Abstract
Retrieving information about genes is a continuous and time-consuming aspect of systems biology. While there are resources that can fulfill this need, they require navigation to different and often complex websites, execution of a search (or searches) and selection of a result before the user can view pertinent information. To streamline this process, we developed a browser extension called GIX (Gene Information eXtension; https://gene-info.org) to allow users to retrieve gene information directly from a website without navigating to another page.
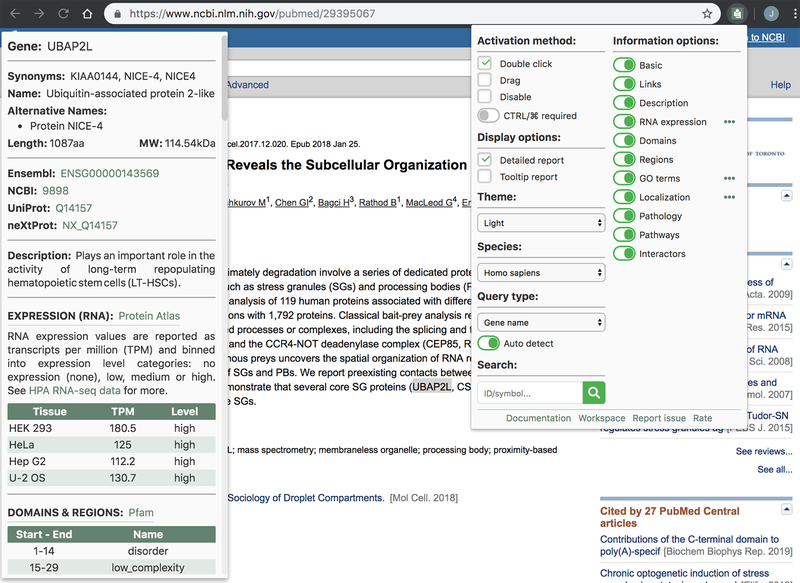
GIX is a browser extension for Google Chrome and Mozilla Firefox that allows users to retrieve customizable gene-product information—especially as it relates to proteins and their expression and functions—directly from a website. Simply double- clicking on (or, alternatively, mouse-dragging) a gene name or supported accession number (Ensembl, Entrez, neXtProt, RefSeq or UniProt) will open an information panel on the current page (Fig. 1). This panel will include gene synonyms, the full gene name, alternative names, the size and molecular weight of its canonical protein product, the UniProt description, protein domains and regions, Gene Ontology terms, protein localization, RNA tissue expression, associated diseases or phenotypes, pathways, protein interactors (from BioGRID and IntAct) and links to external resources (Ensembl, NCBI and UniProt for all species, and organism-specific databases including dictyBase, FlyBase, MGI, neXtProt, PomBase, SGD, TAIR, WormBase, Xenbase and ZFIN). GIX also offers an alternative tooltip mode that simply provides links to these external resources. The extension is fully customizable, allowing users to control the information they see, and it supports queries for Homo sapiens and ten model organisms: Arabidopsis thaliana, Caenorhabditis elegans, Danio rerio, Dictyostelium discoideum, Drosophila melanogaster, Gallus gallus, Mus musculus, Saccharomyces cerevisiae, Schizosaccharomyces pombe and Xenopus laevis. The extension provides a search bar for entering queries manually and an online “workspace” in which the user can paste gene lists from desktop applications for quick queries with GIX. Although double- clicking to retrieve results is not possible on websites with embedded content such as Google Docs or PDFs, querying with the search bar does work on such webpages. GIX collates data from BioGRID1, Compartments2, Gene Ontology3, HUGO Gene Nomenclature Committee4, the Human Protein Atlas5, IntAct6, OMIM7, Pfam8, Reactome9 and UniProt10. The GIX database is updated monthly to incorporate changes from these resources.
Figure 1.
Screenshot of GIX in the Chrome browser. Double-clicking on a gene name/symbol (in this case UBAP2L) opens an information panel (left side) displaying information about the queried gene. The extension has a number of settings that the user can customize by clicking on its icon in the browser toolbar (right side). The screenshot of PubMed is used courtesy of the National Library of Medicine (NLM), and the abstract is reproduced from Mol. Cell, Vol. 69, Youn, J.-Y. et al., High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies, 517–532.e11, 2018, with permission from Elsevier.
ACKNOWLEDGEMENTS
We are grateful to all members of the Gingras lab for feedback on the extension. We acknowledge funding from the Governments of Canada and Ontario through Genome Canada, Ontario Genomics and the Ontario Research Fund (OGI-139 to A.-C.G. and RE08-065 to A.-C.G.), the Canadian Institutes of Health Research (Foundation grant FDN143301 to A.-C.G.) and the National Institutes of Health Office of Research Infrastructure Programs (R01OD010929 to M.T. and K. Dolinski). This research was enabled in part by support provided by Compute Canada (www.computecanada.ca). A.-C.G. is the Canada Research Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics; M.T. is the Canada Research Chair in Systems and Synthetic Biology.
Footnotes
CODE AVAILABILITY
GIX is available for free without restriction at the Chrome Web Store and the Firefox Add-ons site. Download links, documentation, a tutorial video and source code can be found at https://gene-info.org.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Oughtred R et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder JX et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database (Oxford) 2014, bau012 (2014). [DOI] [PMC free article] [PubMed]
- 3.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates B et al. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res 45, D619–D625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uhlen M et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Orchard S et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res 42, D358–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amberger JS, Bocchini CA, Scott AF & Hamosh A OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res 47, D1038–D1043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Gebali S et al. The Pfam protein families database in 2019. Nucleic Acids Res 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabregat A et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res 46, 2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]