Abstract
Carbonylation is one of the most remarkable expressions of the oxidative damage to proteins and the DNPH method the most common procedure to assess protein oxidation in biological samples. The present study was elicited by two hypotheses: i) is malondialdehyde, as a reactive dicarbonyl, able to induce the formation of allysine through a Maillard-type reaction? and ii) to which extent does the attachment of MDA to proteins interfere in the assessment of protein carbonyls using the DNPH method? Human serum albumin (HSA), human hemoglobin (HEM) and β-lactoglobulin (LAC) (5 mg/mL) were incubated with MDA (0.25 mM) for 24 h at 37 °C (HSA and HEM) or 80 °C (LAC). Results showed that MDA was unable to induce oxidative deamination of lysine residues and instead, formed stable and fluorescent adducts with proteins. Such adducts were tagged by the DNPH method, accounting for most of the protein hydrazones quantified. This interfering effect was observed in a wide range of MDA concentrations (0.05–1 mM). Being aware of its limitations, protein scientists should accurately interpret results from the DNPH method, and apply, when required, other methodologies such as chromatographic methods to detect specific primary oxidation products such as allysine.
Keywords: Carbonylation, Allysine, Protein oxidation, Malondialdehyde, DNPH method
1. Introduction
The oxidation of proteins has become a topic of undeniable interest among biochemists given the role of the oxidative damage to proteins in cell function, disease and aging [[1], [2], [3]]. As a posttranslational modification in proteins, oxidation can be part of a precise physiological mechanism (i.e. cellular signaling) or the outcome of uncontrolled oxidative stress. The removal of oxidized proteins responds to a strategy to avoid protein dysfunction and altered physiological processes. Yet, when proteins are severely damaged, they accumulate in cells leading to chronic disorders [2]. Food scientists and nutritionists have also reported the implications of protein oxidation in food systems and they include i) impaired functionality and digestibility ii) altered sensory properties and iii) potential safety concerns as a result of the intake of oxidized proteins and amino acids [4,5].
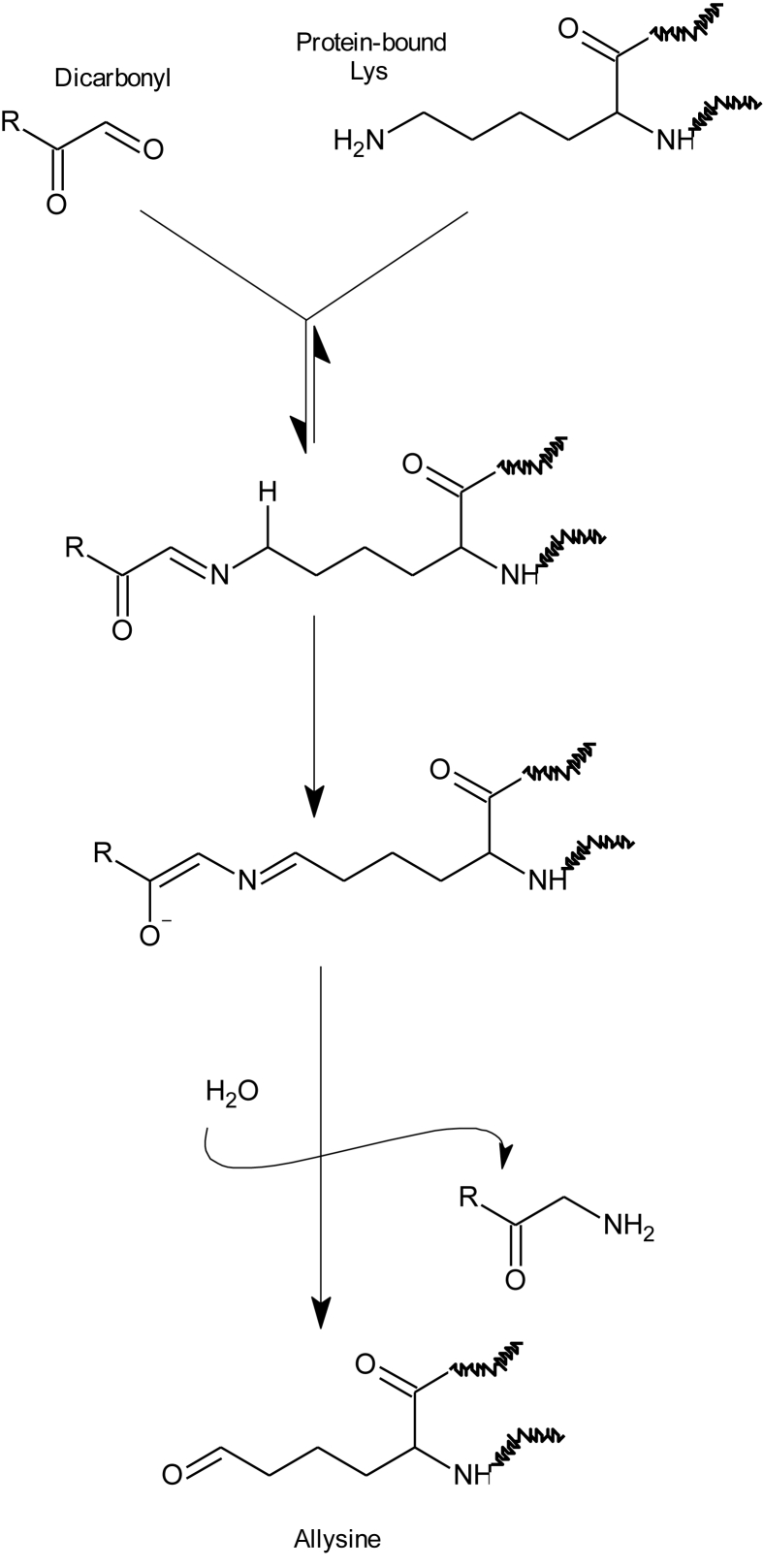
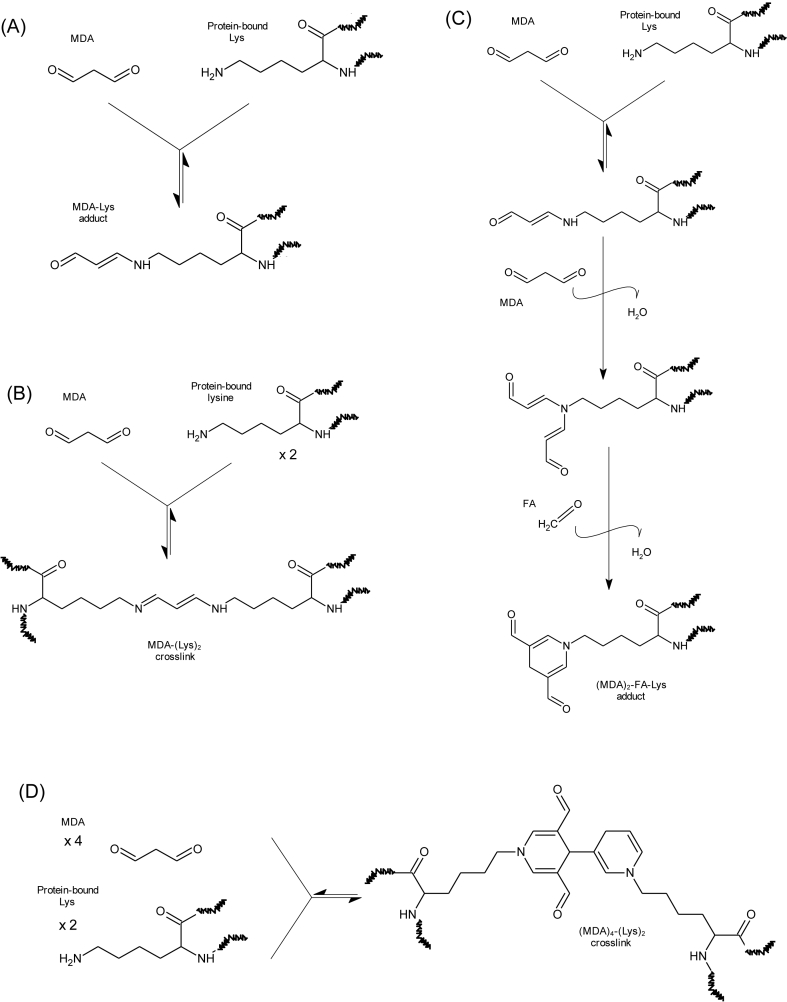
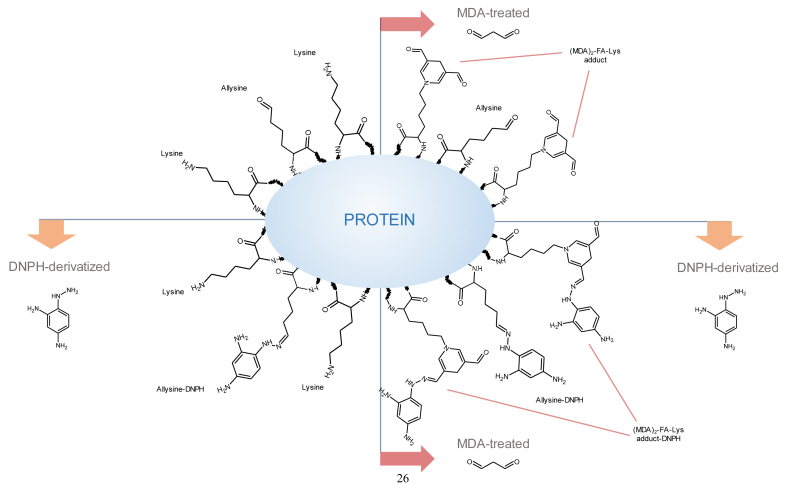
Carbonylation is one of the most remarkable expressions of the oxidative damage to proteins [6]. The on-site formation of carbonyls in proteins (∼primary protein carbonyls) typically occurs as the result of the attack of reactive oxygen species (ROS) to the ε-amino group of susceptible amino acids (lysine, arginine and proline). However, this oxidative deamination mechanism is also triggered by α-dicarbonyls such as glyoxal (GO) and methylglyoxal (MGO), formed from the degradation of reducing sugars [7] (Fig. 1). This Maillard-mediated mechanism has been found to occur in pathological disorders (i.e. diabetes) involving high concentration of circulating glucose [7] and also in food systems in which reducing sugars and their oxidation products have been found to react with ε-amino group of alkaline amino acids in proteins [8,9]. Regardless of the underlying mechanism (ROS-mediated or Maillard-mediated), the oxidative deamination of lysine, one of the most abundant amino acids in proteins, leads to the formation of allysine, a primary protein carbonyl and reliable marker of oxidative stress and disease [6]. Proteins can also be carbonylated by the addition of pre-formed carbonyl groups such as those generated from lipid oxidation (∼secondary protein carbonyls) [10]. Malondialdehyde (MDA) and 4-hydroxy-non-2-enal (4-HNE), among others, form covalent linkages with proteins by reacting, precisely, with ε-amino groups from protein-bound lysines. Some of these adducts (Fig. 2) have been linked to assorted pathological conditions [11,12]. It is, however, ignored, whether MDA and other dicarbonyls from lipid oxidation are able to replicate the Maillard-mediated mechanism of GO and MGO and hence, induce on-site carbonylation of alkaline amino acids (e.g. formation of protein-bound allysine).
Fig. 1.
Formation pathway of allysine in the presence of an α-dicarbonyl in accordance to the Maillard-mediated mechanism proposed by Akagawa et al. (2005).
Fig. 2.
Formation of adducts between MDA, AA and lysine residues (Adapted from Weiβer et al. [24]; and Nakamura et al. [23]).
While all these mechanisms may occur simultaneously, each of them responds to completely different reaction conditions and their implications and consequences for the biological system, whether this is a living organism or food system, may also be different. In light of this complex chemistry, specific methodological approaches are needed to identify which particular mechanism is responsible for the carbonylation of proteins in a given biological sample. Yet, the most common procedure for quantifying protein carbonyls in biological systems does not provide such relevant information. This routine method involves tagging all carbonyl moieties with the dinitrophenylhydrazine (DNPH) reagent [13]. The DNPH reacts with all protein carbonyls, regardless of their formation pathway, and furthermore, lipid-derived carbonyls such as MDA are also tagged. In fact, routine methods for the quantification of MDA in plasma and other biological samples involve using DNPH as a derivatization agent [14].
The hypotheses that elicited the present study were: i) is MDA, as a reactive dicarbonyl, able to induce the formation of allysine through a Maillard-type reaction (Fig. 1) and ii) to which extent does the attachment of MDA to proteins interfere in the assessment of protein carbonyls using the DNPH method?
2. Material and methods
2.1. Chemicals
All chemicals, reagents and proteins used for the present work were purchased from Panreac (Panreac Química, S. A., Barcelona, Spain), and Sigma Chemicals (Sigma- Aldrich, Steinheim, Germany). Water used was purified by passage through a Milli-Q system (Millipore Corp., Bedford, MA). The molarities of all reactants refer to the final concentration in the reaction mixture.
2.2. Synthesis of allysine
N-Acetyl-L-AAS (allysine) was synthesized from N-acetyl-l-lysine using lysyl oxidase activity from egg shell membrane following the procedure described by Akagawa et al. [15]. Briefly, 10 mM N-acetyl-l-lysine was incubated at constant stirring with 5 g egg shell membrane in 50 mL of 20 mM sodium phosphate buffer, pH 9.0 at 37 °C for 24 h. The egg shell membrane was then removed by centrifugation and the pH of the solution adjusted to 6.0 using 1 M HCl. The resulting aldehydes were reductively aminated with 3 mmol p-amino-benzoic acid (PABA) in the presence of 4.5 mmol sodium cyanoborohydride (NaBH3CN) at 37 °C for 2 h with stirring. Then, PABA derivatives were hydrolyzed by 50 mL of 12 M HCl at 110 °C for 10 h. The hydrolysates were evaporated at 40 °C in vacuo to dryness. The resulting allysine-PABA was purified by using silica gel column chromatography and ethyl acetate/acetic acid/water (20:2:1, v/v/v) as elution solvent. The purity of the resulting solution and authenticity of the standard compounds obtained following the aforementioned procedures were checked by using MS and 1H NMR [16].
2.3. Experimental setting
In order to evaluate the ability of MDA to induce carbonylation in proteins, three proteins, namely, human serum albumin (HSA), human hemoglobin (HEM) and β-lactoglobulin (LAC) (5 mg/mL, final concentration) were dissolved in 100 mM phosphate buffer (pH 6.5) and incubated separately with MDA (0.25 mM) at 37 °C (HSA, HEM) and 80 °C (LAC) in an oven at constant stirring for 24 h. Proteins were selected on the basis of their susceptibility to oxidative damage [9,17]. Samples were taken at fixed incubation times (0, 2, 7 and 24 h) for the quantification of total protein carbonyls by the DNPH method, free amino groups, allysine and Schiff bases. The preparation of six experimental units corresponding to the reaction units (HSA-MDA, HEM-MDA and LAC-MDA) and the corresponding controls (proteins without MDA) were replicated three times in corresponding independent assays and all analyses were repeated three times in each same experimental unit (9 measurements per analysis and per treatment to calculate means and standard deviations).
In order to evaluate the dose effect of MDA on the aforementioned changes, HSA (5 mg/mL, final concentration) was dissolved in 100 mM phosphate buffer (pH 6.5), and incubated with increasing concentrations of MDA (0, 0.05, 0.10, 0.25, 1, 5, 10 and 50 mM) at 37 °C in an oven at constant stirring for 24 h. Samples were taken at 24 h for the quantification of total protein carbonyls by the DNPH method, free amino groups, allysine and Schiff bases. The preparation of eight experimental units corresponding to the each MDA concentration, were replicated three times in corresponding independent assays and all analyses were repeated three times in each same experimental unit (9 measurements per analysis and per treatment to calculate means and standard deviations).
2.4. Protein carbonyls by the DNPH method
Total protein carbonyls were determined by means of the dinitrophenylhydrazine (DNPH) method described by Levine et al. [13] with some modifications. Protein suspensions (200 μL) from each experimental unit were precipitated by the addition of 1 mL of cold 10% trichloroacetic acid (TCA), followed by centrifugation at 4 °C at 600 g for 5 min and the supernatants were discarded. Pellets were treated with 1 mL of a 2 M HCl solution with 0.2% DNPH and incubated at room temperature for 1 h. Proteins were subsequently precipitated with 1 mL of cold 10% TCA, followed by centrifugation at 4 °C, 1200 g for 10 min and washed twice with 1 mL of ethanol:ethyl acetate (1: 1 v/v). The pellets were dissolved in 1.5 mL of 20 mM Na3PO4 buffer pH 6.5 added with guanidine hydrochloride to reach 6 M. The amount of carbonyls was expressed in nmoles of protein hydrazones per mg of protein using a molar extinction coefficient of hydrazones (21.0 nM−1 cm−1) with absorbance readings at 370 nm.
2.5. Analysis of free amino groups
Free amino groups were quantified as described by Weigele et al. [18] and Strauss & Gibson [19]. Protein suspensions (850 μL) were added to 2.0 mL 0.05 M sodium tetraborate (pH 8.5) in a 4 mL quartz spectrofluorometer cell. Subsequently, 150 μL of 0.7 mM fluorescamine solution in acetone werte dispensed. The cell was inverted four times, and the resulting fluorescence was measured using 390/485 nm for excitation and emission on a PerkinElmer LS45 Fluorescence spectrometer (Llantrisant, UK). The free amino group concentration was calculated based on a standard curve prepared from lysine diluted in malic acid buffer (pH 5.8). The contribution from malic acid buffer (pH 5.8) was recorded under the same conditions and subtracted from all the samples. The concentration is given as μmol amino groups/mg protein.
2.6. Analysis of Schiff bases by fluorescence spectroscopy
The formation of Schiff bases (SB) was assessed using a LS-55 PerkinElmer fluorescence spectrometer (PerkinElmer, Beaconsfield, UK). Prior to the analysis, reaction mixtures were diluted (1:20) with 8 M urea in 100 mM sodium phosphate buffer, pH 7. SB were excited at 350 nm and the emitted fluorescence recorded at 450 nm. The excitation and emission slit widths were set at 10 nm and the speed of data collection while scanning was of 500 nm per minute. The height of the peaks corresponding to SB spectra was recorded. After taking into consideration the applied dilutions, the results were expressed as arbitrary fluorescence units.
2.7. Analysis of allysine by HPLC
Allysine was identified and quantified upon derivatization with p-amino-benzoic acid (PABA) and subsequent analysis by fluorescent HPLC as reported before by Utrera et al. [20]. Two hundred microliters of protein suspension were dispensed in eppendorf tubes and treated with 1 mL of a cold 10% TCA solution. Each eppendorf was vortexed and then subjected to centrifugation at 2000g for 30 min at 4 °C. The supernatant was removed and the pellet was treated with 1 mL of a cold 5% TCA solution. A new centrifugation was performed at 5000 g for 5 min at 4 °C. The supernatant was removed and the pellets were incubated with the following freshly prepared solutions: 0.5 mL 250 mM 2-(N-morpholino) ethanesulfonic acid (MES) buffer pH 6.0 containing 1% sodium dodecyl sulfate (SDS) and 1 mM diethylenetriaminepentaacetic acid (DTPA); 0.5 mL 50 mM PABA in 250 mM MES buffer pH 6.0 and 0.25 mL 100 mM NaBH3CN in 250 mM MES buffer pH 6.0. The eppendorf tubes were vortexed and then incubated in an oven at 37 °C for 90 min. The samples were stirred every 15 min. After derivatization, samples were treated with a cold 50% TCA solution and centrifuged at 5000 g for 10 min. The pellet was then washed twice with 1 mL of cold 10% TCA and 1 mL of diethyl ether-ethanol (1:1). Finally, the pellet was treated with 0.5 mL of 6 N HCl and kept in an oven at 110 °C for 18 h until completion of hydrolysis. Hydrolysates were dried under nitrogen flow while heated in a thermoblock (40 °C). The generated residue was reconstituted with 200 μL of milliQ water and then filtered through hydrophilic polypropylene GH Polypro (GHP) syringe filters (0.45 μm pore size, Pall Corporation, USA) for HPLC analysis.
A Shimadzu ‘Prominence’ HPLC apparatus (Shimadzu Corporation, Kyoto, Japan), equipped with a quaternary solvent delivery system (LC-20AD), a DGU-20AS on-line degasser, a SIL-20A auto-sampler, a RF-10A XL fluorescence detector (FLD), and a CBM-20A system controller, was used. An aliquot (1 μL) from the reconstituted protein hydrolysates was injected and analyzed in the above mentioned HPLC equipment. Allysine eluted in a Cosmosil 5C18-AR-II RP-HPLC column (5 μm, 150 × 4.6 mm) equipped with a guard column (10 × 4.6 mm) packed with the same material. The flow rate was kept at 1 mL/min and the temperature of the column was maintained at 30 °C. The eluate was monitored with excitation and emission wavelengths set at 283 and 350 nm, respectively. Standard allysine (0.1 μL) was run and analyzed under the same conditions. Identification of derivatized allysine in the FLD chromatograms was carried out by comparing its retention times with that from the standard compound. The peak corresponding to allysine-ABA was manually integrated from FLD chromatograms and the resulting areas plotted against an ABA standard curve with known concentrations that ranged from 0.1 to 0.5 mM. Results are expressed as nmol of allysine per mg of protein.
2.8. Statistical analyses
Data from the analysis of depletion of amino groups and accretion of protein hydrazones, allysine and fluorescent Schiff bases (n = 9 per treatment) in proteins treated with MDA, were collected and subjected to statistical analyses. Normality and homoscedasticity were checked for every data set in order to comply with the ANOVA requirements. In order to assess the effect of MDA addition and incubation time, a two-way analysis of variance (ANOVA) was applied to data (SPSS v. 15.5). In order to assess the effect of MDA dose, a one-way analysis of variance (ANOVA) was applied to data (SPSS v. 15.5). Tukey tests were applied when ANOVA found significant differences among treatments. Correlation coefficients were also calculated to assess potential connections between measurements. The statistical significance was set at p < 0.05.
3. Results and discussion
3.1. Protein hydrazones in MDA-treated protein suspensions
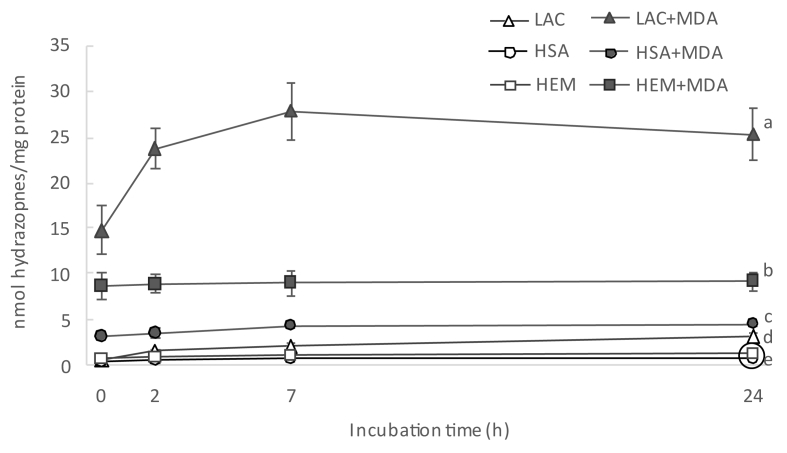
At a first stage, total protein carbonyls, as assessed by the DNPH method, were quantified in proteins incubated with MDA and their corresponding controls (no added MDA). The concentration of protein hydrazones (DNPH-derivatized carbonyls) in LAC, HSA and HEM incubated for 24 h at whether 80 °C (LAC) or 37 °C (human proteins) is displayed in Fig. 3. In control samples, the concentration of protein carbonyls increased from 0.6, 0.35 and 0.74 to 3.18, 0.81 and 1.25 nmol hydrazones/mg protein in LAC, HSA and HEM, respectively. Despite not adding a specific protein oxidation promoter, the carbonylation of proteins took place in control samples as a likely result of the formation of radical species from residual oxygen in the headspace of the vials (∼20.5%). In accordance to calculations made in a similar previous study [17], a low concentration of O2•- in control samples (5 nM; thousand times lower than in Fenton-reaction systems) may be sufficient to initiate the oxidative deamination of alkaline amino acids and hence, to cause the formation of protein carbonyls. Consistently, other authors such as Akagawa et al. [15,21] and Gürbüz et al. [22] reported the carbonylation of HSA and other proteins while incubated at 37 °C in an oxygenated environment. The higher concentration of protein hydrazones in LAC compared to those in human proteins is explained by the severe temperature of incubation to which the former was subjected (compatible with milk-dairy product processing).
Fig. 3.
Concentration of protein hydrazones (means ± standard deviations) during incubation of HSA, HEM and LAC for 24 h.
Footnote: For simplification purposes, only results from statistical analysis at final sampling point are displayed. Different letters denote significant differences (p < 0.05) among experimental units in a post-hoc Tuckey test. Samples grouped within a circle share the same letter.
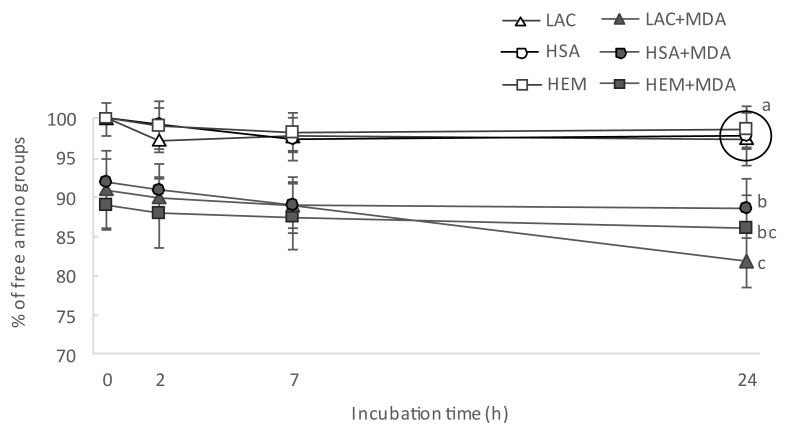
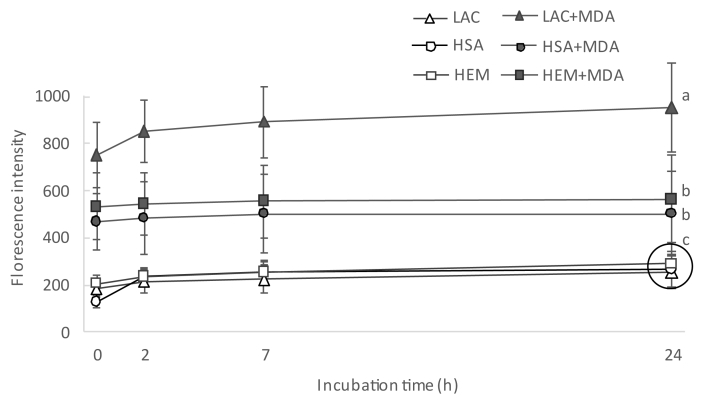
The incubation of proteins with MDA had a remarkable effect on the total amount of hydrazones in all the proteins under study. The notable concentrations of hydrazones in proteins incubated with MDA was already observed at the first sampling, right after MDA addition, and before subjecting the protein suspensions to the incubation temperatures (14.7, 3.2 and 8.7 nmol of hydrazones/mg protein in LAC, HSA and HEM, respectively). The subsequent incubation led to significant and remarkable increases of hydrazones in LAC and significant but more moderate increases of hydrazones in HSA and HEM. The increases in protein hydrazones occurred along with a depletion of free amino groups (Fig. 4), with this decrease being positively and highly correlated with the accretion of hydrazones (r = 0.79; p < 0.05). This timely coincidence provides strength to the plausible implication of MDA in both events. The addition of the carbonyl moieties in MDA to the nucleophilic ε-amino groups in alkaline amino acids in a Michael addition-type reaction could explain the decrease of the latter as compared to the control samples. In literature, this reaction is typically described to take place between MDA and its degradation products, acetaldehyde (AA) and formaldehyde (FA), with lysine to form adducts of assorted stability [23]. The adduct formed between one lysine residue and one MDA molecule (Fig. 2A) is unstable, given the reactivity of the free aldehyde moiety. In fact, the addition of a second lysine residue yields and intra- and/or intermolecular covalent cross-linked 1-amino-3-iminopropene-type MDA-lysine adduct (2B), a final product of higher stability [23]. Other common reactions involve MDA, lysine and FA or AA (2:1:1), which also results in stable products such as 1,4-dihydropyridine-3,5-dicarbaldehyde ([MDA]2-FA-Lys; Fig. 2C) and the reaction of four MDA with two lysines to yield 3,5-diformyl-1,4-dihydropyridine-4-yl-pyridinium (FHP; [MDA]4-[Lys]2), Fig. 2D). To gain further insight into the structures of these adducts, protein suspensions were analyzed for the emission of fluorescence by Schiff base structures by a spectroscopy procedure (Fig. 5). Evident similarities were found between the evolution of the formation of fluorescent Schiff base structures throughout the assay (Fig. 5) and the formation of protein hydrazones (Fig. 3). The positive and significant high correlation calculated between both measurements (r = 0.91; p < 0.05), provides strength to the hypothesis that MDA-lysine adducts not only had one carbonyl available for the addition of DNPH, but also they were able to emit natural fluorescence. These findings strongly incriminate hybrid and complex fluorescent MDA adducts such as (MDA)2-FA-Lys (Fig. 2C) and (MDA)4-(Lys)2 (Fig. 2D) and rule out other linear and simpler adducts such as MDA-Lys (Fig. 2A) and MDA-(Lys)2 (Fig. 2B) [24].
Fig. 4.
Concentration of free amines (means ± standard deviations) during incubation of HSA, HEM and LAC for 24 h.
Footnote: For simplification purposes, only results from statistical analysis at final sampling point are displayed. Different letters denote significant differences (p < 0.05) among experimental units in a post-hoc Tuckey test. Samples grouped within a circle share the same letter.
Fig. 5.
Fluorescence intensity emitted by Schiff bases structures (means ± standard deviations) during incubation of HSA, HEM and LAC for 24 h.
Footnote: For simplification purposes, only results from statistical analysis at final sampling point are displayed. Different letters denote significant differences (p < 0.05) among experimental units in a post-hoc Tuckey test. Samples grouped within a circle share the same letter.
3.2. Formation of allysine in MDA-treated protein suspensions
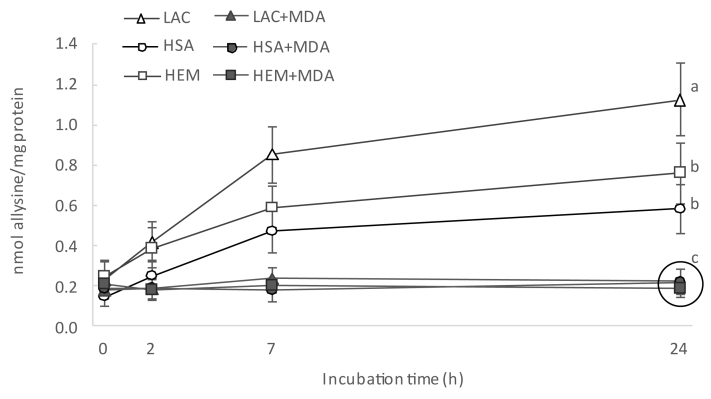
Yet, one of our initial hypothesis was indefinite: is MDA, as a reactive dicarbonyl, able to induce oxidative deamination of lysine residues to yield allysine? This possibility was formulated as feasible given that other small molecular weight dicarbonyls such as GO and MGO have been proven to trigger this Maillard-type mechanism [7]. As displayed in Fig. 1, firstly, the dicarbonyl is added to the ε-amino group in a protein-bound lysine to yield an unstable Schiff base which is comparable to the MDA-Lys adduct depicted in Fig. 2A. Subsequently, the enolization of the second carbonyl moiety leads to the formation of an iminoenaminol which is spontaneously hydrolyzed to release an enaminol and yield allysine. To our knowledge, these last stages have never been described to occur upon the reaction between MDA and lysine residues. However, this reaction would explain all the results reported so far: the depletion of free amino groups in proteins and the subsequent formation of hydrazones through the derivatization of allysine by the DNPH. To elucidate whether our hypothesis was correct or not, protein suspensions incubated with MDA were analyzed for the formation of allysine. Allysine was found to increase in control samples, which is consistent with the DNPH results (Fig. 6). In fact, allysine is the most abundant carbonyl in proteins and accounts between 40 and 70% of total protein hydrazones in HSA, BSA and assorted food proteins [7,15,20,25]. Contradicting our initial hypothesis, allysine concentrations remained unchanged during the incubation of protein suspensions with MDA (Fig. 6). Therefore, under the conditions of the present experiment, MDA is unable to induce the oxidative deamination of lysine residues. Hence, the second aldehyde moiety from MDA would not undergo enolization and instead may fulfil its electrophilic nature by adding to additional amino groups as reported above. Furthermore, the addition of MDA to the ε-amino groups impedes the subsequent oxidative deamination of lysine by any means (radical-mediated or α-dicarbonyl-mediated): the formation of allysine is hindered when MDA is present.
Fig. 6.
Concentration of allysine (means ± standard deviations) during incubation of HSA, HEM and LAC for 24 h.
Footnote: For simplification purposes, only results from statistical analysis at final sampling point are displayed. Different letters denote significant differences among experimental units in a post-hoc Tukey test. Samples grouped within a circle share the same letter.
The findings from the present study confirm that MDA would account for protein hydrazones when the DNPH is used as a routine method for the detection of protein carbonyls. This has relevant consequences from a scientific point of view because the DNPH method is typically used to assess protein carbonylation as the most salient expression of protein oxidation in biological samples. Ever since the DNPH method was applied to quantify protein carbonyls, these compounds have been praised to be the result of a metal-catalyzed and radical-mediated oxidative damage to proteins and hence, indicators of an on-site protein carbonylation [26,27]. In their highly influential paper, Levine et al. [27] performed a detailed description of the method and emphasized its significance to identify oxidatively modified proteins and highlighted γ-glutamyl semialdehyde (GGS, oxidation product from arginine and proline) as the most abundant protein carbonyl. In subsequent revisions of the method, the same authors [13,28] distinguished between primary (∼oxidation of amino acid residues into carbonyls) and secondary protein carbonylation (∼introduction of pre-formed carbonyls into proteins). Nevertheless, in further review papers, GGS and allysine are stated as the main carbonyl products in proteins and the DNPH method emphasized as an accurate technique for their detection [6]. Requena et al. [6] regarded secondary carbonylation as a negligible event limited to rat liver samples in which carbonyls may also be introduced by glycation and lipid peroxidation products. In their recent review, Alomari et al. [29] refer to the ‘Levine’ DNPH method as a valid biomarker of oxidatively stressed proteins. Relevant papers in the field are discriminated between those in which the complex nature of protein carbonyls is emphasized and the validity of the DNPH method as a precise technique for assessing oxidized proteins is questioned [25,[30], [31], [32]] and those in which such complexity is overlooked and protein hydrazones are assumed to mainly reflect primary oxidation of amino acid residues in proteins [33,34]. The present results confirm that such an assumption leads to misleading conclusions. Whenever lipid oxidation is concurrent to the oxidative damage to proteins, MDA and likely, other lipid-derived carbonyls, are introduced in proteins by reacting with ε-amino groups. Furthermore, depending on the oxidation conditions and mechanisms, if lipid oxidation is intense and MDA is profusely formed, the nucleophilic addition of MDA to proteins will hamper the primary oxidation of alkaline amino acids such as lysine, and the subsequent formation of allysine. Fig. 7 aims to depict how MDA impedes the formation of allysine and yet, contribute to the formation of protein hydrazones upon reaction with the DNPH reagent. Nevertheless, the DNPH method specifically reflects the carbonylation of proteins but the biochemical mechanism behind that oxidative modification is so complex and variable that accurate and in-depth scientific discussion are not allowed unless other and more specific methodologies are applied to biological samples.
Fig. 7.
Illustration of protein bound lysines and allysines in the presence and/or absence of MDA and DNPH.
3.3. Dose-effect of MDA on HSA carbonylation
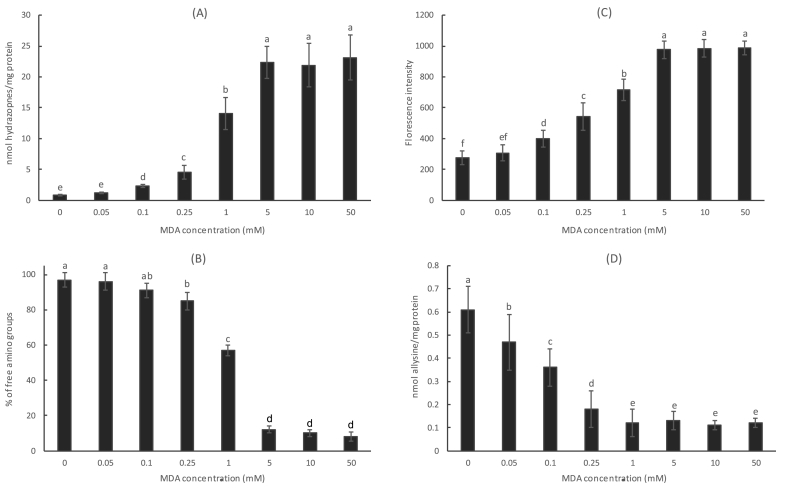
A subsequent assay, aimed to assess the effect of MDA concentration on the events under examination, showed a dose-dependent interfering effect of MDA on allysine formation in the range between 0.05 and 1 mM. Increasing MDA concentration led to accumulative accretion of protein hydrazones (Fig. 8A), subsequent depletion of free amino groups (Fig. 8B) and increasing gain of Schiff base fluorescence (Fig. 8C). Conversely, the formation of allysine declined as the MDA concentration increased, providing strength to the hypothesis of MDA forming adducts with ε-amino groups in protein-bound lysines, and as a result, hindering the oxidative deamination of this amino acid. It is worth highlighting that this interfering effect was observed at biologically relevant MDA concentrations (0.05 mM) [11,12] and the dose-effect was detected at concentrations that may also be relevant in food systems (1 mM). The formation of allysine, already at basal concentrations at 1 mM, was not diminished at higher MDA concentrations. Yet, a complete saturation of amino groups was only observed at 5 mM which is reasonable considering that other amino acids (different from lysine) may also be present and do not yield allysine. Above 5 mM of MDA, no significant changes were observed, with those concentrations being out of the expected range of MDA concentration in biological samples (including food systems) [4].
Fig. 8.
Effect of MDA concentration on accretion of protein hydrazones (A), loss of free amino groups (B), fluorescence gain (C) and concentration of allysine (D) (means ± standard deviations) after incubation with HSA for 24 h.
Footnote: Different letters on top of bars denote significant differences among MDA concentration in a post-hoc Tukey test.
4. Conclusions
The DNPH is a valid method to assess protein carbonylation caused through a variety of pathways and mechanisms, including: i) ROS-mediated oxidation; ii) Maillard-mediated glycooxidation by reducing sugars and their oxidation products and iii) the addition of pre-formed lipid oxidation-aldehydes. While all of them lead to the occurrence of protein carbonyls, the two first mechanisms involve the oxidation of alkaline amino acids and the formation of primary protein carbonyls (such as allysine), and the last mechanism involves the Michael-type addition of pre-formed carbonyls such as MDA. The present study proves that under severe MDA-mediated stress, the formation of primary protein carbonyls is blocked and most protein carbonyls assessed by the DNPH method would reflect the oxidative damage caused by lipid oxidation products on proteins. Being aware of its limitations, protein scientists should accurately interpret results from this commonly used method, and apply, when required, other methodologies such as chromatographic methods to detect specific primary oxidation products such as allysine.
Financial support
Mario Estévez received support from the Spanish Ministry of Economics and Competitiveness (SMEC) through the project AGL2017-84586R. Josué Delgado received support from the Health Council of the Andalusian Regional Government through the project PI-0170-2018.
Acknowledgement
The authors acknowledge the Spanish Ministry of Science, Innovation and Universities (project ID: AGL2017-84586R) and the Health Council of the Andalusian Regional Government (project ID: PI-0170-2018) for funding the present study.
References
- 1.Dean R.T., Fu S., Stocker R., Davies M.J. Biochemistry and pathology of radical-mediated protein oxidation. Biochem. J. 1997;15:1–18. doi: 10.1042/bj3240001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeg S., Grune T. Protein oxidation in aging: does it play a role in aging progression? Antioxidants Redox Signal. 2015;23:239–255. doi: 10.1089/ars.2014.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estévez M., Xiong X.L. Intake of oxidized proteins and amino acids and causative oxidative stress and disease: recent scientific evidences and hypotheses. J. Food Sci. 2017;84(3):387–396. doi: 10.1111/1750-3841.14460. [DOI] [PubMed] [Google Scholar]
- 4.Soladoye O.P., Juarez M.L., Aalhus J.L., Shand P., Estévez M. Protein oxidation in processed meat: mechanisms and potential implications on human health. Compr. Rev. Food Sci. Food Saf. 2015;14:106–122. doi: 10.1111/1541-4337.12127. [DOI] [PubMed] [Google Scholar]
- 5.Estévez M., Luna C. Dietary protein oxidation: a silent threat to human health? Crit. Rev. Food Sci. Nutr. 2017;57:3781–3793. doi: 10.1080/10408398.2016.1165182. [DOI] [PubMed] [Google Scholar]
- 6.Requena J.R., Chao C.-C., Levine R.L., Stadtman E.R. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. U.S.A. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akagawa M., Sasaki T., Suyama K. Oxidative deamination of lysine residue in plasma protein of diabetic rats. Novel mechanism via the Maillard reaction. Eur. J. Biochem. 2002;269:5451–5458. doi: 10.1046/j.1432-1033.2002.03243.x. [DOI] [PubMed] [Google Scholar]
- 8.Villaverde A., Estévez M. Carbonylation of myofibrillar proteins through the Maillard pathway: effect of reducing sugars and reaction temperature. J. Agric. Food Chem. 2013;61:3140–3147. doi: 10.1021/jf305451p. [DOI] [PubMed] [Google Scholar]
- 9.Luna C., Estévez M. Oxidative damage to food and human serum proteins: radical-mediated oxidation vs. glyco-oxidation. Food Chem. 2018;267:111–118. doi: 10.1016/j.foodchem.2017.06.154. [DOI] [PubMed] [Google Scholar]
- 10.Feeney R.E., Blankenhorn G., Dixon B.F. Carbonyl-amine reactions in protein chemistry. Adv. Protein Chem. 1975;29:135–203. doi: 10.1016/s0065-3233(08)60412-x. [DOI] [PubMed] [Google Scholar]
- 11.Houglum K., Filip M., Witztum J.L., Chojkier M. Malondialdehyde and 4-hydroxynonenal protein adducts in plasma and liver of rats with iron overload. J. Clin. Investig. 1990;86:1991–1998. doi: 10.1172/JCI114934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan M.F., Wu X., Tipnis U.R., Ansari G.A.S., Boor P.J. Protein adducts of malondialdehyde and 4-hydroxynonenal in livers of iron loaded rats: quantitation and localization. Toxicology. 2002;173:193–201. [PubMed] [Google Scholar]
- 13.Levine R.L., Williams J.A., Stadtman E.P., Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 14.Malaei R., Ramezani A.M., Absalan G. Analysis of malondialdehyde in human plasma samples through derivatization with 2,4-dinitrophenylhydrazine by ultrasound-assisted dispersive liquid–liquid microextraction-GC-FID approach. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2018;1089:60–69. doi: 10.1016/j.jchromb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Akagawa M., Sasaki D., Kurota Y., Suyama K. Formation of α-aminoadipic and γ-glutamic semialdehydes in proteins by the Maillard reaction. Ann. N. Y. Acad. Sci. 2005;1043:129–134. doi: 10.1196/annals.1333.016. [DOI] [PubMed] [Google Scholar]
- 16.Estévez M., Ollilainen V., Heinonen M. Analysis of protein oxidation markers – α-aminoadipic and γ-glutamic semialdehydes – in food proteins by using LC–ESI- multi-stage tandem MS. J. Agric. Food Chem. 2009;57:3901–3910. doi: 10.1021/jf804017p. [DOI] [PubMed] [Google Scholar]
- 17.Arcanjo N.M.O., Luna C., Madruga M.S., Estévez M. Antioxidant and pro-oxidant actions of resveratrol on human serum albumin in the presence of toxic diabetes metabolites: glyoxal and methyl-glyoxal. Biochim. Biophys. Acta Gen. Subj. 2018;1862(9):1938–1947. doi: 10.1016/j.bbagen.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Weigele M., Debernardo S.L., Tengi J.P., Leimgruber W. A novel reagent for the fluorometric assay of primary amines. J. Am. Chem. Soc. 1972;94:5927–5928. [Google Scholar]
- 19.Strauss G., Gibson S.M. Plant phenolics as cross-linkers of gelatin gels and gelatin-based coacervates for use as food ingredients. Food Hydrocolloids. 2004;18:81–89. [Google Scholar]
- 20.Utrera, M., Morcuende, D., Rodríguez-Carpena, G. & Estévez, M. Fluorescent HPLC for the detection of specific protein oxidation carbonyls – α-aminoadipic and γ-glutamic semialdehydes – in meat systems. Meat Sci., 89, 500-506. [DOI] [PubMed]
- 21.Akagawa M., Sasaki D., Ishii Y., Kurota Y., Yotsu-Yamashita M., Uchida K., Suyama K. New method for the quantitative determination of major protein carbonyls, α-aminoadipic and γ-glutamic semialdehydes: investigation of the formation mechanism and chemical nature in vitro and in vivo. Chem. Res. Toxicol. 2006;19(8):1059–1065. doi: 10.1021/tx060026p. [DOI] [PubMed] [Google Scholar]
- 22.Gürbüz G., Liu C., Jiang Z.-Q., Pulkkinen M., Piironen V., Sontag-Strohm T., Heinonen M. Protein–lipid co-oxidation in emulsions stabilized by microwave-treated and conventional thermal-treated faba bean proteins. Food Sci. Nutr. 2018;6(4):1032–1039. doi: 10.1002/fsn3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura J., Shimomoto T., Collins L.B.…Gold A., Bultman S.J. Evidence that endogenous formaldehyde produces immunogenic and atherogenic adduct epitopes. Sci. Rep. 2017;7:10787. doi: 10.1038/s41598-017-11289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weißer J., Ctortecka C., Busch C.J.…Binder C.J., Bennett K.L. A comprehensive analytical strategy to identify malondialdehyde-modified proteins and peptides. Anal. Chem. 2017;89:3847–3852. doi: 10.1021/acs.analchem.6b05065. [DOI] [PubMed] [Google Scholar]
- 25.Armenteros M., Heinonen M., Ollilainen V., Toldrá F., Estévez M. Analysis of protein carbonyls in meat products by using the DNPH method, fluorescence spectroscopy and liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) Meat Sci. 2009;83:104–112. doi: 10.1016/j.meatsci.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Oliver C.N., Ahn B.W., Moerman E.J., Goldstein S., Stadtman E.R. Aged-related changes in oxidized proteins. J. Biol. Chem. 1987;262:5488–5491. [PubMed] [Google Scholar]
- 27.Levine R.L., Garland D., Oliver C.N., Amici A., Climent I., Lenz A.G., Ahn B.W., Shaltiel S., Stadtman E.R. Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 28.Levine R.L., Wehr N., Williams J.A., Stadtman E.R., Shacter E. Determination of carbonyl groups in oxidized proteins. Methods Mol. Biol. 2000;99:15–24. doi: 10.1385/1-59259-054-3:15. [DOI] [PubMed] [Google Scholar]
- 29.Alomari E., Bruno S., Ronda L.…Bettati S., Mozzarelli A. Protein carbonylation detection methods: a comparison. Data Brief. 2018;19:2215–2220. doi: 10.1016/j.dib.2018.06.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estévez M. Protein carbonyls in meat systems: a review. Meat Sci. 2011;89:259–279. doi: 10.1016/j.meatsci.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Augustyniak E., Adam A., Wojdyla K.…Fedorova M., Griffiths H.R. Validation of protein carbonyl measurement: a multi-centre study. Redox Biol. 2015;4:149–157. doi: 10.1016/j.redox.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber D., Davies M.J., Grune T. Determination of protein carbonyls in plasma, cell extracts, tissue homogenates, isolated proteins: focus on sample preparation and derivatization conditions. Redox Biol. 2015;5:367–380. doi: 10.1016/j.redox.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mesquita C.S., Oliveira R., Bento F.…Rodrigues J.V., Marcos J.C. Simplified 2,4-dinitrophenylhydrazine spectrophotometric assay for quantification of carbonyls in oxidized proteins. Anal. Biochem. 2014;458:69–71. doi: 10.1016/j.ab.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 34.Soglia F., Petracci M., Ertbjerg P. Novel DNPH-based method for determination of protein carbonylation in muscle and meat. Food Chem. 2016;197:670–675. doi: 10.1016/j.foodchem.2015.11.038. [DOI] [PubMed] [Google Scholar]