Abstract
Amputation of the axolotl forelimb results in the formation of a blastema, a transient tissue where progenitor cells accumulate prior to limb regeneration. Connective tissue (CT) – skeleton, periskeleton, tendon, dermis, interstitial fibroblasts – contributes the vast majority of cells that populate the blastema, however, it is unclear how individual CT cells may reprogram their fate in order to rebuild the tetrapod limb. Here we use a combination of Cre-loxP reporter lineage tracking and single-cell (sc) RNA-seq to molecularly track, for the first time, adult CT cell heterogeneity and its transition to a limb blastema state. We uncover a multi-phasic molecular program where CT cell types found in the uninjured adult limb revert to a relatively homogenous progenitor state that participates in inflammation and extracellular matrix disassembly prior to proliferation, establishment of positional information, and ultimately re-differentiation. While the early regeneration transcriptome states are unique to the blastema, the later stages recapitulate embryonic limb development. Notably, we do not find evidence of a pre-existing blastema-like precursor nor limb bud-like progenitors in the uninjured adult tissue. However, we find that distinct CT subpopulations in the adult limb differentially contribute to extending bone at the amputation plane versus regenerating new segments. Together, our data illuminates molecular and cellular reprogramming during complex organ regeneration in a vertebrate.
Among tetrapods, only salamanders show an extraordinary capacity to replace a lost limb (1). The adult axolotl (Ambystoma mexicanum) limb is composed of many different cell types originating from neural, myogenic, epidermal and connective tissue (CT) lineages. Upon limb amputation, cells from nearby the amputation plane accumulate in a unique tissue called the blastema. Cells within the blastema are capable of fully regenerating the missing limb (reviewed in (2)). CT cells, descendants of lateral plate mesoderm, are the most abundant lineage contributing to the blastema (3–5), and encompass bone and cartilage, tendons, periskeleton and dermal and interstitial fibroblasts. The CT cells are a key cell type for deciphering the molecular program of regeneration, since they express factors that guide the regeneration of appropriate limb parts (4, 6–8). Nevertheless, how mature CT produces blastema cells has not been molecularly defined because of the inability to isolate and deconstruct this cell population. It is even unclear whether mature CT cells molecularly “reprogram” into embryonic-like limb progenitors, or whether the CT harbors pre-existing stem cells that selectively seed the blastema.
Here we generated Cre:loxP reporter lines to track CT compartments and uncover compartment-associated contributions to different limb segments. We use single-cell mRNA sequencing (scRNA-seq) to dissect CT heterogeneity in the blastema, as well as the adult, embryonic, and regenerated forelimb, enabling the characterization of CT cell types and lineage relationships as cells regenerate the limb. Notably, CT cells lose their mature phenotypes to form multipotent limb bud-like progenitors in the blastema. The combination of lineage tracking and single-cell transcriptomics resolves the origin and molecular profiles of re-differentiated CT cell types that emerge from the blastema. Our work provides the first molecular view of individual cells that build a blastema and reconstitute a patterned limb skeleton.
A Cre-loxP reporter system tracks connective tissue cells during axolotl limb regeneration
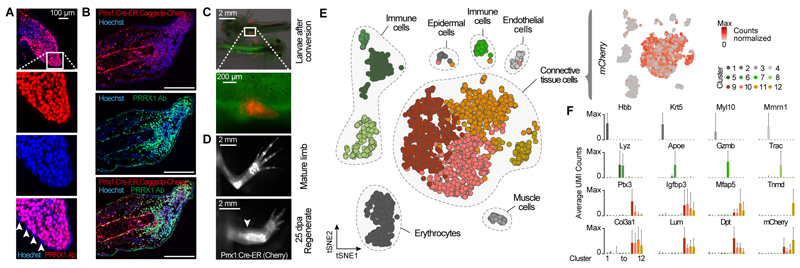
To label and track axolotl CT cells, we generated a germline transgenic line that expresses the tamoxifen-inducible Cre-ERT2 (Cre-ER) gene under the control of the Prrx1 limb enhancer element (Prrx1:Cre-ER;Caggs:lp-Cherry = Prrx1:TFPnls-T2A-Cre-ERT2;CAGGs:lp-GFP-3pA-lp-Cherry) (figs. S1 and S2, Table S1 and S2). Prrx1, a paired-related homeobox gene, is expressed in connective tissue precursors in the developing limb bud and in the limb blastema (9–11). Immunofluorescence staining of PRRX1 protein confirmed specific expression in axolotl limb bud cells (Fig. 1A), blastema cells (Fig. 1B) and adult connective tissue (fig S2). Administration of tamoxifen to Prrx1:Cre-ER;Caggs:lp-Cherry animals at the limb bud stage resulted in an efficient (>80%) genetic labeling of adult limb CT (Fig. 1, C and D; fig. S1E). Notably, after limb amputation, we found that Prrx1-expressing blastema cells express mCherry showing that the transgenic reporter efficiently marks the adult precursors to the blastema cells (Fig. 1B). Examination of 25 day post amputation (dpa) regenerates revealed mCherry-expressing cells in upper and lower arm CT (Fig. 1D; fig. S1, C to F), showing that CT gives rise to new CT during regeneration. Therefore, this new transgenic line provides a system to track CT cells during limb regeneration.
Fig. 1. Tracking and molecular profiling of axolotl limb connective tissue (CT).
(A) Longitudinal section of a limb bud at stage 47 stained with anti-PRRX1 Ab (red) identifies Prrx1 as a pan-CT marker during limb development. Arrowheads indicate absence of PRRX1 staining in the epidermis. (B) Longitudinal section of a blastema 11 days post amputation (dpa) stained with anti-PRRX1 Ab (green). Red: converted cells; Blue: Hoechst = nuclei. Scale bar: 500 μm. (C) Embryos after induction of Prrx1:Cre-ER;CAGGs:lp-Cherry using Tamoxifen (4-OHT) show expression of mCherry only in limb mesenchyme. (D) Fluorescence image of converted cells in uninjured and regenerated limb (conversion at limb bud stage) indicates stable labeling of CT prior to and post regeneration. Arrowhead indicates amputation plane. (E) Left: tSNE plot visualizing single-cell (sc) RNA-seq data of 2,379 single cells (circles) from the adult axolotl upper arm. Gray patches outline related cell types. Right: mCherry expression is detected exclusively in CT cell types. (F) Bar plots showing mean expression of marker genes in each cluster. X-axis represents cell clusters identified in Fig. 1E. Error bars indicate standard deviation. UMI: unique molecular identifier.
We used a high-throughput droplet-based scRNA-seq method (10X Genomics) to sample the cellular diversity in the uninjured adult limb and further validate this transgenic line. We converted cells at the limb bud stage and performed scRNA-seq on the dissociated uninjured adult limb tissue containing labeled and unlabeled cells (2,379 cells; Table S3). Using unbiased clustering, and based on the expression of marker genes, we identified endothelial, epidermal, immune, muscle, red blood, and CT cells (Fig. 1E). mCherry mRNA from Prrx1:Cre-ER;Caggs:lp-Cherry converted cells was only detected in the CT cluster, which included periskeletal, tendon, dermal, and fibroblastic cell subpopulations as identified based on the expression of canonical markers (Fig. 1F). To specifically examine CT heterogeneity, we analyzed 2375 single cell transcriptomes after FACS isolation of labeled Prrx1:Cre-ER;Caggs:lp-Cherry derived CT cells from the adult upper forelimb using tSNE clustering (Fig. 2, A and B; Table S5). We identified 8 distinct clusters that we assigned based on the expression of marker genes as tenocytes (Tnmd), periskeletal cells (Col8a2), actively cycling cells (Ccnb1) as well as 5 fibroblastic CT subpopulations (fCT I-V) (Fig. 2, B and C; Table S6). The two fibroblastic CT populations fCT IV and fCT V could be identified as components of the dermis based on expression of Twist2 and Ptgds. Interestingly, the cycling population contained multiple different cell types including tenocytes, fCT I, fCT III and fCT IV (fig. S4A). We detected only very few skeletal cells (fig. S4B) likely due to their inability to dissociate from the skeletal matrix. Due to the low cell number, skeletal cells fall within the periskeletal cluster in the tSNE plot. Previous live imaging data (12) and our own cell tracking data (fig. S9) show that skeletal cells do not contribute to the regenerate. Since regeneration of bone progresses via endochondral ossification, “skeletal cells” here refers to osteogenic and chondrogenic cells, the proportion of which differs between mature and regenerating bone. In summary, this profiling data validates the specificity of our Prrx1:Cre-ER;Caggs:lp-Cherry - reporter animals, provides a cell atlas and marker set for cell types of the uninjured adult axolotl limb (Table S4) and characterizes the heterogeneity of the upper arm CT (Table S6).
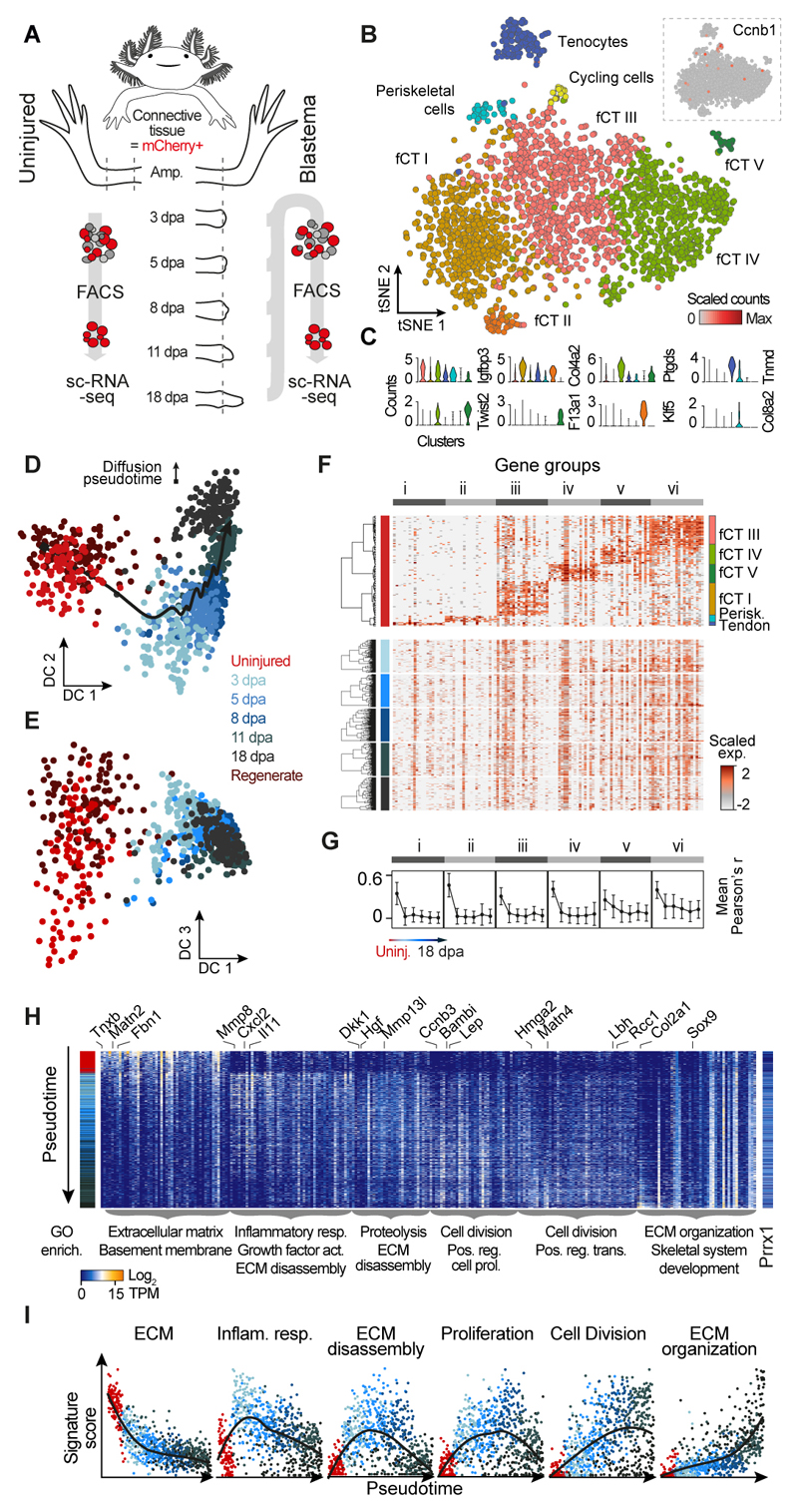
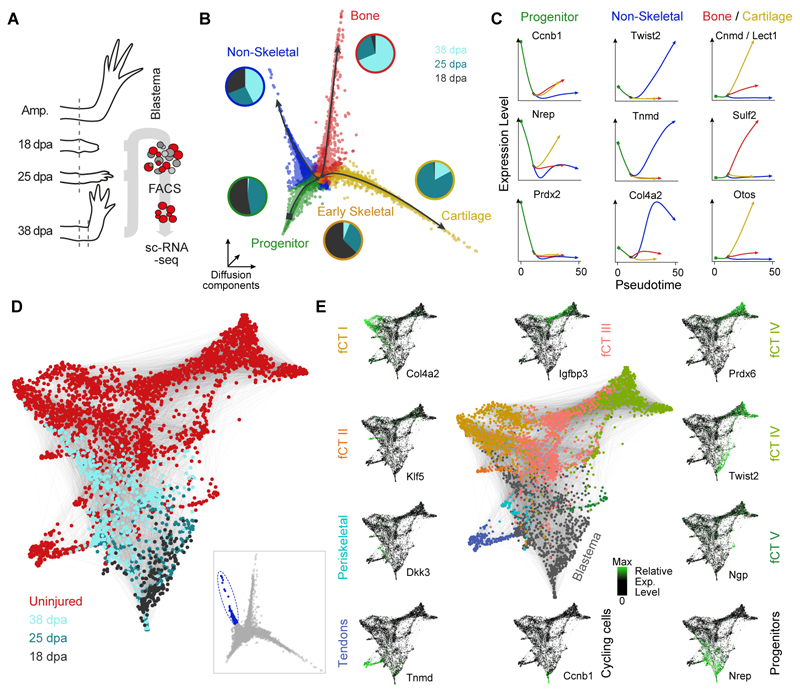
Fig. 2. Blastema formation from axolotl upper arm connective tissue cells involves molecular funneling during regeneration.
(A) Schematic of CT scRNA-seq experiments. ScRNA-seq was performed on FACS sorted mCherry+ CT cells of the uninjured axolotl upper arm (0 days post amputation, dpa) and during regeneration of the upper arm blastema at 3 dpa, 5 dpa, 8 dpa, 11 dpa and 18 dpa using Prrx1:Cre-ER;Caggs:Lp-Cherry animals (conversion at 1 cm size). (B) Cellular heterogeneity of the uninjured upper arm CT based on 2375 single cell transcriptomes. tSNE projection reveals 8 clusters referring to 7 CT subtypes. The 8th cluster contains cycling cells marked by expression of Ccnb1 shown as an inset. fCT: fibroblastic connective tissue. (C) Violin plots showing distribution of expression for selected tSNE cluster marker genes (panel B). The cluster of cycling cells was excluded. Colors refer to cluster colors of tSNE map (panel B). (D) Diffusion map projection (16) describes lineage relationships between uninjured CT cells and cells from all blastema time points as well as cells from a fully regenerated upper arm. DC: Diffusion component. CT cells from limb regenerate cluster with cells from uninjured upper arm tissue. (E) Diffusion component (DC) 3 captures the cell type heterogeneity in the uninjured CT, which is lost in the blastema. (F) Cellular heterogeneity of the mature CT is lost in the blastema. Expression of cell type marker genes (gene groups i to vi) identified for the uninjured CT is shown for each blastema time point as heatmap with genes in columns and cells hierarchically clustered in rows. Transcript levels are scaled across columns, respectively. (G) Mean pairwise correlation (Pearson) between genes of each of the 6 identified gene groups (panel F) across all cells was calculated for each experimental time point. Mean correlation coefficients decrease over the course of blastema formation indicating the loss of cell type heterogeneity in the blastema. Error bars indicate standard deviation. (H) Heatmap visualization of time point-specific marker genes (columns) with cells (rows) ordered by diffusion pseudotime (see also fig. S4I). GO enrichments are provided below the heatmap for each gene group and exemplary genes are shown at the top (see also fig. S5A). Colored sidebar on the left indicates time points. (I) Pseudotemporal expression of different gene signatures across all cells from uninjured upper arm CT to blastema 18dpa. Smoothed conditional means using LOESS are presented.
Blastema formation from axolotl upper arm connective tissue cells involves molecular funneling during regeneration
To understand the molecular pathways involved in CT regeneration, we used a high transcriptome coverage scRNA-seq method (Fluidigm C1) to analyze mCherry+ cells isolated by FACS (Prrx1:Cre-ER;Caggs:lp-Cherry) along a dense time course that captures the major transitions during regeneration. Time points were determined based on the average blastema cell cycle length (53-103 hours) (13, 14), and previous bulk transcriptional dynamics (15), and therefore included the uninjured upper forelimbs (uninjured, 108 cells) as well as blastema stages (3 dpa, 108 cells; 5 dpa, 167 cells; 8 dpa, 121 cells; 11 dpa, 163 cells; 18 dpa, 135 cells), and a fully regenerated upper forelimb (3-12 months post amputation, 128 cells) (Fig. 2A, fig. S3 and Table S7). We first analyzed the uninjured upper forelimb data and could confirm the presence of the CT subpopulations described above, which allowed identification of additional marker genes for each cell type (fig. S4, C and D). We next combined fibroblastic and periskeletal cells from the uninjured CT, which are the major populations contributing to the blastema (12), with all blastema states and the regenerated CT and used a diffusion pseudotime estimate (16) to reconstruct the lineage path from uninjured CT through the blastema state to the regenerated CT (Fig. 2D and fig. S4E). This analysis revealed a general time-dependent progression, with some intermixing between time points. Interestingly, diffusion component 3 visualizes how heterogeneous differentiated CT cells funnel into the relatively homogeneous early blastema state (Fig. 2E) and regenerated CT cells funnel out of the blastema to re-establish the initial cellular heterogeneity (fig. S4F). We confirmed this observation with two alternative analyses. When focusing on the cell type-specific expression patterns found in the uninjured tissue (Fig. 2F), we did not observe a comparable heterogeneity within the blastema cell populations, and instead found that heterogeneity was diminished in all blastema time points (Fig. 2G), up until the re-differentiation of CT cell types (fig. S4G). Further, we calculated for each time point the number of unique genes detected within the top 100 most expressed genes per cell as a measure of intercellular heterogeneity per time point. We detected a decrease in the number of unique genes from the adult uninjured upper limb through the early blastema stages up to 11 dpa blastema, followed by an increase at 18 dpa and in the regenerate (fig. S4H). This is consistent with a progressive decrease in intercellular heterogeneity during blastema formation until 11 dpa with the highest heterogeneity found in the uninjured adult upper arm, the regenerate and in the 18 dpa blastema when cells begin to differentiate. Notably, we do not find blastema-like cells within the uninjured tissue since all cells from the adult upper forelimb cluster separately from blastema-derived cells in a tSNE analysis (fig. S4I). Also, when analyzing the cycling cells in the uninjured tissue together with cycling blastema cells, the cells derived from the uninjured tissue are distinct, since they express signatures of differentiated CT cell types (e.g. Mfap5, Fbn1) and lack expression of blastema markers (e.g. Nrep, Prdx2) (fig. S4, J and K).
Our molecular data taken together with previous cell lineage observations (12) and digital tracking data (17) show little numerical bias in cell contribution during blastema formation allowing us to conclude that blastema formation does not involve the selection of a pre-existing stem cell population. Considering cell cycle length (13, 14) and cell counts pointing to an on average 6-fold expansion of cells from the uninjured to the 11 dpa timepoint (Supplemental Materials and Methods), a “pre-existing” blastema cell would need to comprise at least 20% of the mature CT population to account for the cell expansion. In summary these data indicate that mature CT cells dedifferentiate into a relatively homogeneous pool of progenitor cells when forming the blastema.
Next, we explored the transcriptional changes that ensue during the regenerative process within the blastema (Fig. 2H and fig. S5, A to C). The uninjured CT state is characterized by the expression of many extracellular matrix (ECM) genes that are downregulated during blastema formation (Fig. 2, H and I). We observe an inflammation response (e.g. Il11, Cxcl2) in the early blastema (3-5 dpa) coinciding with the expression of various matrix metalloproteinases (Mmp8/13), indicating extensive ECM remodeling induced by injury. By 8 dpa, cells express genes involved in proliferation (e.g. Ccnb3), and by 11 dpa there is positive regulation of transcription (Hmga2, Lbh) and evidence of re-patterning (Hoxa13, Hoxd12). Re-establishment of ECM (Matn4) is initiated by 11 dpa but peaks at 18 dpa. Notably, genes necessary for skeletal development are detected in the blastema 18 dpa (Hoxd13, Col2a1, Sox9). In addition, the expression of various signaling molecules (Dkk1, Bambi, Lep) associated with regeneration are induced throughout blastema development (15, 18, 19). This data constitutes a molecular map of CT cells as they transition through the limb regenerative process.
Connective tissue reprogramming progresses through a blastema-specific state prior to recapitulating embryonic limb bud development
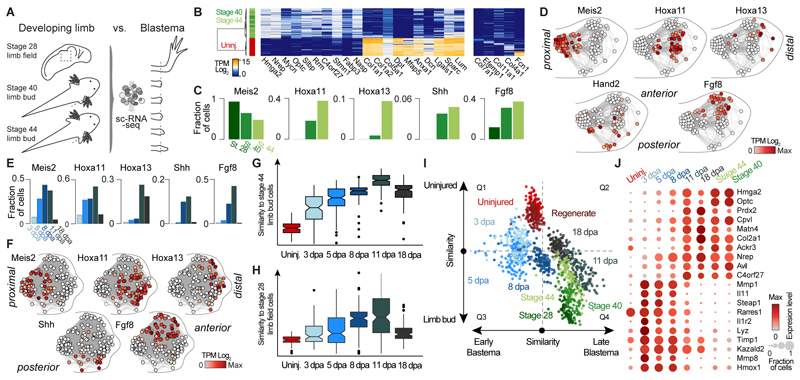
We next explored the extent to which CT regeneration recapitulates limb development. To cover different stages of limb development, we examined the single-cell transcriptomes of stage 28 limb field cells (82 cells) as well as stage 40 (76 cells) and stage 44 (121 cells) limb bud cells (20) (Fig. 3A; Table S7) and identified CT cells based on Prrx1 expression (279 cells). By stage 44 the limb bud contains approximately 1500 cells. We observed that CT precursors in the developing limb are distinct from and less differentiated than adult CT (Fig. 3B and fig. S6A). While cells from the two limb bud stages showed major similarities, cells from the stage 28 limb field were distinct from stage 40 and 44 limb bud cells as shown by the unique expression of genes such as Wnt8a or Hoxd1 (fig. S6B). Interestingly, variation in the expression of spatial patterning genes constituted the major source of heterogeneity in the limb buds (Stage 40 and 44; fig. S6C) whereas patterning genes were generally not yet expressed in stage 28 limb field cells (Fig. 3C). This variation enabled reconstruction of the proximal-distal and anterior-posterior axes through the correlated expression of patterning genes confirming that our data captured the representation of the limb bud cells (Fig. 3D) (21). We investigated when patterning axes start to be established during limb regeneration and found that anterior-posterior markers (Fgf8 and Shh, respectively) and proximal-distal markers (Meis2, Hoxa11 and Hoxa13, respectively) are established between 8-11 dpa (Fig. 3E). In a similar manner as for the limb bud, patterning genes enabled the spatial reconstruction of cell positions in a blastema 11 dpa (Fig. 3F and fig. S6D).
Fig. 3. Connective tissue reprogramming progresses through a blastema-specific state prior to recapitulating embryonic limb bud development.
(A) Overview of scRNA-seq experiments on three axolotl limb bud stages. 279 limb bud CT cells were in silico identified based on Prrx1 expression and their transcriptomes were compared to scRNA-seq data of the blastema cells. (B) Left: Heatmap showing expression of genes (columns) that distinguish mature limb CT cells from limb bud CT cells (rows). Right: Heatmap showing expression of marker genes for uninjured CT cell types (columns) across mature limb and limb bud CT cells (rows). Cells are hierarchically clustered (Pearson) based on expression of all shown genes. (C) Bar graphs show fraction of cells per embryonic stage that express genes involved in proximal-distal patterning (Meis2, Hoxa11, Hoxa13) or in anterior-posterior patterning (Fgf8, Shh). (D) Spatial patterning genes describe most of the heterogeneity found in the limb bud CT (See also fig. S6C). Intercellular correlation network constructed for stage 44 limb bud cells (circles) based on expression of 5 known patterning genes places cells on a hypothetical position within an imaginary limb bud. Note, that Hand2 instead of Shh was used as anterior marker due to the low number of Shh expressing cells (Fig. 3C). (E) Limb bud patterning genes are reactivated during blastema formation. Bar graphs show fraction of cells per blastema time point that express genes involved in proximal-distal patterning (Meis2, Hoxa11, Hoxa13) or in anterior-posterior patterning (Fgf8, Shh). (F) Intercellular correlation network constructed for all blastema 11 dpa cells (circles) based on expression of 5 patterning genes places cells on a hypothetical position within an imaginary limb blastema. (See also fig. S6D). (G) Correlation analysis reveals the highest similarity of limb bud progenitors with blastema 11 dpa cells. Boxplot shows distributions of scaled correlation between single cell transcriptomes at any given time point and the mock bulk transcriptome of stage 44 limb bud CT cells. (H) Correlation analysis reveals the highest similarity of stage 28 limb field cells with blastema 11 dpa cells. Boxplots show distributions of scaled correlation values between single-cell transcriptomes at the different sampled time points and the mock bulk transcriptome of limb field CT cells. (I) Scatterplot showing differential correlation of single cell transcriptomes (dots, color-coded based on time point) with limb bud versus uninjured mature CT transcriptomes (y-axis) and with blastema 5 dpa versus blastema 11 dpa transcriptomes (x-axis). (J) Dotplot visualizing expression of genes shared between blastema 11 dpa and limb bud progenitor cells. Circle size represents the fraction of cells of each time point expressing the gene and color represents the average expression level.
We next quantified the similarity between different blastema stages and limb development. We created mock bulk transcriptomes of each uninjured, regenerate, embryonic, and blastema time point and calculated the correlation (Spearman’s r) of each single cell with each bulk transcriptome. We found that the correlation of blastema cells with stage 40 and stage 44 limb bud peaks at 11 dpa, with a progressively weaker correlation in earlier blastema time points (Fig. 3G and fig. S6, E and F). Notably, the same trend was observed when we compared blastema cells with the stage 28 limb field (Fig. 3H). Additionally, we find that cells in the 3 and 5 dpa blastema are similar to neither uninjured adult cells nor limb bud/limb field cells, suggesting that a cell state emerges in the blastema that is distinctive for limb regeneration (Fig. 3I and fig. S6E). Indeed, within our dataset, we identify many genes that are uniquely expressed in the early blastema stages, whereas 11 dpa cells largely share expression patterns with the cells derived from the developing limb (Fig. 3J and fig. S6, G and H). Our data suggests that the former CT cells within the blastema initiate reprogramming using genetic mechanisms that are distinct from embryonic limb bud development but arrive at a limb bud-like state by day 11.
Tracking different connective tissue subpopulations in the blastema reveals distinct cell sources for proximal versus distal limb regenerate tissue
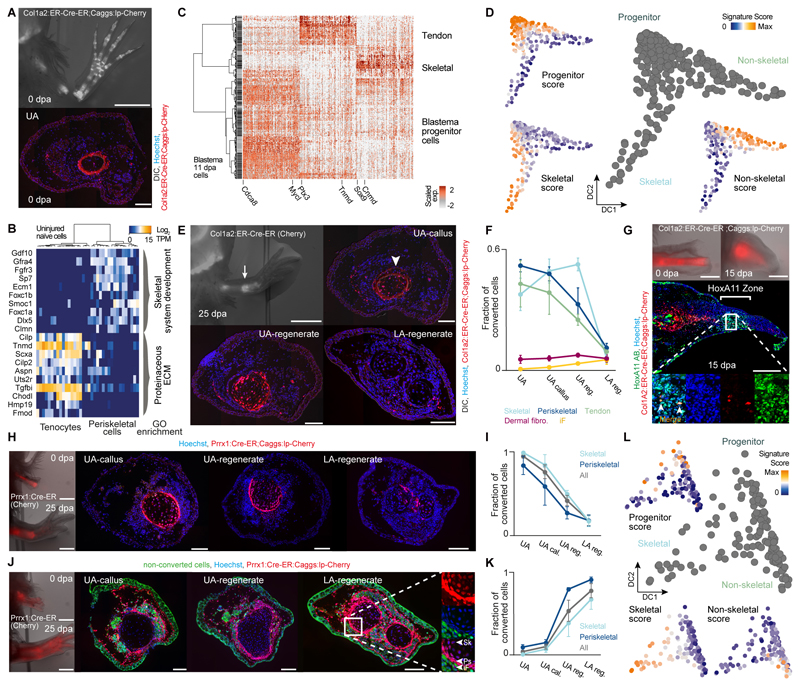
We next sought to analyze how different CT cell subpopulations contribute to re-differentiated cell types in the regenerated limb. We first generated a new transgenic line using the Col1a2 promoter (Col1A2:ER-Cre-ER;Caggs:lp-Cherry = Col1A2:TFPnls-T2a-ERT2-Cre-ERT2;CAGGs:lp-GFP-3pA-lp-Cherry) in which only a subset of CT cells are labeled and tracked (fig. S7). Specifically, conversion of 3 cm larvae leads to limbs with genetic labeling primarily of skeleton, periskeleton, and tendons, but very few dermal fibroblasts and no interstitial fibroblasts, thus comprising a subset of cell types labeled in the Prrx1:Cre-ER;Caggs:lp-Cherry line (Fig. 4A and fig. S7A). ScRNA-seq on 36 sorted, labeled cells from the uninjured adult upper arm revealed periskeletal cells and tenocytes (bone cells were not recovered in the dissociation, but they do not contribute to the blastema), confirming the histological analysis of the labeled cells (Fig. 4B and Table S8). We next performed scRNA-seq on labeled Col1a2:ER-Cre-ER;Caggs:lp-Cherry (“Col1a2”) descendants in the 11 dpa blastema (349 cells), and could identify undifferentiated progenitors as well as immature skeletal and non-skeletal cell lineages (Fig. 4C and Table S8). A pseudotemporal trajectory displayed two branching paths; one transitioning from progenitors (Mycl, Matn4, Nrep) to non-skeletal cells (Tnmd, Fcn2, Aspn; likely precursors to tenocytes and periskeletal cells) and the other to skeletal cells (Cnmd/Lect1, Otos) (Fig. 4D, fig. S6I and Table S10).
Fig. 4. Tracking different connective tissue subpopulations in the blastema reveals distinct cell sources for proximal versus distal limb regenerate tissue.
(A) Top: Fluorescence overlaid with bright-field image of Col1a2:ER-Cre-ER;Caggs:lp-Cherry (conversion at 3 cm size) limbs at 0 dpa overlaid with DIC. Bottom: Upper arm limb cross-section reveals labeling of periskeletal and tendon cells. Hoechst (blue), Col1a2:ER-Cre-ER;Caggs:lp-Cherry (red). (B) Heatmap showing distinct expression profiles for 36 mature limb periskeletal and tendon cells (Col1a2:ER-Cre-ER;Caggs:lp-Cherry labeled) with cells (columns) being hierarchically clustered (Pearson). GO enrichments are shown. (C) Cellular heterogeneity of labeled Col1a2:ER-Cre-ER;Caggs:lp-Cherry descendants in the 11 dpa blastema (349 cells). Heatmap visualizing expression of genes (columns) identified by PCA in 11 dpa blastema cells (rows, hierarchically clustered based on Pearson’s correlation). Transcript levels are scaled across columns. (D) Pseudotemporal trajectory obtained by Diffusion Map projection (26) on genes identified by PCA (Table S10) for 11 dpa blastema cells deriving from labeled Col1a2:ER-Cre-ER;Caggs:lp-Cherry descendants. Blastema progenitors are linked to 2 emerging cell lineages: non-skeletal (likely precursors to periskeletal cells and tenocytes) and skeletal. (23) Scores for developmental, non-skeletal and skeletal signatures projected onto the Diffusion Map are shown. (E) Top left: Fluorescence overlaid with bright-field image of Col1a2:ER-Cre-ER;Caggs:lp-Cherry limbs at 25 dpa. Scale bar: 2 mm. From top right to bottom left to bottom right: Limb cross-sections along the proximo-distal axis (UA: upper arm, LA: lower arm). Hoechst (blue), Col1A2:ER-Cre-ER;Caggs:lp-Cherry (red) overlaid with DIC. Scale bar: 200 μm. (F) Fraction of converted cells in five CT subtypes at different proximo-distal positions after regeneration, (cell number n > 8000, 3 independent limb samples). iF: Interstitial fibroblasts (fCT I-III). (G) Col1a2:ER-Cre-ER;Caggs:lp-Cherry labeled humerus transplantation into unlabeled host. Top: Stereoscopic images of live animals at 0 dpa and 15 dpa. Scale bar: 1 mm. Center: Longitudinal sections of 15 dpa blastema. Converted cells (red), HoxA11 (green), Hoechst (blue). Scale bar: 500 μm. Bottom: Magnified view showing co-localization of mCherry+ cells with HoxA11 antibodies. (H) Prrx1:Cre-ER;Caggs:lp-Cherry labeled humerus transplantation into unlabeled host. Left: Stereoscopic images of live animals at 0 dpa and 25 dpa. Scale bar: 1 mm. From left to right: Cross sections of limbs from UA-callus, UA-regenerate and LA-regenerate. Converted cells (red), Hoechst (blue). Scale bar: 200 μm. (I) Quantification of converted cells in periskeletal, skeletal and aggregate of periskeletal and skeletal subtypes (All) plotted on proximo-distal axis. (J) Unlabeled humerus transplantation into Prrx1:Cre-ER;Caggs:lp-Cherry converted host. Left: Stereoscopic images of live animals at 0 dpa and 25 dpa. Scale bar: 1 mm. From left to right: Cross sections of limbs from UA-callus, UA-regenerate and LA-regenerate. Converted cells (red), Non-converted cells (green), Hoechst (blue). Scale bar: 200 μm. Right: Magnified view showing mCherry+ periskeletal (Ps) and skeletal (Sk) and iF: Interstitial fibroblasts (fCT I-III). (K) Quantification of converted cells in periskeletal, skeletal and aggregate of periskeletal and skeletal subtypes plotted (All) on proximo-distal axis. (L) Pseudotemporal trajectory obtained by Diffusion Map projection (26) on genes identified by PCA (Table S10) for 18 dpa blastema cells of labeled Prrx1:Cre-ER;Caggs:lp-Cherry descendants links limb bud-like blastema progenitors to 2 emerging cell lineages: non-skeletal (precursors to all non-skeletal CT subtypes) and skeletal. (23) Scores for developmental, non-skeletal and skeletal signatures are projected onto the Diffusion Map.
Interestingly, microscopic examination of regenerates revealed a spatial bias in cell contribution. Labeled cells were primarily found in the extension to the existing bone at the amputation site (humerus) with few cells in more distal segments (Fig. 4, E and F). We also found a similar spatial restriction of “Col1a2”-descendants to extending bone in lower arm amputations (Fig. S8B). This is in contrast to the Prrx1:Cre-ER;Caggs:lp-Cherry line in which all proximal and distal positions of the regenerate are labeled. These data provide direct evidence supporting a previous hypothesis that distinct cell sources are used for bone extension versus de novo segment formation (22). The “Col1a2”-derived blastema cells could be intrinsically or extrinsically limited to extending existing bone. We transplanted a humerus from a Col1a2:ER-Cre-ER;Caggs:lp-Cherry donor into an unlabeled host and found that after upper arm amputation some rare mCherry+ cells were found in the distal, Hoxa11-positive region of blastema 15 dpa and showed Hoxa11 staining (Fig. 4G). These observations lead us to conclude that the “Col1a2”-derived blastema cells are not intrinsically limited in their segmental identity, but rather are strongly associated with the callus, and therefore spatially biased towards extending their bone of origin.
We performed multiple transplantation experiments to confirm the above observations and resolve the source of the distal CT. To exclude the possibility that the Col1a2 driver marked only a subset of periskeleton and tendon with spatially-restricted potential, we grafted a humerus from Prrx1:Cre-ER;Caggs:lp-Cherry (“Prrx1”) converted limbs into an unlabeled host limb, replacing the host humerus prior to upper arm amputation (Fig. 4, H and I). Similar to the Col1a2 tracing, mCherry+ cells were found in the callus and the regenerated periskeleton and skeleton just beyond the amputation site with progressive depletion towards the lower arm regenerate. We next performed a complementary graft of unlabeled humerus into “Prrx1”-converted hosts to label non-bone CT. We found nearly complete labeling of tendon, skeleton, and periskeleton as well as dermal and interstitial fibroblasts in the lower arm regenerate indicating that non-skeletal CT cells regenerate the distal CT (Fig. 4, J and K). We separately grafted a muscle fiber bundle containing labeled interstitial fibroblasts from embryonic “Prrx1”-converted animals into unlabeled hosts and found extensive labeling of the distal CT (fig. S10). These data indicated that the fibroblastic CT cells were the major contributors to the distal limb regenerate. To analyze the regeneration of distal limb CT on the molecular level, we performed a lineage reconstruction analysis on the scRNA-seq data of labeled “Prrx1”-descendants in the 18 dpa blastema (Fig. 4L, fig. S6J and Table S10). This analysis revealed a bifurcated path where uncommitted blastema progenitors branch off into a non-skeletal and a skeletal lineage. At the 18 dpa timepoint, differentiated dermal and interstitial fibroblasts were not yet identifiable, consistent with live imaging data showing the late differentiation of these lineages (12). Notably, both “Col1a2”-descendant blastema cells (Fig. 4C, mostly periskeleton-derived) and “Prrx1”-descendant blastema cells (Fig. 4L, mostly fibroblastic CT-derived) had a similar expression profile resembling limb bud progenitors. Taken together, we conclude that cells from multiple CT compartments funnel into an undifferentiated progenitor with the tissue of origin biasing the spatial contribution of the cells.
Connective tissue lineages reemerge through multipotent progenitors
To reconstruct the re-establishment of each CT cell type in the upper arm regenerate, we performed high-throughput droplet-based scRNA-seq of labeled Prrx1:Cre-ER;Caggs:lp-Cherry descendants in the late blastema at three time points including 18 dpa (9939 cells), 25 dpa (9019 cells) and 38 dpa (2861 cells) (Fig. 5A and Table S9). We inferred differentiation trajectories and pseudotemporal cell relationships (23) from a diffusion map embedding (16), and identified a trifurcated path, where multipotent blastema progenitors expressing embryonic limb and cell cycle markers (e.g. Prdx2, Nrep, Ccnb1) branch off to a non-skeletal lineage or a skeletal lineage that then bifurcates into either cartilage or bone (Fig. 5, B and C). We observed temporal differences in the lineage progression as progenitor cells are present only at the earlier two time points (18 and 25 dpa), whereas cells on the non-skeletal branch are found at all three time points. A skeletal precursor state is found mainly at 18 dpa before cartilage and bone start to differentiate at 25 dpa and 38 dpa, respectively (see also fig. S11B). We next analyzed the heterogeneity in the non-skeletal branch and found that the main non-skeletal lineages (periskeletal cells, tenocytes, fibroblastic CT cells) present in the adult uninjured tissue had started to reemerge by 38 dpa (Fig. 5D and fig. S11, C and D). This data suggests that, at the transcriptome level, the progenitor pool between 18 and 25 dpa is still relatively homogeneous, and that these progenitor cells diversify sometime after 25 dpa into diverse CT lineages.
Fig. 5. Following the re-emergence of connective tissue lineages through multipotent progenitors.
(A) Schematic of high-throughput scRNA-seq experiments on late blastema stages using Prrx1:Cre-ER;Caggs:Lp-Cherry converted animals. ScRNA-seq was performed on FACS sorted mCherry+ CT cells of the uninjured axolotl upper arm (0 days post amputation, dpa) and during regeneration of the upper arm blastema at 18 dpa, 25 dpa and 35 dpa using the 10x Genomics Chromium controller. (B) Three-dimensional representation of a Diffusion analysis (16) of blastema time points (18, 25, 38 dpa) identifies 4 branches. Merlot (23) was used to identify end points and branching points (color coded) within the trajectory and to obtain branch-specific gene expression patterns. Pie charts next to the branches show the time point distribution per respective branch. (C) Pseudotemporal expression of marker genes along the branches identified in panel B. Shown markers were used to assign cell types to each branch (blastema progenitors, two skeletal lineages (cartilage and bone) and one non-skeletal lineage). (D) SPRING (27) visualization of late non-skeletal blastema cells (cells with highest pseudotime encircled and highlighted in blue in inset) together with mature CT cells (total 3151 cells) reveals emergence of CT subpopulations identified in the mature tissue. (E) Genes identified as markers for distinct CT subtypes (Tnmd, Col4a2, Twist2) in the mature uninjured tissue highlight the emergence of cell types during the last phases of regeneration while the expression of developmental blastema markers (Nrep) and cell cycle markers (Ccnb1) decrease during final differentiation.
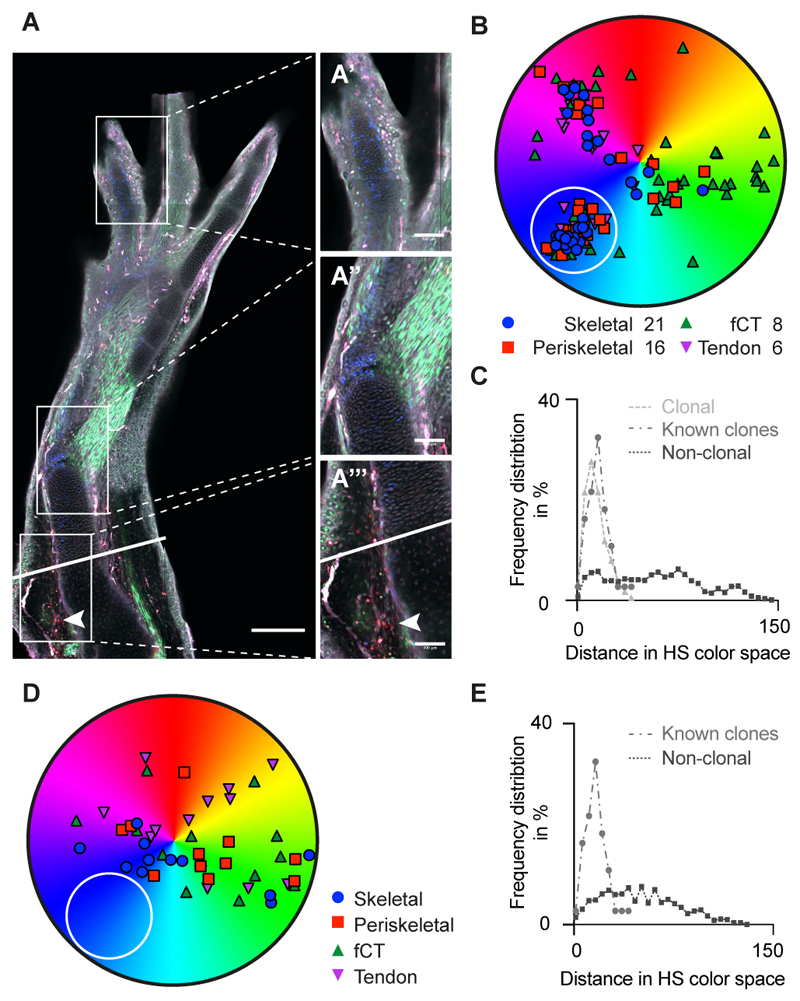
We further sought to validate our molecular analysis by clonal lineage tracing using a Brainbow transgenic animal, which allowed us to determine the spectrum of cell types formed from single CT cells. To establish clonal tracing conditions, recombination was induced in mature limb cells and examined after 7 days. To identify color combinations showing appropriately low occurrence for clonal analysis, clone pairs were identified as infrequently occurring adjacent sisters derived from a cell division, and their distribution in color and hue saturation space was determined (fig S11E). From this data, a rare recombinant type that mapped in the blue color range was identified (fig. S11F). Examination of 9 different source zone equivalents in the mature tissue identified 2, 1, 0, 1, 1, 0, 0, 2, 0 blue cells per zone (fig. S11G). Amputated limbs were allowed to regenerate for 25 days (Fig. 6A) and then examined for the presence of blue clones. Three out of ten limbs contained a blue clone whereas the other seven limbs showed no blue clone, confirming its rare occurrence (Fig. 6, B and D). The frequency distribution in color space of the blue cells in each regenerate was consistent with clonal origin (Fig. 6, C and E) (for more examples see fig. S11, H and I). Assignment of the blue cells from within each limb to connective tissue sub-types revealed contributions of clonal descendants to skeletal, periskeletal, fibroblastic and tendon cells (Fig. 6B). These lineage tracing data confirm for the first time the molecular profiling conclusions that limb blastema cells acquire a limb bud progenitor identity and form a multipotent CT progenitor (24, 25).
Fig. 6. Brainbow clonal analysis confirms multi-lineage potential of connective tissue cells upon limb regeneration.
(A) Representative image of a regenerated limb in a Brainbow axolotl. A presumptive clone of blue cells is observed throughout the limb, from the digit tip (A’), the elbow (A”), and amputation plane (solid line) at the injection site (arrowhead) (A”’). Scale bars: (A) 300 μm; (A’-A”’): 100 μm. (B) Example of HS color distribution of cells from a representative image containing a presumptive clone of blue cells (white circle). Multiple cells of each connective tissue sub-type (Skeletal cells, periskeletal cells, tenocytes and fibroblastic CT cells) are all represented. Similar distributions were observed in 3/10 analyzed samples (for examples see fig. S11H). (C) Frequency distribution in HS color space calculated using the formula (fig. S11E), for known clonally related cells (fig. S11, F and G), presumptive clonally related cells (“blue cells” in white circle, panel B) in a regenerated limb, and non-clonally related cells in a regenerate. Frequency distribution of suspected clonally related cells is indistinguishable from known clones (Kruskal-Wallis analysis and Dunn’s multiple comparison). (D) Example of HS color distribution of cells from a representative image lacking a discreet cluster of blue cells (white circle). Similar distribution was observed for 7/10 analyzed samples (for examples see fig. S11I). (E) Frequency distribution as in C for sample shown in D. Note that due to the lack in identifying a clonally related subset, no presumptive clone could be mapped.
Summary
The molecular understanding of blastema formation had previously suffered from the inability to identify and isolate blastema precursor cells in the adult tissue. We have demonstrated the importance of genetically marked transgenic axolotl strains for isolating blastema precursor cells from adult limb tissue and we have molecularly profiled them using single-cell transcriptomic methods. This profiling has indicated that CT cells express adult phenotypes that are lost upon induction of regeneration and funnel into an embryonic limb bud-like phenotype including multipotency within the CT lineage (24, 25). The molecular reprogrammability of adult cells to cells of embryonic limb potential capable of orchestrating complex limb morphogenesis has clear implications for future prospects in regenerative engineering.
Supplementary Material
One Sentence Summary.
Single-cell transcriptomics and reporter lineage tracking were used to deconstruct cell composition, reconstruct lineage relationships, and trace connective tissue reprogramming during axolotl limb regeneration.
Acknowledgments
We thank B. Borgonovo and D. Drechsel for technical advice; P. Keller for help with Anti-PRRX1 antibodies; A. Heino, B. Gruhl, S. Mögel, A. Wagner, S. Kaudel and H. Görs for excellent animal care and breeding; M. Corriero for Axolotl sketch; T. Engelmaier for help with HE staining; Z. He, G. Parra and J. Kelso for computational support; A. Weihmann and B. Schellbach for sequencing and members of the Tanaka lab, Treutlein lab and S. Pääbo for helpful discussions. We thank the “Core Unit Durchflusszytometrie” (CUDZ) at the College of Veterinary Medicine, University of Leipzig for providing access to FACS. The authors would like to acknowledge financial support from the Max Planck Society (BT), DFG grant TA 274/3-3 (PM, EMT), DFG TA 274/11-1 (DK), and DFG/ANR TA274/13-1 (PM, EMT) and the ERC Advanced Investigator Award (PM, SH and EMT), FZ111 and institutional support from the IMP (EMT). Animal experiments were performed as approved by the State Authorities Saxony, and the Magistrate of Vienna.
Footnotes
Author notes: TG, PM, DK, JGC, EMT and BT designed the study, and wrote the manuscript. PM generated transgenic axolotl lines with assistance from MS and SK. PM performed lineage tracing and immunohistochemistry with help from SH. PM and DK performed grafting experiments. PM and WM performed Brainbow-clonal tracing. TG performed single-cell experiments with assistance from JGC, DK and PM. MG-S performed FACS. TG analyzed single-cell RNA-seq data with help from BT, JGC, JK, PM, DK and EMT. SN provided assistance with Axolotl transcriptome. JC generated HOXA11 antibody.
References and Notes
- 1.Brockes JP, Kumar A. Comparative aspects of animal regeneration. Annu Rev Cell Dev Biol. 2008;24:525–549. doi: 10.1146/annurev.cellbio.24.110707.175336. [DOI] [PubMed] [Google Scholar]
- 2.Nacu E, Tanaka EM. Limb regeneration: a new development? Annu Rev Cell Dev Biol. 2011;27:409–440. doi: 10.1146/annurev-cellbio-092910-154115. [DOI] [PubMed] [Google Scholar]
- 3.Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- 4.Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- 5.Dunis DA, Namenwirth M. The role of grafted skin in the regeneration of x-irradiated axolotl limbs. Dev Biol. 1977;56:97–109. doi: 10.1016/0012-1606(77)90157-9. [DOI] [PubMed] [Google Scholar]
- 6.Carlson BM. The effects of rotation and positional change of stump tissues upon morphogenesis of the regenerating axolotl limb. Dev Biol. 1975;47:269–291. doi: 10.1016/0012-1606(75)90282-1. [DOI] [PubMed] [Google Scholar]
- 7.Nacu E, et al. Connective tissue cells, but not muscle cells, are involved in establishing the proximo-distal outcome of limb regeneration in the axolotl. Development. 2013;140:513–518. doi: 10.1242/dev.081752. [DOI] [PubMed] [Google Scholar]
- 8.Pescitelli MJ, Jr, Stocum DL. The origin of skeletal structures during intercalary regeneration of larval Ambystoma limbs. Dev Biol. 1980;79:255–275. doi: 10.1016/0012-1606(80)90115-3. [DOI] [PubMed] [Google Scholar]
- 9.Nohno T, et al. A chicken homeobox gene related to Drosophila paired is predominantly expressed in the developing limb. Dev Biol. 1993;158:254–264. doi: 10.1006/dbio.1993.1184. [DOI] [PubMed] [Google Scholar]
- 10.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 11.Satoh A, Gardiner DM, Bryant SV, Endo T. Nerve-induced ectopic limb blastemas in the Axolotl are equivalent to amputation-induced blastemas. Dev Biol. 2007;312:231–244. doi: 10.1016/j.ydbio.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Currie JD, et al. Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Dev Cell. 2016;39:411–423. doi: 10.1016/j.devcel.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent CD, Rost F, Masselink W, Brusch L, Tanaka EM. Cellular dynamics underlying regeneration of appropriate segment number during axolotl tail regeneration. BMC Dev Biol. 2015;15:48. doi: 10.1186/s12861-015-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace H, Maden M. The cell cycle during amphibian limb regeneration. J Cell Sci. 1976;20:539–547. doi: 10.1242/jcs.20.3.539. [DOI] [PubMed] [Google Scholar]
- 15.Knapp D, et al. Comparative transcriptional profiling of the axolotl limb identifies a tripartite regeneration-specific gene program. PLoS One. 2013;8:e61352. doi: 10.1371/journal.pone.0061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghverdi L, Buttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 17.Flowers GP, Sanor LD, Crews CM. Lineage tracing of genome-edited alleles reveals high fidelity axolotl limb regeneration. Elife. 2017;6 doi: 10.7554/eLife.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang J, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami Y, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bordzilovskaya NP, Dettlaff TA, Duhon ST, Malacinski GM, editors. Developmental-stage series of axolotl embryos. New York: 1989. pp. 201–219. [Google Scholar]
- 21.Nacu E, Gromberg E, Oliveira CR, Drechsel D, Tanaka EM. FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature. 2016;533:407–410. doi: 10.1038/nature17972. [DOI] [PubMed] [Google Scholar]
- 22.Makanae A, Mitogawa K, Satoh A. Implication of two different regeneration systems in limb regeneration. Regeneration (Oxf) 2014;1:1–9. doi: 10.1002/reg2.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra RG, et al. Reconstructing complex lineage trees from scRNA-seq data using MERLoT. bioRxiv. 2018 doi: 10.1093/nar/gkz706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arques CG, Doohan R, Sharpe J, Torres M. Cell tracing reveals a dorsoventral lineage restriction plane in the mouse limb bud mesenchyme. Development. 2007;134:3713–3722. doi: 10.1242/dev.02873. [DOI] [PubMed] [Google Scholar]
- 25.Pearse RV, 2nd, Scherz PJ, Campbell JK, Tabin CJ. A cellular lineage analysis of the chick limb bud. Dev Biol. 2007;310:388–400. doi: 10.1016/j.ydbio.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 27.Weinreb C, Wolock S, Klein AM. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 2018;34:1246–1248. doi: 10.1093/bioinformatics/btx792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khattak S, et al. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat Protoc. 2014;9:529–540. doi: 10.1038/nprot.2014.040. [DOI] [PubMed] [Google Scholar]
- 29.Bou-Gharios G, et al. A potent far-upstream enhancer in the mouse pro alpha 2(I) collagen gene regulates expression of reporter genes in transgenic mice. J Cell Biol. 1996;134:1333–1344. doi: 10.1083/jcb.134.5.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerney R, Hall BK, Hanken J. Regulatory elements of Xenopus col2a1 drive cartilaginous gene expression in transgenic frogs. Int J Dev Biol. 2010;54:141–150. doi: 10.1387/ijdb.092848rk. [DOI] [PubMed] [Google Scholar]
- 31.Khattak S, et al. Germline Transgenic Methods for Tracking Cells and Testing Gene Function during Regeneration in the Axolotl. Stem Cell Reports. 2014;2:243. doi: 10.1016/j.stemcr.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowoshilow S, et al. The axolotl genome and the evolution of key tissue formation regulators. Nature. 2018;554:50–55. doi: 10.1038/nature25458. [DOI] [PubMed] [Google Scholar]
- 33.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan YA, et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development. 2013;140:2835–2846. doi: 10.1242/dev.094631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gurevich DB, et al. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 2016;353 doi: 10.1126/science.aad9969. aad9969. [DOI] [PubMed] [Google Scholar]
- 36.Picelli S, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 37.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 38.Renaud G, Kircher M, Stenzel U, Kelso J. freeIbis: an efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics. 2013;29:1208–1209. doi: 10.1093/bioinformatics/btt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renaud G, Slon V, Duggan AT, Kelso J. Schmutzi: estimation of contamination and endogenous mitochondrial consensus calling for ancient DNA. Genome Biol. 2015;16:224. doi: 10.1186/s13059-015-0776-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Zhang HM, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Res. 2012;40:D144–149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.