See Kolanko and Malhotra (doi:10.1093/brain/awy282) for a scientific commentary on this article.
Alzheimer's disease is a heterogeneous disorder, and understanding the biological basis for this heterogeneity is key to developing personalized medicine. ten Kate et al. identify atrophy subtypes in patients, and show that these subtypes are present prodromally and are associated with the types of symptoms an individual will develop.
Keywords: Alzheimer’s disease, non-negative matrix factorization, mild cognitive impairment, disease heterogeneity, prognosis
Abstract
Alzheimer’s disease is a heterogeneous disorder. Understanding the biological basis for this heterogeneity is key for developing personalized medicine. We identified atrophy subtypes in Alzheimer’s disease dementia and tested whether these subtypes are already present in prodromal Alzheimer’s disease and could explain interindividual differences in cognitive decline. First we retrospectively identified atrophy subtypes from structural MRI with a data-driven cluster analysis in three datasets of patients with Alzheimer’s disease dementia: discovery data (dataset 1: n = 299, age = 67 ± 8, 50% female), and two independent external validation datasets (dataset 2: n = 181, age = 66 ± 7, 52% female; dataset 3: n = 227, age = 74 ± 8, 44% female). Subtypes were compared on clinical, cognitive and biological characteristics. Next, we classified prodromal Alzheimer’s disease participants (n = 603, age = 72 ± 8, 43% female) according to the best matching subtype to their atrophy pattern, and we tested whether subtypes showed cognitive decline in specific domains. In all Alzheimer’s disease dementia datasets we consistently identified four atrophy subtypes: (i) medial-temporal predominant atrophy with worst memory and language function, older age, lowest CSF tau levels and highest amount of vascular lesions; (ii) parieto-occipital atrophy with poor executive/attention and visuospatial functioning and high CSF tau; (iii) mild atrophy with best cognitive performance, young age, but highest CSF tau levels; and (iv) diffuse cortical atrophy with intermediate clinical, cognitive and biological features. Prodromal Alzheimer’s disease participants classified into one of these subtypes showed similar subtype characteristics at baseline as Alzheimer’s disease dementia subtypes. Compared across subtypes in prodromal Alzheimer’s disease, the medial-temporal subtype showed fastest decline in memory and language over time; the parieto-occipital subtype declined fastest on executive/attention domain; the diffuse subtype in visuospatial functioning; and the mild subtype showed intermediate decline in all domains. Robust atrophy subtypes exist in Alzheimer’s disease with distinct clinical and biological disease expression. Here we observe that these subtypes can already be detected in prodromal Alzheimer’s disease, and that these may inform on expected trajectories of cognitive decline.
See Kolanko and Malhotra (doi:10.1093/brain/awy282) for a scientific commentary on this article.
Introduction
Alzheimer’s disease is a heterogeneous neurodegenerative disorder: patients differ in age of dementia onset, genetic risk factors, clinical presentation, and rate and type of cognitive decline (Lam et al., 2013). Heterogeneity in clinical presentation and trajectories of cognitive decline already manifests in the prodromal stage of Alzheimer’s disease (Vos et al., 2015), i.e. patients with mild cognitive impairment and biomarker evidence of Alzheimer’s pathology (Dubois et al., 2014). Understanding the biological mechanisms that underlie heterogeneity in cognitive symptoms and trajectories of decline is crucial to help clinicians determine prognosis and an important step towards precision medicine in Alzheimer’s disease.
Cognitive subtypes of Alzheimer’s disease dementia (ADD) have been observed in several independent patient cohorts and also in autosomal dominant Alzheimer’s disease, which suggests that clinical heterogeneity has a biological basis (Tang et al., 2016; Scheltens et al., 2017). Post-mortem studies also point towards the existence of biological heterogeneity by showing neuropathological subtypes consisting of typical Alzheimer’s disease, limbic predominant, and hippocampal sparing, which were associated with differences in ante-mortem clinical presentation (Murray et al., 2011). Studying brain atrophy patterns provides an opportunity to examine biological heterogeneity in vivo (Whitwell et al., 2012). Imaging studies classifying patients with ADD based on a priori definitions of heterogeneity have shown that atrophy subtypes can explain part of the variability in clinical and cognitive characteristics, including a medial-temporal atrophy variant with predominant memory dysfunction and a subtype with relative hippocampal sparing with young age of onset and more pronounced non-memory presentation (Möller et al., 2013; Byun et al., 2015; Ferreira et al., 2017; Persson et al., 2017; Risacher et al., 2017). Data-driven methods provide an unbiased approach to detect atrophy subtypes that do not depend on predefined choices, and so may yield a more refined description of heterogeneity in Alzheimer’s disease. A few data-driven (Noh et al., 2014; Hwang et al., 2016; Zhang et al., 2016; Park et al., 2017; Poulakis et al., 2018) or semi-supervised (Dong et al., 2017; Varol et al., 2017) imaging studies have detected atrophy subtypes in patients with probable ADD. However, those studies used clinical criteria without biomarker confirmation to select ADD patients, and therefore the possibility cannot be excluded that subgroups reflect non-Alzheimer pathology. In addition, most previous studies employed clustering methods that rely on determining the global similarity of atrophy patterns amongst individuals, which may not capture particular atrophy patterns that are present in only a subset of individuals. Non-negative matrix factorization (NMF) is a statistical approach that decomposes data into non-negative parts, and allows us to simultaneously cluster features (e.g. cortical atrophy) and individuals into subgroups (Lee and Seung, 1999). A particularly strong aspect of the parts-based approach of NMF is that it enables identifying subsets of atrophy features that are correlated in a subset of individuals, but not in other subgroups: such non-linear associations cannot be detected by methods that depend on global similarity values. As such, this technique is particularly suitable for atrophy subtype discovery.
Furthermore, it remains unclear when heterogeneity in atrophy patterns arises during the disease. We hypothesize that if heterogeneity in atrophy patterns reflects true pathophysiological subtypes of Alzheimer’s disease, then such atrophy subtypes should be detectable in prodromal stages of the disease, and might predict the type of symptoms that a patient will develop (Schuff et al., 2012; Leung et al., 2013). To our knowledge, only one previous study investigated this question, of which the results supported the hypothesis that data-driven derived subtypes may already be present in predementia stages of Alzheimer’s disease (Zhang et al., 2016). But, as the previous study had no replication data available, the robustness of these subtypes remains unclear. Furthermore, it remains unknown whether such subtypes in prodromal Alzheimer’s disease show differences in their rates of clinical and cognitive decline, and whether subtypes show decline in specific cognitive domains.
In the present study, we used a data-driven clustering approach to identify and replicate in two external independent datasets atrophy subtypes in patients with ADD and biomarker evidence of amyloid pathology. We compared patients of the different atrophy subtypes on clinical, biological and cognitive characteristics that are known to be associated with Alzheimer’s disease [i.e. CSF total tau (t-tau) and phosphorylated tau (p-tau), white matter hyperintensities (WMHs) on MRI, and apolipoprotein E (APOE) ε4 genotype (Van Der Flier et al., 2011; Jack et al., 2013; Lee et al., 2016)]. Next, we validated our results further and studied the potential practical use of data-driven derived subtypes, by classifying participants with prodromal Alzheimer’s disease into the atrophy subtypes that best matched with their regional grey matter volumes. We examined whether the atrophy subtypes in this earlier disease stage showed similar clinical and biomarker profiles compared to subjects with dementia, and whether atrophy subtypes would show decline in specific cognitive domains.
Materials and methods
Datasets
We included participants with a good quality 3D T1-weighted structural MRI based on research criteria for ADD, i.e. having a clinical diagnosis of ADD and evidence for amyloid pathology in CSF or on amyloid PET (Supplementary material); or prodromal Alzheimer’s disease, i.e. clinical diagnosis of mild cognitive impairment and evidence of amyloid pathology (Dubois et al., 2014). Participants were included from two large cohorts: the Amsterdam Dementia Cohort (ADC) (van der Flier et al., 2014) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (www.adni-info.org). We first identified atrophy subtypes in a discovery dataset consisting of ADD patients from the ADC who were scanned on the same 3 T scanner (ADCd; n = 299). We then assessed whether we could identify the same clusters in an independent validation set consisting of ADD patients from the ADC scanned on either of two different 3 T scanners (ADCv; n = 181). Finally, we identified clusters in another external validation set consisting of ADD patients from ADNI (n = 227). Participants with prodromal Alzheimer’s disease from ADC (n = 160) and ADNI (n = 443) were grouped into one dataset. As per inclusion criteria for this study, all participants had CSF amyloid-β1-42 or PET amyloid available, and the majority additionally had CSF t-tau and p-tau, as well as WMH, and APOE genotype (Table 1 and Supplementary material). APOE ε4 genotype was dichotomized by the presence of at least one APOE ε4 allele.
Table 1.
Clinical and biomarker characteristics per dementia patient dataset
| ADCd (n = 299) | ADCv (n = 181) | ADNI (n = 227) | P(overall) | ADCd versus ADCv | ADCd versus ADNI | ADCv versus ADNI | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Measure | n missing | Measure | n missing | Measure | n missing | |||||
| Demographics | ||||||||||
| Age, years (%) | 67 ± 8 | 0 (0) | 66 ± 7 | 0 (0) | 74 ± 8 | 0 (0) | <0.0001 | 0.9 | <0.0001 | <0.0001 |
| Sex, female (%) | 149 (50) | 0 (0) | 95 (52) | 0 (0) | 99 (44) | 0 (0) | 0.2 | – | – | – |
| Education, years (%) | 11.2 ± 2.7 | 1 (0) | 11.2 ± 2.9 | 0 (0) | 15.5 ± 3.0 | 0 (0) | <0.0001 | 0.9 | <0.0001 | <0.0001 |
| Global cognition | ||||||||||
| MMSE (%) | 21.8 ± 3.3 | 0 (0) | 22.3 ± 3.2 | 0 (0) | 23.3 ± 2.0 | 0 (0) | <0.0001 | 0.1 | 0.009 | <0.0001 |
| APOE genotype | ||||||||||
| APOE ε4 carrier (%) | 214 (73) | 6 (2) | 123 (69) | 2 (1) | 167 (75) | 3 (1) | 0.4 | – | – | – |
| CSF biomarkersa | ||||||||||
| Amyloid-β1-42, Innotest (%) | 465 ± 97 | 8 (3) | 532 ± 104 | 23 (13) | – | – | n/a | <0.0001 | – | – |
| Amyloid-β1-42, Luminex (%) | – | – | – | – | 130 ± 21 | 23 (10) | n/a | – | – | – |
| Amyloid-β1-42 abnormalb (%) | 291 (100) | – | 158 (100) | – | 204 (100) | – | n/a | – | – | – |
| Total tau, Innotest (%) | 684 ± 398 | 10 (3) | 764 ± 398 | 23 (13) | – | – | n/a | 0.02 | – | – |
| Total tau, Luminex (%) | – | – | – | – | 131 ± 61 | 23 (10) | n/a | – | – | – |
| Total tau, abnormalb (%) | 232 (80) | – | 140 (89) | – | 143 (70) | – | <0.001 | 0.03 | 0.01 | <0.001 |
| P-tau, Innotest (%) | 87 ± 40 | 8 (3) | 88 ± 35 | 23 (13) | – | – | n/a | 0.9 | – | – |
| P-tau, Luminex (%) | – | – | – | – | 59 ± 34 | 95 (42) | n/a | – | – | – |
| P-tau abnormalb (%) | 238 (82) | – | 137 (87) | – | 128 (97) | – | <0.001 | 0.2 | <0.001 | 0.004 |
| Imaging biomarkersc | ||||||||||
| WMH visual rating (%) | 1.1 ± 0.8 | 3 (1%) | 1.1 ± 0.8 | 2 (1%) | – | – | n/a | 0.9 | – | – |
| WMH volume, in ml (%) | – | – | – | – | 5.4 ± 8.0 | 0 (0) | n/a | – | – | – |
Data are presented as n (%) or mean ± standard deviation; n/a = not applicable.
aCSF biomarkers were measured in ADCd/ADCv using Innotest ELISAs (cut-off amyloid-β1-42 < 640 ng/l, t-tau ≥ 375 ng/l, p-tau ≥ 52 ng/l) and in ADNI using Luminex (cut-off amyloid-β1-42 < 192 ng/l, t-tau > 93 ng/l, p-tau > 23 ng/l).
bPercentage abnormal are based on available data (excluding missings).
cWMH were measured in ADCd/ADCv using the visual Fazekas scale (range 0–3) and using automated software in ADNI (unit: ml).
The ADC consists of participants attending the memory clinic of the VU University Medical Centre Amsterdam. The VU University Medical Centre institutional review board approved the ADC study protocol. All participants gave written informed consent for their clinical data to be used for research purposes. Within the ADNI, participants were recruited over 50 sites throughout the USA and Canada. We used data of baseline or screening visits from ADNI phase-1, phase-2, and GO. These data were obtained from the ADNI database (adni.loni.usc.edu). The ADNI study was approved by the Institutional Review Boards of all of the participating institutions. All patients gave written informed consent.
Neuropsychological assessments
In ADC and ADNI the neuropsychological assessment covered similar cognitive domains, although the cohorts differed in the specific tests used (Petersen et al., 2010; van der Flier et al., 2014). To aid comparability between cohorts, we combined test scores into four domains: memory, language, visuospatial, and attention/executive. We grouped the attention and executive domain, as ADNI does not have enough tests available to split these domains. Details of the neuropsychological tests in each domain are presented in the Supplementary material. Before grouping tests in domains, missing values were estimated using multiple imputation (Supplementary material). All test scores were z-transformed within each cohort to remove measuring scale, and inverted when necessary such that lower values represent worse performance. Within each cognitive domain, z-transformed scores were averaged to obtain the composite scores. For prodromal Alzheimer’s disease participants, follow-up z-scores were determined relative to baseline scores.
MRI acquisition and processing
For the ADC, anatomical 3D T1-weighted images were acquired as part of regular patient care on three different MRI 3 T scanners using an 8-channel head coil. Participants in the ADCd were all scanned on a single GE Signa scanner and participants in the ADCv were scanned on one of two scanners: Toshiba Titan 3 T or Philips Ingenuity PET/MRI. Details on acquisition parameters are provided in the Supplementary material. In ADNI, 3D T1-weighted scans were performed on 1.5 T (ADNI-1) or 3 T (ADNI-2 and ADNI-GO) scanners using previously described standardized protocols at each site (Jack et al., 2008).
Structural 3D T1 images were segmented into grey matter, white matter, and CSF using Statistical Parametric Mapping 12 (SPM12) software (Wellcome Trust Centre for Neuroimaging, University College London, UK) running in MATLAB 2011a (MathWorks Inc., Natick, MA, USA). The quality of all segmentations was visually inspected and five were excluded because of erroneous segmentation. Next, we defined 1024 cortical and subcortical anatomical areas in the native space grey matter segmentation of each participant using a brain parcellation that was generated by randomly subdividing the automated anatomical labelling atlas (Tzourio-Mazoyer et al., 2002) into equal-sized regions (Zalesky et al., 2010). This atlas was warped from standard space to subject space using inverted parameters that were calculated by non-linear normalization of subject images to standard MNI space. For each region, grey matter volume was defined as the sum of grey matter estimates across the voxels multiplied by voxel volume, and all volumes were normalized by total grey matter volume.
Cluster analysis
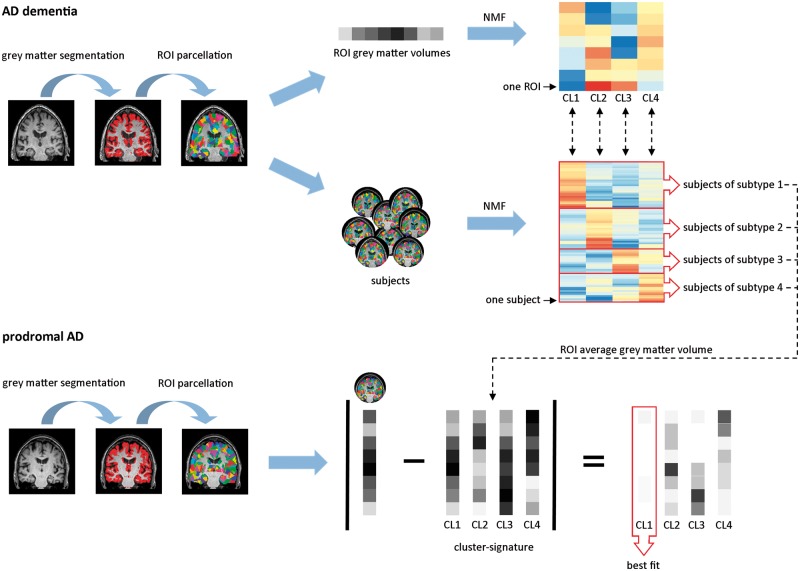
To examine atrophy subtypes, we used NMF in R (v.3.3.1, NMF v.0.20.6) (Gaujoux and Seoighe, 2010), in each of the three ADD patient datasets. NMF is a data-driven dual-clustering approach that identifies clusters of features (in our study atrophy patterns) and participants at the same time (Fig. 1). Participants are grouped into a subtype based on the best fit of their data on the identified atrophy clusters. We characterized each atrophy cluster based on the top 100 cluster-defining features (i.e. regions of interest) in each dataset. Spatial correspondence of atrophy cluster solutions across datasets was assessed with the Dice coefficient (see Supplementary Table 3 and Supplementary Fig. 2 for comparison containing the top 200 features) (Dice, 1945).
Figure 1.
NMF in ADD patients and classification of prodromal Alzheimer’s disease participants. Grey matter segmentations were extracted from structural MRI and parcellated into 1024 equally-sized regions of interest, from which regional grey matter volumes were derived (for illustrative purposes only eight regions of interest are shown). In ADD patients, NMF, a dual-clustering approach, was used to identify clusters of features (in this case atrophy patterns) and participants at the same time. Top right: The regions of interest are clustered into distinct atrophy patterns. Each row represents a region of interest and each column an atrophy cluster. The warmer the colour, the more that region of interest contributes to the atrophy cluster. Middle: Subjects are grouped into subtypes based on the best fit of their region of interest volumes to each of the atrophy clusters. Each row represents one participant and the warmer the colour, the better the fit of that participants’ region of interest volumes to the region of interest volumes of that atrophy cluster. For each of the atrophy clusters, we made a cluster signature by computing the average volume in each of the top cluster-defining regions of interest across all ADD patients classified as that atrophy subtype. We classified prodromal Alzheimer’s disease participants based on the lowest absolute minimal distance between their own region of interest volumes and that of the cluster-signatures. AD = Alzheimer’s disease; CL = cluster; ROI = region of interest.
Classification of prodromal Alzheimer’s disease participants
For each of the identified atrophy clusters, we derived a cluster signature by computing the average grey matter volume in the cluster-defining regions of interest (across datasets) from all ADD participants classified into that subtype (Fig. 1). We then classified participants with prodromal Alzheimer’s disease based on the lowest absolute minimal distance between their own regional grey matter values and each of the cluster signatures.
Statistical analysis
Subtypes were compared on demographic, clinical, neuropsychological, genetic, and biomarker measures with ANOVA, Kruskal Wallis, or chi-square tests where appropriate. We performed comparisons between subtypes for each dataset separately, and pooled across datasets. Prior to pooling, variables that were measured at different scales (i.e. CSF biomarkers, WMH) were z-transformed. For composite neuropsychological scores, results were pooled over imputed datasets using Rubins’s rules as implemented in the R package MICE (van Buuren and Groothuis-Oudshoorn, 2011). Voxel-based morphometry was used to compare patterns of grey matter loss between the atrophy subtypes, and for reference with a control group of participants with normal amyloid markers and normal cognition (264 cognitively normal from ADNI; 88 subjective cognitive decline from ADC) (Supplementary material). In prodromal Alzheimer’s disease participants, we also characterized longitudinal trajectories of cognitive decline in two ways. First, time to dementia (dependent variable) was compared between subtypes (predictor variable) with a Cox proportional hazard model with the ‘survival’ package (version 2.41-3) in R. Second, linear mixed models were used to evaluate baseline cognition and decline over time in the cognitive domains of prodromal Alzheimer’s disease participants classified in the subtypes using the ‘lme4’ package (version 1.1-12) in R. Time from baseline (years), subtype, and their interaction were included as fixed effects. Subject intercepts and slopes were modelled as random effects. We repeated the linear mixed model with covariates age, gender and education. Finally, we analysed point estimate differences between subtypes in cognitive functioning after 3 years follow-up for a subset of prodromal Alzheimer’s disease individuals (n = 352, 63%).
Data availability
The ADNI data used is this study were obtained from the ADNI database (adni.loni.usc.edu). The ADC data used in this study are available from the corresponding author, upon reasonable request.
Results
Dataset characteristics
On average, ADD patients were 69 ± 8 years old, with the ADCd and ADCv participants being ~10 years younger than ADNI participants (P < 0.0001; Table 1). All patients had mild to moderate ADD; participants in ADNI had slightly higher Mini-Mental State Examination (MMSE) scores (mean 23.3 ± 2.0) than participants in ADCd (21.8 ± 3.3) and ADCv (22.3 ± 2.0) (P < 0.001 for ADCd and P = 0.009 for ADCv).
Subtype identification in Alzheimer’s disease dementia participants with NMF
In each of the dementia datasets, four clusters showed an optimal fit of the data, with high stability of the cluster solutions (Supplementary Table 1) and explaining more variance in the data than random partitions (Supplementary Table 2).
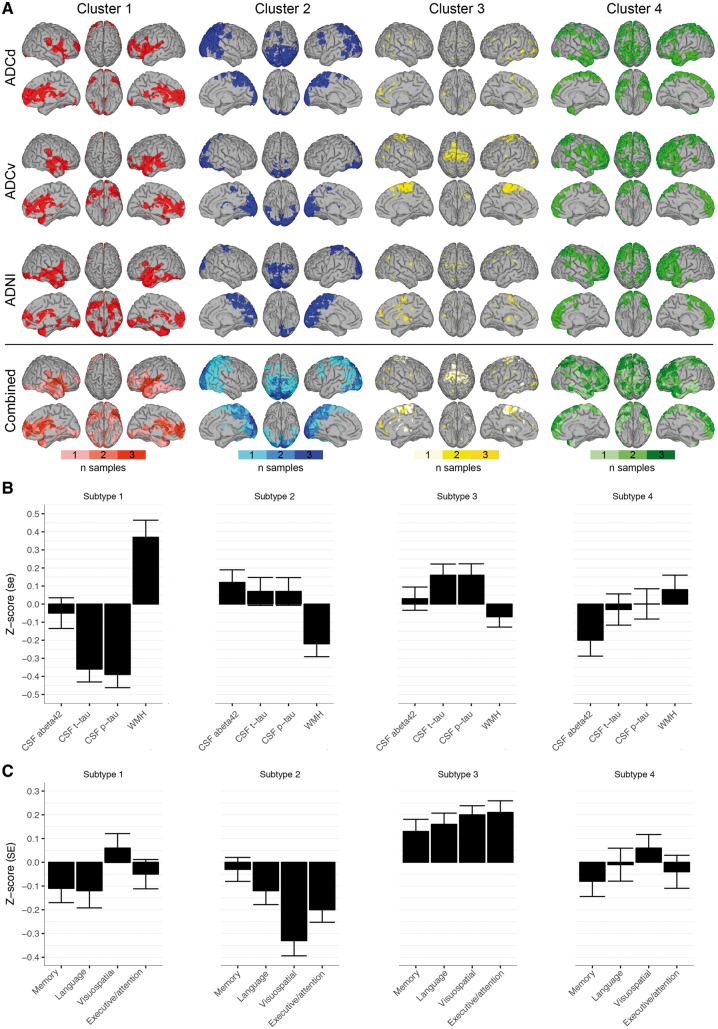
Figure 2 shows the top 100 cluster-defining regions of interest (i.e. the regions of interest that contribute the most to that cluster) for each of the four clusters for each dementia dataset. In each dataset, participants were grouped into subtypes based on the correspondence of their regional grey matter values to one of the four atrophy clusters (Supplementary Fig. 1). On average 19% (range 19–19%) of participants were classified in subtype 1, which showed temporal-dominant cluster regions, also including the insula. On average 28% (range 27–30%) of participants were classified as subtype 2, which showed parieto-occipital dominant cluster regions. On average 34% (range 33–37%) of participants were classified as subtype 3, which showed a dispersed pattern of cluster-defining regions, amongst which was the motor cortex. The fourth subtype, including on average 18% (range 16–21%) of participants, showed most distinctly from the other subtypes involvement of the lateral and medial frontal lobes, and lateral temporal cortex.
Figure 2.
Cluster features across datasets. (A) In each dataset we visualized the top 100 most important cluster-defining features. The bottom row represents the combined important cluster features across datasets: colour bars indicate whether the top 100 cluster-defining features were observed in 1/3, 2/3 or 3/3 datasets. Right hemisphere is displayed on the left side and vice versa. (B) Subtype-specific biomarker profiles: mean ± standard error (SE) of normalized values (z-scores) of CSF levels of amyloid-β1-42 (abeta42), t-tau and p-tau and WMH. (C) Subtype-specific cognitive profiles: mean ± SE of normalized values (z-scores) of neuropsychological composite scores for memory, language, visuospatial and executive/attention domains.
The Dice overlap, assessing the similarity of the cluster-defining regions of interest across datasets, was reasonable-to-good for most of the atrophy clusters ranging from 0.29 to 0.72 (Supplementary Table 3). Since the atrophy clusters showed similar features across datasets (Fig. 2), we pooled participants of each subtype across datasets for further analysis. For the same reason, we pooled cluster-defining features across the three datasets to compute the cluster signatures to classify prodromal Alzheimer’s disease participants.
Atrophy characterization of subtypes
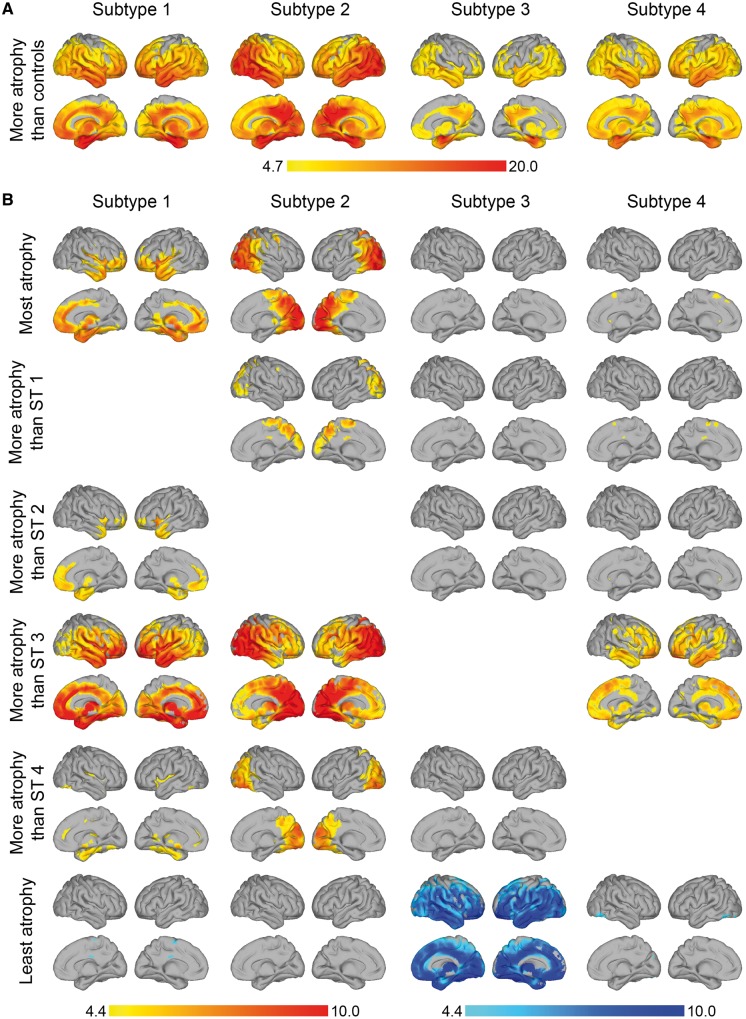
Compared to cognitively normal controls, all subtypes showed widespread atrophy (Fig. 3A and Supplementary Table 4). Pairwise comparisons between the dementia subtypes are presented in Fig. 3B. Subtype 1 had most pronounced medial-temporal atrophy. Subtype 2 had predominant parieto-occipital atrophy. Subtype 3 had the mildest atrophy compared to the other subtypes. Subtype 4 had a diffuse atrophy pattern involving the temporal and frontal lobes, without strong regional differences.
Figure 3.
Atrophy patterns in each subtype. (A) Voxel-based morphometry comparison between atrophy subtypes and control (cognitively normal, amyloid negative) participants. (B) Voxel-based morphometry comparison between atrophy subtypes. Top row: In yellow-red regions where each subtype has most atrophy compared to all other clusters. Rows 2–5: In yellow-red pairwise comparisons between subtypes. Bottom row: In blue regions where each subtype has least atrophy (most grey matter) compared to all other subtypes. Colour bar represents t-statistic. Data are presented at voxel-level PFWE < 0.05. Right hemisphere is displayed on the left side and vice versa. ST1 = subtype 1 (medial-temporal dominant atrophy); ST2 = subtype 2 (parieto-occipital atrophy); ST3 = subtype 3 (mild atrophy); ST4 = subtype 4 (diffuse atrophy).
Cognitive, genetic and biological characterization of atrophy subtypes
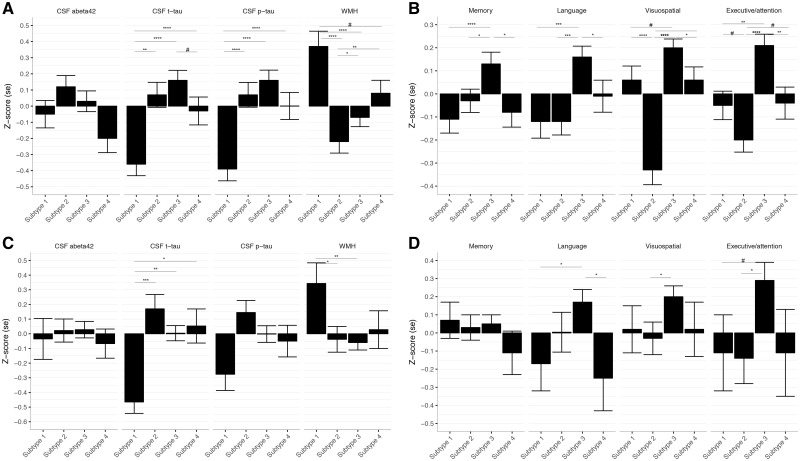
Participants of the first subtype, with medial-temporal predominant atrophy, were the oldest and had unequal sex distribution, with relatively more males (Table 2; see Supplementary Tables 5 and 6 for dataset-specific results). They had the lowest CSF t-tau and p-tau levels, and the highest WMH load (Figs 2B and 4A). They had low memory and language scores (Figs 2C and 4B). Parieto-occipital atrophy subtype participants were young and scored lowest on the MMSE and on visuospatial and executive/attention domains. They had the second highest t-tau and p-tau levels, and the lowest amount of WMH. Mild atrophy subtype participants were youngest, performed best on global cognition (highest MMSE) and in all cognitive domains, and had the highest CSF t-tau and p-tau levels. Finally, the diffuse atrophy subtype participants had intermediate clinical, cognitive and biological features. Participants with this subtype had somewhat lower memory scores and average scores on the other domains. They had intermediate levels of t-tau, p-tau, and WMH. Subtypes showed similar levels of CSF amyloid-β1-42 (overall P = 0.05), but there was a trend towards lowest amyloid-β1-42 concentrations in the diffuse subtype. The subtypes showed similar distributions of APOE ε4 genotype.
Table 2.
Characteristics of dementia subtypes in pooled dataset
| ST1: medial- temporal (n = 136) | ST2: parieto- occipital (n = 198) | ST3: mild (n = 247) | ST4: diffuse (n = 126) | P (overall) | ST1 versus ST2 | ST1 versus ST3 | ST1 versus ST4 | ST2 versus ST3 | ST2 versus ST4 | ST3 versus ST4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | |||||||||||
| Age, years | 73 ± 8 | 68 ± 9 | 67 ± 8 | 70 ± 8 | <0.0001 | <0.001 | <0.0001 | 0.076 | 0.93 | 0.048 | 0.004 |
| Sex, female (%) | 46 (34) | 91 (46) | 140 (57) | 66 (52) | 0.0002 | 0.04 | <0.001 | 0.004 | 0.03 | 0.31 | 0.50 |
| Education, years | 13.1 ± 3.7 | 12.5 ± 3.5 | 12.4 ± 3.3 | 12.6 ± 3.5 | 0.32 | – | – | – | – | – | – |
| Global cognition | |||||||||||
| MMSE | 22.4 ± 3.0 | 21.7 ± 3.0 | 22.9 ± 2.9 | 22.3 ± 2.8 | 0.0002 | 0.043 | 0.073 | 0.54 | <0.0001 | 0.19 | 0.015 |
| APOE genotype | |||||||||||
| APOE ε4 carrier (%) | 95 (70) | 135 (68) | 184 (74) | 90 (71) | 0.44 | – | – | – | – | – | – |
Data are presented as n (%) or mean ± SD. P-values are based on chi-square, ANOVA or Kruskall-wallis tests when appropriate. ST1 = subtype 1 (medial-temporal dominant atrophy); ST2 = subtype 2 (parieto-occipital atrophy); ST3 = subtype 3 (mild atrophy); ST4 = subtype 4 (diffuse atrophy).
Figure 4.
Biomarker and cognitive profile comparisons between subtypes. Data are presented as mean ± SE normalized values (z-scores). P-values are based on ANOVA tests. For neuropsychology, composite scores are presented. Abeta42 = amyloid-β1-42. (A) Biomarker profile comparisons between subtypes in ADD. (B) Cognitive profile comparisons between subtypes in ADD. (C) Biomarker profile comparisons between subtypes in prodromal Alzheimer’s disease. (D) Cognitive profile comparisons between subtypes in prodromal Alzheimer’s disease. Subtype 1 = medial-temporal atrophy; subtype 2 = parieto-occipital atrophy; subtype 3 = mild atrophy; subtype 4 = diffuse cortical atrophy.
Classification of prodromal Alzheimer’s disease participants
Clinical, biological and cognitive characteristics of prodromal Alzheimer’s disease participants can be found in Supplementary Table 7. Most prodromal Alzheimer’s disease participants were classified into the mild atrophy subtype (55%; Table 3), followed by the parietal-occipital cortical atrophy (26%), medial-temporal predominant atrophy (11%) and diffuse cortical (8%). In the prodromal stage, subtypes largely showed similar characteristics as observed in the dementia stage (Table 3, Fig. 4C and D), apart from CSF tau levels, which were highest for the parieto-occipital subtype (instead of second highest in the dementia datasets). Some prodromal Alzheimer’s disease subjects (n = 124, 21%) showed a similar match to multiple subtypes. When repeating analyses taking these subjects as an additional ‘less well matching’ group, we observed similar clinical and biological characteristics for the four subtypes. Subjects with prodromal Alzheimer’s disease who were less well-matching had intermediate clinical and biomarker profiles (data not shown).
Table 3.
Characteristics of prodromal Alzheimer’s disease participants after subtype classification
| ST1: medial- temporal (n = 68) | ST2: parieto- occipital (n = 155) | ST3: mild (n = 329) | ST4: diffuse (n = 51) | P (overall) | ST1 versus ST2 | ST1 versus ST3 | ST1 versus ST4 | ST2 versus ST3 | ST2 versus ST4 | ST3 versus ST4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Demographics | |||||||||||
| Age, years | 76 ± 6 | 73 ± 7 | 70 ± 8 | 74 ± 7 | <0.0001 | 0.006 | <0.0001 | 0.53 | 0.009 | 0.48 | 0.002 |
| Sex, female (%) | 18 (29) | 45 (38) | 142 (48) | 20 (44) | 0.0034 | 0.089 | <0.0001 | 0.14 | 0.053 | 0.95 | 0.36 |
| Education, years | 15.5 ± 3.4 | 14.7 ± 3.8 | 14.8 ± 3.3 | 14.8 ± 3.7 | 0.49 | – | – | – | – | – | – |
| Global cognition | |||||||||||
| MMSE | 27.4 ± 1.8 | 27.5 ± 1.9 | 27.3 ± 2 | 27.3 ± 1.8 | 0.85 | – | – | – | – | – | – |
| APOE genotype | |||||||||||
| APOE ε4 carrier (%) | 38 (56) | 101 (65) | 218 (66) | 29 (57) | 0.14 | – | – | – | – | – | – |
Data are presented as n (%) or mean ± SD. P-values are based on chi-square, ANOVA or Kruskall-Wallis tests when appropriate. ST1 = subtype 1 (medial-temporal dominant atrophy); ST2 = subtype 2 (parieto-occipital atrophy); ST3 = subtype 3 (mild atrophy); ST4 = subtype 4 (diffuse atrophy).
Atrophy subtypes and cognitive decline in prodromal Alzheimer’s disease
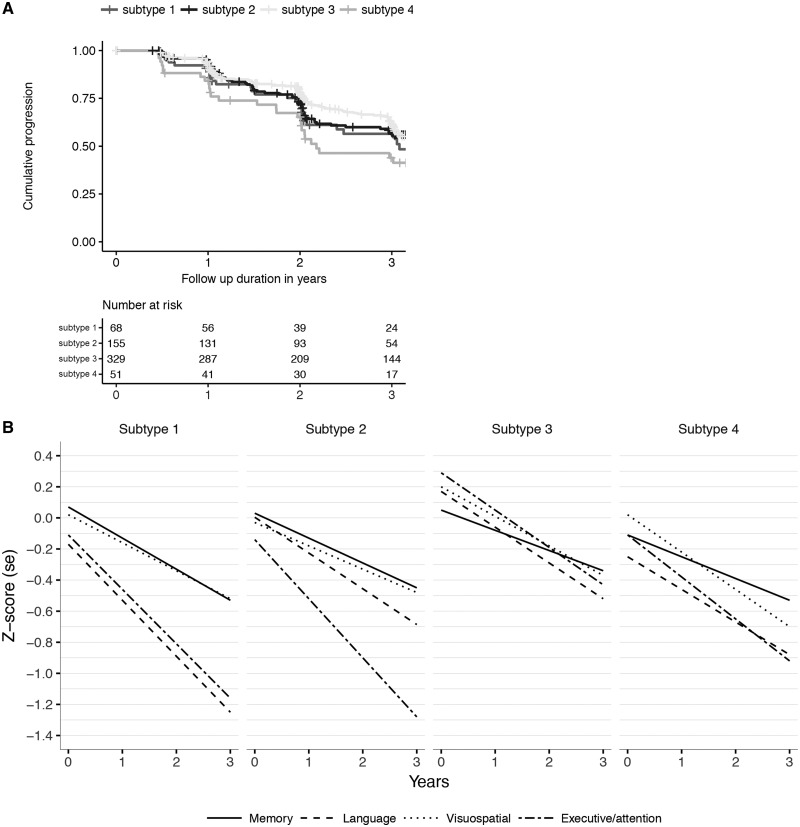
Prodromal Alzheimer’s disease participants of the diffuse cortical subtype had a higher probability of progressing to dementia compared to prodromal participants of the mild atrophy subtype (hazard ratio: 1.50; 95% confidence interval: 1.01–2.2, P = 0.046) within a mean follow-up period of 2.6 ± 1.6 years (range 0.4–10.0) (Fig. 5A). The other two subtypes had intermediate hazard ratios, which did not statistically differ from the former subtypes. Linear mixed models showed that the medial-temporal and diffuse subtypes had lower baseline scores in the language domain, compared to the mild atrophy subtype (Figs 4C and 5B). Prodromal Alzheimer’s disease participants of the parieto-occipital subtype had worse baseline scores in the executive/attention and visuospatial domains compared to participants in the mild atrophy subtype. No baseline group differences in global cognition (MMSE) or in the memory domain were observed (Supplementary Table 8 and Fig. 5B). All prodromal Alzheimer’s disease participants showed a longitudinal decline in global cognition (MMSE) and in each of the cognitive domains (Fig. 5B). Prodromal Alzheimer’s disease participants in the medial-temporal subtype showed steeper longitudinal decline on the MMSE and in the memory domain compared to participants in the mild atrophy subtype, even though there were no significant differences at baseline. Prodromal Alzheimer’s disease participants in the parieto-occipital atrophy subtype showed the steepest decline over time on the executive/attention domain. Effect size remained largely similar after additionally correcting for age, gender and education (Supplementary Tables 8 and 9). Point estimates at 3 years follow-up illustrated that the medial-temporal subtype showed about half a standard deviation worse language impairment than the parieto-occipital atrophy subtype (−1.03 ± 0.30 versus −0.57 ± 0.20, P = 0.008; Supplementary Table 10), and the parieto-occipital atrophy subtype showed worse impairment in attention/executive functioning than the mild and diffuse subtypes (all P < 0.05).
Figure 5.
Disease progression in prodromal Alzheimer’s disease for each of the four atrophy subtypes. (A) Progression curves for time to dementia onset within 3 years. (B) Decline over time in cognitive functioning in memory, language, visuospatial functioning and executive/attention plotted per subtype. Subtype 1 = medial-temporal atrophy; subtype 2 = parieto-occipital atrophy; subtype 3 = mild atrophy; subtype 4 = diffuse cortical atrophy.
Discussion
In this study we identified four atrophy subtypes in participants with ADD using an unbiased data-driven clustering approach in three independent datasets that differed in patient populations (e.g. memory clinic versus multicentre research cohort, geographical location) and imaging acquisition protocols (single scanner versus multiple scanners). Atrophy subtypes showed distinct clinical, neuropsychological, and biomarker characteristics. Moreover, atrophy subtypes were already apparent in the prodromal stage of Alzheimer’s disease, and were associated with decline in subtype-specific cognitive domains.
In line with previous studies, we found a medial-temporal dominant subtype with the worst memory and language performance, a parieto-occipital subtype in which patients had poor visuospatial and executive functioning, and a diffuse cortical atrophy subtype, with intermediate cognitive scores (Van Der Flier et al., 2011; Noh et al., 2014; Dong et al., 2017; Hwang et al., 2016; Zhang et al., 2016; Park et al., 2017; Scheltens et al., 2017; Varol et al., 2017; Poulakis et al., 2018). We extend the literature further by showing that atrophy subtypes can be robustly detected across three independent patient datasets, suggesting that these reflect true pathophysiological subtypes of Alzheimer’s disease. In line with this, the atrophy subtypes showed distinct clinical and biomarker profiles. The medial-temporal subtype was the oldest and had the highest amount of WMH, suggesting concomitant small vessel disease. This subtype might be similar to the limbic-predominant subtype identified in a previous neuropathological study that was also older and showed more vascular pathology on autopsy (Murray et al., 2011). The parieto-occipital cluster might reflect the hippocampal sparing subtype defined with neuropathology, as participants of this subtype were also younger and more often showed an atypical presentation (Murray et al., 2011). In addition to these three subtypes, we found a fourth subtype that was characterized by medial temporal and cortical atrophy compared to control participants, but this atrophy was relatively mild compared to the other Alzheimer’s disease subtypes. Two other studies previously identified a mild atrophy subtype (Dong et al., 2017; Poulakis et al., 2018), but because those studies did not include evidence of amyloid pathology in their clustering analyses, it could not be excluded whether mild atrophy may have reflected a non-Alzheimer pathology. In addition, Dong et al. (2017) included participants with mild cognitive impairment without evidence of amyloid pathology in their clustering analyses, and so it remained unclear whether that mild atrophy may have reflected an earlier disease stage. Since we only included ADD participants with biomarker evidence of underlying amyloid pathology, we minimized the possibility that subtypes reflect non-ADD, and we are able to show that this mild atrophy cluster is a true subtype of ADD. Furthermore, the mild atrophy cluster showed the highest CSF p-tau and t-tau levels. This finding is unexpected, since CSF tau has been hypothesized to increase prior to brain atrophy (Jack et al., 2013), and both tau and atrophy are associated with disease progression. Current classification schemes for disease staging allow us to use CSF tau proteins and atrophy on MRI interchangeably as markers for neuronal injury (Vos et al., 2015). Our results suggest, however, that CSF tau and atrophy may reflect different pathological processes. Future research in which both in vivo MRI and post-mortem data are available is needed to examine how our atrophy subtypes relate to neuropathological characteristics.
We observed atrophy subtypes further in participants with prodromal Alzheimer’s disease. This finding supports the idea that different aetiologies may cause disease heterogeneity in Alzheimer’s disease and that this is already apparent in the predementia stage. One other study could also detect atrophy subtypes in the predementia stage of Alzheimer’s disease (Zhang et al., 2016); however, although that study observed differences in rate of decline, it did not find a clear early distinction in the development of specific symptoms, which may have been due to the number of subtypes studied (three versus four, the latter providing a separation of a posterior and frontal atrophy pattern). In the current study, all prodromal Alzheimer’s disease individuals showed decline in cognitive functioning, which is in line with the notion that individuals with mild cognitive impairment and abnormal amyloid are at considerable increased risk to show clinical progression to dementia (Vos et al., 2015) We observed differences in the rate of decline amongst subtypes, as well as the type of cognitive domain that showed more pronounced decline. Although the differences in cognitive decline are subtle, recognizing this phenotypic variation in atrophy subtypes in participants with prodromal Alzheimer’s disease might aid clinicians in providing a more accurate prognosis for individual patients. Our results might in the future also have an implication for clinical trial design in prodromal Alzheimer’s disease. The existence of atrophy subtypes that show specific trajectories of cognitive decline may require subtype-specific outcome measures tailored to the expected rate of decline in different cognitive domains.
In the prodromal Alzheimer’s disease group, relatively more participants were classified in the third, mild atrophy subtype compared to the ADD group. Various explanations can be postulated for this finding. First, prodromal Alzheimer’s disease subjects have generally less atrophy than ADD patients, and although within the ADD datasets it can be argued that the ‘mild atrophy’ reflects a true subtype of ADD, this is more difficult for prodromal subjects for whom mild atrophy may also reflect their earlier disease stage. Second, the prodromal Alzheimer’s disease participants studied here may potentially be (in part) a different population than the ADD subjects examined. For example, the prodromal participants are the same age or slightly older than the ADD patient datasets. Longitudinal studies are needed to examine whether and how atrophy patterns of these subtypes will change during the course of Alzheimer’s disease.
This study has several potential limitations. First, we did not have pathological data available and so the possibility of misdiagnosis cannot be excluded. However, we used biomarker evidence for amyloid aggregation, which provides strong in vivo support for the presence of underlying Alzheimer’s pathology (Blennow et al., 2015). Second, loss to follow-up may have resulted in an underestimation of cognitive trajectories in prodromal Alzheimer’s disease, in that participants who were followed for more than 2 years were slightly younger, had higher MMSE scores and lower CSF tau levels compared to participants who were followed for a shorter period of time (Supplementary Table 11). Third, an alternative interpretation of our atrophy clusters might be that these represent differences in disease stage, as subtypes showed differences in MMSE scores. If that were the case then the subtype showing the lowest MMSE scores (parieto-occipital subtype) would be expected to show the most severe atrophy, as well as highest tau levels, and worse performance in all cognitive domains. In contrast, our subtypes showed distinct demographics, biomarker profiles, and cognitive symptoms, suggesting that these atrophy subtypes exist in parallel and do not merely reflect staging. Fourth, some ADD patients did not have a high cluster membership probability (Supplementary Fig. 1). It is plausible that some of these participants are a combination of more than one subtype, which is not captured by discretizing subtype membership. Future research should further investigate more refined subtype definitions based on continuous membership probability values. Finally, apart from WMHs, we were not able to examine the influence of other co-morbid disease, as this information is not readily available from the subjects of the ADC. This would be an interesting topic for future research.
In conclusion, we have robustly identified four different atrophy subtypes amongst patients with Alzheimer’s disease using a data-driven clustering approach. The subtypes showed distinct demographic, cognitive, and biomarker profiles. Understanding the causes of this heterogeneity is an important step towards precision medicine for Alzheimer’s disease.
Supplementary Material
Glossary
Abbreviations
- ADC(d/v)
Amsterdam Dementia Cohort (discovery/validation dataset)
- ADD
Alzheimer’s disease dementia
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- MMSE
Mini-Mental State Examination
- NMF
non-negative matrix factorization
- WMH
white matter hyperintensity
Funding
Research of the VUmc Alzheimer Centre is part of the neurodegeneration research program of Neuroscience Amsterdam. The VUmc Alzheimer Centre is supported by Stichting Alzheimer Nederland and Stichting VUmc funds. The clinical database structure was developed with funding from Stichting Dioraphte. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
This work has received support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking (EMIF grant: 115372) (M.t.K. and P.J.V.) and ZonMW Memorabel grant programme (73305056 and 733050824) (B.M.T. and P.J.V.). F.B. was supported by the NIHR biomedical research centre at UCLH. Funding sources were not involved in data collection, data analysis, interpretation, or writing of the manuscript.
Competing interests
The authors report no competing interests.
References
- Blennow K, Mattsson N, Schöll M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol Sci 2015; 36: 297–309. [DOI] [PubMed] [Google Scholar]
- Byun MS, Kim SE, Park J, Yi D, Choe YM, Sohn BK, et al. Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer’s disease. PLoS One 2015; 10: e0142756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L. Measures of the amount of ecologic association between species. Ecology 1945; 26: 297–302. [Google Scholar]
- Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain 2017; 140: 735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K, et al. Advancing research diagnostic criteria for Alzheimer’s disease: the IWG-2 criteria. Lancet Neurol 2014; 13: 614–29. [DOI] [PubMed] [Google Scholar]
- Ferreira D, Verhagen C, Hernández-Cabrera JA, Cavallin L, Guo CJ, Ekman U, et al. Distinct subtypes of Alzheimer’s disease based on patterns of brain atrophy: longitudinal trajectories and clinical applications. Sci Rep 2017; 7: 46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010; 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kim CM, Jeon S, Lee JM, Hong YJ, Roh JH, et al. Prediction of Alzheimer’s disease pathophysiology based on cortical thickness patterns. Alzheimers Dement 2016; 2: 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr., Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013; 12: 207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27: 685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam B, Masellis M, Freedman M, Stuss DT, Black SE. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res Ther 2013; 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature 1999; 401: 788–91. [DOI] [PubMed] [Google Scholar]
- Lee S, Viqar F, Zimmerman ME, Narkhede A, Tosto G, Benzinger TLS, et al. White matter hyperintensities are a core feature of Alzheimer’s disease: Evidence from the dominantly inherited Alzheimer network. Ann Neurol 2016; 79: 929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Bartlett JW, Barnes J, Manning EN, Ourselin S, Fox NC, et al. Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology 2013; 80: 648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller C, Vrenken H, Jiskoot L, Versteeg A, Barkhof F, Scheltens P, et al. Different patterns of gray matter atrophy in early- and late-onset Alzheimer’s disease. Neurobiol Aging 2013; 34: 2014–22. [DOI] [PubMed] [Google Scholar]
- Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol 2011; 10: 785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y, Jeon S, Lee JM, Seo SW, Kim GH, Cho H, et al. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology 2014; 83: 1936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Na HK, Kim S, Kim H, Kim HJ, Seo SW, et al. Robust Identification of Alzheimer’s Disease subtypes based on cortical atrophy patterns. Sci Rep 2017; 7: 43270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson K, Eldholm RS, Barca ML, Cavallin L, Ferreira D, Knapskog AB, et al. MRI-assessed atrophy subtypes in Alzheimer’s disease and the cognitive reserve hypothesis. PLoS One 2017; 12: e0186595–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease neuroimaging initiative (ADNI): clinical characterization. Neurology 2010; 74: 201–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulakis K, Pereira JB, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, et al. Heterogeneous patterns of brain atrophy in Alzheimer’s disease. Neurobiol Aging 2018; 65: 98–108. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Anderson WH, Charil A, Castelluccio PF, Shcherbinin S, Saykin AJ, et al. Alzheimer disease brain atrophy subtypes are associated with cognition and rate of decline. Neurology 2017; 89: 2176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens NME, Tijms BM, Koene T, Barkhof F, Teunissen CE, Wolfsgruber S, et al. Cognitive subtypes of probable Alzheimer’s disease robustly identified in four cohorts. Alzheimers Dement 2017: 13: 1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Tosun D, Insel PS, Chiang GC, Truran D, Aisen PS, et al. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol Aging 2012; 33: 845–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Ryman DC, McDade E, Jasielec MS, Buckles VD, Cairns NJ, et al. Neurological manifestations of autosomal dominant familial Alzheimer’s disease: a comparison of the published literature with the Dominantly Inherited Alzheimer Network observational study (DIAN-OBS). Lancet Neurol 2016; 15: 1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–89. [DOI] [PubMed] [Google Scholar]
- van Buuren S, Groothuis-Oudshoorn K. Mice: multivariate imputation by chained equations inR. J Stat Softw 2011; 45: 67. [Google Scholar]
- van Der Flier WM, Pijnenburg YAL, Fox NC, Scheltens P. Early-onset versus late-onset Alzheimer’s disease: the case of the missing APOE ε4 allele. Lancet 2011; 10: 280–8. [DOI] [PubMed] [Google Scholar]
- van der Flier WM, Pijnenburg YAL, Prins N, Lemstra AW, Bouwman FH, Teunissen CE, et al. Optimizing Patient Care and Research: the amsterdam dementia cohort. J Alzheimers Dis 2014; 41: 313–27. [DOI] [PubMed] [Google Scholar]
- Varol E, Sotiras A, Davatzikos C, Initiative FTADN. HYDRA: Revealing heterogeneity of imaging and genetic patterns through a multiple max-margin discriminative analysis framework. Neuroimage 2017; 145: 346–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SJB, Verhey F, Frölich L, Kornhuber J, Wiltfang J, Maier W, et al. Prevalence and prognosis of Alzheimer’s disease at the mild cognitive impairment stage. Brain 2015; 138: 1327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell J, Dickson D, Murray M, Petersen R, Jack C, Josephs K. MRI in pathologically-defined hippocampal sparing and limbic predominant atypical variants of Alzheimer’s disease. Alzheimers Dement 2012; 8: P160–1. [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yücel M, Pantelis C, et al. Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage 2010; 50: 970–83. [DOI] [PubMed] [Google Scholar]
- Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, Yeo BTT, et al. Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer’s disease. Proc Natl Acad Sci USA 2016; 113: E6535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ADNI data used is this study were obtained from the ADNI database (adni.loni.usc.edu). The ADC data used in this study are available from the corresponding author, upon reasonable request.