Abstract
Background
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) colonizes the gastrointestinal tract of intensive care unit (ICU) patients, and CRPA colonization puts patients at increased risk of CRPA infection. Prior studies have not examined relationships between the microbiota, medications, and CRPA colonization acquisition.
Methods
Data and perirectal swabs were obtained from a cohort of ICU patients at the University of Maryland Medical Center. Patients (N = 109) were classified into 3 groups by CRPA colonization-acquisition status and antimicrobial exposure. We conducted 16S ribosomal RNA gene sequencing of an ICU admission swab and ≥1 additional swab and evaluated associations between patient characteristics, medications, the gastrointestinal microbiota, and CRPA colonization acquisition.
Results
ICU patients had low levels of diversity and high relative abundances of pathobionts. Piperacillin-tazobactam was prescribed more frequently to patients with CRPA colonization acquisition than those without. Piperacillin-tazobactam was associated with low abundance of potentially protective taxa (eg, Lactobacillus and Clostridiales) and increased risk of Enterococcus domination (odds ratio [OR], 5.50; 95% confidence interval [CI], 2.03–14.92). Opioids were associated with dysbiosis in patients who did not receive antibiotics; potentially protective Blautia and Lactobacillus were higher in patients who did not receive opioids. Several correlated taxa, identified at ICU admission, were associated with lower risk of CRPA colonization acquisition (OR, 0.58; 95% CI, .38–.87).
Conclusions
Antibiotics differed in their impact on the microbiota, with piperacillin-tazobactam being particularly damaging. Certain bacterial taxa (eg, Clostridiales) were negatively associated with CRPA colonization acquisition. These taxa may be markers of risk for CRPA colonization acquisition and/or serve a protective role.
Keywords: microbiota, hospital-acquired infection, antibiotic resistance, antimicrobial stewardship, carbapenem-resistant Pseudomonas
Carbapenem-resistant Pseudomonas aeruginosa (CRPA) can colonize intensive care patients and cause infection. Antibiotics differ in their impact on the gastrointestinal microbiota; piperacillin-tazobactam is particularly damaging. Decreases in abundance of several bacterial taxa were associated with increased risk of CRPA colonization and acquisition.
The gastrointestinal (GI) microbiota provides colonization resistance (ie, the prevention of pathogen colonization and/or inhibition of pathogen overgrowth) [1]. A significant subset of hospitalized patients experience collapse of their GI microbiota [2, 3]. Dysbiosis and concomitant loss of colonization resistance put patients at increased risk of colonization and infection by antibiotic-resistant pathogens [4, 5].
Antibiotics are major contributors to dysbiosis in the GI microbiota [6, 7]. However, data to guide clinicians in the selection of antibiotics that minimize microbiota disruption remain limited [8, 9]. Studies of the impact of antibiotics on the microbiota often focus on healthy individuals who differ from hospitalized patients and thus may not represent the appropriate study group [6, 10]. The clinical spectrum (eg, broad or narrow) of a given antibiotic differs from its impact on the GI microbiota, which is determined by concentrations achieved in the GI tract and susceptibility of the microbiota [9]. Moreover, the pharmacokinetics of antibiotics may differ in intensive care unit (ICU) patients compared to healthy individuals [11]. ICU patients are exposed to other medications (eg, opioids), which may also disrupt the microbiota [2, 12].
Pseudomonas aeruginosa is frequently resistant to antibiotics and readily colonizes the GI tract of hospitalized patients [13]. Approximately 28% of ICU patients colonized with carbapenem-resistant P. aeruginosa (CRPA) develop infection due to their colonizing strain during their ICU admission [14]. Thus, prevention of CRPA colonization acquisition represents an important target for interventions to reduce infection and spread of CRPA in the hospital. We reasoned that greater understanding of relationships between the microbiota, medications, and CRPA colonization acquisition would help inform the selection of antibiotics that minimize microbiota disruption and advance development of tools for patient monitoring for risk of CRPA colonization and infection. Thus, our study goals were to (1) describe the GI microbiota of ICU patients; (2) examine the impact of antibiotics and other medications on the GI microbiota; and (3) identify taxonomic markers associated with CRPA colonization acquisition in the GI tract.
METHODS
Study Design and Participants
The institutional review board of the University of Maryland, Baltimore, approved this study. We used samples and data from a longitudinal cohort of adult patients admitted to the medical ICU (MICU) and surgical ICU (SICU) at the University of Maryland Medical Center (UMMC). The UMMC is a 767-bed tertiary care facility in Baltimore. The MICU and SICU have private rooms and provide care to patients with acute and/or life-threatening medical conditions and patients undergoing solid organ transplantation and other surgeries, respectively.
Perirectal swabs were obtained at ICU admission, weekly, and at ICU discharge as part of an ongoing active surveillance program for vancomycin-resistant enterococci (VRE). Rayon swabs (BBL CultureSwab, Becton Dickinson) were cultured for VRE and imipenem-resistant P. aeruginosa (to identify CRPA colonization acquisition) as described previously [14]. After culture, the perirectal swabs were frozen and stored at –80°C.
Study patients entered the ICU negative for CRPA and were classified into 3 groups: Group 1 included patients with CRPA colonization acquisition during their ICU stay; group 2 included patients without CRPA colonization acquisition; and group 3 included patients who did not receive systemic antibiotics and did not have CRPA colonization acquisition during their ICU stay (Supplementary Figure 1). We analyzed 3 swabs from patients in group 1: (1) the ICU admission swab; (2) the swab obtained closest to the date prior to the first CRPA-positive swab; and (3) the first swab with CRPA detected. We evaluated 2 swabs from each patient in groups 2 and 3. For group 2 patients, we analyzed the ICU admission swab and a second swab temporally matched to swab 2 from patients in group 1. For patients in group 3, we analyzed the ICU admission and discharge swabs. Samples were collected between 5 September 2001 and 21 March 2009.
Data were obtained from the UMMC Central Data Repository and medical records review. For each study patient, we collected demographic data such as sex, age, and race. Clinical data included antibiotics and other medications prescribed during the ICU stay, comorbidities, and ICU admission and discharge dates. Discharge International Classification of Diseases, Ninth Revision codes were used to classify patients as immunocompromised and to obtain the comorbidities for calculating Charlson and Elixhauser comorbidity indices [15, 16]. Antibiotics and antibiotic–inhibitor combinations were analyzed individually and in clinically relevant groups. Ampicillin-sulbactam, cefotetan, clindamycin, imipenem-cilastatin, metronidazole, and piperacillin-tazobactam were grouped as antianaerobic. Cefepime, ciprofloxacin, gentamicin, imipenem-cilastatin, and piperacillin-tazobactam were grouped as antipseudomonal. Linezolid and vancomycin were grouped as anti–methicillin-resistant Staphylococcus aureus (MRSA).
Sequencing and Analysis of 16S Ribosomal RNA Gene Sequences
DNA was extracted from swabs using the PureLink Microbiome DNA Purification Kit (Invitrogen, Carlsbad, California). Extracted DNA and negative reagent controls were used for polymerase chain reaction (PCR) amplification of 16S ribosomal RNA hypervariable region V4 [17]. Samples were PCR amplified in duplicate. Pooled PCR product and reagent controls were sequenced at the Yale Center for Genome Analysis on the Illumina MiSeq using a paired-end 250 bp protocol with a PhiX control.
We used Btrim software to sort, trim, and filter low-quality sequence reads [18]. USEARCH software was used for dereplication and removing chimeras. Samples were clustered at 97% identity using UPARSE-OTU. Next, we created operational taxonomic unit (OTU) tables, which were normalized to 10000 reads per sample [5, 19]. USEARCH software was used to calculate diversity indices and define taxonomic groupings [20]. Taxonomic classification was assigned at the genus level and/or the lowest taxonomic level possible at 80% confidence.
Statistical Analysis
Statistical analyses were conducted using SAS version 9.3 and R 3.0.1 software. Diversity indices calculated for each sample were the Shannon (natural log), Jost1 (exp[Shannon]), Simpson, and the Simpson reciprocal index (1/Simpson). The Shannon diversity index provides a measure abundance and evenness within a sample [21]. The Jost1 index provides an estimate of the effective number of species [22]. The Simpson diversity index describes the probability that 2 randomly selected sequence reads will be members of the same OTU and gives more weight to abundant taxa [21]. The Simpson reciprocal index provides an estimate of the effective number of abundant taxa.
We used linear discriminant analysis effect size (LefSe), with 3 as the threshold for significance, to identify differentially present OTUs in each of 2 populations (eg, patients exposed to a given antibiotic [yes/no]) [23]. Principal components analysis (PCA) was used to identify groups of correlated taxa. After excluding rare OTUs (ie, a proportion < 0.001 of the microbiota in ≥90% of the samples), remaining OTU proportions were normalized with an arcsine square-root transformation prior to inclusion in the PCA [24]. We specified an eigenvalue of 1 and an orthogonal rotation. OTUs with a loading value of at least ±0.6 were retained for that factor [25].
We evaluated unadjusted associations between patient characteristics or exposures of interest (eg, taxa differing in abundance between groups) and outcomes (eg, CRPA colonization acquisition) by χ2 test, Fisher exact test, and repeated-measures analysis of variance (ANOVA) as appropriate. Clinical and microbiota-specific data were incorporated in a series of regression models to predict outcomes of interest.
RESULTS
Perirectal swabs and data were collected from 109 ICU patients including 41 in group 1, 45 in group 2, and 23 in group 3 (Table 1). The 3 patient groups did not significantly differ by age, sex, race, Charlson comorbidity index, Elixhauser comorbidity index, or time between collection of swabs 1 and 2 (P > .05 for each). The proportion of patients who were immunocompromised was similar in groups 1 and 2. Fewer patients in group 3 were immunocompromised compared with patients in group 1 or 2.
Table 1.
Characteristics of the Study Population
| Characteristic | All Patients (N = 109) |
Group 1a (n = 41) |
Group 2b (n = 45) |
Group 3c (n = 23) |
P Valued |
|---|---|---|---|---|---|
| Age, y, mean (SD) | 53.5 (13.6); range, 18–89 | 52.8 (14.8) | 54.8 (11.9) | 52.0 (14.7) | .66 |
| Sex | .82 | ||||
| Female | 46 (42.2) | 17 (41.5) | 18 (40.0) | 11 (47.8) | |
| Male | 63 (57.8) | 24 (58.5) | 27 (60.0) | 12 (52.2) | |
| Racee | .38 | ||||
| Black | 53 (49.5) | 22 (55.0) | 21 (46.7) | 10 (45.4) | |
| White | 50 (46.7) | 15 (37.5) | 23 (51.1) | 16 (54.6) | |
| Other | 4 (3.7) | 3 (7.5) | 1 (2.2) | 0 | |
| Immunocompromised | .04 | ||||
| No | 71 (65.1) | 23 (56.1) | 28 (62.2) | 20 (87.0) | |
| Yes | 38 (34.9) | 18 (43.9) | 17 (37.8) | 3 (13.0) | |
| Charlson score, median (IQR) | 1 (2); range, 0–9 | 1 (2) | 1 (2) | 2 (2) | .94 |
| 0 | 24 (22.0) | 9 (22.0) | 11 (24.4) | 4 (17.4) | |
| 1 | 33 (30.3) | 13 (31.7) | 12 (26.7) | 8 (34.8) | |
| 2 | 19 (17.4) | 7 (17.1) | 7 (15.6) | 5 (21.7) | |
| 3–4 | 20 (18.4) | 9 (22.0) | 8 (17.8) | 3 (13.0) | |
| ≥5 | 13 (11.9) | 3 (7.3) | 7 (15.6) | 3 (13.0) | |
| Total Elixhauser score, median (IQR) | 4 (2) (range, 1–7) | 4 (3) | 4 (2) | 4 (3) | .87 |
| 1–2 | 25 (22.9) | 11 (26.8) | 7 (15.6) | 7 (30.4) | |
| 3 | 22 (20.2) | 7 (17.1) | 11 (24.4) | 4 (17.4) | |
| 4 | 21 (19.3) | 9 (22.0) | 9 (20.0) | 3 (13.0) | |
| 5 | 27 (24.8) | 10 (24.4) | 11 (24.4) | 6 (26.1) | |
| ≥6 | 14 (12.8) | 4 (9.8) | 7 (15.6) | 3 (13.0) | |
| Time from swab 1 to swab 2, d, median (IQR)f | 8 (9); range, 2–35 | 7 (14) | 9 (11) | 7 (4) | .35 |
| ≤7 | 53 (48.6) | 21 (51.2) | 19 (42.2) | 13 (56.5) | |
| 8–14 | 29 (26.6) | 7 (17.1) | 14 (31.1) | 8 (34.8) | |
| 15–21 | 12 (11.0) | 5 (12.2) | 6 (13.3) | 1 (4.4) | |
| ≥22 | 15 (13.8) | 8 (19.5) | 6 (13.3) | 1 (4.4) | |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: IQR, interquartile range; SD, standard deviation.
aGroup 1: Patients who entered the intensive care unit (ICU) culture negative for carbapenem-resistant Pseudomonas aeruginosa (CRPA) and had CRPA colonization acquisition during their ICU stay.
bGroup 2: Patients who entered the ICU culture negative for CRPA and did not have CRPA colonization acquisition during their ICU stay.
cGroup 3: Patients selected because they did not receive systemic antibiotics during their ICU stay, nor had CRPA colonization acquisition during their ICU stay.
d P values: F test for age; χ2 test for all other variables.
eMissing data for group 1 (n = 1) and group 2 (n = 1).
fFor group 1 patients, the median time between swabs 2 and 3 was 7 days (IQR, 3 days), with a range of 1–23 days.
Composition of the GI Microbiota of ICU Patients
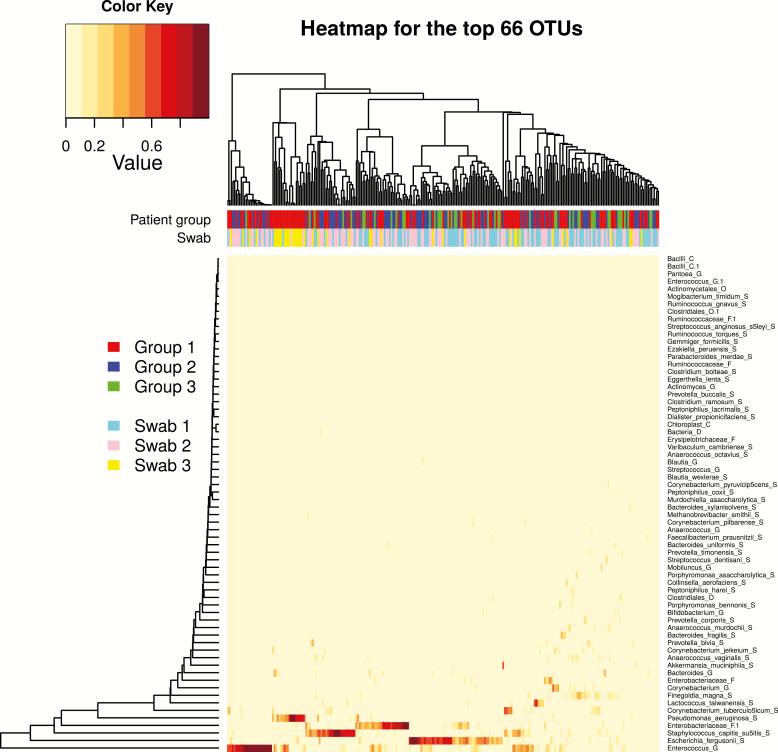
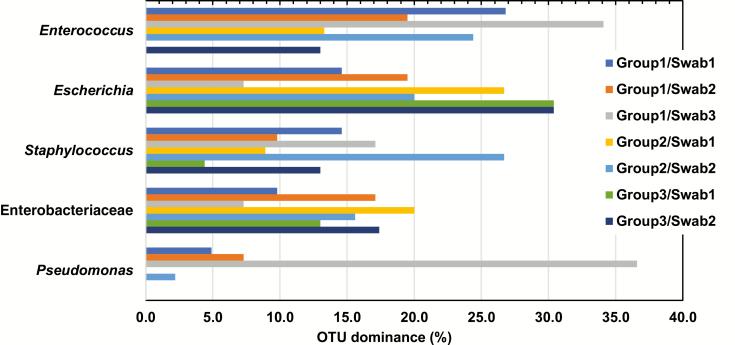
We identified 2047 OTUs in 259 swabs from 109 patients. Hierarchical clustering was used to group GI communities (Figure 1), which clustered based on high relative abundances of Enterococcus (OTU1), Escherichia (OTU2), Staphylococcus (OTU3), Enterobacteriaceae (OTU4), or Pseudomonas (OTU5), or contained a mix of other taxa. The GI microbial communities did not cluster by swab or patient group. A subset of study patients, even some who did not receive antibiotics while in the ICU, had a GI microbiota that was dominated (defined as ≥30% of reads of a single OTU) by at least 1 of 5 pathobionts, Enterococcus, Escherichia, Staphylococcus, Enterobacteriaceae, or Pseudomonas (Figure 2). We examined patterns of dominance in patient groups and swabs (Table 2). Patients in group 2 were 3.7 times more likely to have Staphylococcus dominance on swab 2 if they had Staphylococcus dominance on swab 1. Group 1 patients were >11 times more likely to have Pseudomonas dominance on swab 3 if they had Pseudomonas dominance on swab 1. There were no other significant patterns of dominance observed.
Figure 1.
Heatmap of the most abundant gastrointestinal microbiota taxa (n = 66 of 2047 identified) with a proportion >0.001 for ≥90% of 259 perirectal swab samples from 109 intensive care unit patients. The color keys for patient group and swab are indicated on the left. Abbreviation: OTU, operational taxonomic unit.
Figure 2.
Proportion of samples with domination, defined as ≥30% of reads of a single operational taxonomic unit (OTU), by each of the 5 most frequently identified OTUs by patient and swab. Abbreviation: OTU, operational taxonomic unit.
Table 2.
Odds Ratios and 95% Confidence Intervals From Repeated-Measures Logistic Regression Analyses Predicting Dominance, Defined as ≥30% of Reads of a Single Operational Taxonomic Unit (OTU), on Swab 2 (or Swab 2 or 3 for Group 1) Compared to Swab 1 for Each of the 5 Most Frequent OTUs
| Patient Group | Swab No. | OTU Name | ||||
|---|---|---|---|---|---|---|
| Enterococcus | Escherichia | Staphylococcus | Enterobacteriaceae | Pseudomonas | ||
| Group 1 | Swab 1 (Ref) | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Swab 2 | 0.66 (.30–1.48) | 1.41 (.54–3.69) | 0.63 (.21–1.91) | 1.90 (.62–5.81) | 1.54 (.35–6.70) | |
| Swab 3 | 1.41 (.67–2.99) | 0.46 (.12–1.78) | 1.20 (.37–3.95) | 0.73 (.25–2.12) | 11.25 (2.46–51.42) | |
| Group 2 | Swab 1 (Ref) | 1.00 | 1.00 | 1.00 | 1.00 | … |
| Swab 2 | 2.10 (.80–5.51) | 0.69 (.31–1.54) | 3.73 (1.21–11.49) | 0.74 (.32–1.72) | … | |
| Group 3 | Swab 1 (Ref) | … | 1.00 | 1.00 | 1.00 | … |
| Swab 2 | … | 1.00 (.37–2.68) | 3.22 (.61–16.91) | 1.40 (.32–6.20) | … | |
Data are presented as odds ratio (95% confidence interval). Values in bold represent statistically significant odds ratios. Logistic regression analyses were not possible for OTUs where none of the members in a patient group had the dominant OTU on the reference swab (swab 1)—that is, Enterococcus for group 3 and Pseudomonas for groups 2 and 3.
Abbreviation: OTU, operational taxonomic unit.
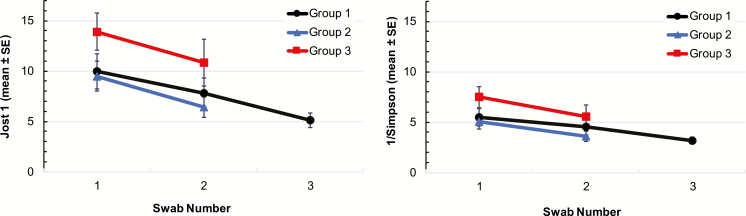
Alpha-diversity declined while patients were in the ICU (Figure 3). Descriptive statistics for α-diversity indices by patient group and swab are shown in Supplementary Table 1. Repeated-measures ANOVA indicated that diversity, as determined by each of the 4 indices, significantly declined in the time between collection of swabs 1 and 2 (Supplementary Table 2). The mean effective number of distinct taxa, Jost1, ranged from a high of 13.9 (standard deviation [SD], 8.8) in group 3, swab 1 to a low of 5.1 (SD, 4.5) in group 1, swab 3 (Figure 3; Supplementary Table 1). The mean effective number of very abundant taxa, the Simpson reciprocal index, ranged from a high of 7.5 (SD, 5.0) in group 3, swab 1 to a low of 3.2 (SD, 2.2) in group 1, swab 3 (Figure 3; Supplementary Table 1). The Shannon and Simpson diversity indices differed by patient group (Supplementary Table 2). Collectively, these data suggest that the effective number of taxa in the GI microbiota is low in ICU patients, even upon admission to the ICU.
Figure 3.
Mean α-diversity indices: Jost1 (effective number of distinct taxa, left panel), Simpson reciprocal index (effective number of abundant taxa, right panel). Abbreviation: SE, standard error.
Impact of Antibiotics and Other Medications on the Microbiota
We quantified prescriptions of antibiotics, proton pump inhibitors, opioids, and steroids between the swab 1 and swab 2 collection dates for each patient (Table 3). Vancomycin was the most frequently prescribed antibiotic. Piperacillin-tazobactam, antianaerobic antibiotics, and antipseudomonal antibiotics were prescribed more frequently to patients in group 1 than in group 2 (P < .05 for each). Piperacillin-tazobactam is included in the antianaerobic and antipseudomonal categories and may be driving some of these associations. Groups 1 and 2 did not significantly differ in frequency of prescriptions for antianaerobic (P = .13) and antipseudomonal (P = .22) antibiotics when piperacillin-tazobactam was not included in these categories.
Table 3.
Medication Use Among Patient Groups
| Medication | Patient Group | Group 1 (n = 41) | Group 2 (n = 45) | P Valuea | Group 3 (n = 23) | P Valueb |
|---|---|---|---|---|---|---|
| Antibiotics | Groups 1 and 2 (n = 86) | |||||
| Any antibiotic | 71 (82.6) | 34 (82.9) | 37 (82.2) | .93 | 0 | … |
| Antibiotic group | ||||||
| Antianaerobic | 57 (66.3) | 32 (78.0) | 25 (55.6) | .04 | … | … |
| Antipseudomonal | 53 (61.6) | 31 (75.6) | 22 (48.9) | .01 | … | … |
| Anti-MRSA | 40 (46.5) | 22 (53.7) | 18 (40.0) | .28 | … | … |
| Antibiotic class | ||||||
| β-lactamc | 60 (70.0) | 31 (75.6) | 29 (64.4) | .35 | … | … |
| Quinoloned | 30 (34.9) | 11 (26.8) | 19 (42.2) | .18 | … | … |
| Cephalosporine | 28 (32.6) | 12 (29.3) | 16 (35.6) | .65 | … | … |
| Macrolidef | 9 (10.5) | 5 (12.2) | 4 (8.9) | .73 | … | … |
| Specific antibiotic | ||||||
| Vancomycing | 38 (44.2) | 20 (48.8) | 18 (40.0) | .52 | … | … |
| Piperacillin-tazobactamc,h,i | 35 (40.7) | 23 (56.1) | 12 (26.7) | .008 | … | … |
| Gatifloxacind | 26 (30.2) | 10 (24.4) | 16 (35.6) | .35 | … | … |
| Imipenem-cilastatinc,h,i | 18 (20.9) | 10 (24.4) | 8 (17.8) | .60 | … | … |
| Metronidazoleh | 16 (18.6) | 10 (24.4) | 6 (13.3) | .27 | … | … |
| Ceftriaxonec,e | 10 (11.6) | 5 (12.2) | 5 (11.1) | .88 | … | … |
| Clindamycinh | 10 (11.6) | 5 (12.2) | 5 (11.1) | .88 | … | … |
| Cefazolinc,e | 9 (10.5) | 3 (7.3) | 6 (13.3) | .49 | … | … |
| TMP-SMX | 9 (10.5) | 4 (9.8) | 5 (11.1) | .84 | … | … |
| Gentamicini | 8 (9.3) | 6 (14.6) | 2 (4.4) | .14 | … | … |
| Ampicillin-sulbactamc,h | 7 (8.1) | 2 (4.9) | 5 (11.1) | .44 | … | … |
| Cefepimec,e,i | 7 (8.1) | 4 (9.8) | 3 (6.7) | .70 | … | … |
| Linezolidg | 6 (7.0) | 5 (12.2) | 1 (2.2) | .10 | … | … |
| Azithromycinf | 5 (5.8) | 2 (4.9) | 3 (6.7) | .72 | … | … |
| Cefotetanc,e,h | 5 (5.8) | 4 (9.8) | 1 (2.2) | .19 | … | … |
| Ciprofloxacind,i | 4 (4.6) | 1 (2.4) | 3 (6.7) | .62 | … | … |
| Erythromycinf | 4 (4.6) | 3 (7.3) | 1 (2.2) | .34 | … | … |
| Ampicillinc | 3 (3.5) | 0 | 3 (6.7) | .24 | … | … |
| Nafcillinc | 3 (3.5) | 0 | 3 (6.7) | .24 | … | … |
| Cefotaximec,e | 2 (2.3) | 0 | 2 (4.4) | .50 | … | … |
| Penicillin G potassiumc | 2 (2.4) | 2 (4.9) | 0 | .22 | … | … |
| Rifampin | 2 (2.3) | 0 | 2 (4.4) | .50 | … | … |
| Doxycycline | 1 (1.2) | 0 | 1 (2.2) | .34 | … | … |
| Rifaximin | 1 (1.2) | 0 | 1 (2.2) | .34 | … | … |
| Other medications | Groups 1–3 (n = 109) | |||||
| PPIs | 91 (83.5) | 33 (80.5) | 37 (82.2) | 21 (91.3) | .51 | |
| Opioids | 80 (73.4) | 31 (75.6) | 34 (75.6) | 15 (65.2) | .61 | |
| Steroids | 38 (34.9) | 16 (39.0) | 17 (37.8) | 5 (21.7) | .33 |
Data are presented as No. (%) unless otherwise indicated. Data in boldface text represent statistically significant P values.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; PPI, proton pump inhibitor; TMP-SMX, trimethoprim-sulfamethoxazole.
aFisher exact test for group 1 vs group 2.
bχ2 test for groups 1, 2, and 3.
cß-lactams: ampicillin, ampicillin-sulbactam, cefazolin, cefepime, cefotaxime, cefotetan, ceftriaxone, imipenem-cilastatin, nafcillin, penicillin G, potassium, piperacillin-tazobactam.
dQuinolones: ciprofloxacin, gatifloxacin.
eCephalosporins: cefazolin, cefepime, cefotaxime, cefotetan, ceftriaxone.
fMacrolides: azithromycin, erythromycin.
gAnti-MRSA: linezolid, vancomycin.
hAntianaerobic: imipenem, piperacillin-tazobactam, ampicillin-sulbactam, cefotetan, clindamycin, metronidazole.
iAntipseudomonal: imipenem, piperacillin-tazobactam, cefepime, ciprofloxacin, gentamicin.
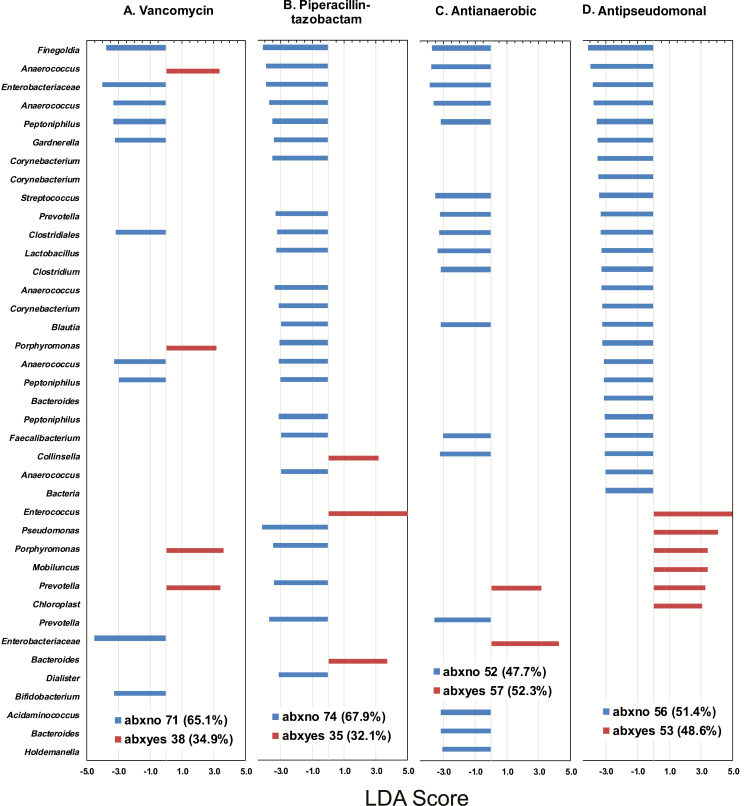
With patients categorized based on antibiotic prescriptions (regardless of patient group), LefSe was used to identify differences in the relative abundance of OTUs on swab 2. The mean proportions of selected taxa, identified by LefSe as differentially abundant by prescription of vancomycin or piperacillin-tazobactam, are shown by patient group and swab (Supplementary Figure 2). Several potentially protective bacteria were more abundant in patients who did not receive specific antibiotics. Bifidobacterium was more abundant in patients who did not receive vancomycin (Figure 4). Relative abundances of Lactobacillus, Blautia, Faecalibacterium, and other potentially beneficial taxa were significantly higher in patients who did not receive piperacillin-tazobactam, antianaerobic antibiotics, or antipseudomonal antibiotics (Figure 4).
Figure 4.
Linear discriminant analysis effect size scores ≥3.0 for comparisons between No. (%) among 109 intensive care unit patients exposed or not exposed to a given antibiotic. A, Vancomycin. B, Piperacillin-tazobactam. C, Antianaerobic antibiotics (ampicillin-sulbactam, cefotetan, clindamycin, imipenem-cilastatin, metronidazole, and piperacillin-tazobactam). D, Antipseudomonal antibiotics (cefepime, ciprofloxacin, gentamicin, imipenem-cilastatin, and piperacillin-tazobactam). Genus names (eg, Anaerococcus) that appear more than once represent different OTUs that are members of the same genus. Abbreviations: abxno, not exposed to antibiotic; abxyes, exposed to antibiotic; LDA, linear discriminant analysis.
LefSe also indicated that Enterococcus was more abundant in patients who received piperacillin-tazobactam (Figure 4). Enterococcus domination in swab 2 was associated with receipt of piperacillin-tazobactam (odds ratio [OR], 5.50; 95% confidence interval [CI], 2.03–14.92). Certain antibiotics may select for overgrowth of specific pathobionts and/or may remove commensals that compete with these taxa.
We compared taxa present on swab 2 in patients who did and did not receive steroids and opioids between swab 1 and 2 using LefSe (Supplementary Figure 3). These analyses were restricted to patients who did not receive antibiotics in the ICU (group 3); proton pump inhibitors were not evaluated because only 2 patients did not receive them. Patients who did not receive opioids had higher levels of potentially protective Blautia and Lactobacillus than those who received opioids. Patients who did not receive steroids had higher levels of Bifidobacterium than those who did not receive steroids.
Antibiotics, Microbiota Composition, and CRPA Colonization-Acquisition
We identified groups of correlated taxa associated with CRPA colonization acquisition by comparing group 1 patients (acquired colonization) with patients without CRPA colonization acquisition (groups 2 and 3 combined). PCA identified 4 independent factors in ICU patients’ GI microbiota (Supplementary Table 3). In logistic regression predicting colonization by CRPA using the 4 factors, high levels of factor 1 taxa on swab 1 were associated with lower odds of CRPA colonization acquisition (OR, 0.58; 95% CI, .38–.87). Factor 1 contained Peptoniphilus (3 OTUs), Clostridiales (2 OTUs), Prevotella, Mogibacterium, Varibaculum, Actinomycetales, Dialister, Ezakiella, Porphyromonas, Finegoldia, and Murdochiella. These data indicate that levels of factor 1 taxa, which can be identified at ICU admission, may serve as a marker for risk of CRPA colonization acquisition. Factor 1 taxa may also have a protective role in CRPA colonization acquisition.
We examined the impact of combined use of 2 commonly prescribed and mutually exclusive antibiotic groups, antianaerobic and anti-MRSA, between the collection dates for swab 1 and swab 2 on CRPA colonization acquisition. This analysis was restricted to patients in groups 1 and 2. The category of antipseudomonal antibiotics was not analyzed because of the overlap in antibiotics in the antianaerobic and antipseudomonal categories. Compared to having no antibiotics from either of these groups, risk of CRPA colonization acquisition was associated with having been prescribed both antianaerobic and anti-MRSA antibiotics (OR, 3.06; 95% CI, 1.00–9.40).
DISCUSSION
Our data demonstrate that ICU patients have a GI microbiota characterized by high levels of pathobionts such as Enterococcus, Escherichia, Staphylococcus, Enterobacteriaceae, and/or Pseudomonas. Antibiotics were differentially associated with pathobiont dominance and lower relative abundances of specific potentially protective taxa. High relative abundances of factor 1 taxa at ICU admission were associated with a lower risk of CRPA colonization acquisition.
Healthy individuals have approximately 160 different taxa in their GI microbiota [26]. In our 109 patients, the effective number of species was low; the mean Jost1 ranged from 5 to 14 effective species per patient across all groups. In addition, 61 of 109 patients (55.9%) were already experiencing dominance by at least 1 of the 5 most frequent pathobionts at ICU admission. These data have implications for selective digestive decontamination strategies for infection control, which seek to reduce the burden of pathogenic colonizing bacteria while preserving the protective microbiota [27, 28]. For some patients, it may be too late to protect the anaerobic microbiota by selective digestive decontamination upon ICU admission. Our analysis of patterns of pathobiont dominance suggests that pathobiont domination shifts over time in the ICU; these data are consistent with findings from Zaborin et al [2]. Pathobiont dominance is of concern because it has been shown to place patients at increased risk for infection [29].
Scientists have proposed reducing the use of certain broad-spectrum antibiotics to decrease colonization with, and spread of, antibiotic-resistant pathogens in hospitalized patients [10, 30]. Piperacillin-tazobactam was commonly prescribed to patients in our study, and use was higher in patients with CRPA colonization acquisition. Piperacillin-tazobactam was associated with Enterococcus dominance and lower abundances of potentially protective taxa such as Faecalibacterium, Blautia, and Lactobacillus. Faecalibacterium species have been evaluated in probiotics [31, 32]. Blautia has been identified as protective for Clostridium difficile colonization and infection [33]. Abundance of Lactobacillus was shown to be higher in hospitalized patients who did not acquire colonization by multidrug-resistant organisms [5]. While patients were also exposed to other medications and antibiotics, these data suggest that piperacillin-tazobactam negatively impacts the GI microbiota. These data are consistent with a recent study, which showed that piperacillin-tazobactam was associated with dysbiosis in the GI microbiota and increased mortality in stem cell transplant recipients [34]. Opioids were prescribed to 73.3% of ICU patients and also likely contribute to dysbiosis in the GI microbiota; potentially protective Blautia and Lactobacillus were higher in group 3 patients who were not prescribed opioids.
Some bacteria species co-aggregate based on similar antibiotic susceptibilities or metabolic requirements [35]. Abundance of several taxa associated with factor 1 from the PCA (eg, Clostridiales, Anaerococcus, Finegoldia, and Peptoniphilus) differed by antibiotic use and declined while patients were in the ICU. Low abundance of Clostridiales has been linked to higher risk of C. difficile infection [36]. Factor 1 taxa may be important markers of microbiota disruption and/or help provide colonization resistance against CRPA. It is also important to note that while taxa such as Anaerococcus, Finegoldia, and Peptoniphilus may play a protective role in the GI microbiota, they have also been sporadically identified in infections and chronic wounds [37].
Our study had several limitations. We did not have data regarding medications or swabs prior to ICU admission. Some of our patients had relatively long ICU stays (Table 1) and may not be representative of all ICU patients. These data indicate that many patients are experiencing dysbiosis upon ICU admission and perhaps all ICU patients should be monitored for microbiota disruption. High levels of factor 1 taxa in swab 1 were associated with lower odds of CRPA colonization acquisition, which suggests admission swabs may provide a useful tool for patient monitoring. We used perirectal swabs to capture the GI microbiota, and the composition may differ from stool samples. Perirectal swabs are frequently used in infection control and have excellent sensitivity for detection of GI tract colonization with pathobionts [38]. Group 1 patients had a longer total time at risk for CRPA acquisition than other patient groups (Kruskal-Wallis test, P = .001; Supplementary Table 4). However, patients in the 3 groups did not significantly differ in terms of the time between collection of swab 1 and swab 2.
Our data suggest that piperacillin-tazobactam contributes to microbiota disruption and indicate that low relative abundances of specific taxa may be related to risk of CRPA colonization acquisition. When possible, antimicrobial therapy should be tailored based on colonization status and/or when infection is suspected in CRPA-colonized patients. Experimental studies are needed to determine which commensals are causally related to colonization resistance. CRPA can colonize patients at non-GI sites [39]. Future studies should examine relationships between the skin and respiratory microbiota and CRPA colonization acquisition at other body sites. Greater understanding of these relationships will facilitate (1) the development of microbiota disruption indices that could be used for monitoring patients at risk of CRPA colonization and infection; and (2) the development of strategies to minimize infection and spread of CRPA in the hospital.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors gratefully acknowledge Martina Wade, Yale School of Public Health, for expert technical assistance in the laboratory, and the Yale Center for Genome Analysis for high-throughput sequencing services.
Disclaimer. The findings and conclusions in this report are solely those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant number 5K24AI079040 to A. D. H.) and the Centers for Disease Control and Prevention’s investments to combat antibiotic resistance (grant numbers 200-2016-91963 and BAA 2016-N-1781 to M. M. P. and J. K. J.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-v Lekkerkerk. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg 1971; 69:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaborin A, Smith D, Garfield K, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio 2014; 5:e01361–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McDonald D, Ackermann G, Khailova L, et al. Extreme dysbiosis of the microbiome in critical illness. Msphere 2016; 1:e00199–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013; 13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Araos R, Tai AK, Snyder GM, Blaser MJ, D’Agata EMC. Predominance of Lactobacillus spp. among patients who do not acquire multidrug-resistant organisms. Clin Infect Dis 2016; 63:937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One 2010; 5:e9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donskey CJ, Chowdhry TK, Hecker MT, et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 2000; 343:1925–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Gunzburg J, Ghozlane A, Ducher A, et al. Protection of the human gut microbiome from antibiotics. J Infect Dis 2017: 217:628–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruppé E, Burdet C, Grall N, et al. Impact of antibiotics on the intestinal microbiota needs to be re-defined to optimize antibiotic usage. Clin Microbiol Infect 2018; 24:3–5. [DOI] [PubMed] [Google Scholar]
- 10. Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis 2006; 43(Suppl 2):S62–9. [DOI] [PubMed] [Google Scholar]
- 11. Gonçalves-Pereira J, Póvoa P. Antibiotics in critically ill patients: a systematic review of the pharmacokinetics of β-lactams. Crit Care 2011; 15:R206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Imhann F, Bonder MJ, Vich Vila A, et al. Proton pump inhibitors affect the gut microbiome. Gut 2016; 65:740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris AD, Jackson SS, Robinson G, et al. Pseudomonas aeruginosa colonization in the intensive care unit: prevalence, risk factors, and clinical outcomes. Infect Control Hosp Epidemiol 2016; 37:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson JK, Smith G, Lee MS, et al. The role of patient-to-patient transmission in the acquisition of imipenem-resistant Pseudomonas aeruginosa colonization in the intensive care unit. J Infect Dis 2009; 200:900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987; 40:373–83. [DOI] [PubMed] [Google Scholar]
- 16. Elixhauser A, Steiner C, Harris DR, Coffey RM. Comorbidity measures for use with administrative data. Med Care 1998; 36:8–27. [DOI] [PubMed] [Google Scholar]
- 17. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kong Y. Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011; 98:152–3. [DOI] [PubMed] [Google Scholar]
- 19. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 21. Li K, Bihan M, Yooseph S, Methé BA. Analyses of the microbial diversity across the human microbiome. PLoS One 2012; 7:e32118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chao A, Chiu CH, Jost L. Phylogenetic diversity measures based on Hill numbers. Philos Trans R Soc Lond B Biol Sci 2010; 365:3599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. MBio 2011; 2:e00245–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qin J, Li R, Raes J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Price R, MacLennan G, Glen J; SuDDICU Collaboration Selective digestive or oropharyngeal decontamination and topical oropharyngeal chlorhexidine for prevention of death in general intensive care: systematic review and network meta-analysis. BMJ 2014; 348:g2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liberati A, D’Amico R, Pifferi S, Torri V, Brazzi L, Parmelli E. Antibiotic prophylaxis to reduce respiratory tract infections and mortality in adults receiving intensive care. Cochrane Database Syst Rev 2009; CD000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quale J, Landman D, Saurina G, Atwood E, DiTore V, Patel K. Manipulation of a hospital antimicrobial formulary to control an outbreak of vancomycin-resistant enterococci. Clin Infect Dis 1996; 23:1020–5. [DOI] [PubMed] [Google Scholar]
- 31. Matsumoto T, Ishikawa H, Tateda K, Yaeshima T, Ishibashi N, Yamaguchi K. Oral administration of Bifidobacterium longum prevents gut-derived Pseudomonas aeruginosa sepsis in mice. J Appl Microbiol 2008; 104:672–80. [DOI] [PubMed] [Google Scholar]
- 32. Brahe LK, Le Chatelier E, Prifti E, et al. Dietary modulation of the gut microbiota—a randomised controlled trial in obese postmenopausal women. Br J Nutr 2015; 114:406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shono Y, Docampo MD, Peled JU, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Science Transl Med 2016; 8: 339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 2011; 9:233–43. [DOI] [PubMed] [Google Scholar]
- 36. Vincent C, Stephens DA, Loo VG, et al. Reductions in intestinal Clostridiales precede the development of nosocomial Clostridium difficile infection. Microbiome 2013; 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murphy EC, Frick IM. Gram-positive anaerobic cocci–commensals and opportunistic pathogens. FEMS Microbiol Rev 2013; 37:520–53. [DOI] [PubMed] [Google Scholar]
- 38. Lautenbach E, Harris AD, Perencevich EN, Nachamkin I, Tolomeo P, Metlay JP. Test characteristics of perirectal and rectal swab compared to stool sample for detection of fluoroquinolone-resistant Escherichia coli in the gastrointestinal tract. Antimicrob Agents Chemother 2005; 49:798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen R, Babushkin F, Cohen S, et al. A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob Resist Infect Control 2017; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.