusion preserves the health of most of the cells and maintains a confluent monolayer. After 3 days of 1 nM, 3 nM, and 10 nM paclitaxel infusion, the confluency was maintained, and no dead cells were observed. The spindlelike phenotype was also preserved. However, with a higher concentration of paclitaxel infusion, the quantity of spindlelike cells decreased, so that almost no such cells can be found in Fig. 7(g) (3-day of 300 nM infusion). With 1 μM drug treatment [Fig. 7(h)], some holes and gaps as well as dead cells could be found on the monolayer. The cell density also decreased, as shown in Fig. 7(i), which indicates that the drug killed some of the cells and the infusion removed most of the dead cells. The coverage of each cell became much larger, because there was more space for each live cell. Such a phenotype was typical, indicating that paclitaxel-induced senescence of EC reported by previous studies.42
FIG. 7.
(a)–(h) Confluent EC in the device subjected to a 3-day treatment of different concentrations of paclitaxel. (i) Relative viability of EC after 3-days of exposure to different concentrations of paclitaxel treatment.
Drug effect on HCT116 spheroids
As mentioned before, HCT116 spheroids with an average diameter of 150 μm were seeded into the devices as well as to other control environments. Through 3 days of drug-free media treatment, both in ultralow attachment well plates and in the EC-loaded devices, spheroids grew vigorously with a remarkable increase in both the diameter and the quantity of dead cells on the spheroid surface, as shown in Fig. 8. Although, due to the hindrance of the Matrigel in the channel, spheroids in EC-loaded devices were smaller than the ones freely cultured in well plates. However, through 3 days of 100 nM drug treatment, a significant difference in spheroid condition was found among the well plate, EC-free device, and EC-loaded device. The quantitative results of volume change and relative viability of the tumor spheroids can be found in Figs. 9 and 10, respectively. A more intuitive comparison of HCT116 viability among the three devices can be found in Fig. S1 in the supplementary material. In the well plate, there were very limited live cells left, and the whole structure was filled with dead cells and debris. Because most of the cells were dead, the 3D spherical shape of the spheroid could not be maintained and collapsed. In the EC-free device, there were many dead cells and a limited amount of live cells after 3 days of treatment. Compared to the situation in the well plate, there were more live cells and less dead cells, indicating that both the membrane and the Matrigel acted as hindrances to drug delivery. In the situation of the EC-loaded device, compared to the other two experimental conditions, more live cells and fewer dead cells were observed after 3 days of treatment. The introduction of an EC layer dramatically hindered the delivery of the drug from the apical channel to the tumor spheroids, especially at the beginning. Tumor cells are able to influence the EC monolayer permeability,43,44 by releasing certain types of chemokines (such as TNF-α) which loosen the cell-cell interactions of EC. As some of the EC were affected by the drug (Fig. 7), openings on the monolayer were formed which allowed the drug to pass through. The 3D structure of the spheroids in both EC-free and EC-loaded situations was maintained, thanks to the support of the Matrigel. The half inhibitory concentration (IC50) values of paclitaxel for the three different environments are 34.8 ± 5.8 nM for the well plate, 56.5±13.2 nM for the EC-free device, and 158±32 nM for the EC-loaded device. According to the drug test results in our previous work,36 the IC50 value of paclitaxel for the 2D cell culture of HCT116 cells is 26.3±12.5 nM, which is much smaller than the one obtained by our device. Therefore, the drug-screening platform with a combination of an EC layer, Matrigel, and tumor spheroids better mimics an in vivo environment compared to the monolayer cell culture method and tumor spheroid cell culture method. Combining the drug effect on EC as shown in Fig. 7(i), we suggest a paclitaxel concentration in blood that reaches the tumor should be in the range of 100–300 nM. We believe this platform has a promising potential for the antitumor drug-screening industry.
FIG. 8.
Confocal images of HCT116 spheroids subjected to 3-day 100 nM paclitaxel (Pac) treatment at different conditions. All the images were split into green and red channels to more vividly show the live and dead cells.
FIG. 9.
The volume change of HCT116 spheroids in EC-loaded channels monitored by OCT subjected to different concentrations of paclitaxel for up to 72 h (*P < 0.05, 1-way ANOVA and Tukey post hoc test).
FIG. 10.
The relative viability of HCT116 cells in EC-loaded channels subjected to different concentrations of paclitaxel for 72 h in well plates (a), in EC-free channels (b), and in EC-loaded channels (c). All the three figures share the same legend in the middle (*P < 0.05, 1-way ANOVA and Tukey post hoc test).
CONCLUSIONS
An in vitro biomimetic antitumor drug-screening platform was presented. The platform consisted of two layers with two channels and a semipermeable membrane. An EC monolayer on the membrane enabled the apical channel to function as a blood vessel, which showed both permeability and resistance to several molecules, including drugs. Additionally, direct seeding of tumor spheroids more closely mimicked the in vivo nature of tumors, while the Matrigel served as a scaffold for the tumor spheroids, which functioned as ECM. The combination of these displayed a more biomimetic microenvironment of a blood vessel and adjacent tissue. The drug efficacy has been expressed by two manners: the viability of tumor cells, which can be quantified by dead/live staining using confocal microscopy, and the volume change of the tumor spheroids, which can be quantified via OCT scanning. Since both the cell culture in the microfluidic device and the two characterization techniques can be automated, we believe this work can dramatically increase the efficiency on both speed and cost on antitumor drug screening.
SUPPLEMENTARY MATERIAL
See the supplementary material for the comparison of the HCT116 cell’s relative viability among the EC-loaded device, EC-free device, and well plate with 300 nM Pac treatment for 72 h.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grants (Nos. R01HL131750 and R01EB025209), the National Science Foundation (NSF) grants (Nos. PFI:AIR-TT 1701136 and DBI-1455613), and the Pennsylvania Infrastructure Technology Alliance (PITA) program.
REFERENCES
- 1.J. McKim, Jr., Comb. Chem. High Throughput Screen. 13, 188–206 (2010). 10.2174/138620710790596736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta P. B., Onder T. T., Jiang G., Tao K., Kuperwasser C., Weinberg R. A., and Lander E. S., Cell 138, 645–659 (2009). 10.1016/j.cell.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greeley J., Jaramillo T. F., Bonde J., Chorkendorff I., and Nørskov J. K., Nat. Mater. 5, 909–913 (2006). 10.1038/nmat1752 [DOI] [PubMed] [Google Scholar]
- 4.Haycock J. W., 3D Cell Culture: Methods and Protocols (Humana Press, 2011), pp. 1–15. [Google Scholar]
- 5.Carletti E., Motta A., and Migliaresi C., Methods in Molecular Biology (Humana Press, 2011), pp. 17–39. [DOI] [PubMed] [Google Scholar]
- 6.Tibbitt M. W. and Anseth K. S., Biotechnol. Bioeng. 103, 655–663 (2009). 10.1002/bit.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., and de Boer J., Trends Biotechnol. 31, 108–115 (2013). 10.1016/j.tibtech.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 8.Kohno N., Ohnuma T., and Truog P., J. Cancer Res. Clin. Oncol. 120, 293–297 (1994). 10.1007/BF01236386 [DOI] [PubMed] [Google Scholar]
- 9.Wartenberg M., Frey C., Diedershagen H., Ritgen J., Hescheler J., and Sauer H., Int. J. Cancer 75, 855–863 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Borgström P., Gold D. P., Hillan K. J., and Ferrara N., Anticancer Res. 19, 4203–4214 (1999). [PubMed] [Google Scholar]
- 11.Jiang X., Xin H., Gu J., Xu X., Xia W., Chen S., Xie Y., Chen L., Chen Y., Sha X., and Fang X., Biomaterials 34, 1739–1746 (2013). 10.1016/j.biomaterials.2012.11.016 [DOI] [PubMed] [Google Scholar]
- 12.Green S. K., Francia G., Isidoro C., and Kerbel R. S., Mol. Cancer Ther. 3, 149–159 (2004). [PubMed] [Google Scholar]
- 13.Frankel A., Buckman R., and Kerbel R. S., Cancer Res. 57, 2388–2393 (1997). [PubMed] [Google Scholar]
- 14.Kathuria H., Kochhar J. S., Fong M. H. M., Hashimoto M., Iliescu C., Yu H., and Kang L., J. Vis. Exp. 105, e52914 (2015). 10.3791/52914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edmondson R., Broglie J. J., Adcock A. F., and Yang L., Assay Drug Dev. Technol. 12, 207–218 (2014). 10.1089/adt.2014.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kijanska M. and Kelm J., In Vitro 3D Spheroids and Microtissues: ATP-Based Cell Viability and Toxicity Assays (Eli Lilly & Company and the National Center for Advancing Translational Sciences, 2004). [PubMed] [Google Scholar]
- 17.Kim S.-A., Lee E. K., and Kuh H.-J., Exp. Cell Res. 335, 187–196 (2015). 10.1016/j.yexcr.2015.05.016 [DOI] [PubMed] [Google Scholar]
- 18.Zanoni M., Piccinini F., Arienti C., Zamagni A., Santi S., Polico R., Bevilacqua A., and Tesei A., Sci. Rep. 6, 19103 (2016). 10.1038/srep19103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longati P., Jia X., Eimer J., Wagman A., Witt M.-R., Rehnmark S., Verbeke C., Toftgård R., Löhr M., and Heuchel R. L., BMC Cancer 13, 95 (2013). 10.1186/1471-2407-13-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong X., Lin C., Cheng J., Su J., Zhao H., Liu T., Wen X., and Zhao P., PLoS One 10, e0130348 (2015). 10.1371/journal.pone.0130348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurent J., Frongia C., Cazales M., Mondesert O., Ducommun B., and Lobjois V., BMC Cancer 13, 73 (2013). 10.1186/1471-2407-13-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland R. M., Science 240, 177–184 (1988). 10.1126/science.2451290 [DOI] [PubMed] [Google Scholar]
- 23.Fischbach C., Chen R., Matsumoto T., Schmelzle T., Brugge J. S., Polverini P. J., and Mooney D. J., Nat. Methods 4, 855–860 (2007). 10.1038/nmeth1085 [DOI] [PubMed] [Google Scholar]
- 24.Li X. J., Valadez A. V., Zuo P., and Nie Z., Bioanalysis 4, 1509–1525 (2012). 10.4155/bio.12.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu A., Loutherback K., Lambert G., Estévez-Salmerón L., Tlsty T. D., Austin R. H., and Sturm J. C., Proc. Natl. Acad. Sci. U.S.A. 110, 16103–16108 (2013). 10.1073/pnas.1314385110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao K., Li L., He L., Hinkle K., Wu Y., Ma J., Chang L., Zhao X., Perez D. G., Eckardt S., McLaughlin J., Liu B., Farson D. F., and Lee L. J., Small 10, 1015–1023 (2014). 10.1002/smll.201300116 [DOI] [PubMed] [Google Scholar]
- 27.Patra B., Peng C.-C., Liao W.-H., Lee C.-H., and Tung Y.-C., Sci. Rep. 6, 21061 (2016). 10.1038/srep21061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas A., Tan J., and Liu Y., Microvasc. Res. 94, 17–27 (2014). 10.1016/j.mvr.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas A., Wang S., Sohrabi S., Orr C., He R., Shi W., and Liu Y., Biomicrofluidics 11, 024102 (2017). 10.1063/1.4977584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas A., Daniel Ou-Yang H., Lowe-Krentz L., Muzykantov V. R., and Liu Y., Biomicrofluidics 10, 014101 (2016). 10.1063/1.4936672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhl C. G., Muzykantov V. R., and Liu Y., Biomicrofluidics 12, 014101 (2018). 10.1063/1.5000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhl C. G. and Liu Y., Lab Chip 19, 1458–1470 (2019). 10.1039/C8LC01250D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz S. B., Trends Pharmacol. Sci. 13, 134–136 (1992). 10.1016/0165-6147(92)90048-B [DOI] [PubMed] [Google Scholar]
- 34.Wood A. J. J., Rowinsky E. K., and Donehower R. C., New Engl. J. Med. 332, 1004–1014 (1995). 10.1056/NEJM199504133321507 [DOI] [PubMed] [Google Scholar]
- 35.Giannakakou P., Robey R., Fojo T., and Blagosklonny M. V., Oncogene 20, 3806–3813 (2001). 10.1038/sj.onc.1204487 [DOI] [PubMed] [Google Scholar]
- 36.Shi W., Kwon J., Huang Y., Tan J., Uhl C. G., He R., Zhou C., and Liu Y., Sci. Rep. 8, 6837 (2018). 10.1038/s41598-018-25203-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood S. C., Tang X., and Tesfamariam B., J. Cardiovasc. Pharmacol. 55, 276–285 (2010). 10.1097/FJC.0b013e3181d263f7 [DOI] [PubMed] [Google Scholar]
- 38.Michailidou M., Brown H. K., Lefley D. V., Evans A., Cross S. S., Coleman R. E., Brown N. J., and Holen I., J. Vasc. Res. 47, 481–493 (2010). 10.1159/000313876 [DOI] [PubMed] [Google Scholar]
- 39.Yu Y., Shang L., Guo J., Wang J., and Zhao Y., Nat. Protoc. 13, 2557–2579 (2018). 10.1038/s41596-018-0051-4 [DOI] [PubMed] [Google Scholar]
- 40.Yu Y., Fu F., Shang L., Cheng Y., Gu Z., and Zhao Y., Adv. Mater. 29, 1605765 (2017). 10.1002/adma.201605765 [DOI] [PubMed] [Google Scholar]
- 41.Huang Y., Wang S., Guo Q., Kessel S., Rubinoff I., Chan L. L.-Y., Li P., Liu Y., Qiu J., and Zhou C., Cancer Res. 77, 6011–6020 (2017). 10.1158/0008-5472.CAN-17-0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ota H., Eto M., Ako J., Ogawa S., Iijima K., Akishita M., and Ouchi Y., J. Am. Coll. Cardiol. 53, 2298–2305 (2009). 10.1016/j.jacc.2009.01.072 [DOI] [PubMed] [Google Scholar]
- 43.Yuan F., Dellian M., Fukumura D., Leunig M., Berk D. A., Torchilin V. P., and Jain R. K., Cancer Res. 55, 3752–3756 (1995). [PubMed] [Google Scholar]
- 44.Clauss M., Gerlach M., Gerlach H., Brett J., Wang F., Familletti P. C., Pan Y. C., Olander J. V., Connolly D. T., and Stern D., J. Exp. Med. 172, 1535–1545 (1990). 10.1084/jem.172.6.1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee M. R., Flotte T., Gregory K., and Puliafito C. A., Science 254, 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y., Zou J., Badar M., Liu J., Shi W., Wang S., Guo Q., Wang X., Kessel S., Chan L. L., Li P., Liu Y., Qiu J., and Zhou Z., J. Visual. Exp. 144, e59020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for the comparison of the HCT116 cell’s relative viability among the EC-loaded device, EC-free device, and well plate with 300 nM Pac treatment for 72 h.