B-cell precursor acute lymphoblastic leukemia (B-ALL), the most common childhood malignancy, comprises genetically, biologically and clinically heterogeneous disease entities.1,2 Given the concepts of risk-adapted or targeted therapies, their precise delineation is becoming increasingly important.1–3 The recently discovered IGH-DUX4 or, less commonly, ERG-DUX4 rearranged subtype of B-ALL accounts for 5-7% of the cases and is characterized by the expression of C-terminally truncated DUX4 isoforms, a highly distinctive gene expression signature and a profound deregulation of ERG.4–6 Somatic ERG deletions, which occur in roughly half of DUX4+ patients,4,6 are indicative of a favorable outcome and attenuate the negative prognostic effect of adverse factors such as IKZF1 deletions alone7,8 or in combination with other deletion events.9 Given that ERG deletions are secondary events driven by the overexpression of DUX4,6 they delineate only a subset of DUX4+ cases and it remains to be determined whether only those patients with ERG deletions or the entire cohort of DUX4+ patients has a superior outcome. However, a reliable identification of DUX4+ leukemia currently requires gene expression profiling or next-generation sequencing approaches,4–6 which are not yet feasible either for large-scale screening studies or in a diagnostic setting for many study centers. Herein we show that expression of the cell surface antigen CD371 (CLL-1), encoded by CLEC12A and easily detectable by flow cytometry,10,11 is a unique feature of DUX4-rearranged B-ALL and identifies virtually all DUX4+ cases.
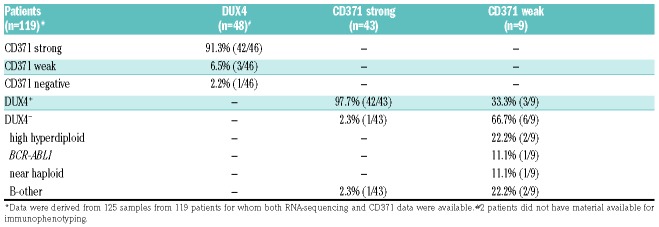
Our evaluation of the immunophenotype of DUX4+ leukemia provides solid evidence that CD371 cell surface expression is a highly specific surrogate marker to identify this otherwise difficult to ascertain genetic subgroup. This notion is based on the finding that of 46 DUX4+ cases, 42 showed strong and three weak CD371 antigen expression (Table 1), while all other genetic subtypes were basically negative (Online Supplementary Tables S1-3).
Table 1.
Summary of DUX4+ and CD371+ patients detected in the cohorts subjected to RNA-sequencing.
In order to detect DUX4+ cases we performed RNA-sequencing of two independent cohorts of patients with childhood or young adolescent leukemia (Online Supplementary Methods; Online Supplementary Tables S1 and S2). The Austrian cohort (cohort 1) consisted of 101 bone marrow samples from 92 patients (n=80 diagnostic, n=3 relapse samples, n=9 diagnosis/relapse matched pairs). It comprised mostly B-other cases (n=65) lacking sentinel genetic alterations as well as representative cases of all major genetic subtypes (Online Supplementary Table S1). Initially, most samples were subjected to RNA-sequencing without prior knowledge of their immunophenotypic details, but later on nine more CD371+ samples with unknown genetic subtype and available material were sequenced. A second cohort (cohort 2) from the Czech Republic consisted of 55 primarily B-other cases, analyzed in parallel for CD371 expression (Online Supplementary Table S2).
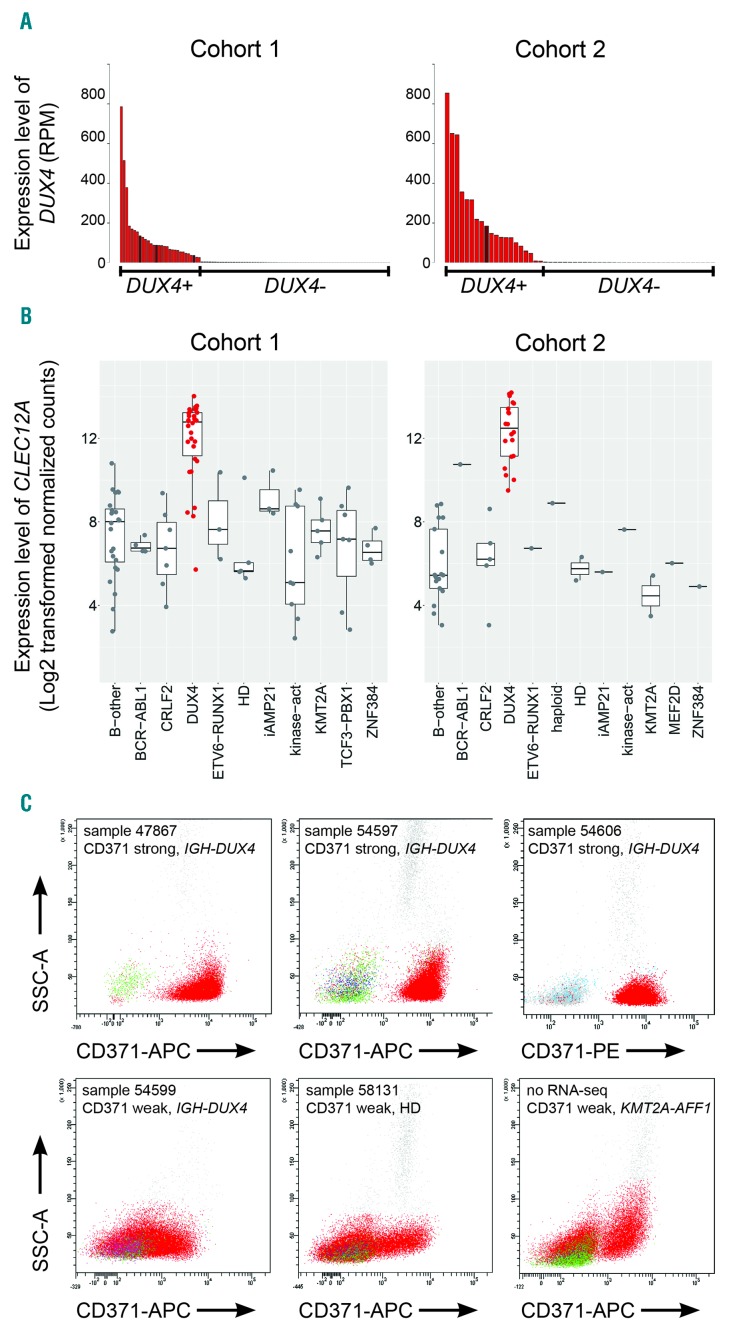
DUX4+ samples were identified by the analysis of DUX4 expression levels (Figure 1A), the presence of DUX4 fusion transcripts (Online Supplementary Tables S1 and S2) and their distinctive gene expression profile4–6,12,13 (Online Supplementary Methods; Online Supplementary Figure S1). Of 48 cases classified as DUX4+, 44 harbored IGH-DUX4 and one a DUX4-ZNF384 fusion, while three lacked any DUX4 fusion (Online Supplementary Tables S1 and S2); but all displayed the gene expression signature typical of DUX4+ leukemia (Online Supplementary Figure S1). As determined by single nucleotide polymorphism array analysis (Online Supplementary Methods) and genomic multiplex polymerase chain reaction,8 32% (15/47) and 65% (13/20) of the patients, respectively, showed ERG deletions (Online Supplementary Tables S1 and S2; Online Supplementary Figure S2), confirming that with either method they are detectable only in a subset of DUX4+ cases.4,6 About one-third of DUX4+ cases displayed IKZF1 deletions (Online Supplementary Methods; Online Supplementary Tables S1 and S2; Online Supplementary Figure S2), which is comparable to the frequency detected in ERG-deleted leukemia.7,8
Figure 1.
Identification of DUX4-positive leukemia. (A) The number of reads that mapped to DUX4 cDNA (NM_001293798.2) per million total mapped reads assigned to RefSeq entries (RPM) for cohort 1 (left panel) and cohort 2 (right panel) are shown. DUX4+ cases are represented in red; cases with elevated DUX4 expression but lacking detectable IGH-DUX4 fusion transcripts are represented in brown. (B) Boxplots showing the expression levels of CLEC12A depicted as log2-transformed normalized counts calculated by DESeq2 in cohort 1 (101 samples from 92 patients) and cohort 2 (55 patients) (left and right panel, respectively). (C) Representative FACS plots of primary bone marrow cells from patients with B-cell acute lymphoblastic leukemia (B-ALL) stained for CD371-APC or CD371-PE; blast cells are depicted in red. B-other, B-ALL cases lacking any sentinel alteration; CRLF2, cases with P2YR8-CRLF2 or IGH-CRLF2 rearrangement; DUX4, cases with expression of DUX4; haploid, masked near haploid; HD, high hyperdiploid; iAMP21, intrachromosomal amplification of chromosome 21; kinase-act, cases harboring a kinase activation fusion gene; KMT2A, KMT2A fusion gene; MEF2D, MEF2D fusion gene; ZNF384, ZNF384 fusion gene; SSC: side scatter; APC: allophycocyanin; PE: phycoerythrin; RNA-seq: RNA-sequencing.
While in the ALL-BFM 2000 clinical trial ERG-deleted patients were more frequently allocated to the intermediate-risk group and had a favorable outcome,8 in the ALL-BFM 2009 study, as a consequence of more refined minimal residual disease stratification, DUX4+ patients were more commonly treated in the high-risk arm14 (Online Supplementary Tables S1 and S2). However, to date, the follow-up of the latter cohort of patients is still too short to draw any conclusions on whether a poor initial treatment response in DUX4+ leukemia eventually results in a poor outcome and whether DUX4+ patients might benefit further from therapy intensification or should rather be spared from additional cytotoxicity.
DUX4+ leukemia is characterized by high expression of distinct genes including CLEC12A6,13 encoding the cell surface protein CD371. Notably, in DUX4-rearranged leukemia DUX4 binds to the CLEC12A locus, suggesting a direct transcriptional regulation.6 CD371 is predomi nantly expressed on myeloid cells and, as a potential myeloid cancer stem cell marker, is considered a target for antibody-based or chimeric antigen receptor T-cell therapies.11 Furthermore, CD371 expression is associated with ‘switch’ ALL,10,14 which has a propensity to switching to monocyte-like cells upon treatment with corticosteroids.15
The previously reported overexpression of CLEC12A in DUX4+ leukemia6,13 was confirmed in both cohorts analyzed by RNA-sequencing (Figure 1B), and hence, we analyzed whether this correlates with CD371 cell surface antigen positivity. Immunophenotyping by flow cytometry and classification into strong or weak antigen expression of the respective samples with available material was performed according to the AIEOP-BFM consensus guidelines (Online Supplementary Methods).10 CD371 expression was determined using phycoerythrin- or allophycocyanin-conjugated mouse anti-human CD371 (clone 50C1; BD Biosciences or BioLegend).
Of 46 DUX4+ cases with immunophenotypic data, 91.3% (42/46) were CD371strong, 6.5% (3/46) CD371weak and 2.2% (1/46) CD371neg (Table 1; Figure 1C; Online Supplementary Figure S2). The last case harbored the exceptional DUX4-ZNF384 fusion but showed an expression signature similar to that of DUX4+ leukemia (Online Supplementary Figure S1). Vice versa, 97.7% (42/43) of all CD371strong cases were DUX4+ and only one single outlier (2.3%; 1/43) did not show either a DUX4 or any other fusion gene that might explain the phenotype.
In most of the nine samples classified as CD371weak only a subfraction of leukemic blasts (10-38%) was antigen positive (Figure 1C). These cases were genetically more diverse and comprised three DUX4+, two high hyperdiploid, one BCR-ABL1, one masked near haploid and two B-other cases, one of which was only analyzed at relapse (Table 1; Online Supplementary Tables S1 and S2). There was no clear difference in the staining pattern between CD371weak DUX4+ samples and those with other genetic subtypes (Figure 1C).
To further substantiate the specificity of CD371 antigen expression for DUX4+ leukemia, we genetically sub-typed 258 consecutive cases from Austrian BFM cohorts prospectively analyzed for CD371 expression (Online Supplementary Methods; Online Supplementary Table S3). Forty-eight of these patients, mostly DUX4+ and B-other cases, overlapped with cohort 1. Of 14 CD371strong cases all 12 analyzed by RNA-sequencing (no material was available for 2) were classified as DUX4+. Conversely, among 240 CD371neg cases a single DUX4+ case was found, corresponding to the one with a DUX4-ZNF384 fusion. The few CD371weak samples again showed variable genotypes (n=1 DUX4+; n=2 KMT2A-AFF1+; n=1 high hyperdiploid). Hence, in the rare cases of weak CD371 expression, exclusion of the major genetic subtypes is required to rule out false interpretation as DUX4+ leukemia.
Considering that KMT2A-rearranged leukemia shows phenotypic heterogeneity with varying degrees of myeloid-lineage-associated antigen expression and that CD371 is primarily expressed on myeloid cells, an international prospective study is necessary to exclude occasional CD371strong expression in this rare B-ALL entity.
CD2 antigen expression, previously described in 35-45% of ERG-deleted cases,7,8 was strongly associated with DUX4+ leukemia and present in roughly 75% of the patients, while basically absent in all other genetic subtypes (Online Supplementary Tables S1 and S2; Online Supplementary Figure S2). The higher frequency of CD2+ cases is most likely attributable to an enrichment of our DUX4+ cohort for ‘switch’ ALL cases commonly expressing CD371 and CD2.14,15 In addition, the classification of the European Group for Immunophenotyping of Leukemias (EGIL) applied in the two previous studies7,8 and the AIEOP-BFM consensus guidelines (Online Supplementary Methods)10 consider cutoffs of 20% and 10%, respectively, to call a sample positive. Except for one single case with an unknown genetic subtype, all CD2 and CD371 double-positive cases were DUX4+. Accordingly, CD2 expression alone will detect only a proportion of DUX4+ cases but is also a strong indicator and in particular in combination with CD371 antigen expression may further underpin DUX4 positivity.
Taken together our data provide compelling evidence that strong CD371 antigen expression in B-ALL is pathognomonic of DUX4+ leukemia. The remarkable finding that one single cell surface protein simply analyzed by flow cytometry may serve as a surrogate marker to identify DUX4+ leukemia will considerably facilitate the detection and further investigation of this disease entity as well as the determination of the prognostic relevance of DUX4 positivity rather than ERG deletions alone,7,8 which are present in only a subset of the cases.4,6
Although the international BFM-FLOW network does not consider immunophenotyping as a general gateway for genetic assessments,10 we explicitly recommend the implementation of CD371 staining in the upfront flow cytometry-based analysis of childhood and adolescent B-cell leukemia for the identification of the DUX4+ subtype.
Acknowledgments
We thank the Vienna BioCenter Core Facilities (VBCF; www.viennabiocenter.org/facilities) Next Generation Sequencing Unit for RNA-sequencing and all those people who are conducting the routine diagnostic work-up of leukemia samples, which builds the essential basis for any research project.
Footnotes
Funding: this study was supported by the St. Anna Kinderkrebsforschung, the Charles University, grant Primus/MED/28 and the Ministry of Health of the Czech Republic, grant n. 15-28525A. Instruments and infrastructure were supported by the Ministry of Education, Youth and Sports, Czech Republic NPU I n. LO1604.
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Iacobucci I, Mullighan CG. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35(9):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwab C, Harrison CJ. Advances in B-cell precursor acute lymphoblastic leukemia genomics. HemaShpere. 2018;2(4):e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor D, Enshaei A, Bartram J, et al. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2018;36(1):34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilljebjorn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasuda T, Tsuzuki S, Kawazu M, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48(5):569–574. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, McCastlain K, Yoshihara H, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48(12):1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clappier E, Auclerc MF, Rapion J, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28(1):70–77. [DOI] [PubMed] [Google Scholar]
- 8.Zaliova M, Zimmermannova O, Dorge P, et al. ERG deletion is associated with CD2 and attenuates the negative impact of IKZF1 deletion in childhood acute lymphoblastic leukemia. Leukemia. 2014;28(1):182–185. [DOI] [PubMed] [Google Scholar]
- 9.Stanulla M, Dagdan E, Zaliova M, et al. IKZF1plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol. 2018;36(12):1240–1249. [DOI] [PubMed] [Google Scholar]
- 10.Dworzak MN, Buldini B, Gaipa G, et al. AIEOP-BFM consensus guidelines 2016 for flow cytometric immunophenotyping of pediatric acute lymphoblastic leukemia. Cytometry B Clin Cytom. 2018;94(1):82–93. [DOI] [PubMed] [Google Scholar]
- 11.Morsink LM, Walter RB, Ossenkoppele GJ. Prognostic and therapeutic role of CLEC12A in acute myeloid leukemia. Blood Rev. 2019;34:26–33. [DOI] [PubMed] [Google Scholar]
- 12.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–143. [DOI] [PubMed] [Google Scholar]
- 13.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novakova M, Vakrmanova B, Slamova L, et al. Switching towards monocytic lineage and discordancy between flow cytometric and PCR minimal residual disease results Is a hallmark feature of DUX4 rearranged B-cell precursor acute lymphoblastic leukemia Blood. 2018;132(Suppl 1):2825. [Google Scholar]
- 15.Slamova L, Starkova J, Fronkova E, et al. CD2-positive B-cell precursor acute lymphoblastic leukemia with an early switch to the monocytic lineage. Leukemia. 2014;28(3):609–620. [DOI] [PubMed] [Google Scholar]