Abstract
Purpose:
Develop an automated exome analysis workflow that can produce a very small number of candidate variants yet still detect different numbers of deleterious variants between probands and unaffected siblings.
Methods:
97 outbred nuclear families from the Undiagnosed Diseases Program/ Network included single probands and the corresponding unaffected sibling(s). SNP chip and exome analyses were performed on all, with proband and unaffected sibling considered independently as the target. The total burden of candidate genetic variants was summed for probands and siblings over all considered disease models.
Results:
Exome analysis workflow include automated programs for: ethnicity-matched genotype calling, salvage pathway for mendelian inconsistency, compound heterozygous recessive detection, BAM file regional curation, population frequency filtering, pedigree-aware BAM file noise evaluation, and exon deletion filtration. This workflow relied heavily on BAM file analysis. A greater average pathogenic variant number was found compared to unaffected siblings. This was significant (p<0.05) when using published recommended thresholds, and implies that causal variants are retained in many probands’ lists.
Conclusion:
Using Mendelian and non-Mendelian models, this agnostic exome analysis shows a difference between a small group of probands and their unaffected siblings. This workflow produces candidate lists small enough to pursue with laboratory validation.
Keywords: exome, diagnosis, agnostic exome analysis, rare diseases, Undiagnosed Diseases Network
INTRODUCTION
Clinical exome analysis has demonstrated utility in cryptic human disease diagnosis, especially with the advancement of low-cost, high-throughput sequencing technology. However, diagnosis rates for unknown disease phenotypes remain modest, approximately 25%1,2. One challenge involves the uncertainty in accepting or rejecting findings that lack an efficient validation method3 that is rapid enough for medical practice4.
Another challenge is related to a typical human genome’s known genetic burden, which includes several deleterious changes, present from conception7, that allow for evolutionary adaptability in response to selective environmental pressures. One pursuit to address this issue would be to strive towards sequencing and analyzing as much of the genomic space as possible, and to adequately determine true deleteriousness for a given genetic variant.
We developed and employed a menu of techniques to maximize completeness and enhance the accuracy of estimates of deleteriousness. These fully automated methods, several of which examine BAM files directly rather than relying on vcfs, remove the human bias associated with manual curation. We applied the programs to two cohorts of phenotypically disparate siblings enrolled in the NIH Undiagnosed Diseases Program (UDP) with an agnostic approach; no prior assumptions were made about disease models or phenotypes. In one cohort, the probands of a sib pair produced lists of variant candidates. In the other cohort, the probands’ normal siblings were analyzed as if they were the affected individuals. The goal was to determine if these analytical programs would identify a difference in the number of deleterious candidate disease-causing variants between the two groups.
MATERIALS AND METHODS
Family Cohort
This clinical research was approved by the NHGRI Institutional Review Board (IRB). Our cohort represented a broad spectrum of disease, and was comprised of 97 families enrolled in the NIH UDP between 2009 and 2015. The families were enrolled in one of two IRB-approved clinical protocols (76-HG-0238 “Diagnosis and Treatment of Inborn Errors of Metabolism and Other Genetic Disorders”; 15-HG-0130). Clinical and Genetic Evaluation of Patients with Undiagnosed Disorders through the Undiagnosed Diseases Network. Only families with a single affected individual were selected, because these are the most difficult families in which to identify candidate deleterious variants. Families with non-paternity, consanguinity, mosaicism, uniparental isodisomy, or copy number variations larger than 150kb were excluded. This was done to ensure that the comparison evaluated only outbred families with single probands. All probands had different unique phenotypes. Of the 97 probands, 50 were female and 51 were children; of the 113 siblings, 61 were female. Each unaffected sibling was medically determined by the intramural NIH UDP to not have serious medical issues or diseases relative to their affected sib.
Exomes and analysis
Exome sequencing was performed by the NIH Intramural Sequencing Center with the same chemistry used for all family members8,9. The resulting variant call files were converted to the VarSifter format, and annotated with population cohort data and variant metadata (Supplement Tables S1 and S2). This analysis covered the entire exome, including coding and non-coding variants, but excluding mitochondrial mutations. No manual curation of variants was performed. Each step in this analysis was run entirely in Java. Mendelian consistency was assumed, except for de novo mutations. Nonpaternity was excluded for all siblings in both groups by SNP chip analysis. The code developed for the exome analyses was written as an automated pipeline of methods already published; the pipeline had previously performed successfully for more than 8 years in a semi-manual curation process10,11. Improvements to the process are described in detail in Supplement B, along with the GitHub repository URL for the open source commented code. The probands’ data have already been submitted to the database of Genotypes and Phenotypes (dbGAP)12.
Analytical Hierarchies
Variants were excluded using a hierarchy of levels of associated certainties. The code eliminates variant candidates using an ordered sequence of discriminating properties starting with the greatest certainty, and finishes with the discriminant technique that has the greatest uncertainty. The hierarchy is: Mendelian phasing, population statistics, BAM curation, and predictions of deleteriousness. For de novo detection, neither parent had evidence of the variant in the sibling being analyzed, nor was there any evidence for that variant in the other sibling. For skipped exon (CNC) detection, variants were processed that had zero coverage in the proband (or sibling), but were otherwise well-sequenced in all control samples. The exon was then called null only if reads were absent in the proband over the local region of the evaluated exon.
The substrate for the workflow is described in Manual 0.
Statistical COMPARISON for differences in the mean number of discovered variants
For comparison of deleterious variants in probands and unaffected siblings, we calculated the sums of all variants meeting the selection criteria for each person at CADD20. See Results, “Applying the Workflow to UDP Quartets”.) De novo and homozygous recessive deleterious variants (DVs) were counted as one each and the compound heterozygous pairs were counted as a single DV. There could only be one count per locus from any model of inheritance, and there were no instances of any locus with more than one DV coming from different models of inheritance. 16 families had two unaffected siblings, so the variant counts for each type of inheritance model were averaged between the two unaffected siblings to equivalently compare the DV counts in the quintet families with the 81 quartet families. Equal numbers of males were compared for X-linked variants in the proband and sibling groups. A single-tailed t-test with heteroskedastic distributions was applied to the two groups’ summed counts to test this single hypothesis: The proband group had a larger unique unshared genetic burden than the sibling group, as manifested by a larger count of all ascertainable DVs.
RESULTS
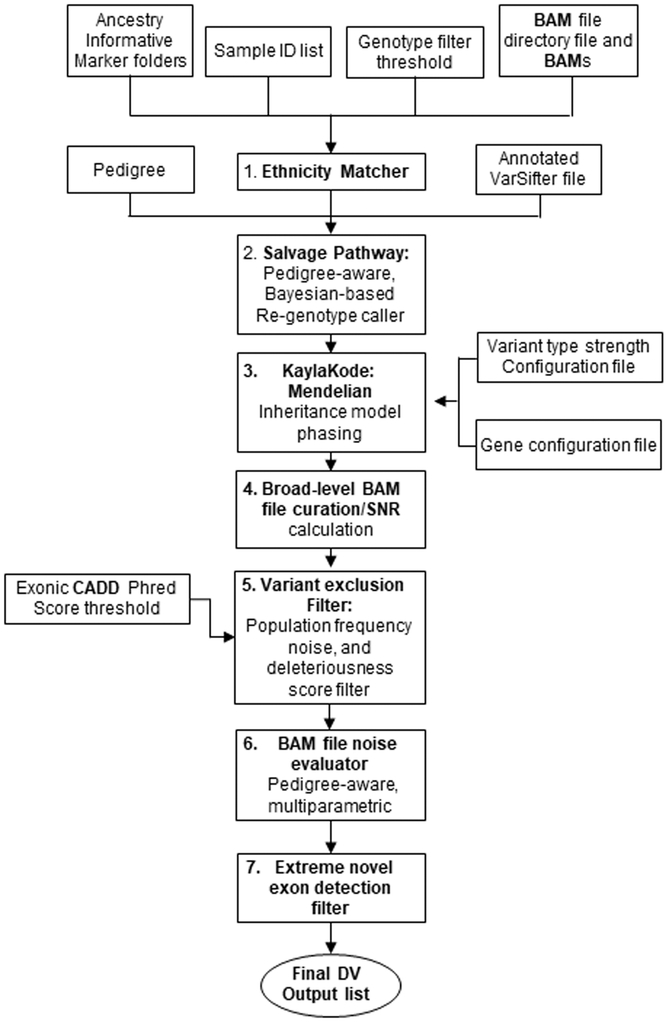
The workflow programs for exome analysis were run using data in a flat file with single annotation metadata per column, and involve the series of procedures (Fig. 1) described below.
Figure 1: Workflow overview of the agnostic exome analysis pipeline.
1. Ethnicity Matching (Mendelian Inconsistent Variant Re-genotyping) (Manual 1)
To identify false negative parental genotypes in apparent de novo cases, the ethnicities of the parents were determined from ancestry informative markers in linkage equilibrium for both parents, then used for ethnicity-matched prior probability re-genotyping at those loci (Manual 1). There was excellent correlation between the Ethnicity Matcher calls and ethnicities recorded in medical records (Supplement Table S3). Variants interpreted as inherited from a parent were entered with the rest of the inherited variants into the compound heterozygous recessive analysis (Supplement Manual 3).
2. Salvage Pathway Module (Manual 2)
This computationally complex module is used only to resolve the several thousand spots in the flat file where siblings appear Mendelian inconsistent relative to their parents. Each inconsistency is resolved by regenotyping using the population prior probability, a chosen mutation rate, and the inheritance information of the quartet in a Mendelian inheritance prior, along with all the reads in every family member’s BAM files to produce a better genotype call at these locations (only).
3. Mendelian Inheritance Model Phasing (KaylaKode) (Manual 3)
This module evaluates the flat file, using various Mendelian models, population frequencies and predictions of deleteriousness to determine gene loci boundaries and phasing of “half of a pair” variants using parental genotypes.
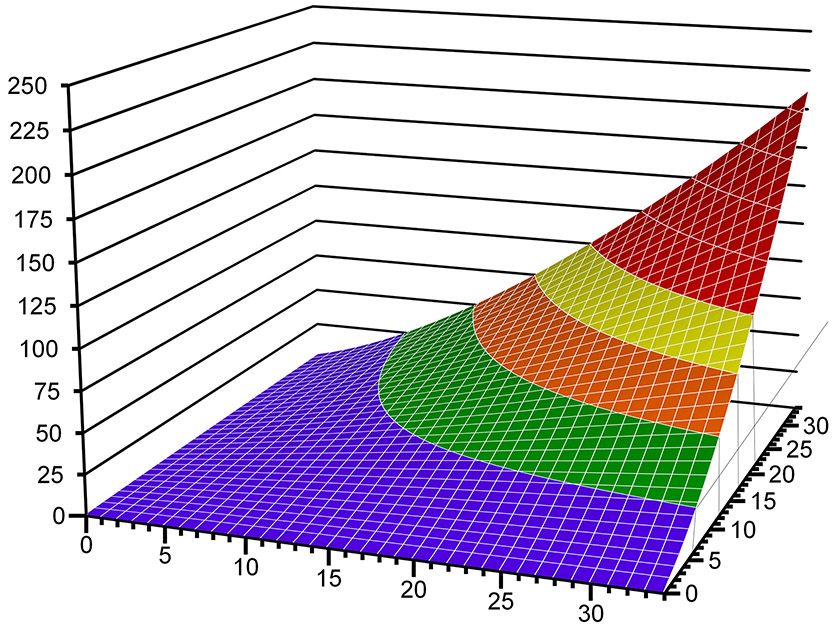
For compound heterozygous recessive detection, the available set of variants within each locus was tested for pairing using a strategy that allows for the possibility of one or both alleles to be noncoding. An unpublished scoring system, Virtual Mendelian Model (VMM), for compound heterozygous recessive pairs of phased variants was developed for this analysis. It uses the formula:
PHRED is the Phred scaled CADD score for each of the two variants in a heterozygous pairing (Fig. 2). This weighting strongly favors two moderately deleterious individual variants over a pair containing one very deleterious variant and one benign variant, which would more likely represent merely the carrier state for a recessive disease model.
Figure 2. VMM scoring function.
The x- and y-axes represent the two CADD (Phred) scores and the z-axis is the VMM score. The longer wavelength colors denote regions more likely to be truly deleterious for the pair of variants.
The pseudoautosomal region of the X-chromosome was excluded from the X-linked analysis and included in the autosomal recessive analysis. The full process and threshold scores for the various inheritance models are provided in the Module 3 code, which is available through open source products.
Steps 4–6 involve various BAM file curation steps. These modules intensely evaluate the local regions of the BAM file pileup for all family members in a quartet only at the candidate variant loci identified by the simple variant analysis module (KaylaKode). They perform an unbiased analysis of the BAM file to reject regions with strong evidence that they are misaligned, mismapped, or have a large number of base called errors.
4. Broad-level BAM file curation (Manual 4)
After each variant is annotated with its Mendelian inheritance state and population frequency, its locus is algorithmically inspected in the BAM file pileup region for all reads containing the variant position, with the goal of filtering out bad BAM file regions, i.e., artifacts of the sequencing/alignment process. A measure of how variant variants are distributed within a local BAM file region (150 bases on either side of the variant) is provided by a parameter called the Signal-to-Noise Ratio (SNR). This term, calculated based upon the Spatial Distribution Score and the Mismatch Density, is explained by a heuristically derived formula (Manual 4, page 6). A second parameter, called “Error”, is defined as 0.25/SNR. The SNR and Error terms contribute to many different decisions in the programs of Modules 4 and 5. For example, if the Error is greater than 2%, then the region of interest is considered too noisy and the variant is excluded. If the Error is greater than 1% with an average read depth across the family of 4 or less, then that region is also excluded. The influence of the Error term varies with the filter and with the inheritance model, and can be discerned from the code.
5. Filtering variants based on population frequency (Manual 5)
Variant exclusion is based on several different human population sequencing statistics. These criteria exclude any variant that has an estimated minor allele frequency ≥2% at a 95% confidence level using cumulative Poisson statistics for the number of variant alleles genotyped, and the number of total samples genotyped at that location for each specific population or subpopulation. The population datasets included the UDP internal cohort (n=1310), ClinSeq13 cohort (n=938), 1000 Genomes14, UK10K15, ExAC16, and the gnomAD genome and exome cohorts. Candidate variants that passed all filters but were in the loci NEB, TTN, or OBSCN, and any KRT, OR, or TAS genes were excluded due to very high false positive rates at these loci9,10; this comprised the complete list of specific, gene-based variants that were excluded. All other loci including non-coding loci were retained. Additional population frequency filter criteria, customized for each specific Mendelian inheritance model, i.e., CM (compound heterozygous), DN (de novo), XL (X-linked), Hemi (hemizygous), and HR (homozygous recessive), are listed in in Manual 5, Tables 3 and 4.
6. Pedigree-aware, multi-parametric BAM file noise evaluation (Manual 6)
This module uses a variety of programs to identify and eliminate “apparent de novos” that are really false positives and constitute “noise”. For a region to be considered cleanly sequenced, three criteria had to be met for analysis at the BAM file level. First, both the mapping and base-call qualities within the candidate region had to be sufficiently high (Supplement Manual 6). Second, the genotype call at the position of the candidate variant had to fit the de novo variant model. That is, a candidate variant position was excluded when variant base calls were made in reads aligned in apparently unaffected family members’ BAM files at the same position. Finally, because each person has two parents, at most two haplotypes are expected in any given region (assuming no polyploidy or mosaicism). Thus, variants within regions that appear to contain multiple (>2) haplotypes in the proband were excluded. In addition, the region had to have at least 8 reads in the pileup, the variant position needed to have been sequenced in at least half the UDP internal cohort (n≥655), and the variant could not be seen in anyone else in the same cohort, since each phenotype was unique to a single proband.
7. Extreme Novel Exon Deletion (CNC; Called/No Coverage) (Manual 7)
CNC, or Called position in a jointly called variant file where the proband has No Coverage, refers to an event unique to one person when virtually everyone else behaved in an orthodox manner at that position. A CNC is distinguishable from the situation in which a unique result occurred but few if any other people were measured at that spot; in that case, it is unknown whether the unique result is truly uncommon or just apparently uncommon because no one looked for it in a large control population. In a jointly called variant call file, using either the vcf or VarSifter format, there is both coverage and genotype information for all samples in which one single person had any form of variant. In those places, there is “free information” on whether someone else also has no coverage, and if everyone remaining has coverage. If everyone except one person has coverage, then the position is well sequenced. Consequently, an individual who had zero reads at that spot represents an extreme novel event. One trivial explanation is that the zero read depth is “on the edge” of everyone’s coverage; many people have only minimal coverage, but this one person, by chance, had zero. However, if the next least amount of coverage involves a large number of reads, and if the other family members have good read depths, then the zero read depth reflects a deleted region; this is typically a skipped or deleted exon. Details of this filter are provided in Supplement Manual 7.
Applying the Workflow to UDP Quartets
The agnostic exome analysis was performed on 97 probands and 113 unaffected siblings in an identical manner to determine all unshared deleterious variants 26,29. The output was an annotated flat file, reconfigured in a text editor manually to an unannotated vcf format. The final lists of variants for each proband and each unaffected sibling are presented in the Appendix, and the workflow code has been submitted to GitHub.
We first examined the claim that a CADD score threshold of 20 (for exonic variants) for differentiating deleterious from non-deleterious variants would distinguish the proband from the sibling groups. The predicted model was that at low thresholds the sample size would be underpowered to show a difference between the two groups due to an overwhelming burden of minor genetic variants. Also, there would be no difference at very high deleteriousness scores between the groups since the causal variants would likely be removed at high thresholds, and both groups would only contain residual noise. This model predicts that the most significant differences would only occur in the region where truly deleterious changes are scored, i.e., in the range of a threshold CADD exonic score of Phred>20, but not when using thresholds significantly below or above that score.
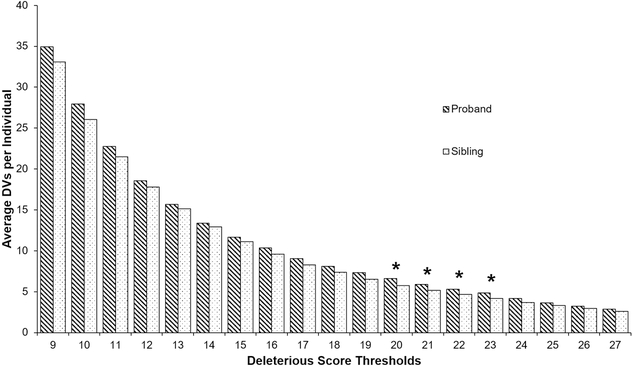
To test the single hypothesis that this pattern is correct, thresholds for exon variants from 9–27 (with intronic threshold and VMM scaled as well) were used in the analysis. Deleterious cutoff scores for intronic variants were set at 75% of the exonic deleterious cutoff scores15. The VMM cutoff was calculated as VMM=30+2×(CADDexonic–20). Only variants equal to or above these CADD or VMM values were included in the final list of candidate variants for the analyzed sibling. The full list of exonic/intronic CADD and VMM cutoff combinations is in Supplement, Table S4. Using the agnostic exome analysis workflow, only CADD thresholds around 20 yielded a significant difference in the number of DVs between the probands and siblings (Fig. 3).
Figure 3: Difference of deleterious variants (DVs) between probands (N=97) and matched siblings (N=113) at a range of deleterious score thresholds.
Deleterious score thresholds represent the exonic CADD thresholds used for each single hypothesis testing. Bars show the average number of DVs per individual, calculated from the combined total compound heterozygous, de novo, homozygous recessive, hemizygous, X-linked, and extreme novel exon deletion variants for each individual. Values above the bars are significance values from a single-tailed t-test with heteroskedastic distributions for the null hypothesis that there is no difference in DV counts between a group of probands and their unaffected siblings. Asterisks mark deleterious score thresholds at which the difference between groups had a p-value less than 0.05.
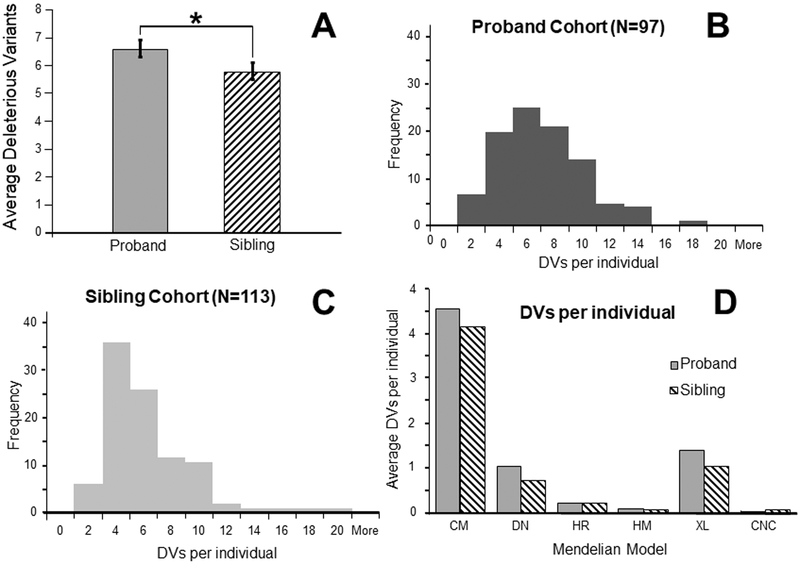
At an exonic CADD Phred threshold score of ≥20, the absolute number of DVs in the proband group (6.6) was significantly different from that of the sibling group (5.8) at p<0.05 (Fig. 4A). The distribution of DVs in each individual at CADD20 is shown for probands and siblings, respectively, in Fig. 4B, 4C. The proband group has a broader distribution of DV counts, while the unaffected sib group has a narrower distribution. This demonstrates that the proband group has a greater proportion of cases with more DVs than the sibling group, not simply a few probands with a large excess of DVs that are skewing the average from the median. There is also no evidence of multimodal distribution at CADD20. Finally, the difference in DVs between the two groups at CADD20 was not confounded by different BAM file sizes (Supplement Fig. S1) or by skewed number of DVs in either the proband group or the sibling group related to a specific ethnicity (Supplement Fig. S2).
Figure 4: Comparison of deleterious variants (DVs) between probands (N=97) and matched siblings (N=113) at each deleterious score threshold.
A. Bars show the average number of DVs per individual, calculated from the combined total compound heterozygous, de novo, homozygous recessive, hemizygous, X-linked, and extreme novel exon deletion variants for each individual at CADD20. * = p<0.05 for a single-tailed t-test with heteroskedastic distributions for the null hypothesis that there is no difference in DV count between a group of probands and a group of their unaffected siblings. B. Histogram of proband DVs at an exonic CADD threshold of 20. C. Histogram of sibling DVs at an exonic CADD threshold of 20. D. Comparison of distribution between probands and their unaffected siblings of average DV counts within six Mendelian states at an exonic CADD threshold of 20, as filtered from the variant exclusion pipeline. : 4.6 DVs and 4.2 DVs (for probands and unaffected sibs, respectively); : 1.0, 0.7; : 0.2, 0.2; : 0.1, 0.1; = 1.4, 1.0; : 0.0, 0.1.
The distribution of inheritance states at CADD20 is shown in Figure 4D. Both groups have almost equivalent quantities of homozygous recessive, hemizygous, and CNC variants. The main difference between the groups results from three categories: excess de novo variants, excess X-linked variants, and an even greater excess of compound heterozygous variants in the proband group. These three disease inheritance models were also the largest overall categories in both the proband and unaffected sib groups.
Of our 97 UDP cases, 36 had an identified variant that was either previously confirmed as causal or is still being validated. These probands’ DV lists (at CADD20) contained the gene previously associated with the diagnosis in 33 (90.1%) of the cases. The 3 cases whose DV lists did not contain the supposedly causal gene involved situations not previously considered for this analysis: incomplete parental penetrance of a variant, a variant in the form of a parental macro-duplication, and inaccuracies in deleteriousness scoring due to a real cryptic splice site that has a very low CADD score in an intronic position.
DISCUSSION
We developed a suite of exome sequence analysis programs (Fig. 1), and tested this workflow by analyzing the genetic variants of 97 nonconsanguineous UDP quartets and quintets. The exome analyses were automated, agnostic, and free of human bias. The average number of DVs for multiple deleteriousness score cutoffs was measured by analyzing the sequence data twice, first by presuming the proband is the affected sib and then by presuming that an unaffected sib is affected and the proband is the unaffected control.
The comparison of these two groups rejected the null hypothesis that there is no difference in the means of these groups, with a p<0.05 at CADD20, the published suggested threshold for deleteriousness for CADD17. Probands on average had nearly one extra variant compared with their sibs (Fig. 4A). This suggests that the distribution of DVs in the proband group is a superposition, one distribution equivalent to that of the healthy group, and the other with an excess of DVs. DVs in the healthy sibling group reflect the genetic burden of these individuals, whereas the “excess” DVs in the probands reflect the additional, causal variants within the proband group (Supplement Fig. S3). In addition, a contingent of the variants found in both the probands and the healthy sibs are “Background”, i.e., false positives based upon technical sequencing mistakes or because they have no pathogenic consequences.
The excess DV counts between the proband and unaffected sibling groups being close to 1.0 at CADD20 is possibly fortuitous, but also supports the speculation that the maximum number of causal genes in most of the individuals in the proband group is not likely to be a large number—much less than the underlying equivalent genetic burden carried by both groups and scored as deleterious. The UDP probands likely have a mixture of non-genetic, mono-genetic and oligo-genetic disorders, consistent with their rarity and the severity of the typical UDP phenotype. However, most are likely monogenic; indeed, all 36 of the working molecular diagnoses are monogenic, and most of those variants were captured in their respective proband’s very small DV list (typically 2–10) at the published deleterious score threshold. (See Appendix.)
Most genetic mutations are benign18, and agnostic genome analysis must filter them out to create a short list of candidate variants that can be intensely considered. Excessively stringent filtering could exclude true pathogenic variants, while over-relaxed filtering will leave too many false positive variants to make validation practical. One cause of excessive false positive variants is the existence of small regions of the genome refractory to designating a reference sequence; this was true for the original reference hg1819,20, and its remedy continues to be pursued.21,22. An example is the HLA region, typically removed by quality control filtering when using globally determined genotype quality scores and applying universal coverage cutoff thresholds. The analysis of such regions would benefit from the use of pedigree inheritance information or ethnicity-derived prior probabilities in Bayesian-based genotype calling; the workflow’s Ethnicity Matcher begins to address this issue. In addition, the VMM determines deleteriousness of compound heterozygous pairs of variants, adding to current sequence analyzing efforts23-25. Finally, the CNC analysis identified missing or skipped exons that provided diagnosis for 3 UDP cases in our cohort.
Other studies have found that there are more deleterious variants in affected probands than their control sibs under ideal analysis2,26,27. However, those studies were limited to a single disease and/or type of Mendelian model (i.e., de novo)28-30. In addition, previous analyses involving case/controls have included tens of thousands of exome sequences. The present study included six different Mendelian inheritance models, diverse diseases, and a much smaller cohort of ~100 families. If there were no distinct differences until the n was much greater (e.g., 10,000), then the difference would not be meaningful.
Compound heterozygosity is the most common recessive disease inheritance model for outbred populations, such as our cohort30. Likewise, de novo dominant variants are well described as major causes of severe genetic disorders31. Indeed, these types of expected pathogenic variants were predominant in our cohort, especially in the excess seen in the proband group (Fig. 4D). Note that any variants not adhering to one of the seven included inheritance models would be missed and would dilute the proband group’s excess variant count, reducing the likelihood of finding a statistical difference between the groups, yet a difference was seen in this comparison.
A major limitation in clinical exome analysis is the relatively low signal-to-noise ratio due to a high initial preponderance of false positive candidate variants when beginning this type of analysis. This results from sequencing errors, low read depth coverage, low complexity sequence regions, and errors in experimental and analytical design11,32-34. Our previous exome analysis returned an average of 88 DVs for quartet families for the proband. In comparison, at CADD20, our new analytical pipeline returned an average of 6.6 DVs, assuming the proband is the patient, and 5.8 DVs, assuming the unaffected sibling is the patient. These final candidate DVs were not based on gene expression or phenotype. Similarly, the selection criteria were not based on mutation type, and a large fraction of these variants were noncoding region variants. All criteria were applied agnostically and equally to both groups without human interpretation. The only non-agnostically excluded variants were those found in the 6 genes or gene families known to commonly misalign and yield false positive results and were carried through from the previously published analysis methods9,10.
Our findings also suggest that it is possible to obtain a justifiable difference per sibling pair at a CADD score threshold associated with actual pathogenicity. If performed on the population at large, this analysis can potentially be used as a diagnostic tool to infer who belongs in a higher risk pool for rare genetic diseases. However, these tools have a poor discrimination power for any specific individual, given the specificity of the selective pressure on deleterious variants and the current accuracy of deleteriousness scoring. Repeating this type of analysis could indicate if predictions of deleteriousness have substantially improved.
This study indicates that it is possible to produce a relatively short list of potentially pathogenic variants by unbiased and automated agnostic exome or genome analysis. These lists likely contain the causal variant of a rare genetic disorder, which should aid in evaluating claims about causation based on agnostic genome-wide analyses and in decisions on resource-intensive laboratory validation.
Supplementary Material
ACKNOWLEDGMENTS
Supported by the Intramural Research Programs of the National Human Genome Research Institute and the NIH Common Fund, Office of the Director, National Institutes of Health.
Footnotes
These authors contributed equally to this work.
Conflict of Interest Notification Page
This clinical research was approved by the NHGRI Institutional Review Board (IRB) and was part of NIH projects HG000215–07 and HG200352–02. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. There are no conflicts of interest to report.
REFERENCES
- 1.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. New England Journal of Medicine. 2013;369(16):1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu X, Petrovski S, Xie P, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genetics in Medicine. 2015;17(10):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JY, Clark P, Londin E, Sponziello M, Kricka LJ, Fortina P. Clinical exome performance for reporting secondary genetic findings. Clinical chemistry. 2015;61(1):213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donnell-Luria AH, Miller DT. A Clinician’s perspective on clinical exome sequencing. Human genetics. 2016;135(6):643–654. [DOI] [PubMed] [Google Scholar]
- 5.Childs B Genetic medicine: a logic of disease. JHU Press; 2003. [Google Scholar]
- 6.Badano JL, Katsanis N. Beyond Mendel: an evolving view of human genetic disease transmission. Nature Reviews Genetics. 2002;3(10):779–789. [DOI] [PubMed] [Google Scholar]
- 7.Muller HJ. Our load of mutations. American journal of human genetics. 1950;2(2):111. [PMC free article] [PubMed] [Google Scholar]
- 8.Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2011;28(4):599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gahl WA, Markello TC, Toro C, et al. The national institutes of health undiagnosed diseases program: insights into rare diseases. Genetics in Medicine. 2011;14(1):51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams DR, Sincan M, Fuentes Fajardo K, et al. Analysis of DNA sequence variants detected by high‐throughput sequencing. Human mutation. 2012;33(4):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gahl WA, Mulvihill JJ, Toro C, et al. The NIH Undiagnosed Diseases Program and Network: applications to modern medicine. Molecular genetics and metabolism. 2016;117(4):393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature genetics. 2007;39(10):1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biesecker LG, Mullikin JC, Facio FM, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome research. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium GP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium UK. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature genetics. 2014;46(3):310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M The neutral theory of molecular evolution. Cambridge University Press; 1983. [Google Scholar]
- 19.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. [DOI] [PubMed] [Google Scholar]
- 20.Chaisson MJ, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nature reviews Genetics. 2015;16(11):627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics. 2016;17(6):333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain M, Koren S, Miga KH, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nature Biotechnology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014;312(18):1870–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature Reviews Genetics. 2011;12(11):745–755. [DOI] [PubMed] [Google Scholar]
- 26.Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Human molecular genetics. 2001;10(21):2319–2328. [DOI] [PubMed] [Google Scholar]
- 27.Sohail M, Vakhrusheva OA, Sul JH, et al. Negative selection in humans and fruit flies involves synergistic epistasis. Science. 2017;356(6337):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature genetics. 2010;42(9):790–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roach JC, Glusman G, Smit AF, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328(5978):636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome biology. 2016;17(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kircher M, Kelso J. High‐throughput DNA sequencing–concepts and limitations. Bioessays. 2010;32(6):524–536. [DOI] [PubMed] [Google Scholar]
- 33.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC bioinformatics. 2014;15(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du C, Pusey BN, Adams CJ, et al. Explorations to improve the completeness of exome sequencing. BMC medical genomics. 2016;9(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.