TP53 mutations are associated with adverse prognosis in both myelodysplastic syndromes (MDS) and in acute myeloid leukemia (AML).1,2 Clinical trials focused on therapeutic options for this group of high-risk patients require a rapid, inexpensive technique to identify patients with TP53 mutations and to closely monitor clinical responses. While sequencing-based assessment of TP53 mutation status is now widely available, the costs and turnaround times may not be optimal for prompt therapeutic selection, clinical trial enrollment, and monitoring of response. Using routine immunohistochemistry (IHC), a rapid, inexpensive and readily available test, we found that expression levels of TP53 were a reliable proxy for both the presence of TP53 mutations and mutation clearance during clinical responses. Across samples with a wide range of TP53 variant allele frequencies (VAF), we show that TP53 IHC has both high sensitivity (86%) and high specificity (90%) to detect TP53 mutations in bone marrow (BM) cores and has a high concordance (r=0.7114, R2=0.5061) with mutation levels across a range of post-therapy responses.
TP53 mutations in AML frequently produce dominant-negative proteins, which may stabilize mutant TP53 due to lack of negative feedback mechanisms, such as TP53-regulation of the ubiquitin ligase MDM2 that targets TP53 for degradation.3,4 Unlike the wild-typeTP53 protein, which is quickly degraded, mutant TP53 protein accumulates in the cell nucleus and is detectable by TP53 IHC. Detection of increased TP53 protein in acute leukemia has also been reported using flow cytometry,5 but this requires intracellular permeabilization and adequate aspirate, and is not a widely used clinical assay. In contrast, TP53 IHC is a commonly used clinical test that can be run on routine BM core or clot sections.
We evaluated TP53 staining by IHC in serial samples from MDS and AML patients (Online Supplementary Table S1) treated on a previously published, prospective, open-label study of 10-day decitabine therapy (clinicaltrials.gov identifier: 01687400).6 Formalin-fixed paraffin-embedded bone marrow (BM) trephine biopsies were available from TP53-mutated (n=21) and TP53-wild-type (n=51) patients. Three additional cases with truncating TP53 mutations were subsequently evaluated from patients collected on unrelated protocols.7,8 All patients consented to genome sequencing analysis and were treated in accordance with the Declaration of Helsinki. TP53 mutation status was established by paired tumor/normal high coverage next-generation sequencing. Serial BM samples obtained during therapy were available for 16 of the TP53-mutated cases (n=44 samples, median number of samples per patient = 4). TP53 and TP53/lineage marker IHC was performed using standard automated protocols [TP53 antibody clones D07 (all cases) and BP53-11 (selected cases), CD3 (2GV6), and CD34 (QBEnd/10), Ventana Medical Systems, Tucson, AZ, USA; CD61 (2f2), E-cadherin (EP700Y), and myeloperoxidase (polyclonal); Cell Marque, Rocklin, CA, USA]. Quantitative scoring of TP53 IHC slides was performed independently by three board-certified hematopathologists (MBR, EJD and YSL) by manually counting any nuclear staining in 500 BM hematopoietic cells. Statistical analysis was performed using GraphPad Prism (San Diego, CA, USA) and SPSS (Chicago, IL, USA).
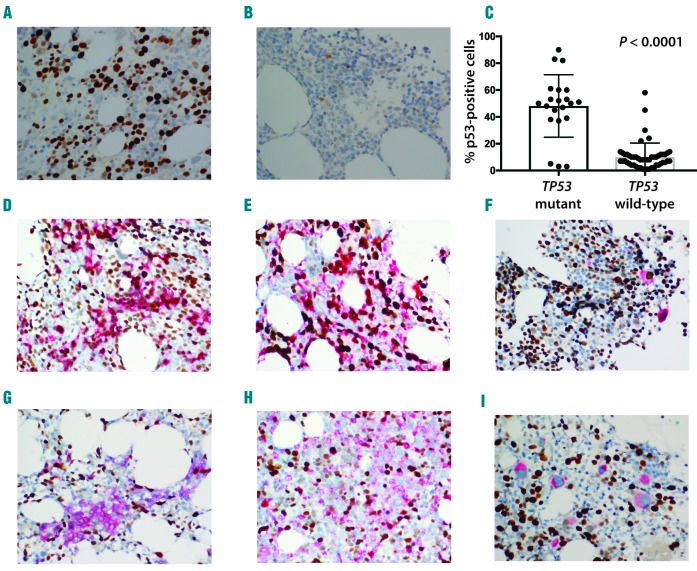
Similar to previous reports,9,10 we found significantly increased nuclear TP53 staining in TP53-mutated cases (48±5%) as compared to TP53-wild-type (10±1.5%) cases (P<0.0001) (Figure 1A-C). We also found significant TP53 overexpression in cases with complex cytogenetics and chromosome 5 and 17 abnormalities (Online Supplementary Appendix and Online Supplementary Figure S1A-C). A high degree of concordance in assessing TP53 counts was noted between observers (R2 =0.92 and 0.94) (Online Supplementary Figure S1D). A receiver operating characteristic (ROC) curve was constructed using different TP53 expression levels in order to establish the best TP53 cut-off value (Online Supplementary Figure S1E). The area under the ROC curve when using TP53 expression to detect TP53 mutations was 0.872 and a cutoff value of >20% TP53-positive cells produced sensitivity of 86% and specificity of 90% compared to sequencing, in close agreement with a previously published study (area under ROC curve of 0.873, cut off value >10% of TP53-positive cells, with sensitivity of 75% and specificity of 91%).10
Figure 1.
TP53 expression is increased in TP53-mutated cases but does not consistently associate with specific bone marrow (BM) lineage. Representative micrographs of TP53 staining of TP53-mutated (A) and TP53 wild-type case (B). Significantly higher TP53 staining (% positive cells) in mutated versus wild-type TP53 cases (C). Co-expression of TP53 in different BM lineages (brown nuclear staining: TP53; red staining: lineage marker). High (D) and low (G) co-expression of TP53 in E-cadherin-positive erythroid elements. High (E) and low (H) co-expression of TP53 in myeloperoxidase-positive myeloid elements. High (F) and low (I) co-expression of TP53 in CD61-positive megakaryocytes.
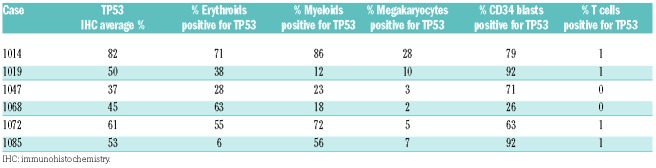
Co-staining with lineage markers was performed to determine whether mutated TP53 protein accumulated preferentially in specific BM lineages. No consistent pattern of TP53 overexpression was identified in erythroid (Figure 1D-G), myeloid (Figure 1E-H), or megakaryocytic (Figure 1F-I) elements (Table 1). Increased expression of TP53 was seen more consistently in CD34-positive (+) blasts (Table 1 and Online Supplementary Figure S1G), but cases with CD34-negative (−) blasts or no increase in blasts limit the usefulness of CD34/TP53 IHC. No significant TP53 overexpression was identified in T cells (Table 1 and Online Supplementary Figure S1H).
Table 1.
Expression of TP53 in different bone marrow lineages.
Previous studies10,11 reported occasional BM samples with TP53 mutations that showed no corresponding elevation of TP53 expression. In our cohort, we identified three cases with TP53 mutations that showed no significant increase in TP53 IHC (3-5% TP53+ cells). Staining discrepant cases with a different anti-p53 antibody clone (BP53-11) produced similar results (7-13% TP53+ cells). We reviewed each of these three cases. The first two cases had truncating variants (patient 1065: p.T81fs*, Online Supplementary Figure S2B; patient 1101: splice site e6-1). An additional three cases with truncating TP53 mutations p.R213*, p.Q167*, p.Q317*7,8 were tested and also showed no significant increase in TP53 expression (0%, 7% and 9%, respectively), as expected. Thus, nonsense and other truncation mutations will likely be a source of false-negative cases. However, these are a minority among TP53 mutations. We reviewed TP53 variants in the Catalogue of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk) among patients with hematopoietic, non-lymphoid malignancies, and 12.6% of catalogued 1752 TP53 variants are predicted to produce a truncated protein likely undetectable by TP53 IHC (5.9% nonsense substitutions, 6.7% frame-shift insertions and deletions). An additional 10% of TP53 mutations are at splice site junctions or in the 5’/3’untranslated region, and these could variably impact protein expression, with a subset likely producing a false-negative IHC result. Within published cohorts of patients, the incidence of mutations likely undetectable by IHC is similar or somewhat higher (14-31%), with higher proportions of truncating mutations reported in studies with lower case numbers (see extended discussion in Online Supplementary Appendix).
The third case from our cohort (patient 1002, Online Supplementary Figure S2C) was associated with TP53 p.G245S with a VAF of 39.6%. Although the pre-treatment sample showed low TP53 IHC staining, subsequent samples showed higher IHC staining and then IHC reduction that paralleled mutation clearance, suggesting that the initial discrepancy may be due to pre-analytic factors such as time to fixation or fixation time of the original BM specimen.12 We also observed two cases with elevated TP53 IHC in the absence of TP53 mutations (Online Supplementary Appendix).
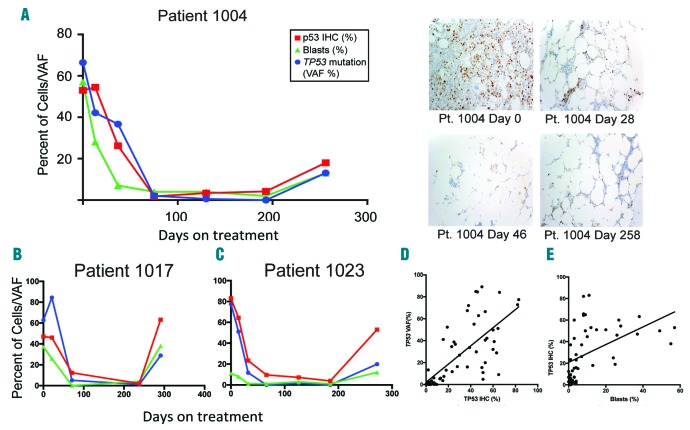
To determine whether TP53 IHC results changed in response to therapy and correlated with clinical response measures, we evaluated serial samples of patients treated with decitabine. Sixteen cases with TP53 mutations had been evaluated previously with serial BM sequencing and noted mutation reduction to VAF level below 5% in con cordance with morphological responses.6 Relapse following initial clinical response to decitabine treatment was common and was associated with return of the original TP53 clone.6 Excluding the above-mentioned discrepant cases with truncating mutations and a sample with technical issue, changes in percentage of IHC TP53+ cells closely paralleled the changes in TP53 mutation VAF levels: decreasing during initial therapy and rising with outgrowth of TP53 clone during relapse (Figure 2A-C and Online Supplementary Figure S2A and D-N). Comparison of pre-treatment TP53 VAF and percentage of TP53 IHC (Online Supplementary Appendix and Online Supplementary Figure S1F) showed no significant correlation, with even low TP53 VAF samples (VAF <20, n=3) showing high TP53 IHC. In contrast, there was a statistically significant positive correlation between percentage of TP53+ cells and TP53 VAF (r=0.7114, R2=0.5061; P<0.0001) (Figure 2D) during decitabine therapy. In cases with elevated blast count at diagnosis, there was also significant positive correlation between percentage of TP53-positive cells and blast count (r=0.4821; P=0.0003) (Figure 2E).
Figure 2.
During decitabine therapy, changes in percentage of immunohistochemistry (IHC) TP53-positive cells parallel closely the changes in TP53 mutation variant allele frequency (VAF) levels. Correlation between TP53 VAF, TP53 IHC and blast percentages and representative micrographs of TP53 staining in serial bone marrows from TP53-mutated patient (patient 1004) at different time points during decitabine therapy (A). Correlation between TP53 VAF, TP53 IHC and blast percentages in two more TP53-mutated patients treated with decitabine (B and C). Significant positive association between TP53 positivity and TP53 VAF during decitabine therapy (D). Significant positive association between TP53 positivity and blast % during decitabine therapy (E).
TP53 mutation is consistently associated with inferior overall survival in MDS and AML, and resistance to anthracycline- or cytarabine-based induction chemotherapy.2,13 In contrast to standard chemotherapy, recent studies have shown that presence of adverse-risk karyotypes or TP53 mutations does not negatively impact sensitivity to hypomethylating therapy,6,14,15 although the extent of observed clinical responses has been variable. In this clinical trial cohort, presence of TP53 mutation strongly correlated with favorable response to decitabine therapy,6 and, similarly, patients with elevated expression of TP53 achieved clinical remission significantly more frequently than patients with TP53 baseline expression (95% vs. 59%; P<0.002). Therefore, a rapid, cost-effective strategy will be relevant to enroll high-risk TP53-mutated patients on future clinical trials. We show that TP53 IHC is a reproducible and rapid tool for detection of TP53 mutation status, monitoring response to therapy, and monitoring for relapse in TP53-mutated AML and MDS patients. We note that each individual laboratory may need to establish its own threshold, and a small subset of cases may produce false-negative results due to truncating mutations or pre-analytical variables. As TP53 IHC is integrated as a rapid screening assay in routine diagnostic evaluations and follow up of MDS and AML, correlation with initial diagnostic sequencing will still be needed to exclude low VAF or truncating mutations. However, p53 IHC can be used as a reliable surrogate for TP53 mutation status in most patients, and may be particularly useful in patients for whom molecular testing is not available.
Acknowledgments
The authors would like to thank Sharon Heath, Kierstin Luber, Megan Janke, Paige Schnoebelen, Theresa Fletcher, Megan Haney, and Shannon Kramer for assistance in patient enrollment, sample collection, and clinical data processing. We thank Greg Malnassy, Nichole Helton, and the Washington University Tissue Procurement Core for sample storage and sample preparation.
Footnotes
Funding: support was provided by an NCI AML-SPORE (P50 CA171963) and the Genomics of AML Program Project grant (P01 CA101937).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Kulasekararaj AG, Smith AE, Mian SA, et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br J Haematol. 2013;160(5):660–672. [DOI] [PubMed] [Google Scholar]
- 2.Rücker FG, Schlenk RF, Bullinger L, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9):2114–2121. [DOI] [PubMed] [Google Scholar]
- 3.Kern SE, Pietenpol JA, Thiagalingam S, Seymour A, Kinzler KW, Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992;256(5058):827–830. [DOI] [PubMed] [Google Scholar]
- 4.Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. [DOI] [PubMed] [Google Scholar]
- 5.Cavalcanti CB, Scheiner MA, Simoes Magluta EP, Vasconcelos FC, Klumb CE, Maia RC. p53 flow cytometry evaluation in leukemias: correlation to factors affecting clinical outcome. Cytometry B Clin Cytom. 2010;78(4):253–259. [DOI] [PubMed] [Google Scholar]
- 6.Welch JS, Petti AA, Miller CA, et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N Engl J Med. 2016;375(21):2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncavage EJ, Jacoby MA, Chang EG, et al. Mutation Clearance after Transplantation for Myelodysplastic Syndrome. N Engl J Med. 2018;379(11):1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGraw KL, Nguyen J, Komrokji RS, et al. Immunohistochemical pattern of p53 is a measure of TP53 mutation burden and adverse clinical outcome in myelodysplastic syndromes and secondary acute myeloid leukemia. Haematologica. 2016;101(8):e320–e323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-pol S, Ma L, Ohgami RS, Arber DA. Immunohistochemistry for p53 is a useful tool to identify cases of acute myeloid leukemia with myelodysplasia-related changes that are TP53 mutated, have complex karyotype , and have poor prognosis. Mod Pathol. 2016;30(3):1–11. [DOI] [PubMed] [Google Scholar]
- 11.Saft L, Karimi M, Ghaderi M, et al. P53 Protein Expression Independently Predicts Outcome in Patients With Lower-Risk Myelodysplastic Syndromes With Del(5Q). Haematologica. 2014;99(6):1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel KB, Moore HM. Effects of preanalytical variables on the detection of proteins by immunohistochemistry in formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med. 2011;135(5):537–543. [DOI] [PubMed] [Google Scholar]
- 13.Wattel E, Preudhomme C, Hecquet B, et al. P53 Mutations Are Associated With Resistance To Chemotherapy and Short Survival in Hematologic Malignancies. Blood. 1994;84(9):3148–3157. [PubMed] [Google Scholar]
- 14.Blum W, Garzon R, Klisovic RB, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc Natl Acad Sci. 2010;107(16):7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cashen AF, Schiller GJ, O’Donnell MR, DiPersio JF. Multicenter, phase II study of decitabine for the first-line treatment of older patients with Acute Myeloid Leukemia. J Clin Oncol. 2010; 28(4):556–561. [DOI] [PubMed] [Google Scholar]