Abstract
A number of well-established servers perform “free” docking of proteins of known structures. In contrast, template-based docking can start from sequences if structures are available for complexes that are homologous to the target. Based on the results of the CAPRI-CASP structure prediction experiments, template-based methods yield more accurate predictions if good templates can be found, but generally fail without such templates. However, free global docking, or focused docking around even poor quality template-based models, can still generate acceptable docked structures in these cases. Based on the analysis of a benchmark set, free docking of heterodimers yields acceptable or better predictions in the top 10 models for around 40% of structures. However, it is likely that a combination of template-based and free docking methods can perform better for targets that have template structures available. Another way of improving the reliability of predictions is adding experimental information as restraints, an option built into several docking servers.
Keywords: Protein-protein interaction, method development, optimization methods, machine learning, sampling, scoring, template-based, homology modeling
Computing the structures of protein complexes has been one of the central but challenging problems in computational structural biology [1]. Even for relatively rigid proteins it is difficult to explore the 6D rotational-conformational space of mutual orientations potentially sampled by a pair of proteins as they interact through complementary patches on their surfaces. Predicting the association of proteins is further complicated by flexibility. Proteins are not static objects; they constantly interconvert between conformers of varying energies [1].
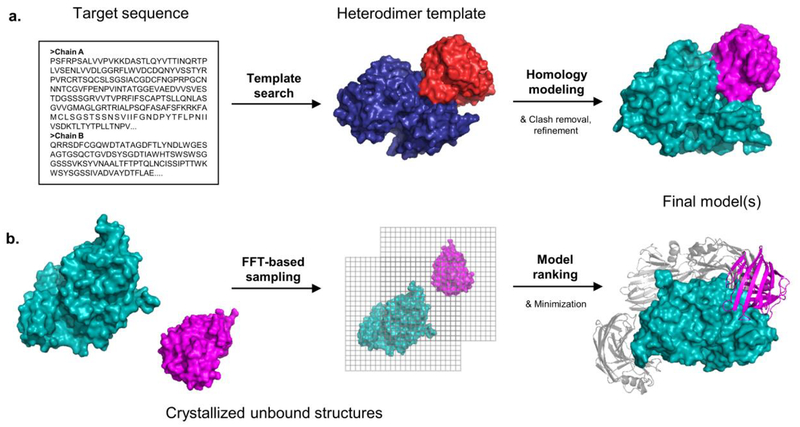
In spite of the complexity of the problem, a variety of docking methods, including some easy-to-use servers, are currently available for predicting the structures of protein-protein complexes. The choice of the method used depends on the nature of the docking problem. “Free” docking methods can be used if X-ray structures are available for all proteins to be docked or for their very close homologs. However, the number of structures of protein complexes has been increased in the Protein Data Bank (PDB). Knowledge of complex structures makes prediction of related protein complexes amenable to template-based and homology modeling methods, even when the structures of component proteins are not available (Figure 1).
Figure 1.
General comparison of template-based and free docking methods for an example heterodimer target. (a) A template-based method begins with the target sequences, using a template search to identify an existing heterodimer template from the PDB (Protein Data Bank). Homology modeling is used to map the target sequence onto the template structure. The model is refined to its final form. (b) In free docking the two components must be individually crystallized. Billions of protein conformations are evaluated, often through the use of an FFT-based algorithm. Final models of the heterodimer are ranked and minimized.
In this review we highlight four observations that we think are relevant to choosing the best method for predicting the structure of a protein-protein complex. First, we investigate the performance of some of the best known and frequently used “free” docking methods that have been tested on the latest update of a widely used protein-protein docking benchmark [2] and in the latest rounds of the CAPRI (Critical Assessment of Predicted Interactions) community-wide protein docking experiment [3]. We also assess the impact of a machine learning-based ranking algorithm on prediction quality. Second, we discuss the shift toward template-based docking, primarily considering the prediction of homo-oligomers, by looking at the results of the latest structure prediction experiments called CASP11-CAPRI [4], and CASP12-CAPRI [5]. Third, we investigate the combination of template-based and free docking algorithms. As will be shown, template-based methods usually yield higher quality predictions if good templates are available, which is frequently the case for homo-oligomeric targets. However, free docking is still useful if no good templates can be found. We also suggest that, depending on template quality, switching between template-based and free methods is likely to be useful for predicting the structures of heterooligomers, as it has already been implemented in some docking servers [6,7]. Fourth, we emphasize the role of additional information from site-directed mutagenesis, cross-linking, SAXS, and other experiments for improving the reliability of docking results.
Testing free docking methods on a recent benchmark
The protein-protein benchmark set, collected by the Weng lab [8–11], has become well established for testing docking methods. The benchmark consists of non-redundant, high-quality structures of protein–protein complexes along with the unbound structures of their components. The most recent addition includes fifty-five new complexes, creating Version 5 of the benchmark, which now contains 230 entries. The developers of Version 5 also tested four “free” docking servers, ZDOCK [12], pyDock [13], SwarmDock [14,15], and HADDOCK (High Ambiguity Driven DOCKing) [16,17]. ZDOCK [12] and pyDock [13] are rigid-body docking algorithms based on the use of fast Fourier transforms (FFTs), with pyDock built on the earlier FTDock method [18]. SwarmDock [14,15] is a flexible docking method which uses a population-based memetic algorithm to optimize parameters characterizing the orientation, position, and conformations of protein subunits. The algorithm combines a modified particle swarm optimization global search to identify broad low-energy regions of parameter space, and an adaptive local search for refinement. HADDOCK [16,17] is a semi-flexible docking protocol which uses experimental information and bioinformatics interface predictions to drive docking. Conformational changes are accounted for through simulated annealing and flexible explicit solvent refinement [19]. The performance of the programs was evaluated using the criteria established by the evaluation team of the CAPRI docking experiment [3] and essentially implemented in the DockQ program [20]. Overall the success rates (at least one acceptable prediction for a benchmark case) ranged between 5% and 16% in the top 1 prediction and 20–38% in the top 10 predictions. The performances of the different docking algorithms were similar and correlated with each other [2], with SwarmDock providing the best results, closely followed by ZDOCK and the other two servers.
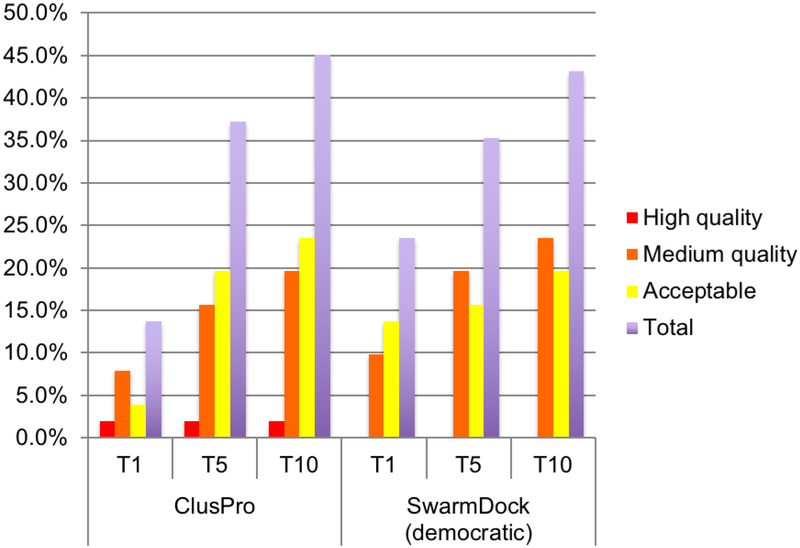
Three of the above servers, i.e., ZDOCK [12], pyDock [13], and SwarmDock [14,15], plus the SDOCK program [21] were more recently improved by adding the rescoring scheme called IRaPPA (Integrative Ranking of Protein–Protein Assemblies) [22]. IRaPPA characterizes decoys using physicochemical descriptors, calculated by the server CCharPPI [23], and combines a large selection of metrics using ranking support vector machines (R-SVMs) to obtain a consensus ranking by a voting method [22]. Models were trained using complexes from Benchmark 4 [11], and the method was evaluated for its ability to select near-native solutions using the new complexes added in Benchmark 5 [2]. All methods substantially improved [22], and the new SwarmDock version (called “democratic” due to the voting scheme used for the ranking) remained the best performer [22]. We were able to run the server on 51 of the 55 targets added to Benchmark 5 (see Figure 2), and obtained the success rates 23.5%, 35.3%, and 43.1%, respectively, in the top 1, top 5, and top 10 predictions. These numbers are close to the ones reported in the IRaPPA paper [22] for the top 1 and top 10 predictions.
Figure 2.
Comparison of ClusPro and the “democratic” SwarmDock servers on their prediction accuracy of 51 targets added to Benchmark 5. Four of the 55 targets in the benchmark were not included since either ClusPro or SwarmDock were not able to yield results.
We also tested our protein docking server ClusPro [24] by predicting the structures of the complexes added in Benchmark 5 [2]. ClusPro uses the FFT-based program PIPER [25], but substantially differs from ZDOCK and pyDOCK because the 1000 lowest energy structures generated by PIPER are clustered using pairwise RMSD as the distance measure, and centers of the largest clusters rather than the lowest energy structures are selected as the most likely models of the complex [24,26]. We used the standard electrostatics-favored ClusPro parameter set for enzyme-inhibitor and “other” type of complexes [24], and the antibody parameterization for antibody-antigen pairs [27] without further training, and obtained acceptable models for 13.7%, 37.3%, and 45.1% of the complexes in the top 1, top 5 and top 10 predictions. According to these results, ClusPro obtained acceptable solution for one more target and thus performed slightly better than the “democratic” SwarmDock server in the top 10 predictions (45.1% versus 43.1%) and in the top 5 predictions (37.3% versus 35.3%), but its success rate was substantially worse in the top 1 predictions (13.7% versus 23.5%) (Figure 2). Thus, while ClusPro remains competitive with the other docking programs if the top 5 or top 10 models are considered, which are the usual evaluation methods in CAPRI, the machine learning-based scoring function IRaPPA improved the discrimination of near-native docked structures. However, the cost of descriptor calculations for IRaPPA greatly increases the computation time when compared to standard docking runs [22]. In addition, further testing in CAPRI and other prediction experiments are needed in order to detect potential overtraining that can occur in machine learning with relatively small training sets. We note that a recent analysis found ClusPro more stable than other methods for docking unbound protein structures [28], most likely due to the final selection based on cluster size rather than scoring function value [29].
Comments on results of the latest CAPRI experiment
At the 6th meeting of the CAPRI (Critical Assessment of Predicted Interactions) community-wide protein docking experiment predictions were evaluated for 12 protein-protein complexes [3]. The five best performing servers were the already discussed ClusPro, SwarmDock, HADDOCK, and PyDock, plus the server LzerD [30]. LZerD uses 3D Zernike descriptors based on a mathematical series expansion of the protein surface. The best “human” predictor group of Guerois used the InterEvDock program that was recently developed into a server [6,31]. The newest version, InterEvDock2, also performs automatic template-based docking, and thus can be used without structural information on the input proteins [6]. The server was extensively tested but not on the Weng benchmark set [2], and hence its performance cannot be directly compared to that of the other servers discussed here. The best success rates (acceptable or better in the top 10 predictions) at CAPRI6 were 41.66% and 58.33%, respectively, for servers and the Guerois group [3].
Shifting toward template-based docking: The CASP-CAPRI experiments
Focus on template-based methods substantially increased by the addition of protein complex prediction to the CASP (Critical Assessment of Techniques for Protein Structure Prediction) experiment [4,5], because the predictions had to be based on sequences rather than structures. The targets in CASP11-CAPRI included 23 homo-oligomers (18 dimers and 5 tetramers), and 2 heterodimers [4]. The best “human” predictors used template-based methods and submitted 15 or 16 acceptable models. The best servers, HADDOCK and ClusPro, also had to use template-based models, but the final predictions came from re-docking, similarly resulting in 16 acceptable models [17]. However, the purely template-based methods yielded 12 to 14 medium quality models, whereas only 8 or 9 such models were obtained by servers that used the combined approach. In CASP12-CAPRI the targets were 12 homo-oligomers (3 dimers, 6 trimers, and 3 tetramers), and 3 hetero-complexes (2 dimers and 1 tetramer). Among these, 5 homo-oligomers and one heterodimer were considered easy targets, with good templates available in the PDB. Accordingly, the best template-based predictors submitted 6 acceptable or better predictions. In addition, one docking group (Grudinin) and the ClusPro server also obtained acceptable predictions for one heterodimer. However, the purely template-based methods again produced a substantially higher number of medium quality models for the other 6 targets with good templates.
It is now clear that CASP11-CAPRI and CASP12-CAPRI together had only 15 targets that are validated homodimers with structures available in the PDB (Table 1). Among these targets 12 were considered easy, whereas three (T72, T86, and T116) were difficult due to the lack of good templates [4,5]. We attempted to identify templates for all these targets using HHPred [32] with default settings and built homology models of the dimers using MODELLER [33] from the templates found. The monomers from the template-based models were submitted for global and focused docking using ClusPro, where focused docking means restricting the conformational search to a box around the template-based model. Table 1 shows the numbers of acceptable or better and medium quality or better models derived using the three different protocols. Template-based modeling yielded acceptable or medium quality models for all easy targets. Subsequent global docking of the models substantially reduced the number of medium quality models (from 32 to 10). No template-based models were generated for any of the 3 difficult targets, but subsequent global docking yielded an acceptable model for T72. Focused docking retained only the structures that had their geometric centers within a box with 3Å sides around the center of the ligand in the template-based model. This strategy increased the number of acceptable predictions, but reduced the number of medium quality ones (from 32 to 22).
Table 1.
Number of models by template-based docking (A), global free docking (B), and focused free docking (C).
| CAPRI ID | PDB ID | A | B | C |
|---|---|---|---|---|
| T69 | 4Q34 | 1* | 1* | 1* |
| T72 | 4Q69 | 0 | 1* | 0 |
| T75 | 4Q9A | 3*/2** | 2* | 3*/2** |
| T79 | 5A49 | 2*/2** | 2* | 2*/2** |
| T80 | 4PIW | 10*/6** | 10*/1** | 10*/3** |
| T85 | 4WJI | 8*/5** | 8*/3** | 8*/4** |
| T86 | 4U13 | 0 | 0 | 0 |
| T87 | 4WBT | 9*/4** | 9* | 9*/2** |
| T90 | 4XAU | 10*/4** | 10*/3** | 8*/3** |
| T91 | 4URJ | 6*/2** | 4* | 5*/1** |
| T92 | 4W66 | 2* | 3* | 10* |
| T93 | 4XRR | 8*/5** | 9*/2** | 8*/3** |
| T94 | 4W9R | 1* | 0 | 1* |
| T116 | 5IDJ | 0 | 0 | 0 |
| T119 | 5YVS | 9*/2** | 9*/1** | 8*/2** |
| TOTAL | 69*/32** | 68*/10** | 73*/22** |
Acceptable or better predictions
Medium or better predictions
As shown above, docking of monomers from the template-based models generally does not improve accuracy, but can add acceptable models if the templates are not very good. The problem is that determining template quality is not always simple. A recent case in CASP13-CAPRI, T137, which was canceled due to an early release of its crystal complex (pdb 6d2v) demonstrated this difficulty. When the target sequence was submitted to HHPred, the top 10 homodimer templates produced high probabilities (> 0.95). Homology models generated from these templates all agreed in their general predicted interface. While these findings suggested that the templates were most likely good, all had less than < 22% sequence identity with the target. Comparison to the X-ray structure revealed that none of the predicted models were acceptable. However, focused docking around these models was able to generate several acceptable predictions from five of the 10 templates.
Template-based modeling of heterodimers
Since almost all targets in CASP-CAPRI were homo-oligomers, it is interesting to explore how the template-based approach would work for heterodimers. We considered the already discussed 55 complexes added in Version 5 of the benchmark [2], ran HHPred with default settings on each chain, and then checked for matching templates. Since HHpred recommends investigating any templates with a probability of 50% or greater, this threshold was used for filtering. Templates released after the target complex release date were removed. Even with the very permissive acceptance condition of 50%, no templates were found for 26 of the 55 targets. The remaining 29 targets had templates, in 21 cases more than one. While we did not further study the quality of these templates, based on the results shown above it is likely that their availability would improve the quality of docking results for some of the 29 targets. Next, we checked how free docking (specifically the ClusPro server) performs for the 26 targets without any acceptable template that would be thus considered difficult for template-based docking. Results have shown that free docking is still very useful for heterodimers as it provided acceptable or better models in the top 10 ClusPro predictions for 10 of 26 targets (38.4%). The best results were obtained for antibody-antigen complexes (5 out of 6), followed by enzyme-inhibitor pairs (4 out of 8). Docking complexes in the “others” category [2] was less successful, with acceptable models only for a single target. Therefore, an approach combining template-based docking for targets with good templates and free docking to the rest would likely increase the overall success rate beyond the roughly 40% that was seen for free docking applied to all targets. However, it is not yet clear what fraction of hetero-oligomers have acceptable targets in general. While for the set considered here this fraction is 47% (26/55), Kundrotas et al. [34] suggest that it can be substantially higher, although they also note that only about one-third of these templates are of good quality. According to Mosca et al., the availability of good templates also depends on the organism [35], but is generally below 20% of all complexes.
Docking with additional information
Free docking generates a large ensemble of potential conformations (Figure 1), but selecting near-native ones is frequently difficult due to the moderate accuracy of scoring functions [29]. This second step can be substantially improved by accounting for prior experimental information, even when the latter is fairly limited. For example, selection of the interface can be facilitated by results from site-directed mutagenesis experiments, whereas cross-linking yields direct distance restraints. The HADDOCK program and server explicitly employs such information based on biochemical/biophysical interaction data to drive the docking process [16]. Other docking servers, including ClusPro [36], ZDOCK [37], and pyDock [38] were more recently enhanced to take advantage of such restraints. Another source of information, increasingly used in docking, is small angle X-ray scattering (SAXS). Accordingly, SAXS data can be directly incorporated into docking by several servers, including HADDOCK [39], pyDock [40], and ClusPro [41]. For ClusPro an ultra-fast filtering implementation of the approach is also available [42].
Conclusions
As demonstrated by validation on a recent benchmark, the best “free” docking servers find acceptable models among the top 10 predictions for around 40% of the targets. Re-ranking the predictions by a machine learning-based scoring method increased the number of near-native structures among the top 1 predictions from about 10% to over 20%. The inclusion of docking in the joint CASP-CAPRI experiments has led to increased visibility for template-based methods utilizing homology modeling of the complexes. Indeed, if good templates are available, template-based docking produces substantially higher quality predictions than free docking. However, free docking is still needed if no good templates are available, and it also offers an opportunity to include prior information to enhance the quality of predictions. Most targets in CASP-CAPRI experiments were homo-oligomers with good templates, and hence the results provided limited information for directly comparing template-based and free docking. Therefore we tested the availability of templates for the 55 heterodimer targets added to the well-established protein docking benchmark set, and found that no templates were available for almost 50% of the complexes. Free docking applied to these targets revealed acceptable or better models for about 40% of these complexes without templates. Thus, template-based docking of targets with good templates and free docking the targets with only poor or no templates is likely to increase the success rates beyond 40%. At this point the only other way to improve the reliability of docking results is to account for experimental information which provides additional restraints, an option already included in several well-known docking servers.
Acknowledgements.
This investigation was supported by grants R35-GM118078 and R21-GM127952 from the National Institute of General Medical Sciences and NSF DBI 1759472 and NSF AF 1759277 from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest. Acpharis Inc. offers commercial licenses to PIPER, the docking program in the ClusPro server. Sandor Vajda and Dima Kozakov own stock in the company. However, the PIPER program and the use of the ClusPro server are free for academic and governmental use.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nussinov R, Papin JA, Vakser I: Computing the dynamic supramolecular structural proteome. PLoS Comput Biol 2017, 13:e1005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vreven T, Moal IH, Vangone A, Pierce BG, Kastritis PL, Torchala M, Chaleil R, Jimenez-Garcia B, Bates PA, Fernandez-Recio J, et al. : Updates to the integrated protein-protein interaction benchmarks: Docking benchmark version 5 and affinity benchmark version 2. J Mol Biol 2015, 427:3031–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lensink MF, Velankar S, Wodak SJ: Modeling protein-protein and protein-peptide complexes: CAPRI 6th edition. Proteins 2017, 85:359–377.• This is the sixth report evaluating the performance of methods for predicting the atomic resolution structures of protein complexes offered as targets to the community-wide initiative on the Critical Assessment of Predicted Interactions (CAPRI). Models were predicted for 8 protein–peptide and 12 protein–protein complexes. Models of acceptable quality or better were obtained for 14 of the 20 targets, including medium quality models for 13 targets and high quality models for 8 targets, indicating progress of computational methods. It is suggested that the progress stems from better integration of different modeling tools with docking procedures.
- 4.Lensink MF, Velankar S, Kryshtafovych A, Huang SY, Schneidman-Duhovny D, Sali A, Segura J, Fernandez-Fuentes N, Viswanath S, Elber R, et al. : Prediction of homo- and hetero-protein complexes by protein docking and template-based modeling: a CASP-CAPRI experiment. Proteins 2016, 10.1002/prot.25007.•• The paper presents the evaluation of CAPRI Round 30, the first joint CASP-CAPRI experiment, which brought together experts from the protein structure prediction and protein–protein docking communities. The targets included 23 homo-oligomers (18 dimers and 5 tetramers), and 2 heterodimers. The best “human” predictors used template-based methods and submitted 15 or 16 acceptable models. The best servers, HADDOCK and ClusPro, also had to use template-based models, but the final predictions came from re-docking, similarly resulting in 16 acceptable models. However, the purely template-based methods yielded 12 to 14 medium quality models, whereas only 8 or 9 such models were obtained by servers that used the combined approach.
- 5.Lensink MF, Velankar S, Baek M, Heo L, Seok C, Wodak SJ: The challenge of modeling protein assemblies: the CASP12-CAPRI experiment. Proteins 2018, 86 Suppl 1:257–273.•• This is the evaluation of the 2nd joint CAPRI-CASP experiment. The targets were 12 homo-oligomers (3 dimers, 6 trimers, and 3 tetramers), and 3 hetero-complexes (2 dimers and 1 tetramer). Among these, 5 homo-oligomers and one heterodimer were considered easy targets, with good templates available in the PDB. Accordingly, the best template-based predictors submitted 6 acceptable or better predictions. In addition, one docking group and the ClusPro server also obtained acceptable predictions for one heterodimer. The purely template-based methods again produced a substantially higher number of medium quality models for the other 6 targets with good templates. As in the 1st joint CAPRI-CASP experiment, there were too few heterocomplex targets to assess the relative performances of free and template-based docking methods.
- 6.Quignot C, Rey J, Yu J, Tuffery P, Guerois R, Andreani J: InterEvDock2: an expanded server for protein docking using evolutionary and biological information from homology models and multimeric inputs. Nucleic Acids Res 2018, 46:W408–W416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan Y, Zhang D, Zhou P, Li B, Huang SY: HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017, 45:W365–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Mintseris J, Janin J, Weng Z: A protein-protein docking benchmark. Proteins 2003, 52:88–91. [DOI] [PubMed] [Google Scholar]
- 9.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z: Protein-Protein Docking Benchmark 2.0: an update. Proteins 2005, 60:214–216. [DOI] [PubMed] [Google Scholar]
- 10.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z: Protein-protein docking benchmark version 3.0. Proteins 2008, 73:705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang H, Vreven T, Janin J, Weng Z: Protein-protein docking benchmark version 4.0. Proteins 2010, 78:3111–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce BG, Wiehe K, Hwang H, Kim BH, Vreven T, Weng Z: ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014, 30:1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pons C, Solernou A, Perez-Cano L, Grosdidier S, Fernandez-Recio J: Optimization of pyDock for the new CAPRI challenges: Docking of homology-based models, domain-domain assembly and protein-RNA binding. Proteins 2010, 78:3182–3188. [DOI] [PubMed] [Google Scholar]
- 14.Torchala M, Moal IH, Chaleil RA, Fernandez-Recio J, Bates PA: SwarmDock: a server for flexible protein-protein docking. Bioinformatics 2013, 29:807–809. [DOI] [PubMed] [Google Scholar]
- 15.Moal IH, Chaleil RAG, Bates PA: Flexible protein-protein docking with SwarmDock. Methods Mol Biol 2018, 1764:413–428. [DOI] [PubMed] [Google Scholar]
- 16.de Vries SJ, van Dijk M, Bonvin AM: The HADDOCK web server for data-driven biomolecular docking. Nature protocols 2010, 5:883–897. [DOI] [PubMed] [Google Scholar]
- 17.Vangone A, Rodrigues JP, Xue LC, van Zundert GC, Geng C, Kurkcuoglu Z, Nellen M, Narasimhan S, Karaca E, van Dijk M, et al. : Sense and simplicity in HADDOCK scoring: Lessons from CASP-CAPRI round 1. Proteins 2017, 85:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moont G, Gabb HA, Sternberg MJ: Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins 1999, 35:364–373. [PubMed] [Google Scholar]
- 19.de Vries SJ, Bonvin AM: CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PLoS One 2011, 6:e17695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu S, Wallner B: DockQ: A Quality Measure for Protein-Protein Docking Models. PLoS One 2016, 11:e0161879.•• CAPRI evaluators made substantial contributions by developing the protocol for determining the quality of predicted structures and defining the acceptable, medium, and high accuracy categories. The protocol and these categories are now widely accepted. However, no software was released to perform the evaluation. The importance of the DockQ program, described in this paper and distributed by the authors, is that it enables individual research group to determine the accuracy of their methods.
- 21.Zhang C, Lai L: SDOCK: a global protein-protein docking program using stepwise force-field potentials. J Comput Chem 2011, 32:2598–2612. [DOI] [PubMed] [Google Scholar]
- 22.Moal IH, Barradas-Bautista D, Jimenez-Garcia B, Torchala M, van der Velde A, Vreven T, Weng Z, Bates PA, Fernandez-Recio J: IRaPPA: information retrieval based integration of biophysical models for protein assembly selection. Bioinformatics 2017, 33:1806–1813.•• The paper describes a rescoring scheme called IRaPPA (Integrative Ranking of Protein–Protein Assemblies). IRaPPA characterizes decoys using physicochemical descriptors, calculated by the server CCharPPI, and combines a large selection of metrics using ranking support vector machines (R-SVMs) to obtain a consensus ranking using a voting method. The new scoring was implemented in the servers ZDOCK, pyDock, SwarmDock, and SDOCK. Machine learning based models were trained using complexes from version 4 of the protein-protein docking benchmark, and evaluated on the new complexes added in version 5 of the benchmark. While all methods were substantially improved, further testing in CAPRI and other prediction experiments are needed in order to detect potential overtraining that can occur in machine learning with relatively small training sets.
- 23.Moal IH, Jimenez-Garcia B, Fernandez-Recio J: CCharPPI web server: computational characterization of protein-protein interactions from structure. Bioinformatics 2015, 31:123–125.• The CCharPPI server calculates up to 108 parameters, including models of electrostatics, desolvation and hydrogen bonding, as well as interface packing and complementarity scores, empirical potentials at various resolutions, docking potentials and composite scoring functions. Although the server is freely available for non-commercial academic use, one can only submit a limited number of structures, and the software is not available for download. Thus, in spite of its potential utility, in the current form the method cannot be used for training machine learning based models for improving the discrimination of docked structures.
- 24.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S: The ClusPro web server for protein-protein docking. Nat Protoc 2017, 12:255–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozakov D, Brenke R, Comeau SR, Vajda S: PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 2006, 65:392–406. [DOI] [PubMed] [Google Scholar]
- 26.Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, Xia B, Hall DR, Kozakov D: New additions to the ClusPro server motivated by CAPRI. Proteins 2017, 85:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenke R, Hall DR, Chuang GY, Comeau SR, Bohnuud T, Beglov D, Schueler-Furman O, Vajda S, Kozakov D: Application of asymmetric statistical potentials to antibody-protein docking. Bioinformatics 2012, 28:2608–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogues H, Gaudreault F, Corbeil CR, Deprez C, Sulea T, Purisima EO: ProPOSE: Direct exhaustive protein-protein docking with side chain flexibility. J Chem Theory Comput 2018, 14:4938–4947. [DOI] [PubMed] [Google Scholar]
- 29.Vajda S, Hall DR, Kozakov D: Sampling and scoring: A marriage made in heaven. Proteins-Structure Function and Bioinformatics 2013, 81:1874–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson LX, Kim H, Esquivel-Rodriguez J, Roy A, Han X, Shin WH, Zhang J, Terashi G, Lee M, Kihara D: Human and server docking prediction for CAPRI round 30–35 using LZerD with combined scoring functions. Proteins 2017, 85:513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Vavrusa M, Andreani J, Rey J, Tuffery P, Guerois R: InterEvDock: a docking server to predict the structure of protein-protein interactions using evolutionary information. Nucleic Acids Res 2016, 10.1093/nar/gkw340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soding J, Biegert A, Lupas AN: The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 2005, 33:W244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb B, Sali A: Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Protein Sci 2016, 86:2 9 1–2 9 37. [DOI] [PubMed] [Google Scholar]
- 34.Kundrotas PJ, Zhu ZW, Janin J, Vakser IA: Templates are available to model nearly all complexes of structurally characterized proteins. Proceedings of the National Academy of Sciences of the United States of America 2012, 109:9438–9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosca R, Ceol A, Aloy P: Interactome3D: adding structural details to protein networks. Nature Methods 2013, 10:47–U127. [DOI] [PubMed] [Google Scholar]
- 36.Xia B, Vajda S, Kozakov D: Accounting for pairwise distance restraints in FFT-based protein-protein docking. Bioinformatics 2016, 10.1093/bioinformatics/btw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vreven T, Schweppe DK, Chavez JD, Weisbrod CR, Shibata S, Zheng C, Bruce JE, Weng Z: integrating cross-linking experiments with ab initio protein-protein docking. J Mol Biol 2018, 430:1814–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallara C, Jimenez-Garcia B, Romero M, Moal IH, Fernandez-Recio J: pyDock scoring for the new modeling challenges in docking: Protein-peptide, homo-multimers, and domain-domain interactions. Proteins 2017, 85:487–496. [DOI] [PubMed] [Google Scholar]
- 39.Karaca E, Bonvin AM: On the usefulness of ion-mobility mass spectrometry and SAXS data in scoring docking decoys. Acta Crystallogr D Biol Crystallogr 2013, 69:683–694. [DOI] [PubMed] [Google Scholar]
- 40.Jimenez-Garcia B, Pons C, Svergun DI, Bernado P, Fernandez-Recio J: pyDockSAXS: protein-protein complex structure by SAXS and computational docking. Nucleic Acids Res 2015, 43:W356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia B, Mamonov A, Leysen S, Allen KN, Strelkov SV, Paschalidis IC, Vajda S, Kozakov D: Accounting for observed small angle X-ray scattering profile in the protein-protein docking server cluspro. Journal of Computational Chemistry 2015, 36:1568–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ignatov M, Kazennov A, Kozakov D: ClusPro FMFT-SAXS: Ultra-fast filtering using small-angle X-ray scattering data in protein docking. J Mol Biol 2018, 430:2249–2255. [DOI] [PubMed] [Google Scholar]