Abstract
ZEB1 and ZEB2 are structurally related E-box binding homeobox transcription factors that induce epithelial to mesenchymal transitions during development and disease. As such, they regulate cancer cell invasion, dissemination and metastasis of solid tumors. In addition, their expression is associated with the gain of cancer stem cell properties and resistance to therapy. Using conditional loss-of-function mice, we previously demonstrated that Zeb2 also plays pivotal roles in hematopoiesis, controlling important cell fate decisions, lineage commitment and fidelity. In addition, upon Zeb2 overexpression, mice spontaneously develop immature T-cell lymphoblastic leukemia. Here we show that pre-leukemic Zeb2-overexpressing thymocytes are characterized by a differentiation delay at beta-selection due to aberrant activation of the interleukin-7 receptor signaling pathway. Notably, and in contrast to Lmo2-overexpressing thymocytes, these pre-leukemic Zeb2-overexpressing T-cell progenitors display no acquired self-renewal properties. Finally, Zeb2 activation in more differentiated T-cell precursor cells can also drive malignant T-cell development, suggesting that the early T-cell differentiation delay is not essential for Zeb2-mediated leukemic transformation. Altogether, our data suggest that Zeb2 and Lmo2 drive malignant transformation of immature T-cell progenitors via distinct molecular mechanisms.
Introduction
T-cell development starts with the migration of bone marrow-derived progenitor cells into the thymus. There, these newly arrived early T-cell progenitors rapidly lose their multipotent character and gradually reprogram into the T-cell lineage. T-cell commitment occurs through an orderly process that is tightly regulated by interplay between key signaling pathways and transcription factors.1 Once committed, immature T-cell progenitors undergo successive and dynamic stages of differentiation, including positive selection for the T-cell receptor complex in the cortex, as well as negative selection for removal of potential self-responsive cells in the medulla.2 Alterations in this process can lead to the development of T-cell acute lymphoblastic leukemia (T-ALL). During malignant transformation, a clonal expansion of immature T cells is selected for via the gradual accumulation of advantageous epigenetic changes and genetic mutations.3,4
E-proteins, E2A and HEB, play pivotal roles in early T-cell commitment, but also at later stages of T-cell differentiation.5,6 These widely expressed basic helix-loop-helix (bHLH) transcription factors cooperate with more tissue/lineage-restricted bHLH proteins, such as LYL1 and SCL/TAL1, to form heterodimers that recognize a single 5′-CANNTG-3′ E-box motif in their target promoters and regulatory elements. As such, E-proteins regulate essential T-cell fate-determining factors including Rag1, Notch1, and the interleukin-7 receptor (Il7r). Only in the presence of the LIM-domain-only (LMO) proteins and LMO-binding protein 1 (LDB1) can bHLH heterodimers form larger multiprotein complexes with a second bHLH heterodimer or with other transcription factors, such as GATA proteins. LMO proteins do not have intrinsic DNA-binding capacity and act solely as essential scaffolding proteins for this multiprotein complex formation to bridge bipartite DNA motifs, e.g. two E-boxes.7,8
Activation of LMO1 and LMO2 genes, which results from chromosomal aberrations such as translocations, deletions or insertions in regulatory elements and promoters, has been recurrently observed in patients with TALL.9–11 Mouse models overexpressing Lmo1 and Lmo2 have demonstrated that both are potent oncogenic drivers within the T-cell lineage.12,13 From 5 months of age, CD2-LMO2tg mice spontaneously develop T-ALL with an immature Lyl1+ expression profile.14 Unlike the thymus of normal mice, which is continously replenished by progenitors from the bone marrow, the pre-leukemic thymus of CD2-LMO2tg transgenic mice is self-sustaining.15,16 Similar to other long-lived pre-leukemic stem cells, Lmo2-overex-pressing thymocyte precursors retain the ability to differentiate into the full spectrum of mature daughter cells, but in addition, their stem cell properties allow clonal expansion and subsequent acquisition of extra oncogenic driver mutations, eventually leading to the onset of a fully transformed leukemia. The pre-leukemic self-renewal capacity of CD2-LMO2tg thymocytes is restricted to the CD4−CD8-double-negative (DN) precursor T cells, more specifically the CD4−CD8−CD44−CD25+ (DN3) subpopulation, and strictly depends on the expression of the bHLH protein LYL1.17
Zinc finger E-box binding homeobox transcription factors, ZEB1 and ZEB2, recognize a similar bipartite E-box motif in their target promoters and regulatory elements.18,19 and as such regulate epithelial-to-mesenchymal transition in the context of progression of solid tumors. In addition, ZEB expression has been correlated with the acquisition of cancer stem cell properties.20,21 Using loss-of-function mouse models, it was previously demonstrated that both ZEB proteins are also essential hematopoietic transcription factors that play pivotal roles at various cell fate decision check points during hematopoiesis,22–24 including the T-cell lineage.24–27 In addition, we recently showed that Zeb2 overexpression can result in spontaneous development of T-ALL with an immature Lyl1+ expression profile21 and a latency similar to that in CD2-LMO2tg/+ mice,14 suggesting a common oncogenic mechanism of action. In these mouse T-ALL, Zeb2 overexpression drives increased expression of Il7r and aberrant activation of the IL7R-JAK/STAT signaling pathway.21 Activating IL7R mutations are also recurrently found in T-ALL patients.28 Interestingly, overexpression of gain-of-function mutant variants of IL7R in p19(Arf)−/− mouse hematopoietic progenitors resulted in a similar T-ALL formation with an immature Lyl1+ expression profile, and high levels of Lmo2.29 Based on the high Lmo2 expression in the IL7R mutant tumors and phenotypic similarities with the CD2-LMOtg mouse models, the authors suggested that T-ALL initiation in both models might act via converging downstream signaling pathways that result in aberrant pre-leukemic thymocyte self-renewal.29
In this study, we used transgenic mouse models to further analyze the effects of Zeb1 and Zeb2 overexpression on pre-leukemic T-cell differentiation. In contrast to Zeb1, Zeb2 overexpression resulted in a partial cell-autonomous differentiation delay and accumulation of a DN3 precursor T-cell population, similar to what has been described in the CD2-LMO2tg and IL7R mutant mouse models. However, Zeb2 overexpression was not associated with gain of pre-leukemic self-renewal capacity. Finally, using a late-acting Cre line, we demonstrated that the early T-cell differentiation defects are not essential for Zeb2-mediated T-ALL initiation. Collectively, our data indicate that Zeb2 and Lmo2 drive a similar immature T-ALL subtype, but via distinct oncogenic mechanisms.
Methods
Animal experimentation and handling
All experiments were performed according to the regulations and guidelines of the ethics committee for care and use of laboratory animals of Ghent University and Monash University.
For thymocyte transplantation experiments, donor thymi were dissected under aseptic conditions. Single cells were prepared in cold phosphate-buffered saline using a 40 μM cell strainer. Cell concentrations were measured using a Burker cell counter chamber. Thymocytes (1x 107) were intravenously injected into 6- to 10-week old syngeneic Ly5.1 recipients that were irradiated with a sublethal (550 Rad) dose 4 h before the transplant. One day before and 14 days after the irradiation, mice were kept on neomycin prophylaxis delivered at a dose of 1.7 mg/mL in acidified (pH 2.5) drinking water.
Histology
Tissue samples were fixed in formalin, embedded in paraffin, sectioned at 6 μm and stained with hematoxylin and eosin (H&E) for histopathological examination as described in detail elsewhere.21
Flow cytometry
Both lobes of thymi were carefully dissected in 1 mL of cold phosphate-buffered saline. Single-cell preparations were made using a 40 μm cell strainer and cell numbers were quantified using a Burker cell counting chamber. Cells were stained and analyzed either by LSRII (BD Biosciences) or FACSAria II (BD Biosciences), and FACSDiva or FlowJo software (BD Biosciences). Cell debris and cell aggregates were gated out and dead cells were discarded using the fixable Viability Dye eFluro506 (eBioscience). The antibodies used for flow cytometry are listed in Online Supplementary Table S1. Intracellular staining was done using a BD Cytofix/Cytoperm kit (BD Bioscience) according to the manufacturer’s guidelines.
In vitro differentiation of fetal hematopoietic progenitors towards the T-cell lineage
The differentiation experiments were performed as described previously,30 plating 1 x 104 Lin−cKit+ hematopoietic progenitor cells per 24-well plates of OP9-DL1 feeders in OP9 culture medium supplemented with 5 ng/mL Flt3 ligand (R&D Systems) and 5 ng/mL IL7 (or 1-0.2 ng/mL IL7 where indicated) (R&D Systems or Peprotech).
OP9-DL1 bone marrow stromal cells30 were maintained in MEMalpha medium (Gibco), supplemented with 20% fetal bovine serum (FCS; Hyclone), penicillin (100 U/mL)-streptomycin (100 μg/mL), 2 mM L-glutamine (Gibco) and incubated at 37°C with 7% CO2 and 95% humidity. E13.5 fetal liver cells were stained with biotin-conjugated lineage cocktail antibodies [Gr-1 (Ly-6G & 6C), Ter119 (Ly-76), CD3ε, and B220 (CD45R); eBioscience], PE-conjugated streptavidin (BD Biosciences, 1:200) and CD117-APC (cKit, Immunosource, 1:200). Lin−cKit+ cells were sorted using a FACSAria II machine (BD Biosciences) and FACSDiva software. Dead cells were discarded via DAPI stain.
Real-time quantitative polymerase chain reactions
Total RNA was isolated using a RNeasy Plus Mini Kit (Qiagen). cDNA was synthesized using the First Strand cDNA Synthesis Kit (Roche) with oligo(dT) primers, starting from equal amounts of RNA. Real-time quantitative polymerase chain reactions (qRT-PCR) were performed using the LightCycler 480 SYBR Green I Master (Roche), monitored on a LichtCycler 480 system (Roche) and analyzed using qBase software from Biogazelle. Gene expression was standardized against the housekeeping genes β-actin, glyceraldehyde-3-phosphate dehydrogenase (Gapdh), RPL13 and TBP. All primers used are listed in Online Supplementary Table S2.
Statistical analysis
Data are presented as the mean ± standard deviation. Comparisons between two data groups were performed using a two-sided Student t-test. *P<0.05, **P<0.01, ***P<0.001 (vs. control).
Results
Overexpression of Zeb2 but not Zeb1 induces spontaneous T-cell acute lymphoblastic leukemia in the mouse
We previously demonstrated that hematopoietic-specific overexpression of Zeb2 from the mouse ROSA26 locus resulted in spontaneous T-ALL development.21 To document the functional similarities and/or differences between the two ZEB family members, we recently generated a similar Zeb1 conditional overexpression mouse model using the same targeting strategies as previously described for Zeb231 (Online Supplementary Figure S1A). In these R26-Zeb1tg mice, an aminoterminal HA-tag Zeb1 cDNA, preceded by a floxed transcriptional stop cassette and followed by an IRES-eGFP, was targeted to the ROSA26 locus. The details on how these mice were generated have been submitted for publication elsewhere.
To compare the oncogenic potential of Zeb1 and Zeb2, we crossed these newly generated R26-Zeb1tg mice to the same Cre line as we used for the R26-Zeb2tg, the Tie2-cre, which targets hematopoietic stem cells and their progeny.32 This resulted in a moderate 2- to 3-fold overexpression of the total Zeb1 mRNA levels in the thymus, similar to that in the R26-Zeb2tg mice (Online Supplementary Figure S1B). These Tie2cre, R26-Zeb1tg/tg mice (henceforth referred to as R26-Zeb1tg/tg) were born in normal Mendelian ratios and no obvious phenotypes were observed at a young age. While we previously reported that mice overexpressing Tie2-cre, R26-Zeb2tg/tg (henceforth referred to as R26-Zeb2tg/tg) spontaneously develop T-ALL starting from the age of 5 months with a penetrance of 53% at 15 months of age, none of the R26-Zeb1tg/tg-overexpressing mice (n=15) showed any signs of leukemia development (Online Supplementary Figure S1C). These data indicate that, in contrast to Zeb2, Zeb1 is not an oncogenic driver of murine T-ALL.
Overexpression of Zeb2 results in thymic hypoplasia and delayed T-cell development
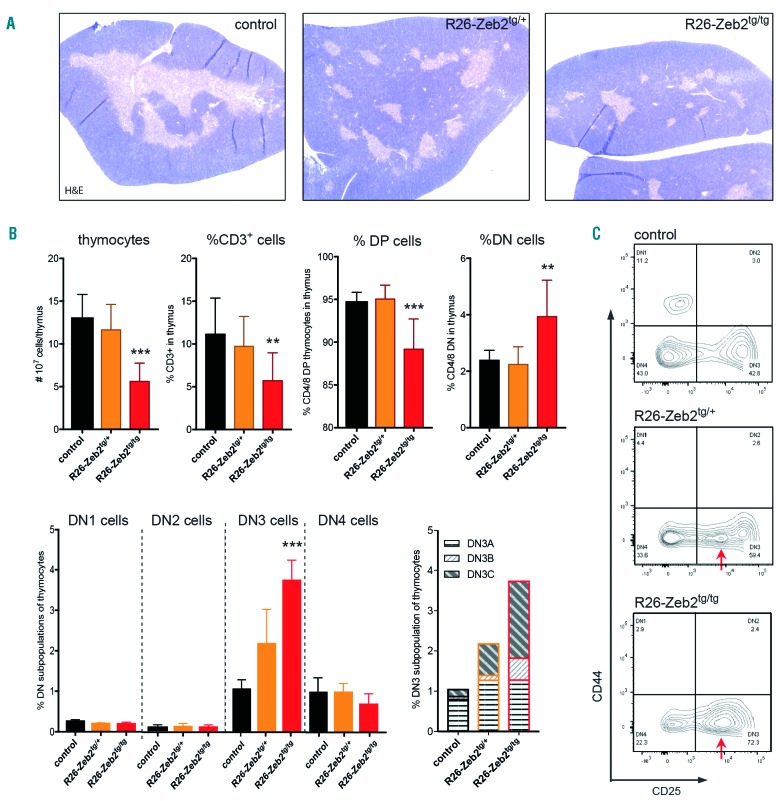
Upon dissection, we noted that juvenile Zeb2-overex-pressing mice, and not Zeb1-overexpressing mice, consistently had smaller thymi compared to their littermate controls. Except for a reduction of the medullary areas, no significant changes were observed in thymus architecture (Figure 1A). Next, detailed flow cytometric analysis was performed on thymi of 8-week old R26-Zeb2tg versus control Cre-negative littermates. Upon Zeb2 overexpression, a significant and dose-dependent decrease in total thymocyte numbers was observed, associated with a significant decrease in percentages and absolute numbers of mature CD3+ and CD4+CD8+ double-positive Thy1+ cell populations, combined with increased percentages of the immature CD4−CD8− double negative (DN) Thy1+ population (Figure 1B). These findings indicate that the thymic hypoplasia in R26-Zeb2tg animals results from a partial block or delay at an early stage during T-cell development. Within the R26-Zeb2tg DN-gated cells, an abnormal population of CD44− cells with intermediate levels of CD25 was observed, which was absent in the control thymus (Figure 1C). These cells are Thy1+, CD49b−, B220− and CD19−, confirming their T-cell lineage identity. Positive staining for CD28 and intracellular TCRβ indicated that these are early post-β-selected cells blocked or delayed at the DN3 to DN4 transition, more specifically at DN3C. In line with this notion, expression analysis of mouse and human T-cell populations (Online Supplementary Figure S2) revealed that endogenous ZEB2 levels are normally downregulated at this DN3-4 transition point.
Figure 1.
Zeb2 overexpression results in a delay in T-cell development at the DN3 stage. (A) Hematoxylin & eosin-stained sections of paraffin-embedded thymi of mice with and without mono- or bi-allelic ROSA26-mediated overexpression. (B) Flow cytometric analysis of thymi of Zeb2-overexpressing mice versus control littermates. Absolute numbers of total thymocytes and percentages of CD3, CD4/8 double-positive (DP) and CD4/8 double-negative (DN) cell populations, DN1-4 and DN3A-C subpopulations are shown. (C) Representative dot plots of CD4/8 DN populations stained with CD25 and CD44. In R26-Zeb2tg/+ and R26-Zeb2tg/tg thymi, an abnormal DN3/4-like population is observed with intermediate CD25 levels (arrows). *P<0.05, **P<0.01, ***P<0.001 (vs control).
Notably, no T-cell differentiation block or delay was seen in the Zeb1-overexpressing mice (Online Supplementary Figure S1D), indicating that Zeb2 overexpression has effects on early T-cell differentiation that differ from those of Zeb1 overexpression.
The Zeb2-mediated differentiation delay is a hematopoietic cell-autonomous phenotype
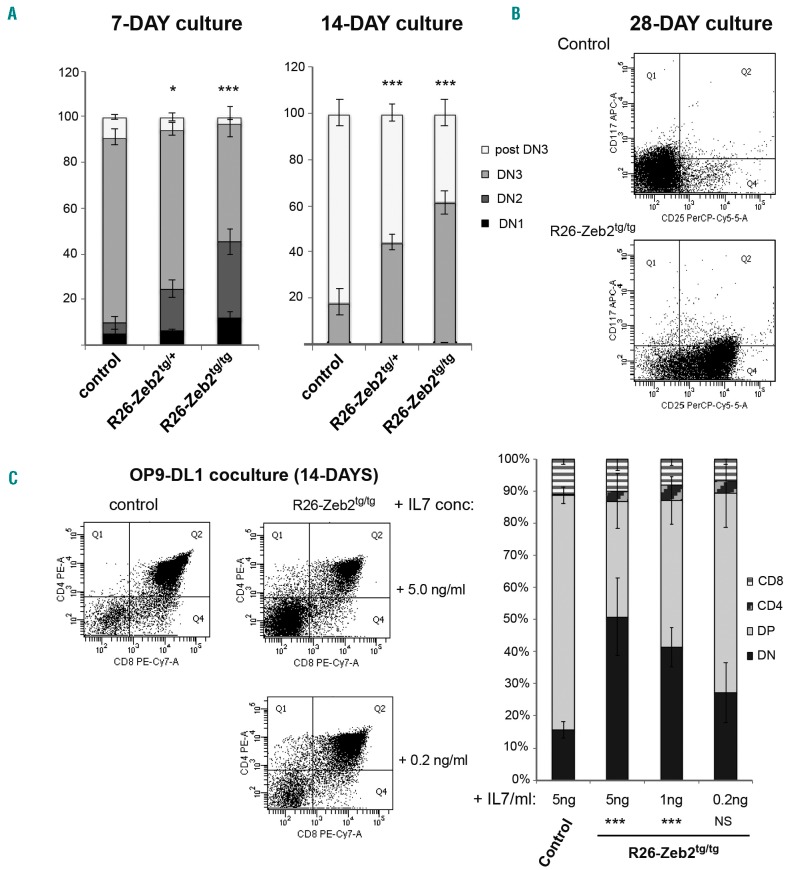
As the Tie2-cre line also targets endothelial cells next to hematopoietic cells, we wanted to exclude that paracrine changes in the thymic microenvironment or architecture cause the delay in T-cell differentiation in R26-Zeb2tg mice. To this end, we performed in vitro T-cell differentiation experiments with purified hematopoietic progenitor cells. Sorted fetal liver hematopoietic progenitors (Lin−, cKit+) from R26-Zeb2tg/tg or Cre-negative control E13.5 embryos were seeded on OP9-DL1 bone marrow stromal cells that express the Notch delta ligand-1,30 in the presence of 5 ng/mL Flt3 ligand and 5 ng/mL IL7. In vitro T-cell development was monitored for 4 weeks. A significant delay in T-cell development was already prominent within 1 week in the Zeb2-overexpressing co-cultures (Figure 2A), exemplified by an increased frequency of cKit+ DN cells. After 2 weeks, an increase of the DN3-like population (CD3−,CD8−,CD44−,CD25int) and a concomitant decrease in more mature cells (CD8+) were also observed, pointing to a hematopoietic cell-autonomous delay or block in early T-cell development upon Zeb2 overexpression (Figure 2A). After 4 weeks of culture, a large proportion of the R26-Zeb2tg/+ cells remained DN3 with intermediate levels of CD25, whereas all T cells had already differentiated beyond the DN stage in the control cultures (Figure 2B).
Figure 2.
Zeb2-mediated T-cell differentiation delay is independent of the thymic mirco-environment. (A,B) Flow cytometric analysis of 1- and 2-week in vitro differentiation cultures of E13.5 fetal liver hematopoietic progenitors on OP9-DL1 feeders. Percentages of DN1 (CD4/8− and CD44+, CD25−), DN2 (CD4/8− and CD44+, CD25+), DN3 (CD4/8−, CD44−, CD25+) and post-DN3 (sCD3 or CD8+) populations show a significant delay in differentiation upon Zeb2 overexpression (A) and the presence of a cell population with intermediate CD25 expression even after 4 weeks of co-culture, as exemplified by a representative dot plot (B). (C) Lowering the concentration of recombinant interleukin 7 (IL7) in OP9-DL1 co-cultures partially rescues the delay in T-cell differentiation of R26-Zeb2tg/tg fetal liver hematopoietic progenitors. Dot plots of representative cultures are shown as are the percentages of T-cell populations following addition of 5 ng/mL, 1 ng/mL and 0.2 ng/mL recombinant IL7. *P<0.05, **P<0.01, ***P<0.001 (vs. control).
Zeb2-mediated increased interleukin-7 receptor signaling is involved in the T-cell differentiation delay
In our previous study, we demonstrated that ZEB2 directly regulates the Il7r promoter.33 Consequently, Zeb2-overex-pressing T-ALL tumors and derived cell lines have high levels of Il7r mRNA and are more responsive to exogenous IL7 levels. As downregulation of IL7 signaling, induced by the pre-TCR signal, is crucial for normal transition of DN3 to double-positive cells,34,35 we hypothesized that the Zeb2-induced delay in T-cell differentiation is due to their inability to downregulate Il7r. We therefore tested whether lowering the amount of recombinant IL7 in the culture medium could overcome the observed delay in T-cell differentiation in vitro. Indeed, reduction of the concentration of recombinant IL7 from 5 ng/mL to 0.2 ng/mL rescued the delay in differentiation, resulting in similar developmental kinetics to that of the control co-cultures (Figure 2C).
ZEB2 overexpression does not induce pre-leukemic thymocyte self-renewal
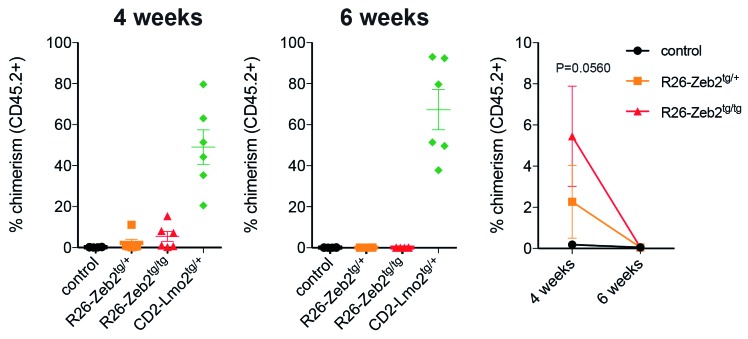
For the following reasons, we subsequently hypothesized that thymocytes would also acquire pre-leukemic self-renewal capacity upon Zeb2 overexpression, similar to what has been observed in CD2-LMO2tg mice: (i) mice with Zeb2 overexpression phenocopy the IL7R gain-of- function mutant and CD2-LMOtg immature T-ALL mouse models; (ii) ZEB2 and LMO2 bind similar bipartite E-box-containing regulatory elements; (iii) ZEB2 expression has been associated with acquisition of cancer stem cell properties in solid tumors; and (iv) Zeb2-overexpressing T-ALL tumors and their derived cell lines have increased leukemia-initiating potential. To test our hypothesis, we backcrossed our R26-Zeb2tg model for ten generations to pure C57BL/6 background (CD45.2+) mice to enable thymocyte transplantation experiments. We injected 1x107 thymocytes from 8-week old R26-Zeb2tg/tg mice or littermate controls into the tail vein of sublethally irradiated syngeneic Ly5.1 (CD45.1+) recipients and used CD2-LMO2tg thymocytes as the positive control. Via flow cytometric analysis of CD45.1 versus CD45.2, we analyzed how many of the donor versus recipient cells contributed to the repopulation of the thymus at 4 and 6 weeks after transplantation. Four weeks after transplantation, no CD45.2+ wildtype thymocytes could be detected, while varying low numbers of CD45.2+ Zeb2-overexpressing thymocytes were consistently seen at this time point. However, and in contrast to the CD2-LMO2tg thymocytes, none of the Zeb2-overexpressing CD45.2 thymocytes could be detected 6 weeks after transplantation (Figure 3). These data demonstrate that Zeb2-overexpressing thymocytes have prolonged survival capacity compared to that of wildtype thymocytes, but that they do not have long-term repopulation or self-renewal capacity.
Figure 3.
Zeb2 overexpression does not induce pre-leukemic thymocyte self-renewal. Percentages of CD45.2+ donor-derived thymocytes that contribute to the repopulation of sublethally irradiated syngeneic CD45.1+ recipients 4 or 6 weeks after transplantation.
The differentiation delay is not essential for the formation of Zeb2-mediated T-cell acute lymphoblastic leukemia
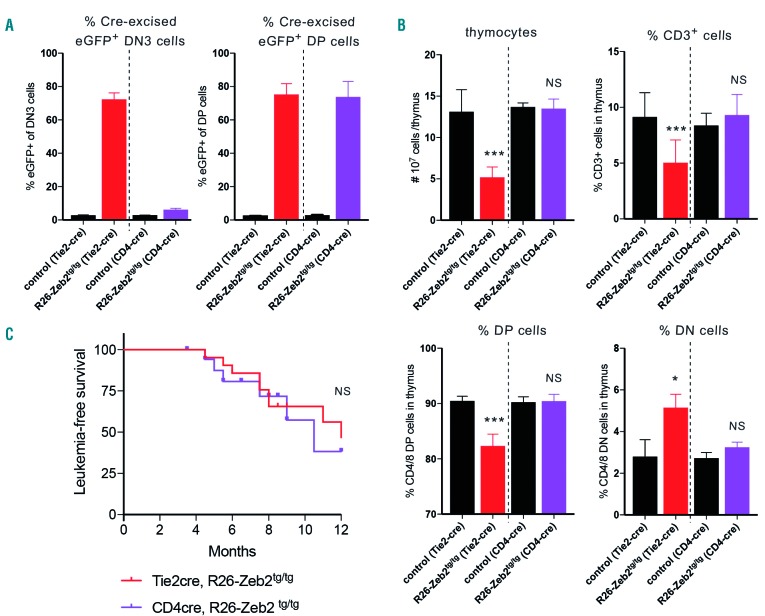
Finally, we wondered whether the observed T-cell differentiation defect in Zeb2-overexpressing mice is essential for Zeb2-mediated T-ALL formation. To this end, we crossed our R26-Zeb2tg mice with the CD4-cre, a late-acting T-cell-specific Cre line that is only active after the above-described DN3C block/delay (Figure 4A). No thymocyte hypoplasia (Figure 4B), or early T-cell differentiation defects could be observed in 8-week old CD4-cre, R26-Zeb2tg/tg mice versus control littermates. Irrespective of the lack of this early T-cell differentiation block, these mice developed leukemia with approximately the same latency and same penetrance (Figure 4C) as observed with the early acting Tie2-cre line. Necropsy and histopatho-logical examination was done on eight Tie2-cre,R26-Zeb2tg/tg and five CD4-cre,R26-Zeb2tg/tg mice, as described in our previous study.21 All leukemic mice were diagnosed with precursor T-cell lymphoblastic leukemia. In general, no immunophenotypic differences were observed between the leukemias developing in the early- versus late-acting Cre line. However, we noted that while all examined Tie2-cre,R26-Zeb2tg/tg mice had large mediastinal masses, two out of the five CD4-cre,R26-Zeb2tg/tg mice had no mediastinal mass. Furthermore, in these two leukemic mice, the neoplastic cells appeared smaller. Since the phenotype in both models is not fully penetrant and we observed large variation in leukemia onset and phenotype, only a limited number (8 vs. 5) leukemic mice could be analyzed fully and we cannot, therefore, draw strong conclusions from our observations.
Figure 4.
Zeb2-mediated T-cell differentiation delay is not essential for Zeb2-induced T-cell acute lymphoblastic leukemia formation. (A) Flow cytometric analysis of the percentages of eGFP+ cells in DN3 versus DP demonstrating differential Cre activity between the early-acting Tie2-cre line and the late-acting CD4-cre line in early T-cell precursors. (B) Flow cytometric analysis of thymi of Zeb2-overexpressing mice versus control littermates upon intercross with either the Tie2-cre or CD4-cre line. Absolute numbers of total thymocytes and percentages of CD3, CD4/8 double-positive (DP) and CD4/8 double-negative (DN) cell populations demonstrate that there is no differentiation delay in the late-acting CD4-cre line. (C) Kaplan-Meier curve for leukemia-free survival of CD4-cre, R26-Zeb2tg/tg (n=18) versus Tie2-cre, R26-Zeb2tg/tg (n=21) mice. *P<0.05, **P<0.01, ***P<0.001 (vs. control); NS, not significant.
Discussion
In various solid cancer types, ZEB expression has been correlated with both poor prognosis and patients’ outcomes. Indeed, ZEB1 and ZEB2 have been demonstrated to increase cancer cell invasion and dissemination via the regulation of epithelial-to-mesenchymal transition. In addition, as for other regulators of epithelial-to-mesenchymal transition, their expression has been associated with the acquisition of cancer stem cell properties and therapy resistance.36
It has been debated whether these structurally very related family members compensate for each other or whether they also have unique functions. The differences in phenotypes between Zeb1 and Zeb2 knockout mice can be explained by their complementary expression patterns.37 Studies with compound Zeb1/Zeb2 double knockouts have shown that they have at least partly overlapping, compensatory functions, but could not rule out that they also have unique functions. Overexpression of ZEB1/ZEB2 in various epithelial cancer cell lines catalyzed similar phenotypes, with overlapping downstream targets.18,38,39 Mild differences between ZEB1/2-mediated epithelial-to-mesenchymal transition were often reported, but these could be attributed to differences in overexpression levels and/or the cellular context. Here, we provide compelling evidence, for the first time, that ZEB1 and ZEB2 can have unique functions. Overexpression in the same cellular context, using the same targeting strategy, expressed from the same endogenous promoter and in a relevant in vivo setting, resulted in clearly distinct phenotypes for ZEB1 and ZEB2. Using luciferase reporter constructs, others have previously shown that ZEB1 and ZEB2 also have opposing effects on TGFβ1-mediated repression of the 3TP and p21 promoter elements.40 At that time, the authors hypothesized that these seemingly opposite effects were mediated by the putative differential recruitment of ZEB1/2-specific co-activators/repressors, such as p300 and P/CAF. This differential recruitment of co-factors may specifically switch ZEB1 from a repressor into an activator. However, others have counteracted this hypothesis by demonstrating that ZEB1 and ZEB2 are equally potent at binding p300 and P/CAF. Furthermore, although the extensive list of ZEB-interacting proteins is growing continuously, no unique interactors have been documented thus far. In this study, we prove that the distinct phenotypes observed are not due to differences in cellular contexts or mRNA (over)expression levels. Unfortunately, no instruments are available to test whether these similar mRNA levels also translate into similar transgene protein levels. Cell-context-dependent differences in post-translational regulation of both ZEB family members may result in more or less of the ZEB1/2 transgene protein and may explain the phenotypic differences observed between the two models.
It is important to mention here that although early T-cell development of R26-Zeb1tg mice is normal and these mice do not spontaneously develop leukemia, this does not exclude that other hematopoietic lineages are not affected by the Zeb1 overexpression.
In the context of T-cell malignancies, ZEB1 and ZEB2 seem to act in an opposite manner. A tumor suppressor role for ZEB1 in T-cell leukemias/lymphomas has previously been suggested,41 based on its expression, the mutations found in patients and the observed spontaneous T-cell lymphoma development in a mouse model expressing a carboxyterminal truncated form of ZEB1. This is in contrast with the oncogenic role we have described for Zeb2 in T-ALL.21 In this study, we demonstrated that pre-leukemic T-cell differentiation is also affected by Zeb2 overexpression. Maintenance of Zeb2 expression results in a partial differentiation block or delay at an early DN3C stage, coinciding with the T-cell developmental stage at which Zeb2 expression is normally downregulated in mice, as well as in humans. This delay could be partially rescued by decreasing IL7R pathway activation, suggesting that the delay is caused by an inability of the Zeb2-overexpressing thymocytes to downregulate IL7R expression. Interestingly, other mouse models have shown a similar block in T-cell differentiation at this key transition point, including mice that overexpress the IL7r.35,42 CD2-LMO2tg animals accumulate a similar aberrant DN3 population at a young age. Notably, our detailed flow cytometric analysis revealed that LMO2-overexpressing cells are delayed slightly earlier in T-cell differentiation compared to the Zeb2-overexpressing thymocytes, at the DN3A stage, and before T-cell rearrangement (data not shown).
As ZEB have been previously associated with the acquisition of cancer stem cell properties, we hypothesized that the cause of spontaneous T-ALL formation in R26-Zeb2tg could be due to acquired self-renewal of pre-leukemic thymocytes, as previously demonstrated for LMO2. This was supported by the fact that both proteins bind similar regulatory elements, and that both factors can drive murine T-ALL development with a similar immature expression profile and increased stem cell properties. Nevertheless, using thymocyte transplantation experiments, we demonstrated that R26-Zeb2tg thymocytes are not able to reconstitute an irradiated thymus and therefore have no pre- leukemic self-renewal capacity. These data indicate different oncogenic mechanism for ZEB2 and LMO2. Upon transplantation, we observed that the survival of R26-Zeb2tg thymocytes was longer than that of wildtype thymocytes, with this difference most probably being associated with increased IL7R signaling. Whether the increase in Il7r expression is involved in tumor initiation in R26-Zeb2tg animals remains to be determined.
Finally, we used a late-acting CD4-Cre line to show that Zeb2 could also transform thymic precursor cells at later stages during their development, suggesting that the delay at the DN3C stage is not essential for leukemic transformation. Interestingly, a forward genetic screening used the same CD4-Cre line to activate the “sleeping beauty transposase” and induce oncogenic transformation specifically at later stages of T-cell development, which resulted in TALL with an immature expression profile.43 These data indicate that cases of immature murine T-ALL can also originate in cells beyond the DN stage. Furthermore, our data suggest that a Zeb2-transformed T-ALL with an immature/stem cell expression profile can originate from a more differentiated cell without self-renewal capacity.
In summary, we conclude that multiple oncogenes such as ZEB2 and LMO2 are able to induce subtypes of immature murine T-ALL via distinct oncogenic mechanisms of action. In addition, Zeb2 can also drive T-cell transformation in more differentiated T-cell precursor cells with similar kinetics.
Acknowledgments
This work was supported by the Australian NHMRC (grant 1047995 to JJH), the Worldwide Cancer Research Fund, the Swiss Bridge Foundation, Stand Up Against Cancer Fund, the Ghent University Special Research Fund and the Fund for Scientific Research-Flanders.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/8/1608
References
- 1.Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186(12):6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat Immunol. 2012;13(2):121–128. [DOI] [PubMed] [Google Scholar]
- 3.Meijerink JP. Genetic rearrangements in relation to immunophenotype and outcome in T-cell acute lymphoblastic leukaemia. Best Pract Res Clin Haematol. 2010;23(3):307–318. [DOI] [PubMed] [Google Scholar]
- 4.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3398–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol. 2001;1(3):193–199. [DOI] [PubMed] [Google Scholar]
- 6.Wang LH, Baker NE. E Proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell. 2015;35(3):269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadman I, Li J, Bash RO, et al. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994;13(20):4831–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wadman IA, Osada H, Grutz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16(11):3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991;88(10):4367–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Vlierberghe P, van Grotel M, Beverloo HB, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108(10): 3520–3529. [DOI] [PubMed] [Google Scholar]
- 11.Goossens S, Van Vlierberghe P. Novel oncogenic noncoding mutations in T-ALL. Blood. 2017;129(24):3140–3142. [DOI] [PubMed] [Google Scholar]
- 12.Fisch P, Boehm T, Lavenir I, et al. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7(12):2389–2397. [PubMed] [Google Scholar]
- 13.Neale GA, Rehg JE, Goorha RM. Ectopic expression of rhombotin-2 causes selective expansion of CD4-CD8- lymphocytes in the thymus and T-cell tumors in transgenic mice. Blood. 1995;86(8):3060–3071. [PubMed] [Google Scholar]
- 14.Smith S, Tripathi R, Goodings C, et al. LIM domain only-2 (LMO2) induces T-cell leukemia by two distinct pathways. PloS One. 2014;9(1):e85883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack MP, Young LF, Vasudevan S, et al. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327(5967):879–883. [DOI] [PubMed] [Google Scholar]
- 16.Cleveland SM, Smith S, Tripathi R, et al. Lmo2 induces hematopoietic stem cell-like features in T-cell progenitor cells prior to leukemia. Stem Cells. 2013;31(5):882–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormack MP, Shields BJ, Jackson JT, et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood. 2013;122(12):2093–2103. [DOI] [PubMed] [Google Scholar]
- 18.Comijn J, Berx G, Vermassen P, et al. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol Cell. 2001;7(6):1267–1278. [DOI] [PubMed] [Google Scholar]
- 19.Remacle JE, Kraft H, Lerchner W, et al. New mode of DNA binding of multi-zinc finger transcription factors: delta EF1 family members bind with two hands to two target sites. EMBO J. 1999;18(18):5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12): 1487–1495. [DOI] [PubMed] [Google Scholar]
- 21.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goossens S, Janzen V, Bartunkova S, et al. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 2011;117(21):5620–5630. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Riedt T, Goossens S, et al. The EMT transcription factor Zeb2 controls adult murine hematopoietic differentiation by regulating cytokine signaling. Blood. 2017;129(4):460–472. [DOI] [PubMed] [Google Scholar]
- 24.Higashi Y, Moribe H, Takagi T, et al. Impairment of T cell development in delta EF1 mutant mice. J Exp Med. 1997;185(8): 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omilusik KD, Best JA, Yu BF, et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8(+) effector and memory T cell populations during infection. J Exp Med. 2015;212(12):2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominguez CX, Amezquita RA, Guan TX, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med. 2015;212(12):2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan T, Dominguez CX, Amezquita RA, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8(+) T cell fates. J Exp Med. 2018;215(4):1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treanor LM, Zhou S, Janke L, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med. 2014;211(4):701–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zuniga-Pflucker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat Immunol. 2004;5(4):410–417. [DOI] [PubMed] [Google Scholar]
- 31.Tatari MN, De Craene B, Soen B, et al. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell Mol Life Sci. 2014;71(18):3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–242. [DOI] [PubMed] [Google Scholar]
- 33.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Kassar N, Lucas PJ, Klug DB, et al. A dose effect of IL-7 on thymocyte development. Blood. 2004;104(5):1419–1427. [DOI] [PubMed] [Google Scholar]
- 35.Munitic I, Williams JA, Yang Y, et al. Dynamic regulation of IL-7 receptor expression is required for normal thymopoiesis. Blood. 2004;104(13):4165–4172. [DOI] [PubMed] [Google Scholar]
- 36.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–591. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi T, Maruhashi M, Van De Putte T, Kondoh H, Huylebroeck D, Higashi Y. Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev Dyn. 2006;235(7):1941–1952. [DOI] [PubMed] [Google Scholar]
- 38.Vandewalle C, Comijn J, De Craene B, et al. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33(20):6566–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eger A, Aigner K, Sonderegger S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14): 2375–2385. [DOI] [PubMed] [Google Scholar]
- 40.Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGF beta/BMP signaling pathway. EMBO J. 2003;22(10): 2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hidaka T, Nakahata S, Hatakeyama K, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112(2): 383–393. [DOI] [PubMed] [Google Scholar]
- 42.Trigueros C, Hozumi K, Silva-Santos B, et al. Pre-TCR signaling regulates IL-7 receptor alpha expression promoting thymocyte survival at the transition from the double-negative to double-positive stage. Eur J Immunol. 2003;33(7):1968–1977. [DOI] [PubMed] [Google Scholar]
- 43.Berquam-Vrieze KE, Nannapaneni K, Brett BT, et al. Cell of origin strongly influences genetic selection in a mouse model of TALL. Blood, 2011;118(17):4646–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]