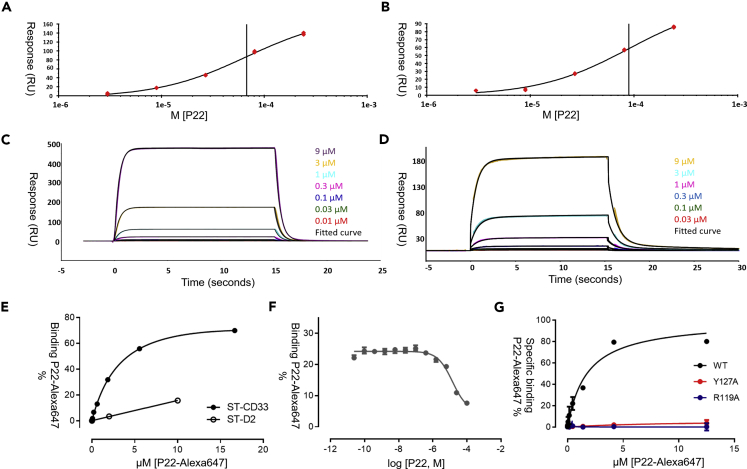
Figure 3.
P22 Binding to CD33 Wild-Type and Mutants
(A) An example of affinity analysis for 1–243 μM (3-fold dilutions) of P22 binding to immobilized CD33C36S. Data obtained in duplicate using single-cycle analysis and fit to a steady-state binding model to determine the equilibrium binding constant.
(B) An example of affinity analysis of P22 binding to immobilized CD33 wild-type (WT) as per example in (A).
(C) Sensorgram data for 0.01–9 μM (3-fold dilutions) of CD33C36S binding to immobilized P22. Data obtained in triplicate using single concentration cycles and fit to a 1:1 binding model for kinetic analysis.
(D) Sensorgram data for 0.03–9 μM (3-fold dilutions) CD33 WT binding to immobilized P22. Data obtained in triplicate using single concentration cycles and fit to a bivalent analyte model for kinetic analysis. Measurements in triplicate showing mean ± SD.
(E) Saturation total binding experiments on either CD33 (black circles) or the dopamine D2 receptor (open circle) showing that P22-Alexa647 binds specifically to CD33. Data presented are representative of three independent experiments performed in quadruplicate for each compound. Data were normalized on P22-Alexa647 estimated Bmax on CD33 and are represented as averages ± SD.
(F) Competition binding experiment on CD33 using 2 μM P22-Alexa647. Data presented are representative of three independent experiments performed in quadruplicate for each compound. Data were normalized on P22-Alexa647 estimated Bmax on CD33 and are represented as averages ± SD.
(G) The specific binding of P22-Alexa647 (total binding – binding in presence of 100 μM non-labeled P22) was evaluated for the WT CD33 and two other single point mutation variants. Binding was completely abolished by the Y127A and R119A variants. Data presented are representative of three independent experiments performed in quadruplicate for each compound. Data were normalized on P22-Alexa647 estimated Bmax on CD33 and are represented as averages ± SD.