Abstract
RUNX1 is a key transcription factor in hematopoiesis and its disruption is one of the most common aberrations in acute myeloid leukemia. RUNX1 alterations affect its DNA binding capacity and transcriptional activities, leading to the deregulation of transcriptional targets, and abnormal proliferation and differentiation of myeloid cells. Identification of RUNX1 target genes and clarification of their biological functions are of great importance in the search for new therapeutic strategies for RUNX1-altered leukemia. In this study, we identified and confirmed that KLF4, a known tumor suppressor gene, as a direct target of RUNX1, was down-regulated in RUNX1-ETO leukemia. RUNX1 bound to KLF4 promoter in chromatin to activate its transcription, while the leukemogenic RUNX1-ETO fusion protein had little effect on this transactivation. KLF4 was also identified as a novel binding partner of RUNX1. RUNX1 interacted with KLF4 through Runt domain and further co-activated its target genes. However, RUNX1-ETO competed with RUNX1 to bind KLF4 through Runt and ETO domains, and abrogated transcription of KLF4. Finally, overexpression experiments indicated that RUNX1 inhibited proliferation and induced apoptosis of t(8;21) leukemia cells via KLF4-mediated upregulation of P57. These data suggest KLF4 dysregulation mediated by RUNX1-ETO enhances proliferation and retards apoptosis, and provides a potential target for therapy of t(8;21) acute myeloid leukemia.
Introduction
RUNX1, also known as AML1 and CBFα, is a critical transcription factor in hematopoiesis, which regulates various kinds of hematopoiesis-related genes, including cytokines, cytokine receptors, microRNA and other transcription factors.1 It is very versatile and also interacts with a number of other hematopoietic regulators, such as CBFβ, PU.1, GATA1, PAX5, and ETS1.2–6 These interaction partners provide RUNX1 with the potential to target genes primarily regulated by other transcription factors, and vice versa. RUNX1 is frequently involved in gene mutations and chromosomal translocations in leukemias, which indicates that the altered function of RUNX1 is closely related with leukemogenesis. Among them, t(8;21)(q22;q22) is one of the most common chromosomal translocations in acute myeloid leukemia (AML), which results in RUNX1-ETO fusion protein. RUNX1-ETO fuses the N-terminus of RUNX1 including only runt domain (RHD) in-frame with the almost entire ETO protein. This leukemogenic fusion protein competes with wild-type RUNX1 in binding to its target genes and recruits a transcriptional co-repressor complex NCoR/SMRT/HDAC to further repress transcription of RUNX1 target genes.7–9 The dominant negative repressive effects of RUNX1-ETO on RUNX1 is considered to be the major pathogenic mechanism of t(8;21) AML, which causes blockage of normal hematopoietic differentiation and accumulation of immature myelocytes.10
A large number of studies on RUNX1 and RUNX1-ETO have been performed to investigate their roles in normal hematopoiesis and leukemogenesis. However, the mechanisms by which RUNX1 and RUNX1-ETO regulate their target genes and interact with binding partners are far from clear. Identification of RUNX1 and RUNX1-ETO novel target genes and interacting proteins are still of great importance in order to develop new therapeutic strategies for t(8;21) leukemia.
Our previous study reported that sodium phenylbutyrate (PB), one of the HDAC inhibitors, could induce t(8;21) leukemia cells to undergo differentiation and apoptosis.11 In another study, we identified PIG7 as a direct target gene of RUNX1 that was responsible for differentiation and apoptosis induction of t(8;21) leukemia cells.12 In addition to PIG7, a series of genes were also up-regulated in the process, including RUNX1, KLF4 and P57. Among them, KLF4, a transcription factor frequently deregulated in a variety of malignant tumors, including several examples in hematologic malignancies, was considered a tumor suppressor. In AML, KLF4 was negatively regulated by HDAC1 and overexpression of KLF4 could markedly repress proliferation of AML cells by blocking cell cycles and inducing the expression of P21 and P27.13 In T-cell acute lymphoblastic leukemia, KLF4 delayed disease progression through directly inhibiting T-cell associated genes, such as NOTCH1, BCL11B, GATA3 and TCF7, and activating the BCL2/BCLXL pathway.14 Another study by Morris et al. reported that KLF4-mediated anti-leukemic effects through regulation of microRNA networks, including MIR-150 and the MYC/MAX/MXD network, providing novel mechanistic support for AML treatment via increasing KLF4 expression.15
In this report, we identified KLF4 as both a novel target gene and a binding partner of RUNX1, which induced the expression of cell cycle inhibitor P57. We further confirmed that overexpression of either of RUNX1, KLF4 or P57 could inhibit proliferation and induce apoptosis of t(8;21) leukemia cell. Our results support the hypothesis that RUNX1, KLF4 and P57 might compose a transcriptional activation cascade in t(8;21) leukemia cells. RUNX1 inhibited proliferation and induced apoptosis of t(8;21) leukemia cells via KLF4-mediated upregulation of P57. We believe that reactivating the “RUNX1-KLF4-P57” signaling pathway might be a potentially potent therapeutic strategy for t(8;21) AML.
Methods
Cell culture
All cell lines used in this study were purchased from ATCC (Manassas, VA, USA). Kasumi-1 and SKNO-1 cells were cultured in RPMI-1640 medium (Gibco-Life Technologies, Grand Island, NY, USA) supplemented with 20% fetal bovine serum (FBS) (Gibco). SKNO-1 was also supplemented with 10 ng/mL recombinant human granulocyte-macrophage colony-stimulating factor (Peprotech, USA). CV-1 and HEK-293T cells were maintained in DMEM (Gibco) supplemented with 10% FBS.
Plasmids construction
Construction of the plasmids for luciferase assays, co-immunoprecipitation assays and overexpression experiments are described in the Online Supplementary Appendix.
Lentiviral preparation and transduction of Kasumi-1 cells
Details of lentiviral preparation and transduction of Kasumi-1 cells are available in the Online Supplementary Appendix.
Real-time polymerase chain reaction and western blot analyses
Real-time quantitative polymerase chain reaction (PCR) and western blot analyses for gene expression are described in the Online Supplementary Appendix.
Co-immunoprecipitation assay
Cells were collected and lysed in Cell lysis buffer for western blotting and immunoprecipitation (IP) (Beyotime, China) on ice for 30 minutes (min). Then the lysates were pre-cleared with protein A/G-Sepharose beads (Santa Cruz, CA, USA) at 4°C for 1h before IP with indicated primary antibodies or anti-IgG antibody (Beyotime) overnight. The protein-antibody complexes were incubated with protein A/G-Sepharose beads for another 4 hours (h). The beads were subsequently washed three times with cold lysis buffer and the bound proteins were separated by SDS-PAGE, followed by western blot assay.
Immunofluorescence analysis
Cell preparation and immunofluorescence staining procedures have been described previously.16 Fluorescence images were taken on a spinning disk confocal microscope (PerkinElmer, USA) by a 100X oil-immersion objective.
Luciferase assay
5×104 CV-1 cells were transfected with 100 ng indicated pGL3 luciferase reporter plasmid, 50 ng control Renilla luciferase plasmid together with different combinations of transcription factor expression plasmid. At 48 h after transfection, cells were harvested and the luciferase activities were analyzed according to the Dual-Luciferase Reporter 1000 Assay System Technical Manual (Promega, USA) on a luminometer (Lumat LB 9507, Berthold Technologies GmbH & Co. KG, Baden-Württemberg, Germany).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed with a Pierce Agarose ChIP Kit according to the manufacturer’s instructions (Thermo Scientific, USA). Each ChIP reaction contained chromatin from 1×107 cells and 2 μg indicated antibody. Following IP, the binding sites of transcription factor to target gene promoter were analyzed by PCR. Anti-RUNX1 antibody (ChIP Grade, Abcam, ab23980) and Rabbit IgG antibody (Beyotime) were used in this study. Primers used in ChIP assays were listed in the Online Supplementary Table S1.
Cell proliferation, cell cycle, cell apoptosis and differentiation analyses
Details are available in the Online Supplementary Appendix.
Statistical analysis
All results were presented as mean with error bars indicating standard deviation of triplicate experiments. Statistical differences were determined by Student t-test using GraphPad Prism (v.5.0). P<0.05 was considered statistically significant; *P<0.05; **P<0.01; ***P<0.001.
Results
Phenylbutyrate up-regulated expressions of RUNX1, KLF4 and P57 in Kasumi-1 cells
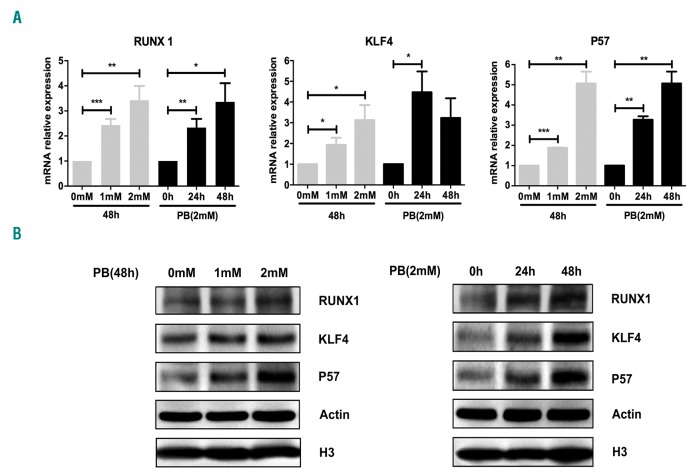
Gene expression profiles of PB-treated Kasumi-1 cells demonstrated that RUNX1, KLF4 and P57 were up-regulated during the process of cell differentiation and apoptosis induced by PB. To confirm the upregulation of these three genes, Kasumi-1 cells were treated with PB at different doses and incubation times. Then cells were harvested, and the mRNA and protein expressions of RUNX1, KLF4 and P57 were evaluated by qRT-PCR and western blot. The results clearly showed that PB up-regulated RUNX1, KLF4 and P57 expressions at both mRNA (Figure 1A) and protein (Figure 1B) level in a time- and dose-dependent manner.
Figure 1.
Upregulation of RUNX1, KLF4 and P57 in t(8;21) leukemia cells by sodium phenylbutyrate (PB) treatment. (A) Relative expressions of RUNX1, KLF4 and P57 mRNA were measured by quantitative real-time polymerase chain reaction in Kasumi-1 cells after treatment with different doses of PB for the time (hours, h) indicated. The gene transcript levels were normalized to those of GAPDH and set to 1 in the control group. (B) Western blot analysis of RUNX1, KLF4 and P57 protein expressions in Kasumi-1 cells after PB treatment. β-actin and H3 were used as internal loading controls.
Identification of KLF4 as a novel target gene of RUNX1 and RUNX1-ETO
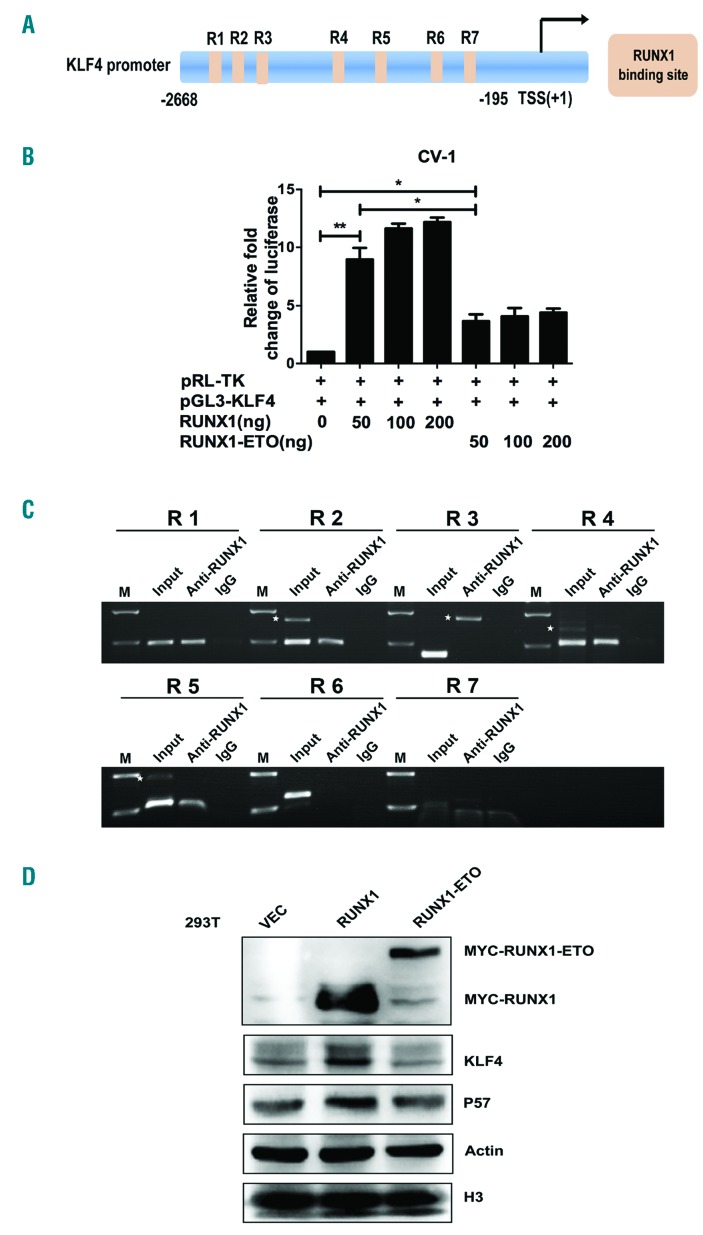
To determine the mechanism of KLF4 upregulation by PB treatment, we first analyzed KLF4 promoter region (bp -2668 to -195) (Homo sapiens chromosome 9, GRCh37) for the transcription factors binding sites by Jaspar database. The result showed that there were seven putative RUNX1-binding sites (R1~R7) within KLF4 promoter region (Figure 2A). We then constructed a pGL3-KLF4 promoter reporter plasmid (pGL3-KLF4) containing RUNX1 binding sites to analyze the transcriptional regulation effects of RUNX1 and RUNX1-ETO on KLF4. The luciferase activity of pGL3-KLF4 was activated by RUNX1 in a dose-dependent manner with a nearly 12-fold increase at the dose of 200 ng (Figure 2B). However, there was a less than 3-fold increase of pGL3-KLF4 luciferase activity by RUNX1-ETO at the doses from 50 ng to 200 ng. To further determine the specific binding sites of RUNX1 and RUNX1-ETO on KLF4 promoter, ChIP assays were performed with a specific anti-RUNX1 antibody in Kasumi-1 cells. The results confirmed the direct binding of RUNX1 to KLF4 promoter region via four predicted binding sites at bp -2617 to -2607 (R1), bp -2312 to -2302 (R2), bp -1136 to -1126 (R4), and bp -764 to -754 (R5) (Figure 2C). Subsequently, western blot analyses were performed to verify the regulation relationship on protein level. 293T cells were transfected with pCMV5-vector, pCMV5-RUNX1 and pCMV5-RUNX1-ETO plasmids, respectively. After 48 h of transfection, cells were harvested for western blot. The results showed that KLF4 was clearly up-regulated by RUNX1 while RUNX1-ETO had almost no effect (Figure 2D).
Figure 2.
KLF4 is a novel target gene of RUNX1 and RUNX1-ETO. (A) Schematic representation of KLF4 promoter fragments fused to a pGL3-Basic Vector. The putative RUNX1 binding sites are indicated by orange boxes as follows: R1 (-2617 to -2607), R2 (-2312 to -2302), R3 (-2014 to -2004), R4 (-1136 to -1126), R5 (-764 to -754), R6 (-399 to -394), and R7 (-263 to -258). Transcription start site (TSS) is indicated by an arrow. Numbers represent base pairs relative to TSS. (B) pGL3-KLF4 promoter reporter plasmid (pGL3-KLF4) was co-transfected with increasing doses of pCMV5-RUNX1 or pCMV5-RUNX1-ETO into CV-1 cells. At 48 h after transfection, the luciferase transcriptional activity of pGL3-KLF4 was measured and normalized to that of Renilla luciferase. (C) Chromatin immunoprecipitation analysis of RUNX1 binding sites to KLF4 promoter region in Kasumi-1 cells. The predicted binding sites R1, R2, R4 and R5 were successfully amplified, both from the input DNA and chromatin immunoprecipitation by an anti-RUNX1 antibody, whereas no amplified product was obtained in the IgG control group. White stars indicate non-specific amplified bandings. (D) 293T cells were transfected with pCMV5-vector, pCMV5-RUNX1 and pCMV5-RUNX1-ETO, respectively. At 48 h after transfection, cells were harvested and the protein levels of KLF4 were assayed by western blot. β-actin and H3 were used as protein loading controls.
KLF4 is also a novel binding partner of RUNX1 and RUNX1-ETO
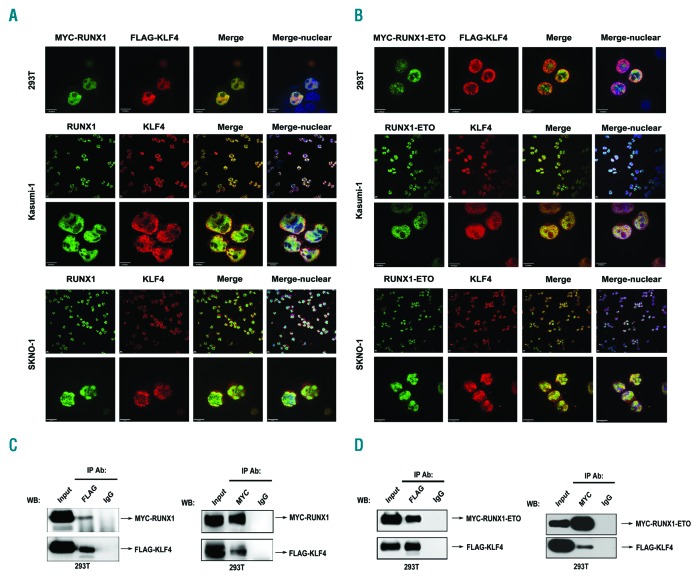
De novo motif analysis of our previous RUNX1 Chip-Seq data demonstrated that KLF4 binding sites were significantly enriched within RUNX1 chip regions, which raised the hypothesis that KLF4 might co-localize and interact with RUNX1. To validate our hypothesis, immunofluorescence confocal imaging analysis and co-immunoprecipitation (Co-IP) assays were performed. Immunofluorescence analysis showed both exogenous and endogenous co-localization of RUNX1 and KLF4 in nuclei (Figure 3A). Co-IP assay further confirmed the interaction between RUNX1 and KLF4 (Figure 3C and Online Supplementary Figure S1A). However, due to the low expression level of KLF4 in Kasumi-1 and SKNO-1 cells, the endogenous interaction between KLF4 and RUNX1 was less evident than that under conditions of exogenous overexpression in 293T cells. The co-localization and protein interaction of fusion protein RUNX1-ETO and KLF4 were also investigated. Similar to wild-type RUNX1, RUNX1-ETO co-localized and interacted with KLF4 in nuclei (Figure 3B and D and Online Supplementary Figure S1B).
Figure 3.
Identification of KLF4 as a novel binding partner of RUNX1 and RUNX1-ETO. (A) KLF4 co-localized with RUNX1 in nuclei. (Top) Co-localization analysis of exogenous MYC-RUNX1 and FLAG-KLF4 in 293T cells at 48 h after transfection by immunofluorescence assay. Anti-MYC (green) and anti-FLAG (red) antibodies were used as primary antibodies; DAPI (blue) was used for nuclear staining. Scale bar represents 10 μm. (Middle and bottom) Co-localization analysis of endogenous RUNX1 and KLF4 in Kasumi-1 and SKNO-1 cells with anti-RUNX1 (green) and anti-KLF4 (red) antibodies; DAPI (blue) was used for nuclear staining. Scale bar represents 10 μm. (B) KLF4 co-localized with RUNX1-ETO in nuclei. (Top) Co-localization analysis of exogenous MYC-RUNX1-ETO and FLAG-KLF4 in 293T cells at 48 h after transfection. Antibodies were used as described in (A). Scale bar represents 10 μm. (Middle and bottom) Co-localization analysis of endogenous RUNX1-ETO and KLF4 in Kasumi-1 and SKNO-1 cells with anti-ETO (green) and anti-KLF4 (red) antibodies. DAPI (blue) was used for nuclear staining. Scale bar represents 10 μm. (C) KLF4 interacted with RUNX1. 293T cells were co-transfected with pCMV5-MYC-RUNX1 and pCMV5-FLAG-KLF4. At 48 h after transfection cells were harvested and cell lysates underwent immunoprecipitation (IP) with anti-FLAG or anti-MYC antibody. Immunoblotting (IB) analysis was performed with the other antibody. (D) KLF4 interacted with RUNX1-ETO. 293T cells were co-transfected with pCMV5-MYC-RUNX1-ETO and pCMV5-FLAG-KLF4. At 48 h after transfection, cells were harvested and cell lysates underwent IP with anti-FLAG or anti-MYC antibody. Immunoblotting analysis was performed with the other antibody. WB: western blotting.
Identification of the specific domains of RUNX1 and RUNX1-ETO mediating interaction with KLF4
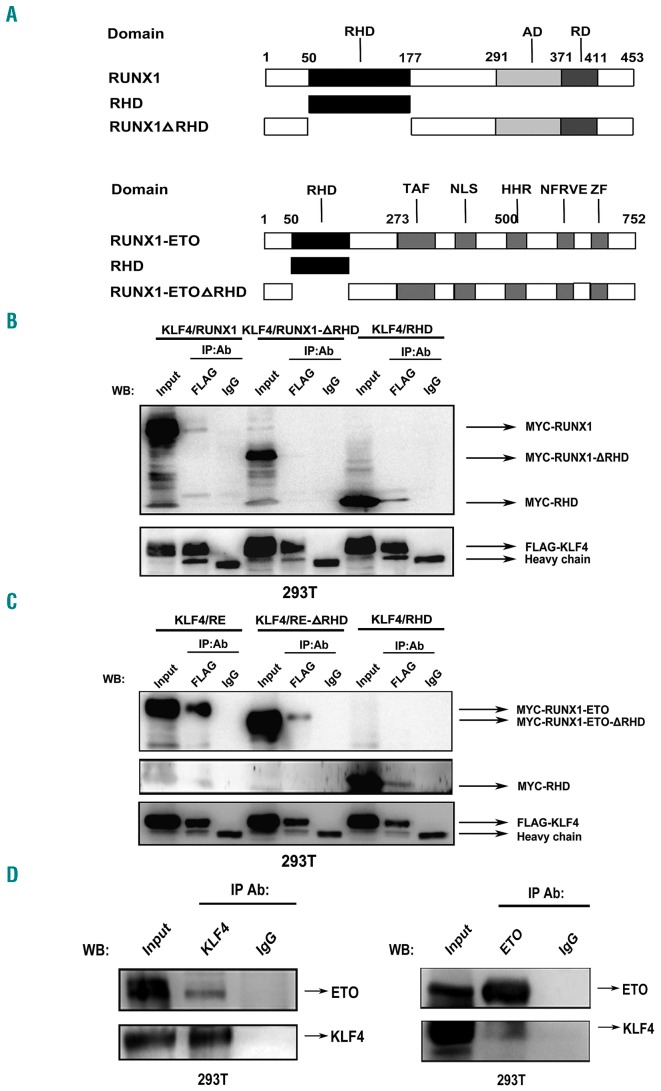
A large number of studies have reported that the runt homology domain (RHD) was responsible for RUNX1 interacting with other transcription factors, such as PU.1, STAT5 and GATA1.3,4,17 To clarify whether RUNX1 interacted with KLF4 through RHD, full-length or truncated mutant (RHD deleted, ΔRHD) of RUNX1 and RHD domain were cloned into a pCMV5 MYC-tagged plasmid, respectively (Figure 4A). After co-transfecting 293T cells with each of the MYC-tagged RUNX1 constructs and FLAG-tagged KLF4 construct, Co-IP assays were performed with anti-FLAG antibody for IP and anti-MYC antibody for immunoblotting (IB). Both the full-length RUNX1 and RHD could directly interact with KLF4, while the truncated mutant RUNX1-ΔRHD failed to interact with KLF4 (Figure 4B). The specific domains of RUNX1- ETO-mediated interaction with KLF4 were also investigated. The results showed that KLF4 not only interacted with RHD, but also with the ETO part of RUNX1-ETO (Figure 4A and C). Further study verified the physical interaction between KLF4 and ETO protein (Figure 4D).
Figure 4.
Identification of the specific domains of RUNX1 and RUNX1-ETO mediating interaction with KLF4. (A) Schematic representation of full-length and truncation mutants of RUNX1 (top) and RUNX1-ETO (bottom). (B and C) FLAG-KLF4 and full-length or truncation mutants of RUNX1 (B) or RUNX1-ETO (C) with MYC tag were co-transfected into 293T cells. At 48 hours (h) after transfection, cell lysates were prepared and underwent immunoprecipitation (IP) with anti-FLAG antibody. Immunoblotting (IB) analyses were performed with anti-MYC antibody. Different combinations of co-transfection with KLF4 and full-length or truncation mutants of RUNX1/RUNX1-ETO were labeled as KLF4/RUNX1, KLF4/RUNX1-ΔRHD, KLF4/RHD, KLF4/RE, KLF4/RE-ΔRHD and KLF4/RHD, respectively. (D) KLF4 and ETO expression plasmids were co-trans-fected into 293T cells. At 48 h after transfection, cells were harvested and cell lysates underwent IP with anti-KLF4 or anti-ETO antibody. IB analysis was performed with the other antibody. WB: western blotting.
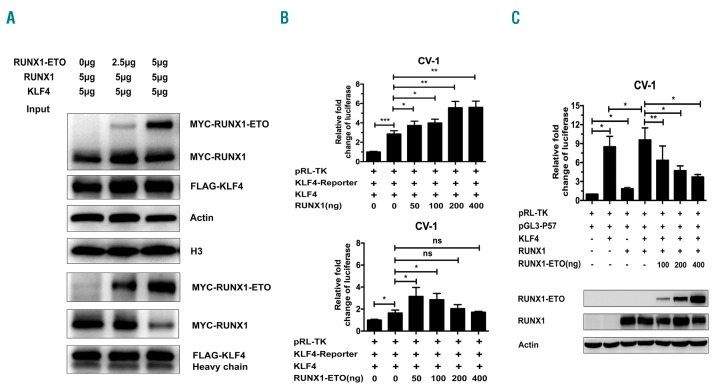
RUNX1-ETO competes with RUNX1 for binding to KLF4
We investigated the physiological interaction between RUNX1 and KLF4. We found that both RUNX1 and RUNX1-ETO interacted with KLF4 (Figure 3) and leukemic fusion protein RUNX1-ETO obstructed the physiological interaction between RUNX1 and KLF4. Fixed doses of pCMV5-FLAG-KLF4 and pCMV5-MYC-RUNX1 were co-transfected with increasing doses of pCMV5-MYC-RUNX1-ETO into 293T cells. At 48 h after transfection, cell lysates were prepared for Co-IP assay. RUNX1-ETO competed with RUNX1 for binding to KLF4 in a dose-dependent manner (Figure 5A). To further investigate the effects of RUNX1 and RUNX1-ETO on KLF4- dependent transactivation of target genes, a luciferase reporter plasmid containing KLF4 binding sites (KLF4-Reporter) was constructed. Luciferase assay was then performed (Figure 5B). The results demonstrated that KLF4 was capable of transactivating its target genes reporter plasmid (KLF4-Reporter) in CV-1 cells. RUNX1 significantly enhanced the transcriptional activation effect of KLF4 in a dose-dependent manner, while RUNX1-ETO could only enhance the effect slightly at low doses (50-100 ng). The above results suggested that RUNX1-ETO might compete with RUNX1 for binding to KLF4 and abrogate transcription of the KLF4 target gene. To confirm this hypothesis, we performed co-transfection of RUNX1 and RUNX1-ETO for luciferase assay using KLF4 target gene P57 promoter reporter. CV-1 cells were transfected with KLF4 or RUNX1 expression plasmid alone or in different combinations of KLF4, RUNX1 and RUNX1-ETO expression plasmids to investigate their regulating effect on P57 promoter reporter. The transactivation of P57 promoter mediated by KLF4 and RUNX1 was abrogated by RUNX1-ETO in a dose-dependent manner (Figure 5C), suggesting that RUNX1-ETO competed with RUNX1 for binding to KLF4 and abrogated transcription of KLF4.
Figure 5.
RUNX1-ETO competes with RUNX1 for binding to KLF4. (A) Fixed dose of pCMV5-FLAG-KLF4 and pCMV5-MYC-RUNX1 were co-transfected with increasing dosages of pCMV5-MYC-RUNX1-ETO into 293T cells. At 48 hours (h) after transfection, cell lysates were prepared and underwent immunoprecipitation (IP) with anti-FLAG antibody. Immunoblotting analyses were performed with anti-MYC antibody. β-actin and H3 served as loading controls for the whole cell lysate. (B) The effects of RUNX1 and RUNX1-ETO on KLF4-dependent transactivation of target genes. CV-1 cells were transfected with fixed dose of KLF4 target genes reporter plasmid (KLF4-Reporter), pCMV5-KLF4 and increasing doses of pCMV5-RUNX1 (top) or pCMV5-RUNX1-ETO (bottom). At 48 h after transfection, transfected cells were harvested for luciferase assay. The luciferase transcriptional activities of KLF4-Reporter were measured and normalized to that of Renilla luciferase. Cells transfected with only KLF4-Reporter were set as control. (C) The transcriptional regulatory effects of RUNX1 and RUNX1-ETO on KLF4 target gene P57 promoter reporter (pGL3-P57). CV-1 cells were transfected with KLF4 or RUNX1 expression plasmid alone or different combinations of KLF4, RUNX1 and RUNX1-ETO expression plasmids together to investigate their regulating effect on pGL3-P57. At 48 h after transfection, transfected cells were harvested for luciferase assay and western blot analysis. The luciferase transcriptional activities of pGL3-P57 were measured and normalized to that of Renilla luciferase. Cells transfected with pGL3-P57 only were set as control.
Biological roles of RUNX1 and KLF4 in t(8;21) leukemia cells
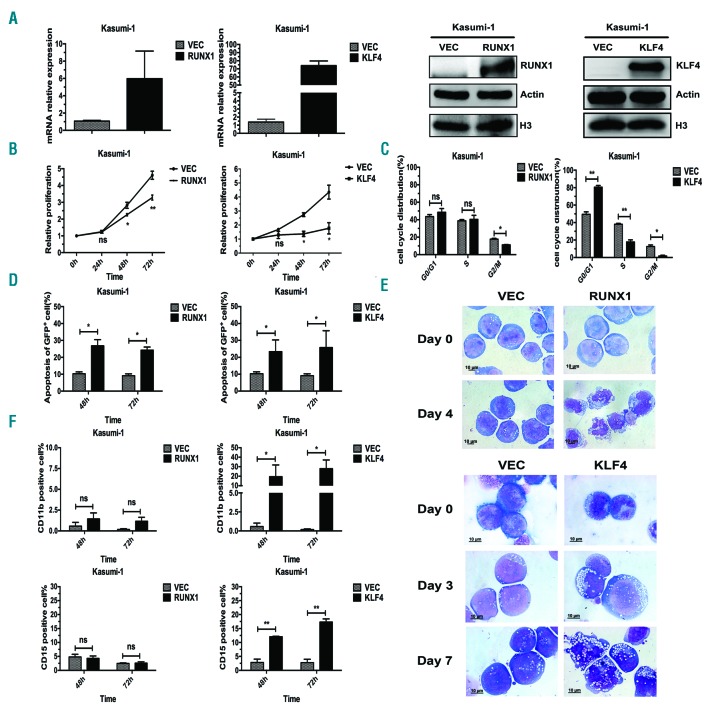
The results seen in Figure 1 suggested that RUNX1 and KLF4 might contribute to the apoptosis and differentiation of Kasumi-1 cells induced by PB. Overexpression experiments were then performed to address their roles in t(8;21) leukemia cells. RUNX1 and KLF4 were over-expressed respectively in Kasumi-1 cells by a pCDH lentivirus system, and the overexpression efficiencies were confirmed at both mRNA and protein levels by qRT-PCR and western blot assay (Figure 6A). 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was performed to evaluate their effects on cell proliferation. The results showed that both of them could markedly inhibit cell proliferation (Figure 6B). Cell cycle analysis was carried out with propidium iodide staining and examined by flow cytometry. KLF4 overexpression blocked cell cycle in G0/G1 phase and reduced S and G2/M cell proportion, while overexpression of RUNX1 had no significant effect on the cell cycle in Kasumi-1 cells (Figure 6C). Apoptosis analysis demonstrated that both RUNX1 and KLF4 promoted apoptosis of Kasumi-1 cells (Figure 6D). Further evidence of cell apoptosis was provided by Wright-Giemsa staining, which displayed typical apoptotic characteristics such as karyopyknosis, reduced ratio of nucleus to cytoplasm, nuclear fragmentation with intact cell membrane, and vacuolar degeneration in pCDH-RUNX1 and pCDH-KLF4 groups compared with that of the control group (Figure 6E). We next examined the effects of RUNX1 and KLF4 overexpression on Kasumi-1 cell differentiation. The expression levels of myeloid cell surface markers CD11b and CD15 were analyzed by flow cytometry at 48 h and 72 h after lentivirus infection. The results showed that there was a big increase in the proportion of CD11b+ and CD15+ cells after infection with pCDH-KLF4, whereas no parallel changes were found in pCDH-RUNX1 group (Figure 6F and Online Supplementary Figure S3).
Figure 6.
Biological effects of RUNX1 and KLF4 overexpression on Kasumi-1 cell proliferation, apoptosis and differentiation. (A) Overexpression of RUNX1 and KLF4 in Kasumi-1 cells were mediated by a pCDH lentivirus system. At 72 hours (h) after infection, the infected cells were sorted by flow cytometry for GFP+ population and the expression levels of RUNX1 or KLF4 were measured by quantitative real-time polymerase chain reaction (left) and western blot (right), respectively. (B) MTS assay was performed to evaluate cell proliferation ability of GFP+ Kasumi-1 cells over-expressing RUNX1 or KLF4 at 72 h after lentivirus infection. (C) Cell cycle distribution of GFP+ Kasumi-1 cells over-expressing RUNX1 or KLF4 was analyzed by flow cytometry with PI staining at 48 h after cell sorting. (D) Cell apoptosis analysis of GFP+ Kasumi-1 cells over-expressing RUNX1 or KLF4 were performed by the flow cytometry at 48 h and 72 h after lentivirus infection. (E) Morphological assessment of GFP+ Kasumi-1 cells over-expressing RUNX1 or KLF4 at indicated days after infection. Cells were stained by Wright-Giemsa staining and observed by oil microscopy after flow sorting (magnification x100). (F) Flow cytometry analysis of cell surface markers CD11b and CD15 of GFP+ Kasumi-1 cells over-expressing RUNX1 or KLF4 at 48 h and 72 h after lentivirus infection.
Identification of P57 as a target gene of KLF4
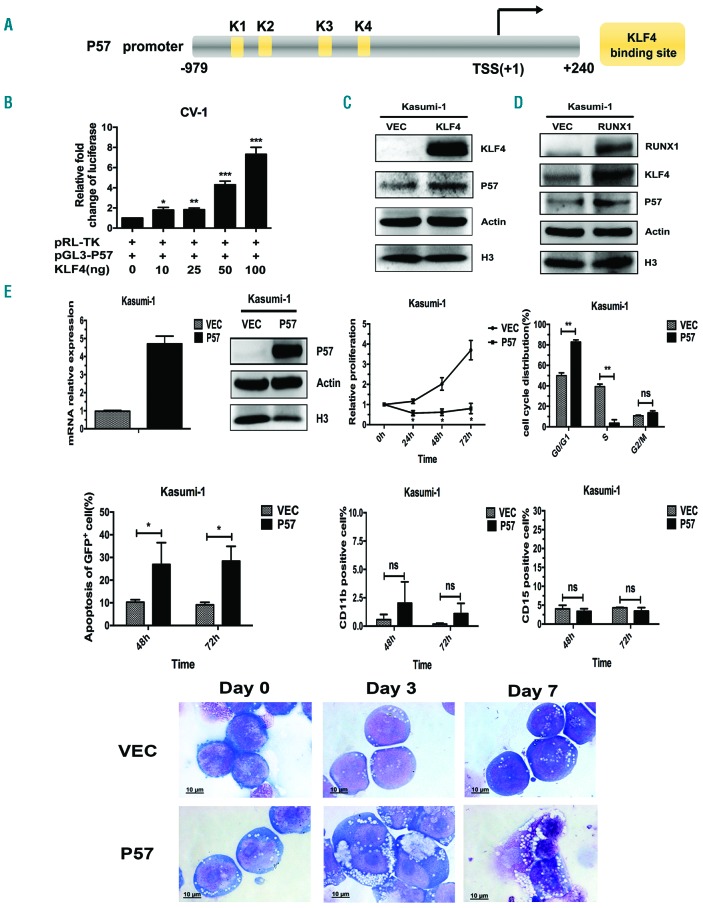
As mentioned in Figure 1, the upregulation of P57 correlated with that of RUNX1 and KLF4 during PB-induced cell differentiation and apoptosis; however, the mechanism of this coherence was unknown. Previously published studies reported that KLF4 could up-regulate P57 in lymphatic endothelial cells and colon cancer cells.18,19 Based on this, we explored whether KLF4 up-regulated the expression of P57 in t(8;21) leukemia cells. Gene promoter analysis showed that there were four KLF4 putative binding sites (K1~K4) within P57 promoter region (bp -979 to +240) (Homo sapiens chromosome 11, GRCh38) (Figure 7A). We then cloned the promoter region of P57 containing KLF4 putative binding sites into a pGL3-Basic Vector to construct the pGL3-P57 plasmid. Next, the pGL3-P57 plasmid was co-transfected with different doses of KLF4 expression plasmid into CV-1 cells and the luciferase activity of pGL3-P57 was evaluated. KLF4 could transactivate pGL3-P57 in a dose-dependent manner with the maximal effect with an almost 8-fold increase at the dose of 100 ng (Figure 7B). Moreover, P57 protein level was found remarkably up-regulated in Kasumi-1 cells after KLF4 overexpression, which further substantiated the regulation relationship (Figure 7C). As discussed (see above), KLF4 was a downstream gene of RUNX1; we therefore concluded that RUNX1, KLF4 and P57 might make up a transcriptional activation cascade in t(8;21) leukemia cells. To further testify the “RUNX1-KLF4-P57” pathway, we over-expressed RUNX1 in Kasumi-1 cells and examined the expression level of putative downstream genes KLF4 and P57. The result showed that both KLF4 and P57 were remarkably up-regulated after RUNX1 overexpression (Figure 7D). Finally, we explored the biological functions of P57 in t(8;21) leukemia cells. Overexpression of P57 mediated inhibition in cell proliferation, blockage in cell cycle, and induction of cell apoptosis, similar to RUNX1 and KLF4 (Figure 7E). However, P57 overexpression had no effects on cell differentiation as assessed by myeloid markers CD11b and CD15 and morphology analysis (Figure 7E and Online Supplementary Figure S3). Taken together, the above results suggested that the “RUNX1-KLF4-P57” pathway, activated by PB, was closely related to cell proliferation and apoptosis in t(8;21) leukemia cells.
Figure 7.
P57 is involved in “RUNX1-KLF4-P57” transcriptional activation cascade, which promotes cell proliferation inhibition and apoptosis induction of t(8;21) leukemia cells. (A) Schematic representation of P57 promoter fragments fused to a pGL3-Basic vector. The putative KLF4 binding sites are indicated by yellow boxes. Transcription start site (TSS) is indicated by an arrow. Numbers represent base pairs relative to TSS. (B) pGL3-P57 promoter reporter plasmid (pGL3-P57) was co-transfected with increasing doses of pCMV5-KLF4 into CV-1 cells. At 48 hours (h) after transfection, the luciferase transcriptional activity of pGL3-P57 was measured and normalized to that of Renilla luciferase. (C and D) Kasumi-1 cells were infected with pCDH lentivirus over-expressing KLF4 (C) and RUNX1 (D), respectively. At 72 h after infection, the protein levels of putative downstream genes P57 and KLF4 were evaluated by western blot assay. (E) Overexpression of P57 in Kasumi-1 cells was mediated by a pCDH lentivirus system. At 72 h after infection, the infected cells were sorted for GFP+ population and the overexpression efficiency was confirmed by quantitative real-time polymerase chain reaction and western blot assay, respectively. Then, the biological effects of P57 on cell proliferation, cell cycle distribution, cell apoptosis and differentiation were evaluated by MTS assay, flow cytometry, and cell morphological analysis (see Figure 6).
Discussion
Dysfunction of RUNX1 through gene mutations and chromosome translocations occurs frequently in various myeloid malignancies. As a transcription factor, RUNX1 exerts its biological effects through transcriptional regulation of its target genes. Thus, identification of RUNX1 target genes and clarification of their roles would offer an intriguing insight into the mechanisms of how perturbed RUNX1 function may result in hematologic malignancies.
For this purpose, we comprehensively analyzed RUNX1 Chip-Seq data with our previous gene expression profiles of PB-treated Kasumi-1 cells. The result revealed a list of putative RUNX1 target genes which had a high potential of driving differentiation and apoptosis of t(8;21) leukemia cells. These targets also served as potential candidates for developing new therapies to treat t(8;21) AML. Among them, p53-induced gene 7 (PIG7), a gene transactivated by RUNX1 through a specific RUNX1-binding site located in the promoter region (bp -1511 to -1503) and promoted apoptosis and differentiation of leukemia cells, has been reported.12 In the present study, we focused our attention on KLF4. On the basis of previously published studies, KLF4 is likely to have similar cellular phenotypes as those observed with RUNX1 in regulating cell proliferation and differentiation. It is reported that RUNX1 mediated inhibition of AML cell survival through repression of VEGF expression, a major mediator of angiogenesis, proliferation, migration, and survival in AML.20 Loss of Runx1 down-regulated p19ARF to accelerate the development of MLL-ENL leukemia.21 Besides, RUNX1 co-operated with PU.1 to activate hematopoietic differentiation genes, such as MCSF, and contributed to the downregulation of the erythroid gene expression program by repressing MIR451 transcription.22,23 Likewise, KLF4 overexpression inhibited cell proliferation of AML cell lines through regulation of microRNA networks and cell cycle inhibitors, and it also played important roles in regulating hematopoietic differentiation, especially myeloid and monocytic differentiation.15,24 Furthermore, a recent study described the role of RUNX1 in inducing intestinal goblet cell differentiation, acting through direct transcriptional activation of KLF4.25 However, the interactions between RUNX1 and KLF4 in AML and its association with leukemia development have remained largely unexplored.
In this study, we demonstrated that KLF4 was directly regulated by RUNX1 through four of seven putative binding sites within the KLF4 promoter region. KLF4 expression could be markedly transactivated by RUNX1 in a dose-dependent manner while leukemogenic fusion protein RUNX1-ETO only had a slight effect. We expected KLF4 expression to be specifically lower in t(8;21) leukemia samples due to haploinsufficiency of RUNX1 in these cells. However, according to an analysis of a publicly available microarray gene expression profiling (GEP) dataset (containing 285 annotated AML cases and 10 healthy cases), KLF4 expression was consistently decreased in the majority of AML cases, and there was no significant difference between t(8;21) and non-t(8;21) groups.26,27 This suggested that KLF4 inactivation might be occurring in non-t(8;21) leukemia cells in distinctly different ways. One mechanism would be DNA hypermethylation, which has already been confirmed by two independent groups in chronic lymphocytic leukemia and adult T-cell leukemia.28,29 Other possibilities remain of great interest for further investigation. In addition to RUNX1, analysis of KLF4 promoter region also revealed putative binding sites correspond to STAT3 and NF-κB (date not shown), both of which are important transcription factors in the hematopoietic system, suggesting that RUNX1 might collaborate with these factors to regulate KLF4 expression. Clarification of the connections between RUNX1, STAT3 and NF-κB is the subject of our ongoing study.
By analyzing RUNX1 Chip-Seq data, we identified several RUNX1 binding sites within the KLF4 promoter region. Furthermore, we found remarkable enrichment of KLF4 consensus motif within RUNX1 chip regions, which suggested the physical interaction between these two factors. Supporting this possibility, both exogenous and endogenous RUNX1 and KLF4 were confirmed to co-localize and interact with each other in nuclei by co-immunoprecipitation and immunofluorescence confocal imaging assay. To the best of our knowledge, this is the first report demonstrating KLF4 both as a direct target and a binding partner of RUNX1. The physical interaction between RUNX1 and KLF4 was mediated through RHD, a critical domain of RUNX1 responsible for DNA binding and commonly involved in protein-protein interaction. RUNX1-KLF4 interaction increased KLF4 transactivation capacity on a promoter reporter driven by KLF4-response elements only (KLF4-Reporter). RUNX1-ETO was also shown to interact with KLF4; however, it had almost no co-activation effect on KLF4-Reporter. The competitive protein-protein interaction experiments showed that RUNX1-ETO disrupted RUNX1-KLF4 interaction in a competitive manner, which might block RUNX1 from co-activating KLF4 target genes. Previous studies described the role of RUNX1 in mediating the interaction between PU.1 and NuAT and BAF families of co-activators. RUNX1-ETO displaced the co-activators from PU.1 and produced a striking switch to PU.1 interaction with co-repressors, such as Dnmt1, Sin3A, Nurd, CoRest, and B-Wich.30 Whether interaction between RUNX1-ETO and KLF4 would abolish KLF4 binding capacity with co-activators or impact its DNA-binding ability are questions that need to be further addressed.
RUNX1 and KLF4 are both up-regulated genes during HDAC inhibitor-induced differentiation and apoptosis of t(8;21) leukemia cells. In this study, we performed overexpression experiments to address their biological roles in t(8;21) leukemia cells. The results showed that both of them contributed to cell proliferation inhibition and cell apoptosis induction. Besides, KLF4 also promoted myeloid differentiation of t(8;21) leukemia cells by up-regulating myeloid markers CD11b and CD15. Furthermore, we reported the role of P57, a novel target gene of KLF4 in limiting proliferation and inducing apoptosis. Thus, RUNX1, KLF4 and P57 might make up a transcriptional activation cascade in regulating t(8;21) leukemia cells survival. To further investigate whether the anti-leukemic effects of the “RUNX1-KLF4-P57” pathway existed only in t(8;21) AML, we performed additional overexpression experiments of RUNX1, KLF4 and P57 in a non-RUNX1-ETO expression cell line, the HL-60 cells. The result showed that overexpression of RUNX1 also up-regulated KLF4 and P57 expression in HL-60 cells (Online Supplementary Figure S2A and B), and overexpression of either RUNX1, KLF4 or P57 (Online Supplementary Figure S2C) could inhibit the proliferation and induce apoptosis in HL-60 cells (Online Supplementary Figure S2D and F), which was similar to the results observed in Kasumi-1 cells. The above results obtained from HL-60 cells suggested that the “RUNX1-KLF4-P57” pathway not only existed in RUNX1-ETO expressing cells but also in non-RUNX1-ETO expressing cells. The effect of RUNX1-ETO on the “RUNX1-KLF4-P57” pathway needs to be further clarified and this is something that we will carry forward in our future studies.
In summary, this study identified KLF4 as a target gene and a binding partner of RUNX1. KLF4 mediated proliferation inhibition and apoptosis induction of t(8;21) leukemia cells through transactivating P57. Restoration of the “RUNX1-KLF4-P57” signaling pathway might be an effective therapeutic strategy for t(8;21) AML.
Acknowledgments
The authors would like to thank the staff for their kindly assistance, especially Wanzhu Yang, Haoyue Liang and Weichao Fu in Core Facility of flow cytometry, State Key Laboratory of Experimental Hematology, Institute of Hematology and Blood Diseases Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/8/1597
Funding
This work was supported by grants from the National Natural Science Foundation of China (81570147, 81430004 and 81800153), Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81421002) and CAMS Initiative Fund for Medical Sciences (2016-I2M-1-001, 2016-I2M-1-007).
References
- 1.Ichikawa M, Yoshimi A, Nakagawa M, Nishimoto N, Watanabe-Okochi N, Kurokawa M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int J Hematol. 2013;97(6):726–734. [DOI] [PubMed] [Google Scholar]
- 2.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. Embo J. 2000;19(12):3004–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol Cell Biol. 1998;18(7):3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101(11):4333–4341. [DOI] [PubMed] [Google Scholar]
- 5.Libermann TA, Pan Z, Akbarali Y, et al. AML1 (CBFalpha2) cooperates with B cell-specific activating protein (BSAP/PAX5) in activation of the B cell-specific BLK gene promoter. J Biol Chem. 1999;274(35): 24671–24676. [DOI] [PubMed] [Google Scholar]
- 6.Kim WY, Sieweke M, Ogawa E, et al. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. Embo J. 1999; 18(6):1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89(3):349–356. [DOI] [PubMed] [Google Scholar]
- 8.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89(3): 341–347. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci U S A. 1998;95(18):10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark Ed). 2012;17:1120–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Saunthararajah Y, Redner RL, Liu JM. Inhibitors of histone deacetylase relieve ETO-mediated repression and induce differentiation of AML1-ETO leukemia cells. Cancer Res. 1999;59(12):2766–2769. [PubMed] [Google Scholar]
- 12.Liu J, Xing H, Chen Y, et al. PIG7, transactivated by AML1, promotes apoptosis and differentiation of leukemia cells with AML1-ETO fusion gene. Leukemia. 2012;26(1):117–126. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, Chen J, Lu C, et al. HDAC1 and Klf4 interplay critically regulates human myeloid leukemia cell proliferation. Cell Death Dis. 2014;5:e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Jiang Z, Li T, et al. Genome-wide analyses identify KLF4 as an important negative regulator in T-cell acute lymphoblastic leukemia through directly inhibiting T-cell associated genes. Mol Cancer. 2015;14:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris VA, Cummings CL, Korb B, Boaglio S, Oehler VG. Deregulated KLF4 Expression in Myeloid Leukemias Alters Cell Proliferation and Differentiation through MicroRNA and Gene Targets. Mol Cell Biol. 2016;36(4):559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Lu W, Li S, et al. Identification of JL1037 as a novel, specific, reversible lysine-specific demethylase 1 inhibitor that induce apoptosis and autophagy of AML cells. Oncotarget. 2017;8(19):31901–31914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa S, Satake M, Ikuta K. Physical and functional interactions between STAT5 and Runx transcription factors. J Biochem. 2008;143(5):695–709. [DOI] [PubMed] [Google Scholar]
- 18.Choi D, Park E, Jung E, et al. ORAI1 Activates Proliferation of Lymphatic Endothelial Cells in Response to Laminar Flow Through Kruppel-Like Factors 2 and 4. Circ Res. 2017;120(9):1426–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ky N, Lim CB, Li J, Tam JP, Hamza MS, Zhao Y. KLF4 suppresses HDACi induced caspase activation and the SAPK pathway by targeting p57(Kip2). Apoptosis. 2009;14(9):1095–1107. [DOI] [PubMed] [Google Scholar]
- 20.Ter Elst A, Ma B, Scherpen FJ, et al. Repression of vascular endothelial growth factor expression by the runt-related transcription factor 1 in acute myeloid leukemia. Cancer Res. 2011;71(7):2761–2771. [DOI] [PubMed] [Google Scholar]
- 21.Nishimoto N, Arai S, Ichikawa M, et al. Loss of AML1/Runx1 accelerates the development of MLL-ENL leukemia through down-regulation of p19ARF. Blood. 2011;118(9):2541–2550. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Vradii D, Gutierrez S, et al. Subnuclear targeting of Runx1 is required for synergistic activation of the myeloid specific M-CSF receptor promoter by PU.1. J Cell Biochem. 2005;96(4):795–809. [DOI] [PubMed] [Google Scholar]
- 23.Kohrs N, Kolodziej S, Kuvardina ON, et al. MiR144/451 Expression Is Repressed by RUNX1 During Megakaryopoiesis and Disturbed by RUNX1/ETO. PLoS Genet. 2016;12(3):e1005946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg MW, Wara AK, Cao Z, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. Embo J. 2007;26(18):4138–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buchert M, Darido C, Lagerqvist E, et al. The symplekin/ZONAB complex inhibits intestinal cell differentiation by the repression of AML1/Runx1. Gastroenterology. 2009;137(1):156–164, 164.e1–3. [DOI] [PubMed] [Google Scholar]
- 26.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. [DOI] [PubMed] [Google Scholar]
- 27.Stirewalt DL, Meshinchi S, Kopecky KJ, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer. 2008;47(1):8–20. [DOI] [PubMed] [Google Scholar]
- 28.Filarsky K, Garding A, Becker N, et al. Kruppel-like factor 4 (KLF4) inactivation in chronic lymphocytic leukemia correlates with promoter DNA-methylation and can be reversed by inhibition of NOTCH signaling. Haematologica. 2016;101(6):e249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasunaga J, Taniguchi Y, Nosaka K, et al. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64(17):6002–6009. [DOI] [PubMed] [Google Scholar]
- 30.Gu X, Hu Z, Ebrahem Q, et al. Runx1 regulation of Pu.1 corepressor/coactivator exchange identifies specific molecular targets for leukemia differentiation therapy. J Biol Chem. 2014;289(21):14881–14895. [DOI] [PMC free article] [PubMed] [Google Scholar]