Abstract
Assessment of measurable residual disease, also called “minimal residual disease,” in patients with acute myeloid leukemia in morphological remission provides powerful prognostic information and complements pretreatment factors such as cytogenetics and genomic alterations. Based on data that low levels of persistent or recurrent residual leukemia are consistently associated with an increased risk of relapse and worse long-term outcomes, its routine assessment has been recommended by some experts and consensus guidelines. In addition to providing important prognostic information, the detection of measurable residual disease may also theoretically help to determine the optimal post-remission strategy for an individual patient. However, the full therapeutic implications of measurable residual disease are uncertain and thus controversy exists as to whether it should be routinely incorporated into clinical practice. While some evidence supports the use of allogeneic stem cell transplantation or hypomethylating agents for some subgroups of patients in morphological remission but with detectable residual leukemia, the appropriate use of this information in making clinical decisions remains largely speculative at present. To resolve this pressing clinical issue, several ongoing studies are evaluating measurable residual disease-directed treatments in acute myeloid leukemia and may lead to new, effective strategies for patients in these circumstances. This review examines the common technologies used in clinical practice and in the research setting to detect residual leukemia, the major clinical studies establishing the prognostic impact of measurable residual disease in acute myeloid leukemia, and the potential ways, both now and in the future, that such testing may rationally guide therapeutic decision-making.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease, with a widely variable likelihood of cure with conventional therapies that depends on both patient-and disease-related characteristics.1 Historically, pretreatment characteristics such as the patients’ age, karyotype, and, more recently, genetic mutations have been the primary determinants of prognosis in patients with newly diagnosed AML. How well the leukemia responds to initial treatment also provides important information about chemosensitivity of an individual’s leukemia that cannot always be predicted from pretreatment characteristics. One measure of treatment response is the achievement of complete remission, defined as bone marrow with <5% blasts with recovery of peripheral blood elements and no evidence of extramedullary disease.2 However, among patients <60 years of age who achieve complete remission with induction and then receive consolidation chemotherapy, the cure rate is only approximately 50% (and significantly lower in older patients), suggesting that morphological assessment of the bone marrow alone is inadequate to discriminate relapse risk.3 Accurate assessment of the risk of relapse is imperative in the treatment of AML, as the decision to pursue consolidation chemotherapy rather than allogeneic hematopoietic stem cell transplantation (HSCT) in first remission is largely decided by the anticipated risk of relapse in the absence of HSCT balanced against the risk of relapse after HSCT as well as the expected transplant-related mortality.4 Although improvements in genomic classification of AML have refined our current risk stratification systems, relapses are unfortunately still common even with risk-adapted treatment.
Assessment of measurable residual disease (MRD), also called “minimal residual disease,” allows for the detection and quantification of lower levels of residual leukemia that can be detected by morphological assessment alone.5,6 In both pediatric and adult acute lymphoblastic leukemia, MRD is routinely used to refine prognostic assessment and also allocate post-remission therapies, particularly in the pediatric population.7–11 Similarly, studies utilizing various methods of MRD determination have consistently shown that MRD is highly prognostic in AML and complements (and sometimes supersedes) historically relevant pretreatment characteristics.12–18 However, many questions remain as to the optimal MRD assay, appropriate timing of MRD assessment, and whether MRD should guide treatment. Herein, we review methods of MRD detection and the prognostic impact of MRD across AML subtypes. We also offer some practical considerations for MRD testing in the clinical setting and review the available data on how MRD can potentially guide post-remission treatment decisions in AML.
Methods of measurable residual disease assessment
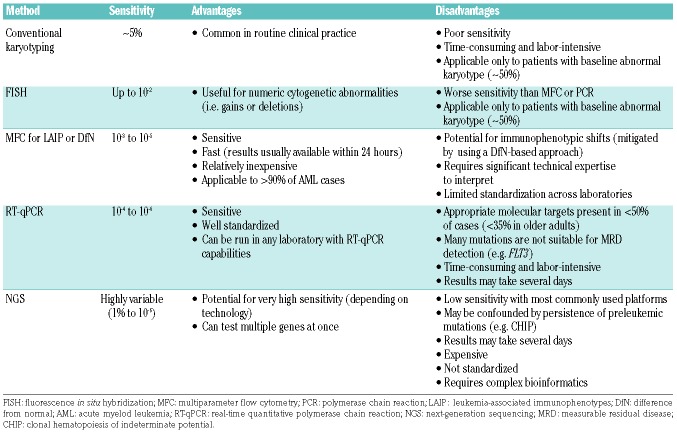
Several methods of MRD assessment are available in the clinical and research settings, each of which has its own advantages and disadvantages. The European LeukemiaNet MRD Working Party has released comprehensive, consensus recommendations regarding their appropriate use and technical limitations.6 These various MRD methods vary in their level of sensitivity, applicability to the vast majority of patients with AML versus only certain subtypes, cost, and level of technical expertise needed to obtain an accurate result, all of which may influence the choice of assay for a particular patient.19 The major differences among MRD technologies are summarized in Table 1.
Table 1.
Methods of measurable residual disease assessment in acute myeloid leukemia.
Karyotype analysis and fluorescence in situ hybridization
In patients with an abnormal pretreatment karyotype, the persistence of an abnormal leukemia-associated karyo type at remission suggests the presence of residual disease. Persistent cytogenetic abnormalities have been associated with worse survival in several studies and may also identify patients who could benefit from HSCT in first remission.20,21 While cytogenetic analysis at remission adds prognostic information in patients who achieve complete remission, the sensitivity of this method is relatively poor, as it can only detect one abnormal metaphase out of 20 (i.e. sensitivity ≈5%). Furthermore, up to 50% of adult patients present with cytogenetically normal AML, further limiting karyotype analysis as a universal MRD marker.1 Fluorescence in situ hybridization may also be used to detect persistent cytogenetic abnormalities with slightly increased sensitivity compared to conventional cytogenetics. However, the maximum sensitivity achieved with fluorescence in situ hybridization is approximately 1%, which is not adequate to detect low levels of clinically relevant MRD.22
Multiparameter flow cytometry
Multiparameter flow cytometry (MFC) uses a panel of fluorochrome-labeled monoclonal antibodies to identify aberrantly expressed antigens on leukemic blasts. This MFC-based MRD analysis may be accomplished through either the tracking of leukemia-associated immunophenotypes in the pretreatment and remission samples or the use of “difference from normal” analysis.22 Leukemia-associated immunophenotypes consist of the aberrant expression of antigens compared to that on normal myeloid precursors, cross-lineage antigen expression (e.g. expression of lymphoid antigens on myeloblasts), over- or under-expression of antigens normally expressed, and aberrant co-expression of antigens normally found in early or late hematopoietic differentiation.6 In contrast, the “difference from normal” approach is used to detect any differences in the remission immunophenotype compared to the highly stereotypical normal immunophenotype distribution.23 The advantage of “difference from normal” analysis is that it does not require knowledge of the diagnostic immunophenotype; furthermore, it may also be less susceptible to immunophenotypic shifts that can occur as a direct result of therapy or due to a shift in clonal architecture.24,25 Workflows that incorporate both of these methods (i.e., a leukemia-associated immunophenotype-based, “difference from normal” approach) may help to further optimize MRD assessment.6 Some studies suggest that addition of leukemia stem cell markers (e.g. CLL1, CD44, CD123, and CD184, among others) to flow MRD antibody panels may add additional prognostic information to standard MFC-based MRD, particularly by identifying those patients at very high risk of relapse (i.e. those who are positive for both MRD and leukemia stem cell markers).26–28
MFC-based MRD assessment can achieve a sensitivity of 10−3 to 10−5, which is dependent on the number of cells analyzed, gating method, and number of antibody colors used; in most cases, a sensitivity of 10−4 is achieved. Compared to real-time quantitative polymerase chain reaction (RT-qPCR), MFC is significantly faster and less labor-intensive. It also has the advantage of being applicable to more than 90% of patients with AML, unlike other methods that rely on specific genetic or molecular targets.22 Despite these advantages, the interpretation of MFC MRD is not standardized in most countries, including the USA, and requires significant technical expertise on the part of the interpreting pathologist, which can lead to inter-laboratory discordance. Because of the complexity of interpreting flow-based MRD, there is interest in the use of machine-based learning artificial intelligence to reduce the potential for bias or other subjective errors in MRD interpretation. Such artificial intelligence-based algorithms are promising and may result in more clinically significant MRD results, although further validation of this technology is needed.29
Polymerase chain reaction
RT-qPCR can be used to monitor recurrent genomic alterations in certain subtypes of AML. To be a useful marker for PCR-based MRD assessment, the target gene fusion or mutation should be stable throughout the disease course and its presence should reflect true persistent disease (rather than preleukemic subclones). Suitable targets that have been evaluated in large studies include PML-RARA in acute promyelocytic leukemia, CBFB- MYH11 and RUNX1-RUNX1T1 in core-binding factor AML, and mutant NPM1.15,30–36 In contrast, other mutations may emerge or disappear at the time of relapse (e.g., mutant FLT37,38) and are therefore generally unreliable MRD markers for assessing meaningful “MRD negativity,” although their persistence likely represents residual disease in most cases. One disadvantage of PCR is that appropriate, validated targets are present in less than 50% of patients with AML, and the incidence of these AML subtypes declines substantially with increasing age at diagnosis.1 To overcome this limitation, attempts have been made to track residual disease using markers that are expressed at significantly higher levels in leukemic blasts than in normal hematopoietic cells. For example, several studies have evaluated monitoring levels of WT1 or EVI1 mRNA transcripts over the course of treatment as a marker of MRD.39–42 While these targets may provide some prognostic information, they are generally not specific enough for residual leukemia to be used in routine practice.6
PCR-based MRD assessment has the advantage of being highly sensitive (sensitivity ≈ 10−4 to 10−6, depending on the input of RNA/DNA) and is generally better standardized than MFC.19 To further refine relapse risk, ultrasensitive digital droplet PCR technologies have been developed in order to detect low levels of residual gene mutations.43 In contrast to standard PCR, digital droplet PCR does not require calibration standards, and is thus faster and more precise and reproducible.44 Due to its absolute quantification of DNA copy numbers, it can also provide information about clonality and subclonality. This technology may have greater sensitivity and be better able to quantify very low levels of MRD than standard PCR22; however, whether this added sensitivity will translate into more accurate prognostic discrimination is yet to be determined.
Next-generation sequencing
Targeted next-generation sequencing (NGS) panels are commonly used at the time of diagnosis to identify prognostic gene mutations or mutations that may be therapeutically targeted (e.g. FLT3 or IDH1/2 mutations). Several studies have also evaluated re-using similar NGS panels at the time of remission to assess the relationship between decline in mutational burden and clinical outcomes.17,45,46 This methodology relies on a similar principal as PCR regarding the tracking of genomic or molecular targets, although NGS is able to target multiple genes at once, or even the entire genome, if desired.
Although there is understandable excitement about the potential for high-throughput NGS-based MRD monitoring in AML, there are several practical considerations that limit its clinical use at present. NGS MRD assessment is relatively expensive, is not standardized, and requires complicated bioinformatics. The interpretation of NGS MRD results is further complicated by the presence of preleukemic clones that may not fully clear even in patients who achieve deep, long-term remissions with chemotherapy, such as mutations associated with clonal hematopoiesis of indeterminate potential (CHIP).47–49 CHIP mutations, particularly DNMT3A, TET2, and ASXL1, commonly persist in patients who do not relapse, suggesting that they should not be routinely used as MRD markers.17,46 Furthermore, at present, the sensitivity of NGS for MRD assessment is generally ~1% with most commonly used platforms, due largely to the intrinsic error rate of sequencing. This lack of sensitivity is often because the assays are designed for identification of mutations in baseline samples (for which high sensitivity is generally not needed) and then repurposed for assessment of MRD (for which adequate sensitivity is imperative). However, depending on the NGS platform used and the amount of input DNA, NGS can theoretically achieve a sensitivity of 10−6, making it an attractive potential option for very sensitive MRD detection. Advances in NGS technologies, including molecular barcoding and duplex sequencing, may improve the sensitivity of NGS and allow for the identification of very low levels of residual leukemia.50,51
Prognostic Impact of measurable residual disease in acute myeloid leukemia
Achievement of MRD negativity has been shown to be a powerful prognostic factor in numerous studies of patients undergoing frontline AML therapy. Based on the consistent impact of MRD on long-term outcomes across multiple studies and AML subtypes, consensus guidelines from the European LeukemiaNet support “complete remission without MRD” as an official AML response criterion.2 There are also ongoing efforts in the USA and elsewhere to validate MRD as a surrogate endpoint for accelerated regulatory approval of novel drugs and combinations. Here we review some of the major studies supporting the clinical use of MRD to provide prognostic and predictive information in AML.
Multiparameter flow cytometry-based measurable residual disease studies
MRD as assessed by MFC is highly prognostic when measured at various time points, including after induction, during and after consolidation, and peri-transplant.12,13,16,18,52–56 While studies have used varied cutoffs to define MRD “negativity,” many have used levels of ≤0.1% to define negativity, both because this level of sensitivity can be reliably achieved with flow-based assays and because this level appears to provide the best discrimination for relapse risk at most time points.13,56 However, it is important to note that some studies have also suggested that even levels of residual leukemia below the 0.1% threshold may be associated with worse outcomes than lack of detectable MRD.12,53 Several studies in younger adults ≤60 years of age with newly diagnosed AML undergoing standard frontline chemotherapy have shown that achievement of MRD negativity is associated with a significantly lower risk of relapse and better survival.13,16,18 The impact of MRD status consistently adds additional prognostic information beyond that provided by pretreatment characteristics, such as cytogenetics and genetic mutations. Notably, most studies have evaluated MRD in younger adults and the data regarding the impact of MRD in patients >60 years are relatively scant. However, the available data suggest that achievement of MRD negativity in the older population may be predictive for lower relapse risk whether the patients are treated with intensive chemotherapy or hypomethylating agents.12,57 Because most older patients with AML will not be candidates for allogeneic HSCT, the potential therapeutic implications of persistent MRD in these patients is less clear than in their younger counterparts.
The National Cancer Research Institute AML17 trial is the largest study of MRD in AML to date and provides some key insights into the impact of MRD in AML.18 This study enrolled 1,874 adults <60 years of age with AML who received standard daunorubicin plus cytarabine-based induction, followed by risk-adapted chemotherapy consolidation, with or without allogeneic HSCT. Patients with MRD positivity (defined as ≥0.1% by MFC assay) after cycle 1 had similar 5-year overall survival as patients who only achieved a partial response (51% vs. 46%, respectively), highlighting the poor outcomes associated with persistent MRD. Given the large size of this population, subgroup analyses were possible, including those among patients with standard-risk AML without NPM1 mutations. Of note, this is a particularly important group to refine prognosis using MRD, since there is controversy as to whether standard-risk patients (as determined by pretreatment characteristics) should routinely undergo HSCT in first remission; it is also a population in which MFC-based MRD assessment plays an integral role given the absence of a reliable molecular MRD target (e.g. mutant NPM1).1,22 Among patients with standard-risk AML without NPM1 mutation, the 5-year overall survival rates for patients who achieved MRD negativity after two cycles and for patients who remained MRD-positive were 63% and 33%, respectively (P=0.003), with the difference being driven by a significantly higher relapse rate in the MRD-positive group. Thus, MRD status after induction or in early consolidation may be able to stratify standard-risk patients according to risk of relapse and, consequently, help to determine the potential benefit from consolidative allogeneic HSCT.
Detectable MRD immediately prior to HSCT was also associated with increased risk of post-HSCT relapse and worse overall survival in several studies.52–55 These observations are further supported by a meta-analysis of 19 studies evaluating pre-HSCT MRD (the majority of which assessed MRD by MFC).58 In one study of patients with AML who underwent HSCT, those in complete remission but with detectable MRD had a similar post-HSCT relapse rate and overall survival as those transplanted when not in morphological remission.54 The 3-year overall survival rates for patients who were in complete remission but MRD-positive versus those with active AML at the time of HSCT were 26% and 23%, respectively, compared to 73% for patients who were MRD-negative. The impact of post-HSCT MRD status is, however, less clear.55,59 While patients with persistent or recurrent MRD after HSCT appear to have relatively poor outcomes, one analysis suggests that pre-HSCT MRD status carries relatively more prognostic information than post-HSCT MRD status.55 In this study, only pre-HSCT MRD was independently associated with long-term outcomes. The 3-year overall survival rate for patients with pre-HSCT MRD that persisted after the transplant was 19% and only slightly higher for those with pre-HSCT MRD that cleared with transplantation (3-year overall survival rate: 29%).
Polymerase chain reaction-based measurable residual disease studies
The prognostic impact of MRD detected by PCR has been shown primarily in acute promyelocytic leukemia, core-binding factor leukemias, and NPM1-mutated AML, as the target alterations in these AML subtypes represent stable, foundational genomic lesions that become undetectable in long-term survivors (with rare exceptions60,61) and persist or re-emerge in the setting of relapsed disease.19 In acute promyelocytic leukemia, the persistence of PML-RARA transcripts strongly predicts for relapse, and patients with persistent transcripts invariably relapse without intervention.30,31,62 Among patients with acute promyelocytic leukemia treated with chemotherapy plus all-trans retinoic acid, initiation of arsenic trioxide for those with positive MRD prevents morphological relapse in the majority of cases.31 However, in this context, it is important to confirm a positive MRD assessment with repeat testing, given the potential for false positives. Consensus guidelines have therefore defined molecular remission as an important therapeutic milestone and recommend molecular assessment in routine clinical practice.63 The link between MRD status and prognosis, as well as an effective salvage therapy for MRD-positive disease, makes acute promyelocytic leukemia a model for MRD-guided therapies, although it is unclear whether routine, universal monitoring is still required in the era of regimens capable of curing >95% of non-high-risk patients.64,65
Core-binding factor AML accounts for 20-25% of cases of AML in younger patients, but <10% in older adults.2,66 PCR-based MRD assessment of CBFB-MYH11 and RUNX1-RUNX1T1 transcripts for patients with inv(16) and t(8;21), respectively, has been consistently associated with risk of relapse across several studies.32,33,67–70 In the largest study of core-binding factor AML and MRD, 278 patients [163 with t(8;21) and 115 with inv(16)] were treated.33 A >3 log reduction in RUNX1-RUNX1T1 in the bone marrow and CBFB-MYH11 copy number >10 in the peripheral blood after induction were the most prognostic measures of MRD for risk of relapse. Levels of MRD above these thresholds were associated with overall survival in the t(8;21) group but not in the inv(16) group, possibly due to better salvage options fpr patients with the latter subtype. Thresholds of MRD were identified that could predict relapse in 100% of patients in both groups and, notably, rising MRD levels on sequential samples predicted morphological relapse, suggesting a potential role for pre-emptive therapy in this population.
Monitoring of mutant NPM1 throughout treatment may also provide important prognostic information in AML.15,34–36 Mutations in NPM1 are present in approximately 30% of younger patients with AML and in up to 60% of cases of AML with normal karyotype.71 Like PML-RARA, CBFB-MYH11 and RUNX1-RUNX1T1, these mutations are stable at diagnosis and relapse, making them ideal targets for molecular MRD assessment.72 In the largest study of NPM1-based MRD (the National Cancer Research Institute AML17 trial), 346 patients with NPM1-mutated AML received intensive chemotherapy and were monitored for mutant NPM1 levels by PCR in the bone marrow and peripheral blood after each cycle of chemotherapy.15 With a median sensitivity of 10−5, MRD negativity in the peripheral blood after two cycles of chemotherapy was achieved by approximately 85% of patients and was the most prognostic MRD measure. Lack of achievement of this MRD milestone was the only independent prognostic factor for death in multivariate analysis [hazard ratio (HR) 4.84; 95% confidence interval (95% CI): 2.57-9.15; P<0.001). When MRD was assessed in the peripheral blood at this time point, the 3-year overall survival rate was 24% for those who were MRD-positive and 75% for those who were MRD-negative (P<0.001). Similar to the experience with acute promyelocytic leukemia and core-binding factor AML, rising levels of mutant NPM1 transcripts on sequential testing reliably predicted relapse. In a separate study, patients with NPM1 transcripts detectable by PCR immediately prior to HSCT had significantly worse 5-year overall survival rates than those who were MRD-negative (40% vs. 89%: P=0.007); furthermore, the overall survival of those in complete remission prior to HSCT but with detectable NPM1 MRD was similar to that of patients transplanted with active disease.73 Together, these studies suggest that PCR-based MRD for mutant NPM1 is prognostic across clinical contexts, both after induction/early consolidation and before HSCT.
Next-generation sequencing-based measurable residual disease studies
Compared to both MFC and PCR, the use of NGS as an instrument for detecting MRD is relatively new and consequently there are fewer studies to guide its use in clinical practice.17,45,46 In a study of 50 patients who underwent paired whole genome or exon sequencing at diagnosis and remission, 48% had persistent leukemia-associated mutations in at least 5% of bone marrow cells (i.e. variant allelic frequency ≥2.5%).45 These patients with persistent mutations had shorter event-free survival (median: 6.0 vs. 17.9 months, P<0.001) and overall survival (median: 10.5 vs. 42.2 months, P=0.004), compared to those of patients with lower post-remission mutation burden. In a study from the MD Anderson Cancer Center of 131 patients undergoing induction for newly diagnosed AML, complete molecular clearance (i.e. absence of detectable mutations by NGS) was independently associated with decreased risk of relapse and better event-free survival even when MFC-based MRD status was also available.46 These results were strengthened by removing mutations associated with CHIP (i.e. DNMT3A, TET2, and ASXL1, or “DTA” mutations) from the analysis, suggesting that persistence of these preleukemic mutations may not be associated with worse outcomes. In the largest study of NGS MRD in AML, 482 patients were evaluated with NGS at diagnosis and remission, 430 (89.2%) of whom had a leukemia-associated mutation detected at baseline.17 Mutations persisted in 51% of patients at variant allelic frequencies ranging from 0.02-47%. CHIP-associated DTA mutations were not associated with relapse. When DTA mutations were excluded from analysis, residual disease, as detected by NGS, was independently associated with risk of relapse, relapse-free survival and overall survival. Importantly, NGS MRD added prognostic information to MFC MRD status. Four-year relapse rates were similar among patients who were MRD-positive by NGS/MRD-negative by MFC and those who were MRD-negative by NGS/MRD-positive by MFC (52.3% vs. 49.8%, respectively), whereas patients who were MRD-positive or MRD-negative by both assays had highly divergent risks of relapse (73.3% vs. 26.7%, respectively). Together, these studies suggest that NGS-based MRD may complement MFC MRD assays, as these two approaches evaluate different targets (i.e. leukemia-associated mutations and aberrant surface protein expression), and also that reduction and/or clearance of mutations associated with CHIP may not be necessary for cure.17,46
Due to the background error rate of commonly used NGS-based technologies, the sensitivity of these assays is generally no better than ~1%, limiting their discrimination for small amounts of residual disease. In a study of patients with AML undergoing HSCT, an error-corrected, molecular barcoded NGS MRD assay was able to detect residual mutations prior to HSCT with a median variant allelic frequency of 0.33% (range, 0.016%-4.91%), suggesting that such an assay can detect lower levels of MRD than can standard NGS MRD assays.51 Among the 96 evaluable patients, 45% were MRD-positive prior to HSCT using this approach. The 5-year cumulative incidence of relapse was 66% for those who were MRD-positive versus 17% for those who were MRD-negative (HR 5.58; P<0.001). On multivariate analysis, MRD remained an independent predictor of relapse and also overall survival (HR 3.0; P=0.004). Further studies of these more sensitive, error-corrected NGS technologies in other contexts are warranted.
Practical considerations for the evaluation of measurable residual disease in clinical practice
While MRD response has been reliably shown to be predictive for better outcomes, there is significant heterogeneity across studies depending on the specific population of patients, regimen used, AML subtype, MRD method and target, timing of MRD assessment, and threshold to define adequate response, among other factors. All of these variables must be considered when interpreting study results and attempting to generalize to a patient in clinical practice. Despite these challenges, we routinely assess MRD in our clinical practice, as this information helps to refine prognostic assessment and, in some cases, informs therapeutic decisions (e.g. identifies patients who may be candidates for MRD-directed clinical trials or HSCT). Consensus recommendations from the European LeukemiaNet support this approach of routine MRD assessment in AML.6 We also routinely discuss the findings with our patients, particularly when this information is used for clinical decision-making. However, despite the strong association of MRD with clinical outcomes, it is imperative to remember (and to convey to the patient) that “MRD-negativity” does not equate to “cure,” as cumulative incidences of relapse of approximately 20-50% are still observed in “MRD-negative” patients, depending on the assay and clinical context. In contrast, while “MRD-positive” patients almost invariably relapse without subsequent therapy, this is also not an absolute certainty. Thus, an acknowledgment of the limitations of our current MRD technologies is required by the clinician to avoid “over-interpreting” MRD information.
When using MRD in clinical practice, it is important to choose a complementary MRD assay tailored to the AML subtype of a particular patient. For patients with an appropriate target (e.g. those with acute promyelocytic leukemia, core-binding factor AML or NPM1-mutated AML), PCR is preferred to MFC since the clinical data in these populations are more robust for the former and it also has a higher sensitivity.2,6,22 When MFC is used, it is imperative that it is performed in a laboratory with extensive experience in interpretation to ensure accuracy of the assessment.
Whether peripheral blood can replace bone marrow for MRD testing is an area of controversy, although it is generally agreed that an accurate and sensitive peripheral blood-based MRD assay would be welcome to both clinicians and patients, given the less invasive nature of this approach. While some studies have suggested similar findings with these two sources of assay material,74,75 most of the available clinical data on the prognostic and predictive impact of MRD were derived from bone marrow specimens. Furthermore, bone marrow likely provides additional sensitivity, with detectable MRD levels approximately 1-log higher than those in the peripheral blood.6 For example, in core-binding factor AML, a “negative” peripheral blood MRD assay could miss positive bone marrow MRD in up to 40% of cases during therapy and 10-15% of cases during follow up.33 Conversely, in a study of PCR-based MRD in NPM1-mutated AML, discrimination for survival was better when peripheral blood was used than when bone marrow was used.15 Thus, the optimal source for MRD assessment is likely dependent on assay, regimen, and time. Future studies of peripheral blood MRD monitoring are needed across MRD technologies and treatment contexts, and should include investigation of the potential use of circulating cell-free DNA as a source for MRD assays similarly to what has been achieved for some solid tumors.76,77 Initial efforts are already underway in AML.78,79
While the optimal prognostic time point of MRD assessment varies slightly based on the specific regimen and other factors, most studies have shown that detectable MRD is associated with worse outcomes regardless of when it is measured. For patients treated with intensive chemotherapy in the frontline setting, we routinely assess for MRD, at a minimum, at the end of induction and consolidation. It is reasonable to assess MRD at the end of the first cycle of consolidation as well, particularly in patients who remained MRD-positive after induction. In several studies, this time point (i.e. after 2 cycles of chemotherapy) is the most discriminatory for relapse and long-term survival.15,18 MRD status before transplantation is also highly associated with risk of post-HSCT relapse and may identify patients who may be candidates for investigational post-HSCT maintenance strategies.52–55
Therapeutic implications of measurable residual disease in acute myeloid leukemia
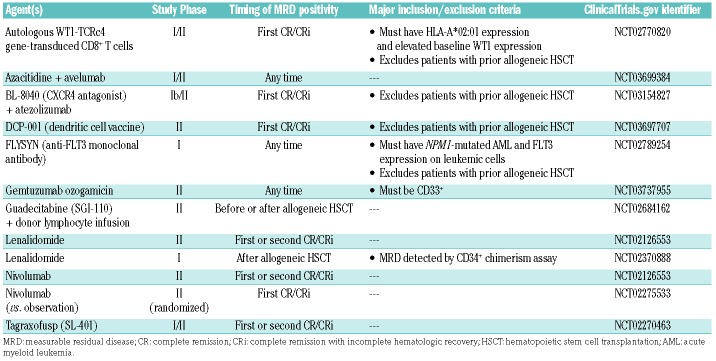
While it is important to obtain MRD information for prognostic purposes, ultimately the goal of developing MRD assays that can more accurately determine risk of relapse in an individual patient with AML is so that post-remission therapies can be tailored accordingly. Currently, risk stratification in AML is largely based on cytogenetic and molecular alterations present at the time of diagnosis; these factors are currently the primary disease-related considerations for deciding to pursue HSCT in first remission.2 In contrast, in acute lymphoblastic leukemia, the decision to pursue HSCT in first remission is determined in large part by MRD status, particularly in the pediatric population.7–9 Furthermore, the CD3-CD19 bispecific T-cell engaging antibody blinatumomab is highly effective in eradicating MRD in patients with acute lymphoblastic leukemia and is approved by the United States Food and Drug Administration for this use.80 While some data support the use of MRD to guide therapeutic decisions in AML, the role of MRD-guided therapies is less clear in AML than in acute lymphoblastic leukemia. With the possible exceptions of allogeneic HSCT and hypomethylating agents, effective treatments for MRD-positive disease have not been established in AML. It has yet to be conclusively shown that eradication of MRD that persists or recurs after initial treatment is associated with improved outcomes in AML; however, given the available evidence (as discussed in detail below), we do frequently use HSCT or hypomethylating agents for patients with MRD-positive disease. Enrollment of these patients in MRD-directed clinical trials with novel agents and combinations is also imperative (Table 2).
Table 2.
Ongoing measurable residual disease-based clinical trials in adults with acute myeloid leukemia.
Measurable residual disease-guided allogeneic hematopoietic stem cell transplantation
While it is common practice to refer patients who are MRD-positive to HSCT, robust data supporting this practice are lacking. Nevertheless, while the outcomes of MRD-positive patients are relatively poor regardless of whether HSCT is or is not performed, several studies suggest that outcomes may be improved with HSCT.14,18,70,81 In a multicenter study of 116 patients with AML harboring t(8;21), patients received either allogeneic HSCT or chemotherapy, with or without autologous HSCT, based on MRD response.70 High-risk patients were defined as those who did not achieve a major molecular response (i.e., a >3-log reduction in RUNX1/RUNX1T1 transcript levels in bone marrow, determined by PCR analysis) after the second consolidation, or who lost the major molecular response within 6 months of achieving it. Among patients in the high-risk group, those who underwent allogeneic HSCT had a lower cumulative incidence of relapse compared to that of the patients who did not undergo allogeneic HSCT (22.1% vs. 78.9%, respectively; P<0.001), which translated into an improvement in overall survival (71.6% vs. 26.7%, respectively; P=0.007). Conversely, the overall survival rate was lower in low-risk patients who underwent allogeneic HSCT (75.7% vs. 100%; P=0.013). These data suggest that MRD could reasonably be used to assign patients with t(8;21) to post-remission therapies. Data for similar approaches in core-binding factor AML with inv(16) are lacking.
The role of allogeneic HSCT in patients with intermediate-risk AML is controversial, and it is possible that MRD may be able to guide post-remission therapeutic decisions in this group.2,4 In a study of younger patients (age 18-60 years) with newly diagnosed NPM1-mutated AML, HSCT was associated with better disease-free survival and overall survival in patients with non-favorable risk disease who had a <4-log reduction in mutant NPM1 in the peripheral blood at the time of remission (disease-free survival: HR, 0.25; 95% CI: 0.06-0.98; P=0.047; overall survival: HR, 0.25; 95% CI: 0.06-0.98; P=0.047), whereas HSCT did not improve outcomes for patients with a >4-log reduction.14 Similarly in the National Cancer Research Institute AML17 study of standard-risk patients without NPM1 mutations, HSCT appeared to preferentially benefit those who remained MRD-positive by MFC after the second cycle of chemotherapy but not those who were MRD-negative, although the interaction between HSCT and MRD status did not reach statistical significance (P=0.16), possibly because of the small number of patients in the cohort who underwent HSCT in first remission.18 Although definitive evidence is lacking, collectively these studies provide some support for the use of MRD to guide post-remission therapies in patients with intermediate-risk AML.
Hypomethylating agents for measurable residual disease-positive acute myeloid leukemia
Although HSCT has historically been the preferred consolidation option for patients with high-risk AML, there is also interest in the use of hypomethylating agents for these patients, including those with detectable MRD. Several studies have suggested that treatment with hypomethylating agents (azacitidine or decitabine) may be beneficial for patients with AML and persistent or recurrent MRD.82–85 The RELAZA2 trial was the largest prospective study to assess this issue.84 One hundred and ninety patients with AML or myelodysplastic syndrome who achieved complete remission after chemotherapy or HSCT were monitored for MRD for 24 months with either PCR for mutant NPM1, leukemia-specific gene fusions (DEK-NUP214, RUNX1-RUNX1T1, or CBFB-MYH11) or donor chimerism in flow cytometry-sorted CD34+ cells (for patients who had undergone prior allogeneic HSCT). Patients who developed detectable MRD received a standard dose of azacitidine for up to 24 cycles. Among the 53 evaluable patients who became MRD-positive and received treatment with azacitidine, 19 (36%) achieved MRD-negativity after 6 months of treatment. Median relapse-free survival and overall survival times for these MRD-positive patients who received azacitidine was 10 months and 31 months, respectively. Importantly, MRD response was associated with improved relapse-free survival (HR, 0.2; 95% CI: 0.1-0.5; P<0.0001) and a trend towards improved overall survival (HR, 0.4; 95% CI: 0.1-1.3; P=0.112). These data suggest that hypomethylating agents may be a reasonable option for patients with MRD-positive AML, particularly those who are not suitable candidates for allogeneic HSCT. Other studies have, however, failed to demonstrate a benefit for the routine use of hypomethylating agents in the maintenance setting.86,87 A phase III randomized study of an oral formulation of azacitidine as maintenance therapy in patients with AML in remission will likely provide further important information in this area (NCT01757535).
Conclusions
Despite substantial heterogeneity across studies of MRD in AML (e.g. variations in AML subtype, MRD methodology and target, cutoff, regimen, and timing of assessment), achievement of MRD negativity has been reliably shown to predict risk of relapse and long-term outcomes. More sensitive assays are still needed, however, as up to 50% of patients who are “MRD-negative” according to commonly applied MRD technologies still relapse. Beyond the important prognostic information that MRD provides, the ultimate goal of assessing MRD is to use this information to appropriately guide decisions regarding post-remission therapies. This approach is, however, strongly dependent on the availability of therapies that are effective in eradicating small amounts of residual leukemia (as with blinatumomab in acute lymphoblastic leukemia). While some studies suggest that allogeneic HSCT or hypomethylating agents may provide benefit for some patients with persistent or recurrent MRD, long-term outcomes with these approaches are largely disappointing. Innovative treatments are therefore needed for these patients. There are several ongoing MRD-directed clinical trials that are evaluating novel immune-based strategies (e.g. checkpoint inhibitors, vaccines, and T-cell-based therapies such as bi-specific T-cell engaging antibodies and chimeric antigen receptor T cells) which may be able to overcome the chemoresistant phenotype associated with MRD-positive disease. Novel combinations with Bcl-2 inhibitors (e.g. venetoclax) may also be promising in this setting. Ultimately, enrollment of high-risk patients into these trials is imperative in order to advance our understanding of how to use MRD information to drive clinical decisions and to improve outcomes of patients with AML.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/8/1532
Funding
Supported by an MD Anderson Cancer Center Support Grant (CA016672).
References
- 1.Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet. 2018;392(10147):593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer RJ, Davis RB, Schiffer CA, et al. Intensive postremission chemotherapy in adults with acute myeloid leukemia. Cancer and Leukemia Group B. N Engl J Med. 1994;331(14):896–903. [DOI] [PubMed] [Google Scholar]
- 4.Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood. 2016;127(1):62–70. [DOI] [PubMed] [Google Scholar]
- 5.Hourigan CS, Karp JE. Minimal residual disease in acute myeloid leukaemia. Nat Rev Clin Oncol. 2013;10(8):460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuurhuis GJ, Heuser M, Freeman S, et al. Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131(12):1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Short NJ, Jabbour E, Albitar M, et al. Recommendations for the assessment and management of measurable residual disease in adults with acute lymphoblastic leukemia: a consensus of North American experts. Am J Hematol. 2019;94(2):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pui CH, Pei D, Coustan-Smith E, et al. Clinical utility of sequential minimal residual disease measurements in the context of risk-based therapy in childhood acute lymphoblastic leukaemia: a prospective study. Lancet Oncol. 2015;16(4):465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vora A, Goulden N, Wade R, et al. Treatment reduction for children and young adults with low-risk acute lymphoblastic leukaemia defined by minimal residual disease (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2013;14(3):199–209. [DOI] [PubMed] [Google Scholar]
- 10.Campana D, Pui CH. Minimal residual disease-guided therapy in childhood acute lymphoblastic leukemia. Blood. 2017;129(14): 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruggemann M, Raff T, Kneba M. Has MRD monitoring superseded other prognostic factors in adult ALL? Blood. 2012;120(23):4470–4481. [DOI] [PubMed] [Google Scholar]
- 12.Freeman SD, Virgo P, Couzens S, et al. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol. 2013;31(32):4123–4131. [DOI] [PubMed] [Google Scholar]
- 13.Terwijn M, van Putten WL, Kelder A, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31(31):3889–3897. [DOI] [PubMed] [Google Scholar]
- 14.Balsat M, Renneville A, Thomas X, et al. Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: a study by the Acute Leukemia French Association Group. J Clin Oncol. 2017;35(2):185–193. [DOI] [PubMed] [Google Scholar]
- 15.Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard-risk AML. N Engl J Med. 2016;374(5):422–433. [DOI] [PubMed] [Google Scholar]
- 16.Ravandi F, Jorgensen J, Borthakur G, et al. Persistence of minimal residual disease assessed by multiparameter flow cytometry is highly prognostic in younger patients with acute myeloid leukemia. Cancer. 2017;123(3):426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378(13):1189–1199. [DOI] [PubMed] [Google Scholar]
- 18.Freeman SD, Hills RK, Virgo P, et al. Measurable residual disease at induction redefines partial response in acute myeloid leukemia and stratifies outcomes in patients at standard risk without NPM1 mutations. J Clin Oncol. 2018;36(15):1486–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for "prime time"? Blood. 2014;124(23):3345–3355. [DOI] [PubMed] [Google Scholar]
- 20.Marcucci G, Mrozek K, Ruppert AS, et al. Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: results from Cancer and Leukemia Group B study 8461. J Clin Oncol. 2004;22(12):2410–2418. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Cortes J, Estrov Z, et al. Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: prognostic significance and the potential role of allogeneic stem-cell transplantation. J Clin Oncol. 2011;29(18):2507–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravandi F, Walter RB, Freeman SD. Evaluating measurable residual disease in acute myeloid leukemia. Blood Adv. 2018;2(11):1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood BL. Principles of minimal residual disease detection for hematopoietic neoplasms by flow cytometry. Cytometry B Clin Cytom. 2016;90(1):47–53. [DOI] [PubMed] [Google Scholar]
- 24.Baer MR, Stewart CC, Dodge RK, et al. High frequency of immunophenotype changes in acute myeloid leukemia at relapse: implications for residual disease detection (Cancer and Leukemia Group B study 8361). Blood. 2001;97(11):3574–3580. [DOI] [PubMed] [Google Scholar]
- 25.Langebrake C, Brinkmann I, Teigler-Schlegel A, et al. Immunophenotypic differences between diagnosis and relapse in childhood AML: implications for MRD monitoring. Cytometry B Clin Cytom. 2005;63(1):1–9. [DOI] [PubMed] [Google Scholar]
- 26.Terwijn M, Zeijlemaker W, Kelder A, et al. Leukemic stem cell frequency: a strong biomarker for clinical outcome in acute myeloid leukemia. PLoS One. 2014;9(9): e107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradbury C, Houlton AE, Akiki S, et al. Prognostic value of monitoring a candidate immunophenotypic leukaemic stem/progenitor cell population in patients allografted for acute myeloid leukaemia. Leukemia. 2015;29(4):988–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeijlemaker W, Grob T, Meijer R, et al. CD34(+)CD38(-) leukemic stem cell frequency to predict outcome in acute myeloid leukemia. Leukemia. 2019;33(5):1102–1112. [DOI] [PubMed] [Google Scholar]
- 29.Ko BS, Wang YF, Li JL, et al. Clinically validated machine learning algorithm for detecting residual diseases with multicolor flow cytometry analysis in acute myeloid leukemia and myelodysplastic syndrome. EBioMedicine. 2018;37:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnett AK, Grimwade D, Solomon E, Wheatley K, Goldstone AH. Presenting white blood cell count and kinetics of molecular remission predict prognosis in acute promyelocytic leukemia treated with all-trans retinoic acid: result of the randomized MRC trial. Blood. 1999;93(12):4131–4143. [PubMed] [Google Scholar]
- 31.Grimwade D, Jovanovic JV, Hills RK, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27(22):3650–3658. [DOI] [PubMed] [Google Scholar]
- 32.Krauter J, Gorlich K, Ottmann O, et al. Prognostic value of minimal residual disease quantification by real-time reverse transcriptase polymerase chain reaction in patients with core binding factor leukemias. J Clin Oncol. 2003;21(23):4413–4422. [DOI] [PubMed] [Google Scholar]
- 33.Yin JA, O’Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120(14):2826–2835. [DOI] [PubMed] [Google Scholar]
- 34.Schnittger S, Kern W, Tschulik C, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114(11):2220–2231. [DOI] [PubMed] [Google Scholar]
- 35.Kronke J, Schlenk RF, Jensen KO, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29(19):2709–2716. [DOI] [PubMed] [Google Scholar]
- 36.Hubmann M, Kohnke T, Hoster E, et al. Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica. 2014;99(8):1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nazha A, Cortes J, Faderl S, et al. Activating internal tandem duplication mutations of the fms-like tyrosine kinase-3 (FLT3-ITD) at complete response and relapse in patients with acute myeloid leukemia. Haematologica. 2012;97(8):1242–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cloos J, Goemans BF, Hess CJ, et al. Stability and prognostic influence of FLT3 mutations in paired initial and relapsed AML samples. Leukemia. 2006;20(7):1217–1220. [DOI] [PubMed] [Google Scholar]
- 39.Smol T, Nibourel O, Marceau-Renaut A, et al. Quantification of EVI1 transcript levels in acute myeloid leukemia by RT-qPCR analysis: a study by the ALFA group. Leuk Res. 2015;39(12):1443–1447. [DOI] [PubMed] [Google Scholar]
- 40.Cilloni D, Renneville A, Hermitte F, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol. 2009;27(31):5195–5201. [DOI] [PubMed] [Google Scholar]
- 41.Gianfaldoni G, Mannelli F, Ponziani V, et al. Early reduction of WT1 transcripts during induction chemotherapy predicts for longer disease free and overall survival in acute myeloid leukemia. Haematologica. 2010;95(5):833–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomdedeu JF, Hoyos M, Carricondo M, et al. Bone marrow WT1 levels at diagnosis, post-induction and post-intensification in adult de novo AML. Leukemia. 2013;27(11): 2157–2164. [DOI] [PubMed] [Google Scholar]
- 43.Parkin B, Londono-Joshi A, Kang Q, Tewari M, Rhim AD, Malek SN. Ultrasensitive mutation detection identifies rare residual cells causing acute myelogenous leukemia relapse. J Clin Invest. 2017;127(9):3484–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanagal-Shamanna R. Digital PCR: principles and applications. Methods Mol Biol. 2016;1392:43–50. [DOI] [PubMed] [Google Scholar]
- 45.Klco JM, Miller CA, Griffith M, et al. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314(8):811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita K, Kantarjian HM, Wang F, et al. Clearance of somatic mutations at remission and the risk of relapse in acute myeloid leukemia. J Clin Oncol. 2018;36(18):1788–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thol F, Gabdoulline R, Liebich A, et al. Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood. 2018;132(16):1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walter RB, Gooley TA, Wood BL, et al. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J Clin Oncol. 2011;29(9):1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter RB, Buckley SA, Pagel JM, et al. Significance of minimal residual disease before myeloablative allogeneic hematopoietic cell transplantation for AML in first and second complete remission. Blood. 2013;122(10):1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Araki D, Wood BL, Othus M, et al. Allogeneic hematopoietic cell transplantation for acute myeloid leukemia: time to move toward a minimal residual disease-based definition of complete remission? J Clin Oncol. 2016;34(4):329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Othus M, Araki D, et al. Pre- and post-transplant quantification of measurable (‘minimal’) residual disease via multiparameter flow cytometry in adult acute myeloid leukemia. Leukemia. 2016;30(7): 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feller N, van der Pol MA, van Stijn A, et al. MRD parameters using immunophenotypic detection methods are highly reliable in predicting survival in acute myeloid leukaemia. Leukemia. 2004;18(8):1380–1390. [DOI] [PubMed] [Google Scholar]
- 57.Boddu P, Jorgensen J, Kantarjian H, et al. Achievement of a negative minimal residual disease state after hypomethylating agent therapy in older patients with AML reduces the risk of relapse. Leukemia. 2018;32(1): 241–244. [DOI] [PubMed] [Google Scholar]
- 58.Buckley SA, Wood BL, Othus M, et al. Minimal residual disease prior to allogeneic hematopoietic cell transplantation in acute myeloid leukemia: a meta-analysis. Haematologica. 2017;102(5):865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shah MV, Jorgensen JL, Saliba RM, et al. Early post-transplant minimal residual disease assessment improves risk stratification in acute myeloid leukemia. Biol Blood Marrow Transplant. 2018;24(7):1514–1520. [DOI] [PubMed] [Google Scholar]
- 60.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82(3):712–715. [PubMed] [Google Scholar]
- 61.Miyamoto T, Nagafuji K, Akashi K, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87(11): 4789–4796. [PubMed] [Google Scholar]
- 62.Diverio D, Rossi V, Avvisati G, et al. Early detection of relapse by prospective reverse transcriptase-polymerase chain reaction analysis of the PML/RARalpha fusion gene in patients with acute promyelocytic leukemia enrolled in the GIMEMA-AIEOP multicenter "AIDA" trial. Blood. 1998;92(3): 784–789. [PubMed] [Google Scholar]
- 63.Sanz MA, Grimwade D, Tallman MS, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113(9):1875–1891. [DOI] [PubMed] [Google Scholar]
- 64.Lo-Coco F, Avvisati G, Vignetti M, et al. Retinoic acid and arsenic trioxide for acute promyelocytic leukemia. N Engl J Med. 2013;369(2):111–121. [DOI] [PubMed] [Google Scholar]
- 65.Abaza Y, Kantarjian H, Garcia-Manero G, et al. Long-term outcome of acute promyelocytic leukemia treated with all-trans-retinoic acid, arsenic trioxide, and gemtuzumab. Blood. 2017;129(10):1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cancer Genome Atlas Research Network. Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boddu P, Gurguis C, Sanford D, et al. Response kinetics and factors predicting survival in core-binding factor leukemia. Leukemia. 2018;32(12):2698–2701. [DOI] [PubMed] [Google Scholar]
- 68.Leroy H, de Botton S, Grardel-Duflos N, et al. Prognostic value of real-time quantitative PCR (RQ-PCR) in AML with t(8;21). Leukemia. 2005;19(3):367–372. [DOI] [PubMed] [Google Scholar]
- 69.Weisser M, Haferlach C, Hiddemann W, Schnittger S. The quality of molecular response to chemotherapy is predictive for the outcome of AML1-ETO-positive AML and is independent of pretreatment risk factors. Leukemia. 2007;21(6):1177–1182. [DOI] [PubMed] [Google Scholar]
- 70.Zhu HH, Zhang XH, Qin YZ, et al. MRD-directed risk stratification treatment may improve outcomes of t(8;21) AML in the first complete remission: results from the AML05 multicenter trial. Blood. 2013;121(20):4056–4062. [DOI] [PubMed] [Google Scholar]
- 71.Falini B, Martelli MP, Bolli N, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117(4):1109–1120. [DOI] [PubMed] [Google Scholar]
- 72.Jain P, Kantarjian H, Patel K, et al. Mutated NPM1 in patients with acute myeloid leukemia in remission and relapse. Leuk Lymphoma. 2014;55(6):1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kayser S, Benner A, Thiede C, et al. Pretransplant NPM1 MRD levels predict outcome after allogeneic hematopoietic stem cell transplantation in patients with acute myeloid leukemia. Blood Cancer J. 2016;6(7):e449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maurillo L, Buccisano F, Spagnoli A, et al. Monitoring of minimal residual disease in adult acute myeloid leukemia using peripheral blood as an alternative source to bone marrow. Haematologica. 2007;92(5):605–611. [DOI] [PubMed] [Google Scholar]
- 75.Zeijlemaker W, Kelder A, Oussoren-Brockhoff YJ, et al. Peripheral blood minimal residual disease may replace bone marrow minimal residual disease as an immunophenotypic biomarker for impending relapse in acute myeloid leukemia. Leukemia. 2016;30(3):708–715. [DOI] [PubMed] [Google Scholar]
- 76.Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 2006;24(26):4270–4276. [DOI] [PubMed] [Google Scholar]
- 77.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15(9):577–586. [DOI] [PubMed] [Google Scholar]
- 78.Assi RE, Albitar M, Ma W, et al. Comparison of somatic mutations profiles from next-generation sequencing (NGS) of cell-free DNA (cfDNA) versus bone marrow (BM) in acute myeloid leukemia (AML). J Clin Oncol. 2018;36(15_suppl):7051–7051. [Google Scholar]
- 79.Nakamura S, Yokoyama K, Shimizu E, et al. Prognostic impact of circulating tumor DNA status post-allogeneic hematopoietic stem cell transplantation in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2018;132(Suppl 1):247. [DOI] [PubMed] [Google Scholar]
- 80.Gokbuget N, Dombret H, Bonifacio M, et al. Blinatumomab for minimal residual disease in adults with B-cell precursor acute lymphoblastic leukemia. Blood. 2018;131(14): 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buccisano F, Maurillo L, Spagnoli A, et al. Cytogenetic and molecular diagnostic characterization combined to postconsolidation minimal residual disease assessment by flow cytometry improves risk stratification in adult acute myeloid leukemia. Blood. 2010;116(13):2295–2303. [DOI] [PubMed] [Google Scholar]
- 82.Sockel K, Wermke M, Radke J, et al. Minimal residual disease-directed preemptive treatment with azacitidine in patients with NPM1-mutant acute myeloid leukemia and molecular relapse. Haematologica. 2011;96(10):1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Platzbecker U, Wermke M, Radke J, et al. Azacitidine for treatment of imminent relapse in MDS or AML patients after allogeneic HSCT: results of the RELAZA trial. Leukemia. 2012;26(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Platzbecker U, Middeke JM, Sockel K, et al. Measurable residual disease-guided treatment with azacitidine to prevent haematological relapse in patients with myelodysplastic syndrome and acute myeloid leukaemia (RELAZA2): an open-label, multi-centre, phase 2 trial. Lancet Oncol. 2018;19(12):1668–1679. [DOI] [PubMed] [Google Scholar]
- 85.Ragon BK, Daver N, Garcia-Manero G, et al. Minimal residual disease eradication with epigenetic therapy in core binding factor acute myeloid leukemia. Am J Hematol. 2017;92(9):845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boumber Y, Kantarjian H, Jorgensen J, et al. A randomized study of decitabine versus conventional care for maintenance therapy in patients with acute myeloid leukemia in complete remission. Leukemia. 2012;26(11): 2428–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blum W, Sanford BL, Klisovic R, et al. Maintenance therapy with decitabine in younger adults with acute myeloid leukemia in first remission: a phase 2 Cancer and Leukemia Group B study (CALGB 10503). Leukemia. 2017;31(1):34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]