Abstract
The recent emergence and re-emergence of porcine epidemic diarrhea virus (PEDV) underscore the urgent need for the development of novel, safe, and effective vaccines against the prevailing strain. In this study, we generated a cold-adapted live attenuated vaccine candidate (Aram-P29-CA) by short-term passage of a virulent PEDV isolate at successively lower temperatures in Vero cells. Whole genome sequencing identified 12 amino acid changes in the cold-adapted strain with no insertions and deletions throughout the genome. Animal inoculation experiments confirmed the attenuated phenotype of Aram-P29-CA virus in the natural host. Pregnant sows were orally administered P29-CA live vaccines two doses at 2-week intervals prior to parturition, and the newborn piglets were challenged with the parental virus. The oral homologous prime-boost vaccination of P29-CA significantly improved the survival rate of the piglets and notably mitigated the severity of diarrhea and PEDV fecal shedding after the challenge. Furthermore, strong antibody responses to PEDV were detected in the sera and colostrum of immunized sows and in the sera of their offspring. These results demonstrated that the cold-adapted attenuated virus can be used as a live vaccine in maternal vaccination strategies to provide durable lactogenic immunity and confer passive protection to litters against PEDV.
Keywords: Attenuated vaccine, cold adaptation, porcine epidemic diarrhea virus, protection, whole genome sequencing
INTRODUCTION
Porcine epidemic diarrhea virus (PEDV) is a member of the genus Alphacoronavirus, belonging to in the family Coronaviridae of the order Nidovirales. PEDV is a large, enveloped virus possessing a 5′ capped, single-stranded positive-sense RNA genome of approximately 28 kb, with a 3′ polyadenylated tail [1,2]. The PEDV genome consists of seven canonical genes, including an open reading frame (ORF) 3, flanked by 5′- and 3′-untranslated region (UTRs). The first two large ORFs comprise two-thirds of the genome and encode the replicase polyproteins, 1a and 1ab, which are then posttranslationally cleaved into 16 mature non-structural proteins (nsp1–16). The last one-third encodes four canonical coronavirus structural proteins in a fixed order, spike (S), membrane (M), envelope (E), and nucleocapsid (N), as well as a single accessory gene encoding ORF3 between S and M [1,2,3]. The virus can be phylogenetically divided into two genotypes comprising two sub-genotypes: genogroup 1 (classical G1a and recombinant G1b) with low-pathogenicity, and genogroup 2 (local epidemic G2a and global epidemic or pandemic G2b) with high-pathogenicity [1,2,4,5,6].
PEDV causes acute gastrointestinal symptoms characterized by a rapid onset of severe watery diarrhea, vomiting, fatal dehydration, and high mortality in newborn and suckling piglets [7]. In 1971, the virus was initially recognized in England and had since been geographically restricted and problematic in Europe and Asia during the past four decades [8]. However, in early 2013, PEDV first emerged in the United States and rapidly spread to the adjacent North and South American countries, causing significant financial losses to their swine industries [9,10,11]. Soon after, the US prototype-like highly virulent G2b PEDV strains almost simultaneously invaded multiple Asian nations, including South Korea, Taiwan, and Japan, resulting in the recurrence of a massive nationwide PED epizootic [5,12,13]. PEDV has now emerged or re-emerged as one of the most devastating porcine viruses, posing a tremendous threat to the global pork business.
The challenges of substantial monetary damage caused by PEDV highlight the exigent necessity for the establishment of optimal immunization strategies. In most cases of porcine enteric viruses that produce clinical disease in newborn piglets before the development of active immunity, the neonates are protected by passive lactogenic immunity derived from the dams [14,15,16]. Prophylactic maternal immunization regimens with oral live viruses, including intentional infection and vaccination of sows during gestation, remain the most effective way to stimulate intestinal mucosal immunity that is subsequently transferred to piglets via mammary secretions [15,17]. Although intentional infection or feedback through controlled oral exposure is extensively used to induce herd immunity, there are several potential risks associated with this practices such as the wide dissemination of other microbial pathogens and the uncertainty of immune induction [1,2,15]. Therefore, numerous research groups are attempting to develop safer and more effective live attenuated PEDV vaccines to induce passive lactogenic immunity as an alternative to intentional infection.
In South Korea, a number of modified live virus (MLV) vaccines for classical G1a PEDV came to be widely used throughout the country; however, they were incapable of controlling the impact of the recent massive G2b outbreaks in the domestic swine industry because of limited cross-protection between two genetic clusters [1,2,4,18]. Considering this efficacy issue, there is a high priority for the development of a next-generation MLV vaccine against G2b epizootic or related strains prevalent in the field, which can replicate to high titer in the gut and also boost lactogenic immune responses without giving rise to clinical illnesses. In general, the attenuated virus utilized to prepare the MLV vaccine can be achieved by traditional cell culture adaptation procedures of the virulent wild-type virus in non-host cell lines, but this process is impeded by difficulties in performing laboratory procedures, such as numerous time-consuming, and repetitive passages in non-host cell lines. In contrast, adaptation to growth at low (< 37°C) temperatures by short-term serial passages in vitro has been frequently used to generate several attenuated DNA and RNA viruses [19,20,21,22]. In this study, we sought to create a cold-adapted attenuated G2b PEDV low-passage strain by progressively decreasing growth temperatures to 32°C in Vero cells and then attempted to evaluate its protective efficacy on neonatal piglets against virulent PEDV challenge.
MATERIALS AND METHODS
Cells, virus, and antibody
Vero cells (ATCC CCL-81) were cultured in alpha minimum essential medium (α-MEM; Invitrogen, USA) with 5% fetal bovine serum (FBS; Invitrogen) and antibiotic-antimycotic solutions (100×; Invitrogen) and maintained at 37°C in a humidified 5% CO2 incubator. The isolation of a highly virulent Korean PEDV G2b strain, KOR/Aram/2014, was conducted from clinical fecal samples on Vero cells in the presence of trypsin (USB, USA). Virus isolation was confirmed by cytopathic effects (CPEs) observation, immunofluorescence assay (IFA), and nucleotide sequencing as described previously [23]. A viral stock was prepared from the 5th passage in cell culture (Aram-P5) and used as the parental and challenge virus in this study. PEDV N protein-specific monoclonal antibody (MAb) was obtained from ChoongAng Vaccine Laboratories (CAVAC; Korea).
Cold adaptation of PEDV
The PEDV strain, Aram-P5, was plaque-purified twice in Vero cells, and the purified virus was serially passaged in Vero cells by gradually reducing the incubation temperature from 37°C to 32°C as described previously with some modifications [24]. Confluent Vero cells grown in 100-mm diameter tissue culture dishes were washed with PBS and inoculated with PEDV Aram-P5 along with trypsin. After incubation at 37°C for 1 h, virus growth medium [BMpro-V medium (Cell Science & Technology Institute, Inc., Japan) supplemented with 5 μg/mL of trypsin] was added. The inoculated cells were maintained at 37°C under 5% CO2 and monitored daily for CPE. When > 70% of the cells showed CPE, the inoculated cells were subjected to three rounds of freezing and thawing. The culture supernatants were then centrifuged for 10 min at 400 × g (Hanil Centrifuge FLETA5, Korea) and filtered through a 0.45-μm pore-size filter (Millipore, USA). The clarified supernatants were aliquoted and stored at −80°C as a viral stock (Aram-P1-CA) for the next passage. In the same manner, Aram-P1-CA was passaged 4 times with step-wise descending temperatures of 1°C per passage in Vero cells up to 32°C. Subsequently, Aram-P5-CA was serially cultivated at 32°C for an additional 45 passages for cold adaptation, and virus stock were produced at 24, 28, 35, and 45 passages that were designated Aram-P29-CA, P33-CA, P40-CA, and P50-CA.
Virus titration
Vero cells were infected with each Aram virus stock in the presence of trypsin as described above. The culture supernatants were collected 48 h post-infection (hpi) when 70% CPE commonly developed. Virus titers were measured by end-point titration in 96-well plates using 10-fold serial dilutions of the supernatant samples in triplicate for each dilution to determine the amount of virus required to produce CPE in 50% of the inoculated Vero cells. The 50% tissue culture infectious dose (TCID50) per mL of virus stock was calculated using the Reed and Muench method [25].
Nucleotide sequence analyses
The full-length genomic sequences of the parental Aram-P5 and cold-adapted Aram-P29-CA strains were determined by traditional Sanger methods. Ten overlapping cDNA fragments spanning the entire genome of each virus strain were amplified and sequenced as described elsewhere [5,23,24]. The 5′ and 3′ ends of the genome of the strain were determined by rapid amplification of cDNA ends (RACE) as described previously [26]. The full-length genomic sequences were deposited in the GenBank database under accession numbers MK559454 through MK559456.
Multiple alignments and phylogenetic analyses
The sequences of 49 fully sequenced S genes and 39 complete genomes of global PEDV isolates were independently used in sequence alignments and phylogenetic analyses. Multiple sequence alignments were generated using the ClustalX 2.0 program [27] and the percentages of nucleotide sequence divergences were assessed using the same software. Phylogenetic trees were constructed from the aligned nucleotide or amino acid sequences using the neighbor-joining method and subsequently subjected to bootstrap analysis with 1000 replicates to determine the percentage reliability values of each internal node of the tree [28]. All phylogenetic trees were generated using Mega 4.0 software [29].
Animal infection experiments and clinical assessments
The four in vivo swine studies described here were independently performed at the CAVAC Animal Facility under the guidelines established by its Institutional Animal Care and Use Committee (IACUC, Approval No. 160129-03). All animals were obtained from a conventional breeding farm with a good health record and either vaccination against PEDV or no known prior PED outbreak and were tested to confirm that they were not infected with any porcine enteric viruses.
Piglet challenge trials was conducted to determine the infectious dose and virulence of a PEDV Aram strain using 3-day-old conventional suckling piglets delivered from commercial crossbred sows (Great Yorkshire × Dutch Landrace). Thirty pigs were divided into six groups (group 1–group 6) of five animals and were fed commercial milk replacer frequently (4 times daily) with ad libitum access to water for the study (7 days). Animals in groups 1–6 were allowed to acclimate for two days and then inoculated orally with 1 mL of 10−2, 10−3, 10−4, 10−5, and 10−6 dilutions of the Aram-P5 virus (105.0 TCID50/mL) and cell culture medium at 5 days of age, respectively. After inoculation, piglets were observed daily for clinical signs, including diarrhea and mortality. Stool samples from pigs in all groups were collected prior to inoculation and daily with 16-inch cotton-tipped swabs and scored for fecal consistency for 5 days post-inoculation (DPI): 0 = normal; 1 = pasty; 2 = semi-liquid; 3 = liquid or watery. Mean pig diarrhea dose (PDD50) and lethal dose (LD50) were determined as the reciprocal of the virus dilution at which 50% of the pigs developed watery diarrhea (score 3) or died at a given time point using the Reed and Muench method.
To assess immunogenicity of cold-adapted derivatives, a total of 14 3-week-old conventional piglets delivered from commercial crossbred sows (Great Yorkshire × Dutch Landrace) were allocated into the cold adapted Aram-P29-CA-inoculated (n = 3), P33-CA-inoculated (n = 3), P-45-CA-inoculated (n = 3), P50-CA-inoculated (n = 3), and sham-inoculated control (n = 2) groups. Animals were immunized orally with a 1 mL dose of 104.5 TCID50/mL of each virus or sham inoculated with cell culture medium. Pre-immune sera were collected at the immunization, and antisera were collected at 1 week intervals for 3 weeks.
Pathogenicity tests were performed using a total of 12 suckling piglets of 3 days of age obtained from commercial crossbred sows (Great Yorkshire × Dutch Landrace). Pigs were randomly assigned to three experimental groups: the parental Aram-P5-inoculated (n = 4), cold-adapted P29-CA-inoculated (n = 4), and sham-inoculated control (n = 4) groups. Animals were fed commercial milk replacer frequently (4 times daily) and had ad libitum access to water for the duration of the study (9 days). After a 2-day acclimation period, piglets (5-day-old) in the virus-inoculated groups received a 1 mL dose of 104.0 TCID50/mL (equivalent to 1000 LD50 as determined in this study) of one of the viruses orally. The sham-inoculated pigs were administered cell culture medium as a placebo. Animals were monitored daily for clinical signs of vomiting, diarrhea, and mortality throughout the experiment. Rectal swabs were collected from all pigs and diluted with PBS to 10% (w/v) suspensions followed by centrifugation. The clarified supernatants were subjected to conventional reverse transcription polymerase chain reaction (RT-PCR) using an i-TGE/PED Detection Kit (iNtRON Biotechnology, Korea) and real-time RT-PCR to detect the presence of PEDV shedding. A clinical significance score (CSS) was determined with the following scoring criteria based on visual examination for 7 DPI used to measure diarrheal severity: 0 = normal and no diarrhea (mean cycle threshold [Ct] values of > 45); 1 = mild and fluidic feces; 2 = moderate watery diarrhea; 3 = severe watery and projectile diarrhea (mean Ct values of < 20); 4 = death. Piglets were necropsied upon death after challenge throughout the study, whereas all surviving pigs from the challenge and control groups were euthanized at 7 DPI for post-mortem examinations.
Swine vaccination and challenge experiments were conducted with a total of 6 commercial crossbred pregnant sows (Great Yorkshire × Dutch Landrace) with the same parity and expected farrowing date. Animals were allocated randomly into three experimental groups: vaccinated group 1 (n = 3), challenge control group 2 (n = 2), and strict negative control group 3 (n = 1). A multiple-dose PED homologous vaccination schedule at 2-week intervals starting prior to farrowing was applied in the present protection study. Three sows in group 1 were orally administered twice with a 1 mL dose of the Aram-P29-CA with 104.5 TCID50/mL at 4 and 2 weeks pre-partum. The remaining sows in groups 2 and 3 were unvaccinated and served as controls. All sows were monitored daily for clinical alterations and adverse effects following vaccination. To mimic the field conditions of nursing piglets, all sows were allowed to farrow naturally and nurse their piglets freely for the duration of the study. Ten 4-day-old suckling piglets per litter (a total of 50 newborn piglets) in groups 1 and 2 were challenged orally with a 1 mL dose of 104.0 TCID50/mL of virulent Aram-P5 virus. The sham-inoculated piglets from group 3 were administered with cell culture medium as a placebo. Animals were observed daily for clinical signs, including vomiting, diarrhea, and mortality. Fecal specimens were taken from all groups and examined for PEDV shedding as described above. A CSS was determined for 7 days post-challenge (DPC) as described above. Blood samples were collected from sows before (at each vaccination), at, after farrowing (up to 2 weeks) and also from 5 representative piglets per litter from each dam at 0 and 7 DPC. Colostrum was collected at the day of farrowing.
Quantitative real-time RT-PCR
Viral RNA was extracted from fecal suspensions prepared as described above using an i-TGE/PED Detection Kit according to the manufacturer's protocol. Quantitative real-time RT-PCR was performed using a One Step SYBR PrimeScript RT-PCR Kit (TaKaRa, Japan) as described elsewhere [24,30,31]. A PEDV isolate with a known infectivity titer was 10-fold serially diluted to generate a standard curve in each PCR plate. The virus concentrations (TCID50/mL) in samples were calculated based on the standard curve. The mean Ct values were calculated based on PCR positive samples, and the mean virus titers were calculated based on all pigs within the group.
Histopathology and immunohistochemistry of the small intestines
Intestinal tissues and other major organs were grossly examined upon necropsy. Small intestinal tissue specimens (< 3-mm thick) collected from each piglet were fixed with 10% formalin for 24 h at RT and embedded in paraffin according to standard laboratory procedures. The formalin-fixed paraffin-embedded tissues were cut in 5–8-μm thick sections on a microtome (Leica, Germany), floated in a 40°C water bath containing distilled water, and transferred to glass slides. The deparaffinized intestinal tissue sections were stained with hematoxylin and eosin (Sigma, USA) to observe histopathological change or were subjected to immunohistochemistry (IHC) for the detection of PEDV antigen using an MAb specific for PEDV N protein as described previously [23,24]. PEDV antigen detection was semi-quantitatively measured throughout the IHC-stained jejunal sections based on the following scoring criteria as described previously [32]: 0 = no signal, 1 = 1%–10% of villous enterocytes within the section showing a positive signal, 2 = 11%–50% of villous enterocytes showing a positive signal, and 3 = greater than 50% of villous enterocytes showing a positive signal.
Virus neutralization
The presence of PEDV-specific neutralizing antibodies in serum and colostrum samples collected from pigs in all groups was determined using a serum neutralization (SN) test using PEDV isolate Aram-P5 as previously described [24]. The neutralization titer was calculated as the reciprocal of the highest dilution of serum that inhibited virus-specific CPE in duplicate wells.
Statistical analysis
All values are expressed as the means ± standard deviation of the means (SDM). All statistical significances were evaluated by a Student's t-test by using GraphPad Prism software version 5.0 (GraphPad Prism Inc., USA). The p values of less than 0.05 were considered statistically significant.
RESULTS
Isolation and characterization of a PEDV Aram strain
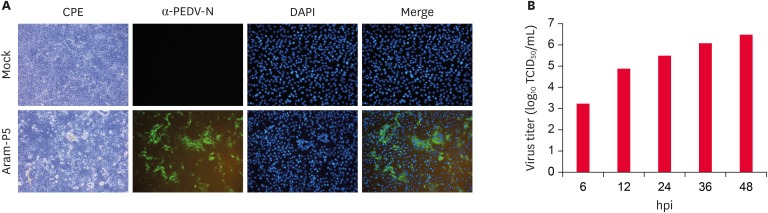
PEDV isolation in cell culture is the first step toward the generation of a live attenuated vaccine. Thus, we initially attempted to isolate PEDV from clinical fecal suspensions and successfully propagated one Korean PEDV strain, designated as Aram, in Vero cells. The Aram strain produced obvious CPE typical of PEDV infection, such as cell fusion and multi-nucleated cell (syncytium) formation, in infected Vero cells, which was verified by detecting PEDV antigens by the IFA using a PEDV N protein-specific MAb (Fig. 1A). Growth kinetics analysis indicated that Aram replicated efficiently in Vero cells, reaching a maximum viral titer of > 106 TCID50 by 36 hpi (Fig. 1B).
Fig. 1.
Cytopathology and growth properties of PEDV G2b isolate Aram. (A) CPE formation in Vero cells infected with Aram-P5 virus. PEDV-specific CPE was observed daily, and cells were photographed at 24 hpi using an inverted microscope at a magnification of 200× (left panels). For immunostaining, infected cells were fixed at 24 hpi and incubated with MAb against the N protein, followed by incubation with Alexa green-conjugated goat anti-mouse secondary antibody. The cells were then examined under a fluorescence microscope at 200× magnification (left panels). (B) One-step growth kinetics for Aram-P5. At the indicated time points post-infection, culture supernatants were harvested from Aram-P5-infected Vero cells, and virus titers were determined.
CPE, cytopathic effects; PEDV, porcine epidemic diarrhea virus; DAPI, 4′,6-diamidino-2-phenylindole; TCID, tissue culture infectious dose.
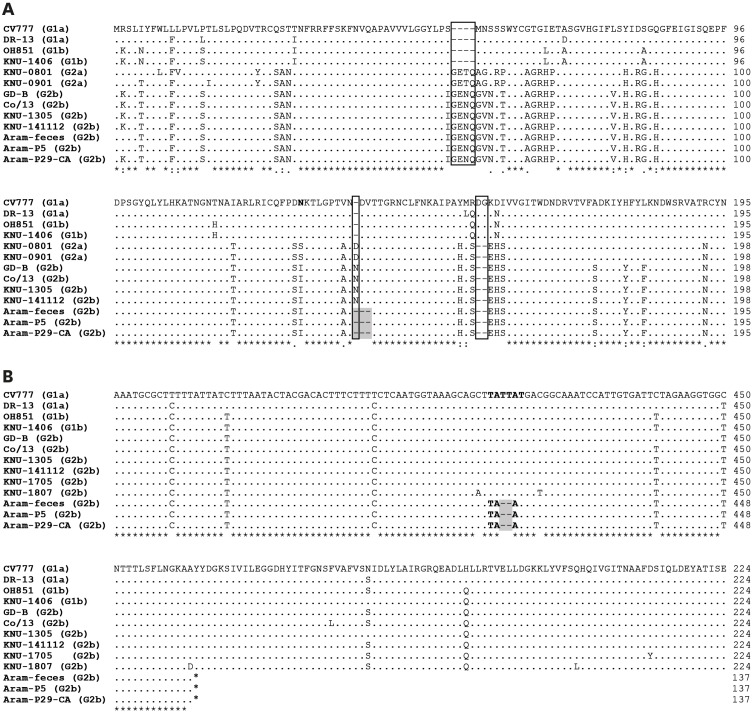
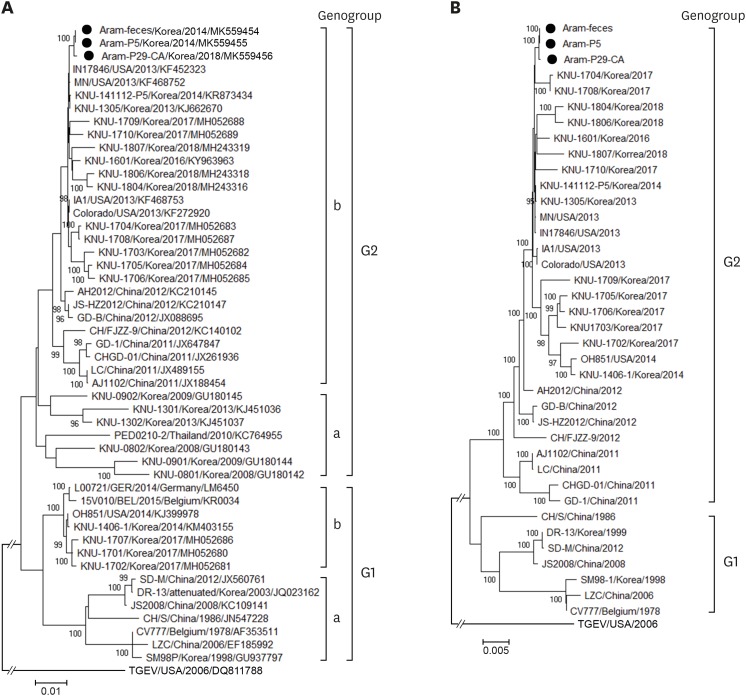
We subsequently determined the entire genomic sequences of the original fecal sample (Aram-feces) and the cell culture-passaged Aram-P5 virus using the Sanger technology. Both the identified genomes were identical in length [28,027-nucleotide (nt)], except for the 3′ poly(A) tails, and showed the typical genomic organization of alphacoronaviruses, consisting of the 292-nt 5′ UTR, the 20345-nt ORF1a/1b (nt 293 to 12601 for 1a and nt 12601 to 20637 for 1b), the 4152-nt S gene (nt 20634 to 24785), the 414-nt ORF3 (nt 24785 to 25198), the 231-nt E gene (nt 25438 to 25668), the 681-nt membrane (M) gene (nt 25676 to 26356), the 1326-nt N gene (nt 26368 to 27693), and the 334-nt 3′ UTR. Compared to the complete genome sequence of the Aram-feces, the Aram-P5 virus had no amino acid (aa) substitution throughout the entire genome, suggesting genetic stability during cell passages. Interestingly, the genome size of the Aram strain was 11-nt shorter than that of most G2b field viruses; this was due to the presence of the unique 9-nt and 2-nt deletions (DELs) at genomic positions 21,051–21,059 and 25,207–25,208, respectively. These DELs are completely absent in the genome sequences of the other global G1 and G2 strains available in the GenBank database. The former 9-nt DEL resulted in a 3-aa DEL at aa positions 140–142 in the N-terminal domain (NTD) of the S protein (Fig. 2A), while the latter 2-nt (thymine residues) DEL at nt positions 414–415 (412TATTAT417) in ORF3 caused premature termination 414-nt upstream from the authentic stop codon, thereby possibly producing a truncated ORF3 without C-terminal 87-aa residues (Fig. 2B). Sequencing analysis of the complete S gene showed that the Aram strains were classified as G2b, sharing 97.4%–99.2% aa identity with the global G2b strains, but only having 91.3%–92.7% and 94.6%–95.3% aa homology with the classical G1a or variant G1b strains, respectively. Likewise, the Aram isolate had a high degree of nt homology with other global G2b PEDVs at the genomic level (98.1%–99.8%). However, it was genetically distinct from the G1a strains, showing relatively low nt identity in the range of 96.2% to 97.2%. Furthermore, the Aram strain possessed the genetic signature of the G2 epidemic strains with S insertions-deletions (S INDELs) within the N-terminal hypervariable region of S compared to the prototype CV777 strain [1,2,4,5] (Fig. 2B). Phylogenetic analysis based on the complete S protein clearly classified the PEDV strains into two distinct genogroup clusters, G1 and G2, with two subgroups in each genogroup (Fig. 3A). The Aram strains belonged to subgroup G2b, as it clustered closely with the contemporary domestic and global G2b isolates. Subsequent whole-genome phylogeny showed the same grouping structure as that of an S gene-based phylogenetic tree (Fig. 3B).
Fig. 2.
Sequence alignments of the N-terminal region of the S protein (A) and the C-terminal region of ORF3 (B) of global PEDV strains. Genetic subgroups of PEDV are indicated in parenthesis on the left. The dashes (–) indicate deleted sequences. Potential N-glycosylation sites in the S protein predicted by the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) are shown in boldface type. The S insertions-deletion genetic signature of G2 epidemic strains, that includes two discontinuous 4-aa and 1-aa insertions at positions 55/56 and 135/136 and one 2-aa DEL at positions 160 and 161 within S compared to the prototype CV777 strain, is shown in solid boxes. The Aram strain-specific DELs in S and ORFs are shaded. An alternative TAA stop codon (boldface) and a corresponding premature termination (asterisk) in nucleotide (top panel) and deduced amino acid (bottom panel) sequences of ORF3 are also indicated.
ORF, open reading frame; PEDV, porcine epidemic diarrhea virus; DEL, deletion.
Fig. 3.
Phylogenetic analyses based on the nucleotide sequences of the spike (S) genes (A) and full-length genomes (B) of the porcine epidemic diarrhea virus strains. A region of the S gene and the complete genome sequence of transmissible gastroenteritis virus were included as the outgroups in each tree. Multiple sequence alignments were performed using ClustalX software and phylogenetic trees were constructed from the aligned nucleotide sequences using the neighbor-joining method. Numbers at each branch are bootstrap values greater than 50% based on 1000 replicates. The names of the strains, countries and dates (year) of isolation, GenBank accession numbers, and genogroups and subgroups are shown. Solid circles indicate the Aram strains described in this study. Scale bars indicate nucleotide substitutions per site.
A PEDV Aram-P5 stock with a known cell culture infectious titer of 105.0 TCID50/mL was serially diluted 10-fold with cell culture medium, giving rise to theoretical infectious titers of 103 to 10−1 TCID50/mL for the 10−2 to 10−6 dilutions, respectively, which were later used to determine the outcomes of infection in 5-day-old piglets (Table 1). Each group (groups 1–5) of five piglets was challenged orally with the 10-fold serially diluted (10−2–10−6) Aram-P5 virus, while control animals in group 6 were fed cell culture medium. All five piglets in the groups 1 to 3 (inoculated with virus dilutions 10−2 to 10−4) and three piglets in group 4 (inoculated with virus dilution 10−5) developed moderate to watery diarrhea (scores 2–3) by 3 DPI and it lasted through the study period (Table 1). By 5 DPI, 100% pigs of group 1, 60% (3/5) pigs of groups 2 and 3, and 20% (1/5) pigs of group 4 died from PED-related clinical signs. In contrast, all five pigs in groups 5 (inoculated with virus dilution of 10−6) and the negative control group remained active and clinically unaffected throughout the 5-day experimental period. The PDD50 and LD50 of the Aram virus were determined as 5.2 log10 PDD50/mL and 4.0 log10 LD50/mL, indirectly corresponding to theoretical cell culture infectious titers of 100.2 TCID50/mL and 101 TCID50/mL, respectively.
Table 1. Summary of pig groups and corresponding numbers, inoculum, and pig diarrhea and death outcomes after inoculation.
| Pig group | Pig numbers | Inoculum dilution* | Calculated inoculum infectious titers (log10 TCID50/mL)† | Diarrhea (percent) | Death (percent) |
|---|---|---|---|---|---|
| Group 1 | 5 | 10−2 | 3 | 5/5 (100) | 5/5 (100) |
| Group 2 | 5 | 10−3 | 2 | 5/5 (100) | 3/5 (60) |
| Group 3 | 5 | 10−4 | 1 | 5/5 (100) | 3/5 (60) |
| Group 4 | 5 | 10−5 | 0 | 3/5 (60) | 1/5 (20) |
| Group 5 | 5 | 10−6 | −1 | 0/5 (0) | 0/5 (0) |
| Group 6 | 5 | Cell culture media | - | 0/5 (0) | 0/5 (0) |
*Each pig was inoculated orally with 1 mL inoculum; †Titers were calculated based on the titer of the original virus pool and dilution times.
Immunogenicity of the cold-adapted Aram strains
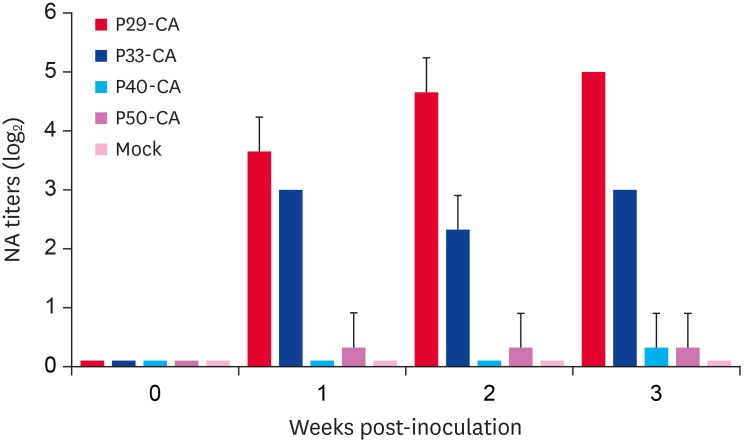
The cold-adapted G2b PEDV Aram-CA strains were generated by a serial passage of Aram-P5 at low temperatures commencing from 37°C to 32°C and additional adaptation for growth up 45 passages at 32°C in Vero cells. We initially aimed to evaluate the immunogenicity of the cold-adapted Aram derivatives in the natural host. Antisera were collected from the pigs at 0-, 1-, 2-, and 3-week after oral administration of each virus and were tested for neutralizing activity against the Aram isolate. The serum samples obtained from pigs immunized with the cold-adapted Aram-P29-CA strain had higher neutralizing antibody titers, when compared with those from pigs in other CA virus-inoculated groups (Fig. 4). Furthermore, the immune sera of pigs inoculated with Aram-P40-CA or -P50-CA strain showed little or no neutralizing activity against PEDV. Since our data indicated that only Aram-P29-CA can elicit robust antibody responses in immunized animals, this cold-adapted virus was selected for subsequent in vitro and in vivo studies.
Fig. 4.
Porcine epidemic diarrhea virus -specific neutralizing antibody responses of cold-adapted strains. Piglets at 3 weeks of age were inoculated orally with each cold-adapted Aram-CA strain. The serum samples were collected at the indicated time points and were tested by virus neutralization assay using Aram-P5. Neutralizing antibody titers for individual samples were presented as a log2 scale. Values are representative of the mean from three independent experiments in duplicate and error bars denote the mean ± standard deviation of the mean.
Phenotypic and genotypic characteristics of the cold-adapted Aram-P29-CA strain
Like the parental virus as shown in Fig. 1A, the cold-adapted Aram-P29-CA strain caused noticeable CPE and IFA staining showing typical syncytia in virus-infected cells at 32°C. However, the Aram-P29-CA virus showed delayed replication (late CPE onset) at 37°C as compared to 32°C, indicating impairment of growth at 37°C. This diminished growth at 37°C was confirmed by comparing growth kinetics in Vero cells at both the temperatures. The Aram-P29-CA strain exhibited efficient growth and had an infectious titer of > 106.5 TCID50/mL at 32°C by 2 dpi, but slower replication rates with a maximum titer of 102.5 TCID50/mL than that of the parental Aram-P5 strain (a 10,000-fold reduction) at 37°C, indicating an adaptation to low temperatures.
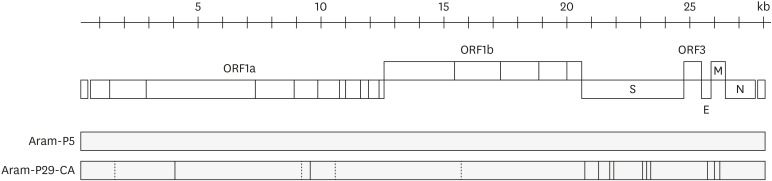
To identify the mutation that occurred during the cold adaptation of PEDV, the genome of the cold-adapted virus was sequenced and compared to that of the wild-type virus. No additional INDELs were found in the genome after cold adaptation, and the genome size of the Aram-P29-CA strain was determined to be 28,027-nt, which was identical to the parental genome size. Two genetic DEL markers of the Aram strain were also completely conserved in S and ORF3 of the cold-adapted Aram-P29-CA strain (Fig. 2). The 5′- and 3′- UTRs of the cold-adapted virus remained unchanged, whereas the coding region contained a total of 16 nt substitutions (Fig. 5). Among these mutations, 12 were non-synonymous, causing changes of 1-aa in nsp3, 1-aa in nsp5, 7-aa in S, 1-aa in E, and 2-aa in M. Details in nt and aa differences between the parental Aram-P5 and cold-adapted P29-CA strains are summarized in Table 2. Further phylogenetic analysis based on the full-length S and the entire genome indicated that the cold-adapted Aram-P29-CA strain was still clustered into subgroup G2b along with the parental and global epizootic strains (Fig. 3).
Fig. 5.
Schematic representation of amino acid differences between the parental Aram-P5 virus and its cold-adapted derivative Aram-P29-CA. The illustration on top represents the organization of the porcine epidemic diarrhea virus genome (approximately 28 kb). The coding region for each non-structural protein in ORFs 1a and 1b and for each structural protein is indicated. The lower panels symbolize the genomes of the parental Aram-P5 strain and its cold-adapted derivative, where the vertical lines denote silent substitutions (dotted) and non-silent aa substitutions (solid) in relation to the aa sequence of Aram-P5.
ORF, open reading frame.
Table 2. Changes of nucleotides and amino acids between the parental Aram-P5 and the cold adaptive P29-CA virus during in vitro serial passages.
| ORFs | Gene | Position | Aram strains | ||
|---|---|---|---|---|---|
| Nucleotide | Amino acid | P5 | P29-CA | ||
| ORF1a | nsp3 | 4007 | 1336 | GAT (D) | GGT (G) |
| nsp5 | 9625 | 3209 | ATT (I) | TTT (F) | |
| ORF2 | S | 5 | 2 | AGG (R) | AAG (K) |
| 628 | 209 | AAC (N) | TAC (Y) | ||
| 1120 | 374 | GTG (V) | TTG (L) | ||
| 1314 | 438 | ATA (I) | ATG (M) | ||
| 2320 | 774 | ACG (T) | GCG (A) | ||
| 2466 | 822 | CAG (Q) | CAT (H) | ||
| 2492 | 831 | GAG (E) | GTG (V) | ||
| ORF4 | E | 209 | 70 | CCT (P) | CTT (L) |
| ORF5 | M | 149 | 50 | GCT (A) | GTT (V) |
| 440 | 147 | GTT (V) | GGT (G) | ||
The bold letters indicate mutated nucleotides based on P5 virus.
ORF, open reading frame; nsp, non-structural protein.
Pathogenicity of the cold-adapted Aram-P29-CA virus in neonatal piglets
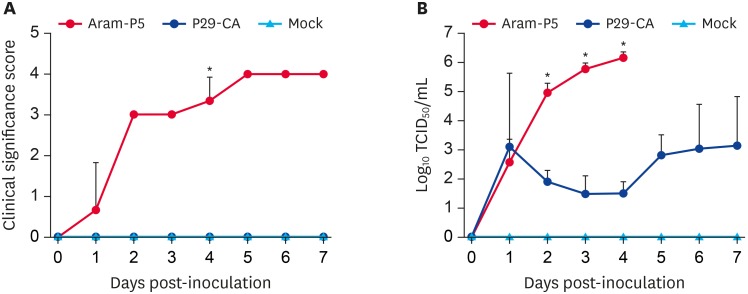
Next, animal inoculation experiments were performed to examine the in vivo phenotypic alterations associated with serial in vitro passage at low temperatures (a cold adaption tool for virus attenuation) of the virulent Aram strain. The pathogenicity of the parental Aram-P5 and its cold-adapted derivative, Aram-P29-CA, was characterized in pigs. Twelve piglets divided into three groups of four animals each were challenged orally with parental Aram-P5 (group 1) or cold-adapted Aram-P29-CA (group 2), and the remaining piglets in a control group received cell culture medium. Clinical signs were recorded daily, and fecal swabs were collected pre- and post-challenge for the duration of the study. During the acclimation period, all piglets were active, showed no clinical symptoms, and had normal fecal consistency that did not contain any PEDV genetic material. After the challenge, none of the negative-control piglets developed clinical presentations typical of PEDV throughout the study. In contrast, Aram-P5-challenged piglets (group 1) displayed clinical signs including loss of appetite and diarrheic feces by 1 DPI (mean CSS ± SDM of 0.67 ± 1.15) and underwent lethal watery diarrhea with vomiting thereafter (mean CSS of > 3.0) (Fig. 6A). All the inoculated animals in group 1 finally died by 5 DPI. Remarkably, the cold-adapted Aram-P29-CA virus-inoculated piglets in group 2 experienced neither PEDV-related clinical symptoms nor mortality throughout the experiment.
Fig. 6.
Clinical significance scores and virus shedding in piglets inoculated with PEDV Aram-P5, -P29-CA, or mock. (A) Clinical significance scores were measured as described in the Materials and Methods section. (B) PEDV titers in rectal swap samples at each time point were determined by a quantitative real-time reverse transcription polymerase chain reaction. The virus titers (log10 tissue culture infectious dose50/mL) are the mean virus titers from all pigs and error bars represent the mean ± standard deviation of the mean. The p values were calculated by comparing the parental (Aram-P5) virus- and cold-adapted (P29-CA) virus-inoculated groups using Student's t-test.
PEDV, porcine epidemic diarrhea virus.
*p < 0.001.
All animals in group 1 were positive for PEDV, as determined by RT-PCR, by 1 DPI with a mean Ct value of 28.55 (equivalent to 102.59 TCID50/mL) and shed significantly higher amounts of PEDV in feces with Ct ranging from 6.80–12.35 (104.97–106.14 TCID50/mL) until death (Fig. 6B). Likewise, fecal shedding of PEDV was detected in all the piglets in group 2 by 1 DPI with the mean Ct values of 24.59 (103.11 TCID50/mL), but thereafter decreased to the lowest levels thereafter followed by a slight increase at 5 DPI. Overall, the quantities of viruses in the feces of animals of group 2 significantly declined compared to those in group 1, with wide Ct ranges of 34.46–24.34 (101.50–103.16 TCID50/mL) until the termination of the study, indicating limited shedding (almost a 3-log reduction) of the P29-CA strain. Negative control piglets remained active without the onset of diarrhea and PEDV fecal shedding throughout the experimental period.
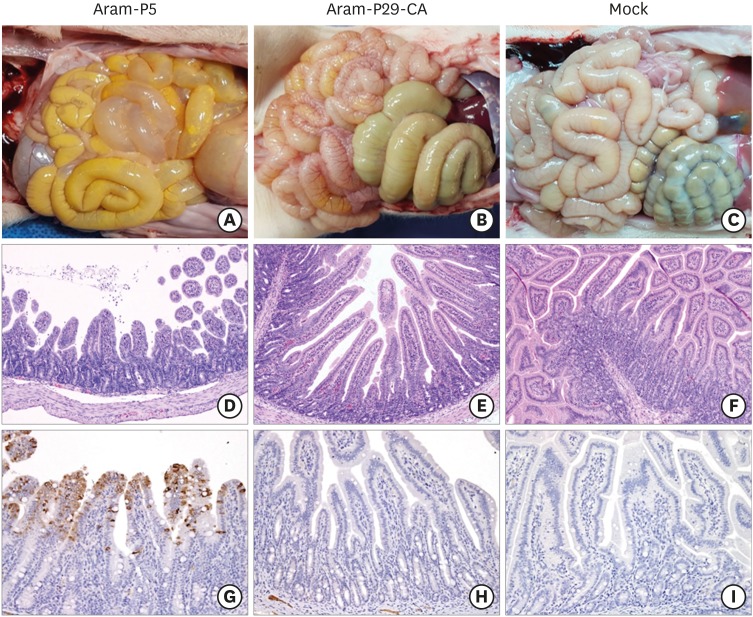
All animals in the parental Aram-P5-infected group were necropsied upon death at 4 or 5 DPI, while piglets in the remaining groups were euthanized at the end of the study for postmortem examinations (Fig. 7). Neither macroscopic nor microscopic intestinal lesions were evident in the negative control piglets (right panels C, F, and I). The virulent Aram-P5-inoculated pigs macroscopically showed archetypal PED-like gross lesions. Their small intestines were dilated with accumulated yellowish fecal fluid and had thin transparent walls as a result of villous atrophy (panel A), whereas the other internal organs appeared normal. In contrast, all animals infected with cold-adapted Aram-P29-CA in group 2 displayed no remarkable visible pathological lesions in their gastrointestinal tracts with normal bowel wall thickness, comparable to those in the negative control group (panel B). Microscopic assessment revealed that the small intestines from all dead piglets in group 1 were characterized by acute viral enteritis, with villous shortening and fusion, involving vacuolation of superficial epithelial cells, in the jejunum (panel D). Furthermore, IHC staining showed intense antigen labeling in the cytoplasm of epithelial cells in atrophied intestinal villi with a immunosignal score of 2.3 ± 0.6 (mean ± SDM) in the jejunum (panel G). However, two out of four cold-adapted virus-inoculated pigs in group 2 exhibited mild villous atrophy in the small intestines, while the remaining pigs showed normal intestinal histopathologies (panel E), analogous to those of the negative control group (panel F). PEDV antigen was rarely present in the small intestines in animals of group 2 with a significantly lower mean jejunal viral antigen score (0.3 ± 0.6) (panel H) than that in group 1 (panel G). Altogether, our results indicate that cold-adapted Aram-P29-CA showed noticeably weakened virulence with an attenuated phenotype in the highly susceptible piglets under experimental conditions.
Fig. 7.
Macroscopic and microscopic small intestine lesions in piglets from three groups. (A-C) Small intestines from representative piglets inoculated with PEDV Aram-P5 (A), -P29-CA (B), or mock (C) were examined for gross lesions. Note that only piglets inoculated with the parental virus typical thin and transparent intestinal walls (panel A). (D-F) Hematoxylin and eosin-stained tissue sections of the jejunum from representative piglets inoculated with PEDV Aram-P5 (D), -P29-CA (E), or mock (F) (100× magnification). Jejunums from piglets infected with the virulent Aram-P5 strain showed acute diffuse, severe atrophic enteritis with villous shortening (panel D). The normal villous epithelium of the jejunums was recorded in pigs inoculated with Aram-P29-CA (panel E), and a mock-inoculated piglet (panel F). (G-I) Detection of PEDV antigen by IHC analysis of jejunum tissue sections from representative piglets inoculated with PEDV Aram-P5 (G), -P29-CA (H), or mock (I) (200× magnification). PEDV antigen signals appear as brown staining and were detected in epithelial cells of the jejunums of all PEDV Aram-P5-inoculated piglets (panel G). No PEDV antigen was detected in the jejunums of piglets inoculated with Aram-P29-CA (panel H), or a mock-inoculated piglet (panel I).
PEDV, porcine epidemic diarrhea virus.
Protective efficacy of the cold-adapted live attenuated Aram-P29-CA vaccine
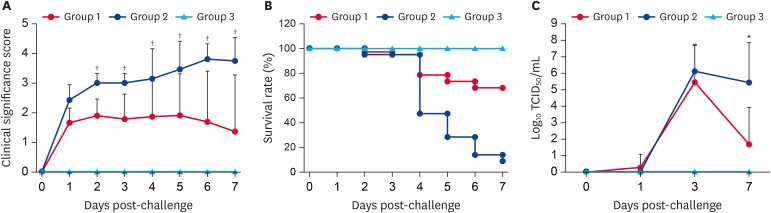
To evaluate the protective effectiveness of a cold-adapted PEDV vaccine, our study employed an oral homologous prime-boost vaccination scheme at 2 weeks apart before farrowing. Three sows in group 1 were orally primed and boosted with live vaccines at 4 and 2 weeks pre-partum, whereas 2 positive (group 2) and 1 negative (group 3) control animals were kept unvaccinated. All sows in the vaccinated and unvaccinated groups experienced neither PED-like clinical symptoms, including PEDV fecal shedding, nor adverse reactions to vaccines. At farrowing, there were no significant differences in reproductive performance between the vaccinated and unvaccinated sows.
Ten piglets per litter were allocated to each sow and were nursed by their dam. At day 4 post-farrowing, all neonates in groups 1 and 2 were orally exposed to the virulent Aram-P5 virus. In contrast, nursing piglets in group 3 were mock-inoculated with MEM. Clinical signs were recorded daily, and fecal swabs were collected before and after the challenge for the duration of the study. During the pre-challenge period, all piglets were healthy and had no clinical symptoms showing normal fecal consistency without PEDV shedding. Following the challenge, PEDV-exposed piglets from unvaccinated sows (group 2) developed clinical symptoms, including anorexia and diarrhea by 1 DPC (mean CSS of > 2.4) and suffered from severe watery diarrhea with vomiting thereafter (Fig. 8A). However, piglets from vaccinated sows (group 1) had mild diarrhea with mean CSS ranges of < 1.36–1.92 throughout the challenge period. These results demonstrated that litters from vaccinated sows significantly mitigated diarrhea after the challenge compared to those from unvaccinated sows.
Fig. 8.
Clinical significance scores, survival rates, and virus shedding in piglets from three experimental groups. Pregnant sows were primed/boosted orally with a live Aram-P29-CA vaccine at 2-week intervals pre-farrowing and their nursing piglets were challenged with virulent G2b PEDV at 4 days of age (A) Clinical significance scores were measured as described in the Materials and Methods section. (B) Survival rate of piglets from vaccinated (group 1), challenge-control (group 2), and negative-control (group 3) sows through 7 DPC. (C) PEDV titers in rectal swap samples at each time point were determined by quantitative real-time reverse transcription polymerase chain reaction analysis. The virus titers (log10 tissue culture infectious dose50/mL) are the mean virus titers from all pigs and error bars represent the mean ± standard deviation of the mean. The p values were calculated by comparing the data from the vaccinated and unvaccinated sow groups after challenge using Student's t-test.
PEDV, porcine epidemic diarrhea virus; DPC, days post-challenge.
*p = 0.001 to 0.05; † p < 0.001.
PEDV-associated mortality occurred in > 50% piglets (11 of 20) from unvaccinated sows by 4 DPI, and additional fatalities occurred until the end of the study, indicating a rate of > 90% mortality in the unvaccinated challenge control group 2 (Fig. 8B). In contrast, 10 of 30 piglets birthed by vaccinated sows (group 1) died from PEDV exposure, showing an approximately 70% survival rate in the piglets. PEDV fecal shedding was detected in all piglets from unvaccinated sows (group 2) by 1 or 2 DPC, and they persistently discharged high quantities (> 105.00 TCID50/mL) of PEDV in feces until death (Fig. 8C). However, all animals from vaccinated sows (group 1) showed PEDV shedding in feces by 3 DPI with a peak mean titer of 105.47 TCID50/mL but amounts of fecal shedding significantly declined thereafter. All piglets from the negative control sow (group 3) remained alive and had no viral shedding in rectal swabs for the duration of the study.
Neutralizing antibody responses of the cold-adapted live attenuated Aram-P29-CA vaccine
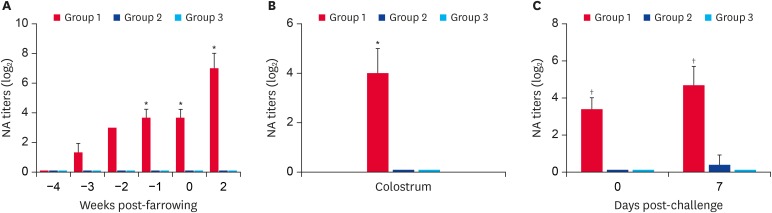
Considering that neutralizing antibodies are associated with protective immunity in neonates against PEDV infection, we tested serum and colostrum samples collected from sows and their neonatal piglets for the presence of anti-PEDV antibodies by an SN assay. As shown in Fig. 9A, all sows developed anti-PEDV neutralizing antibodies after the live prime vaccination. Thereafter, the neutralizing antibody titers in sera of vaccinated sows gradually increased, likely resulting from a homologous boost. All the three sows maintained high neutralizing antibody titers for 2 weeks post-farrowing. Consistent to the results from the serum samples, the neutralizing antibody level was significantly high in the colostrum of vaccinated sows (Fig. 9B). Furthermore, the nursing piglets from each litter in group 1 had pre-challenge neutralizing antibody levels similar to those of their own dam and retained antibodies up to 7 DPI, indicating the protective effect of lactogenic immunity passively acquired from vaccinated sows (Fig. 9C). In contrast, none of unvaccinated sows and their piglets showed anti-PEDV neutralizing antibodies in the serum and colostrum samples (groups 2 and 3). Taken together, these data demonstrate that oral vaccination with the cold-adapted live attenuated virus efficiently elicits potent antibody responses in sows, which are subsequently transferred to the offspring via their milk to provide protection against PEDV.
Fig. 9.
PEDV-specific neutralizing antibody responses in serum (A) and colostrum (B) samples of sows and sera (C) of their corresponding litters from three experimental groups. Pregnant sows were primed/boosted orally with a live Aram-P29-CA vaccine at 4 and 2 weeks pre-farrowing and their nursing piglets were challenged with virulent G2b PEDV at 4 days of age. The samples were collected at the indicated time points and were tested by virus neutralization assay using Aram-P5. Neutralizing antibody titers for individual samples were presented as a log2 scale. Values are representative of the mean from three independent experiments in duplicate and error bars denote the mean ± standard deviation of the mean. The p values were calculated by comparing the data from the vaccinated and unvaccinated sow groups after challenge using Student's t-test.
*p = 0.001 to 0.05; † p < 0.001.
DISCUSSION
A sow does not possess the means to transplacentally transmit maternal immunoglobulins to its fetus; hence, piglets are born without passive immune protection, being highly susceptible to a variety of infectious agents, particularly multiple enteric pathogens, such as PEDV. As a substitute, the passive immunity is dependent on the supplement of maternal-derived immune components via mammary secretions (i.e., colostrum and milk) [15,33]. Therefore, maternal vaccination is the only crucial and effective tool to confer passive protection to newborn piglets through lactogenic immunity against PEDV infection. This objective may be accomplished by the selection of an ideal vaccine candidate, the antigenicity of which is close to that of an epidemic strain, and virulence is minimal or absent in piglets. In South Korea, several G1a PEDV-based live vaccines have been extensively employed to prevent PED for decades. However, owing to partial protection of these historical vaccines against the contemporary G2b strains prevailing since the 2013–2014 pandemic, a demand for the development of new effective vaccines based on the dominant epidemic strain has increased. Although efforts to pioneer a new G2b-based MLV vaccine have been made over the past several years, its success was delayed because of experimental hurdles such as a laborious long-term attenuation procedure in cell culture. It is acknowledged that cold adaptation of animal viruses at serially reduced temperatures for cultivation is the best method to derive a live virus vaccine line, as it is generally accompanied by loss of virulence [21]. In this study, we used a cold adaptation approach with a stepwise lowering of the cultivation temperature in vitro for PEDV attenuation and aimed to determine the immunogenicity and pathogenicity of the cold-adapted strain and its efficacy as an attenuated live vaccine candidate under experimental conditions.
As the pandemic of the G2b PEDV strain ravages the global swine industry, the PEDV field is moving towards the development of novel vaccines and vaccination strategies. To accomplish this task, we first isolated field G2b PEDV that can be efficiently propagated in vitro. The G2b isolate, Aram-P5, exhibited comparable growth characteristics with other G2b strains in regard to cytopathology, infectious titers, and replication kinetics in Vero cells as reported previously [23]. Genetic and phylogenetic analyses showed that the Aram isolates are most closely related to the recent G2b strains prevalent worldwide. However, sequence comparisons with other PEDV strains revealed outstanding genomic features in the Aram strain, which naturally included two independent DELs in S and ORF3, respectively. More interestingly, due to the 2-nt DEL in ORF3, Aram is predicted to encode an alternative ORF3 protein with a large 87-aa excised C-terminus. Although the exact function of PEDV ORF3 remains unknown, a number of attenuated PEDV strains contain their signature DELs in several parts of ORF3, suggesting its critical role in viral pathogenesis [24,34,35]. In particular, Wang et al. [11] reported an attenuated PEDV that encodes a C-terminal truncated ORF3 protein of 91-aa residues, which is form the most analogous to the ORF3 of the Aram strain. However, experimental oral inoculation of newborn pigs with Aram-P5 induced severe clinical presentations and macroscopic and microscopic intestinal lesions typical of acute PEDV infection as reported previously [23]. In general, death rates average 50% in suckling piglets up to one week of age, often approaching 100% in neonates less than 3 days of age, and decrease to 10% thereafter [1,2]. Likewise, our study reproduced 100% mortality in 5-day-old conventional piglets inoculated orally with the 10−2 diluted Aram virus (calculated dose of 103 TCID50 per pig). The data demonstrated that the Aram isolate was highly enteropathogenic in neonatal piglets, implying that the ORF3 product would be irrelevant to PEDV virulence. Since a growing body of evidence proposes that a combination of multiple genetic mutations in S, ORF3, and functional nsps affects the virulence of PEDV, we cannot exclude that the C-terminal defect in ORF3 may be one of molecular and physiological factors that may simultaneously alter pathogenic mechanisms. However, consistent with previous works [24,36], growth kinetics of the Aram strain in cell culture supports the notion that the accessory ORF3 gene is dispensable for PEDV replication in vitro. Collectively, despite a redundant role in virus propagation in vitro, conservation of the complete ORF3 in PEDV field isolates still suggests its importance in causing natural infection in the animal host.
To serve as a novel MLV vaccine strain, we generated an Aram-derived attenuated PEDV strain using a cold adaptation attenuation method. Our approach for viral attenuation was to obtain a cold-adapted strain with impaired growth at physiological temperature (37°C), which can sufficiently induce mucosal immunity in the natural host. The cold-adapted Aram-P29-CA strain was derived by 24 serial passages of the Aram isolate at a low temperature (32°C). The Aram-P29-CA strain developed unsuccessful in vitro infection at the physiological temperature, as a significant decrease in infectious units was observed. However, this virus showed significantly high immunogenicity in orally inoculated pigs. Furthermore, when investigating the phenotype of Aram-P29-CA in highly susceptible 5 day-old piglets, none of the inoculated animals became clinically sick and grossly abnormal. Our experimental data indicate that the cold-adapted Aram-P29-CA virus is attenuated virologically in vitro and clinically in vivo. Subsequently, we sequenced the entire genome of the Aram-P29-CA strain to decode the genetic mutations that emerged during sequential cell passages at 32°C. Upon cold adaptation, a total of 12 non-silent mutations arose in ORFs 1 through 5, except for ORF3, without extra INDELs throughout the genome, whereas the two S and ORF3 DEL signatures of the Aram strain were entirely maintained. Intriguingly, 7-aa substitutions were accumulated in the S protein, among which 4- and 3-aa substitutions were distributed in the N-terminus of S1 and S2 domains, respectively, suggesting that some of these changes are associated with viral attenuation. Since the PEDV S protein is heavily glycosylated, altered glycan motifs may be involved in viral pathogenesis by modifying the protein conformation and function [37]. However, no genetic changes were found in the putative N-glycosylation sites between the parental and cold-adapted strains. As the major antigenic determinant, the S glycoprotein possesses at least four neutralizing domains of PEDV at aa positions 19–220 (NTD/S0), 499–600, 744–774, and 1371–1377 [38]. The aa residues that comprise these domains remained nearly unchanged throughout the cold adaptation, except for one N209Y mutation in the NTD/S0 region. Furthermore, no aa changes were found in additional discrete domains, including a C-terminal domain (residues 477–629) of S1 that can interact with cellular receptor(s) and a S2 fusion domain comprising of a hydrophobic fusion peptide (residues 891–908) and two heptad repeat regions (residues 978–1117 and 1274–1313). Although the polygenic traits are likely associated with PEDV virulence, it is of utmost importance that future work using reverse genetics technology should aim to address whether genetic drift, especially in the S protein, influences the pathogenicity of PEDV. In addition, attenuated viruses often have the propensity to revert to virulent form under certain circumstances, which is a main drawback for their use as an MLV vaccine. Indeed, the Aram-P29-CA strain had an ability to replicate at 37°C since it denoted impaired, but not entirely halted, growth at 37°C, as the viral titer was determined to be 102.5 TCID50/mL. This incompletely abolished growth of Aram-P29-CA at the physiological temperature implies a residual risk of reversion to virulence in vivo. Thus, further safety studies will be necessary to evaluate the phenotypic stability of the attenuated Aram-P29-CA virus in neonatal piglets.
To assess the effectiveness of the cold-adapted live attenuated vaccine, a homologous prime-boost sow immunization trial consisting of oral administration of a double-dose of the P29-CA virus (L/L vaccination) was implemented at intervals of 2 weeks before delivery. Newborn piglets born to vaccinated and unvaccinated sow groups were inoculated orally with the parental virulent PEDV strain. The difference in the burden of diarrheic disease as measured by mortality and morbidity was investigated between the challenged litters from vaccinated and unvaccinated sows. Piglets born to unvaccinated sows presented a much higher mean CSS, as determined by the intensity of diarrhea, than those of vaccinated sows. After 2 DPC, the challenged animals born to vaccinated (group 1) sows had mild-to-moderate diarrhea, whereas the piglets born to unvaccinated (group 2) sows experienced fatal watery diarrhea. Although nearly all the piglets in group 2 died by 7 DPC, group 1 also exhibited more than 30% neonatal mortalities during the experimental period. On the contrary, our previous trial has shown that a multiple-dose parenteral immunization with inactivated killed G2b vaccines (K/K vaccination) offered a more protective efficacy (93% survival rate) in piglets against G2b virus than that in the present study [39]. In contrast to piglets from unvaccinated sows that discharged high amounts of the virus in feces until death, PEDV fecal shedding in animals from vaccinated sows was greatly reduced at 7 DPC and more than 50% of the piglets that survived shed no virus in stools at the end of the trial. Collectively, the maternal L/L vaccination described in the present study is unlikely to be more efficacious than the K/K treatment in terms of the protective rate; however, it could help alleviate diarrheal severity, including the duration and quantity of fecal shedding of PEDV. Although our L/L vaccination of sows did not entirely avert morbidity in piglets upon a PEDV challenge, its advantageous effects on relieving PEDV shedding would lessen the possibility of the environmental contamination in the farrowing barns, thereby breaking the chain of secondary spread of the virus to other animals and herds during epidemics. Furthermore, PEDV transmission via the L/L vaccination is unlikely, as no PEDV genetic material was detected in feces from any of the pregnant sows vaccinated with P29-CA. Moreover, we were able to confirm the presence of satisfactory amounts of anti-PEDV neutralizing antibodies in the sera and colostrum of L/L vaccinated sows at farrowing and post-farrowing stages, as well as in sera obtained from their offspring, indicating the piglet protection by transfer of maternal immunity via mammary secretions from the immunized dams. These data further strengthen the concept that an optimal vaccination regimen is associated with retaining high levels of neutralizing antibodies in the lactogenic secretions of vaccinated sows, which in turn are correlated with colostrum and milk IgA antibody titers to offer protective immunity against PEDV [1,2,15,40].
In summary, this is the first report describing the development of a cold-adapted MLV vaccine based on a virulent G2b PEDV strain. The present study demonstrated the loss of virulence of a highly pathogenic PEDV strain, with 12 genetic mutations throughout the entire genome, after cold adaptation. Vaccination-challenge studies revealed that sows immunized with a homologous oral L/L maternal vaccination regimen provides protective lactogenic immunity to piglets, thereby reducing mortality, morbidity, and fecal shedding of the virus. In South Korea, multiple-dose vaccination programs including L/K/K at 2- or 3-week intervals before delivery have been recommended nationwide in pregnant sows for decades [1,2]. Due to the absence of a new safe and effective G2b live vaccine, controlled oral exposure (feedback) for prime-boost vaccination strategies is mostly implemented as a countermeasure in the field since the 2013–2014 PED epidemics. Despite certain positive contributions of feedback to naïve herds, intentional infection using an undefined inoculum may cause undesirable negative consequences, which include insufficient immunity induction, unexpected virus transmission, amplified viral recombination and diversity; and more importantly, dissemination of other microbial pathogens conceivably evolving to develop into a new disease agent. Once the cold-adapted Aram-P29-CA-based MLV vaccine is supplied in the market, gradual withdrawal of the feedback practice will be imminent. Since oral administration does not have the capacity to ensure that all animals are immunized with a proper dose, this demerit should be ameliorated to easily use the live vaccine under field conditions. With the availability of inactivated G2b PEDV vaccines in the domestic market, customized vaccination programs, such as the application of heterologous oral prime-parenteral boost L/K/K or L/L/K schemes, could be established based on various herd circumstances, including naïve, epidemic, and endemic statuses. It is noteworthy that maternal vaccination has to be implemented in conjunction with additional intervention strategies involving intensive biosecurity, husbandry, and hygiene practices for PED prevention and eradication.
Footnotes
Funding: This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2018R1D1A1B07040334).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Won H, Choi HW, Lee C.
- Data curation: Lee C.
- Formal analysis: Won H, Lee DU, Jang G, Noh YH, Lee C.
- Funding acquisition: Lee C.
- Investigation: Won H, Lee DU, Jang G, Noh YH, Lee SC.
- Methodology: Won H, Lee DU, Jang G.
- Project administration:
- Resources: Yoon IJ, Lee C.
- Software: Lee DU, Jang G.
- Supervision: Yoon IJ, Yoo HS, Lee C.
- Visualization: Won H, Jang G.
- Writing - original draft: Won H, Lee C.
- Writing - review & editing: Lee C.
References
- 1.Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. Porcine epidemic diarrhoea virus. In: Zakaryan H, editor. Porcine Viruses: From Pathogenesis to Strategies for Control. Norfolk: Caister Academic Press; 2019. pp. 107–134. [Google Scholar]
- 3.Lai MM, Perlman S, Anderson LJ. Coronaviridae. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, Straus SE, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1306–1336. [Google Scholar]
- 4.Lee DK, Park CK, Kim SH, Lee C. Heterogeneity in spike protein genes of porcine epidemic diarrhea viruses isolated in Korea. Virus Res. 2010;149:175–182. doi: 10.1016/j.virusres.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S, Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg Infect Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J, Lee KW, Choi HW, Lee C. Immunogenicity and protective efficacy of recombinant S1 domain of the porcine epidemic diarrhea virus spike protein. Arch Virol. 2014;159:2977–2987. doi: 10.1007/s00705-014-2163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvajal A, Argüello H, Martínez-Lobo FJ, Costillas S, Miranda R, de Nova PJ, Rubio P. Porcine epidemic diarrhoea: new insights into an old disease. Porcine Health Manag. 2015;1:12. doi: 10.1186/s40813-015-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mole B. Deadly pig virus slips through US borders. Nature. 2013;499:388. doi: 10.1038/499388a. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 11.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg Infect Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin CN, Chung WB, Chang SW, Wen CC, Liu H, Chien CH, Chiou MT. US-like strain of porcine epidemic diarrhea virus outbreaks in Taiwan, 2013–2014. J Vet Med Sci. 2014;76:1297–1299. doi: 10.1292/jvms.14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki T, Murakami S, Takahashi O, Kodera A, Masuda T, Itoh S, Miyazaki A, Ohashi S, Tsutsui T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect Genet Evol. 2015;36:363–368. doi: 10.1016/j.meegid.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Bohl EH, Gupta RK, Olquin MV, Saif LJ. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect Immun. 1972;6:289–301. doi: 10.1128/iai.6.3.289-301.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langel SN, Paim FC, Lager KM, Vlasova AN, Saif LJ. Lactogenic immunity and vaccines for porcine epidemic diarrhea virus (PEDV): historical and current concepts. Virus Res. 2016;226:93–107. doi: 10.1016/j.virusres.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poonsuk K, Giménez-Lirola LG, Zhang J, Arruda P, Chen Q, Correa da Silva Carrion L, Magtoto R, Pineyro P, Sarmento L, Wang C, Sun Y, Madson D, Johnson J, Yoon KJ, Zimmerman J, Main R. Does circulating antibody play a role in the protection of piglets against porcine epidemic diarrhea virus? PLoS One. 2016;11:e0153041. doi: 10.1371/journal.pone.0153041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goede D, Murtaugh MP, Nerem J, Yeske P, Rossow K, Morrison R. Previous infection of sows with a “mild” strain of porcine epidemic diarrhea virus confers protection against infection with a “severe” strain. Vet Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Lee JM, Jung J, Kim IJ, Hyun BH, Kim HI, Park CK, Oem JK, Kim YH, Lee MH, Lee KK. Genetic characterization of porcine epidemic diarrhea virus in Korea from 1998 to 2013. Arch Virol. 2015;160:1055–1064. doi: 10.1007/s00705-015-2353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buonagurio DA, O'Neill RE, Shutyak L, D'Arco GA, Bechert TM, Kazachkov Y, Wang HP, DeStefano J, Coelingh KL, August M, Parks CL, Zamb TJ, Sidhu MS, Udem SA. Genetic and phenotypic stability of cold-adapted influenza viruses in a trivalent vaccine administered to children in a day care setting. Virology. 2006;347:296–306. doi: 10.1016/j.virol.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Cha TA, Kao K, Zhao J, Fast PE, Mendelman PM, Arvin A. Genotypic stability of cold-adapted influenza virus vaccine in an efficacy clinical trial. J Clin Microbiol. 2000;38:839–845. doi: 10.1128/jcm.38.2.839-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maassab HF, DeBorde DC. Development and characterization of cold-adapted viruses for use as live virus vaccines. Vaccine. 1985;3:355–369. doi: 10.1016/0264-410x(85)90124-0. [DOI] [PubMed] [Google Scholar]
- 22.Wei Y, Qi L, Gao H, Sun H, Pu J, Sun Y, Liu J. Generation and protective efficacy of a cold-adapted attenuated avian H9N2 influenza vaccine. Sci Rep. 2016;6:30382. doi: 10.1038/srep30382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Kim Y, Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015;208:215–224. doi: 10.1016/j.virusres.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, Son KY, Noh YH, Lee SC, Choi HW, Yoon IJ, Lee C. Genetic characteristics, pathogenicity, and immunogenicity associated with cell adaptation of a virulent genotype 2b porcine epidemic diarrhea virus. Vet Microbiol. 2017;207:248–258. doi: 10.1016/j.vetmic.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27:493–497. [Google Scholar]
- 26.Lee YN, Lee C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch Virol. 2013;158:1765–1772. doi: 10.1007/s00705-013-1770-z. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 30.Kim SH, Kim IJ, Pyo HM, Tark DS, Song JY, Hyun BH. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J Virol Methods. 2007;146:172–177. doi: 10.1016/j.jviromet.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagong M, Lee C. Porcine reproductive and respiratory syndrome virus nucleocapsid protein modulates interferon-β production by inhibiting IRF3 activation in immortalized porcine alveolar macrophages. Arch Virol. 2011;156:2187–2195. doi: 10.1007/s00705-011-1116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madson DM, Magstadt DR, Arruda PH, Hoang H, Sun D, Bower LP, Bhandari M, Burrough ER, Gauger PC, Pillatzki AE, Stevenson GW, Wilberts BL, Brodie J, Harmon KM, Wang C, Main RG, Zhang J, Yoon KJ. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet Microbiol. 2014;174:60–68. doi: 10.1016/j.vetmic.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Saif LJ, Jackwood D. Viral Diarrhea of Man and Animals. Bocal Raton: CRC Press; 1990. [Google Scholar]
- 34.Park SJ, Moon HJ, Luo Y, Kim HK, Kim EM, Yang JS, Song DS, Kang BK, Lee CS, Park BK. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. 2008;36:95–104. doi: 10.1007/s11262-007-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Lu W, Chen J, Xie S, Shi H, Hsu H, Yu W, Xu K, Bian C, Fischer WB, Schwarz W, Feng L, Sun B. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012;586:384–391. doi: 10.1016/j.febslet.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Li Z, Zou Y, Wicht O, van Kuppeveld FJ, Rottier PJ, Bosch BJ. Manipulation of the porcine epidemic diarrhea virus genome using targeted RNA recombination. PLoS One. 2013;8:e69997. doi: 10.1371/journal.pone.0069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato T, Takeyama N, Katsumata A, Tuchiya K, Kodama T, Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo . Virus Genes. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okda FA, Lawson S, Singrey A, Nelson J, Hain KS, Joshi LR, Christopher-Hennings J, Nelson EA, Diel DG. The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology. 2017;509:185–194. doi: 10.1016/j.virol.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baek PS, Choi HW, Lee S, Yoon IJ, Lee YJ, Lee S, Lee S, Lee C. Efficacy of an inactivated genotype 2b porcine epidemic diarrhea virus vaccine in neonatal piglets. Vet Immunol Immunopathol. 2016;174:45–49. doi: 10.1016/j.vetimm.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Q, Stone S, Drebes D, Greiner LL, Dvorak CM, Murtaugh MP. Characterization of anti-porcine epidemic diarrhea virus neutralizing activity in mammary secretions. Virus Res. 2016;226:85–92. doi: 10.1016/j.virusres.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]