Abstract
The morula-stage embryo is transformed into a blastocyst composed of epiblast, hypoblast, and trophectoderm (TE) through mechanisms that, in the mouse, involve the Hippo signaling and mitogen-activated kinase (MAPK) pathways. Using the cow as an additional model, we tested the hypotheses that TE and hypoblast differentiation were regulated by the Hippo pathway regulators, yes-associated protein 1 (YAP1) and angiomotin (AMOT), and MAPK kinase 1/2 (MAPK1/2). The presence of YAP1 and CDX2 in the nucleus and cytoplasm of MII oocytes and embryos was evaluated by immunofluorescence labeling. For both molecules, localization changed from cytoplasmic to nuclear as development advanced. Inhibition of YAP1 activity, either by verteporfin or a YAP1 targeting GapmeR, reduced the percent of zygotes that became blastocysts, the proportion of blastocysts that hatched and numbers of CDX2+ cells in blastocysts. Moreover, the YAP1-targeting GapmeR altered expression of 15 of 91 genes examined in the day 7.5 blastocyst. Treatment of embryos with an AMOT targeting GapmeR did not affect blastocyst development or hatching but altered expression of 16 of 91 genes examined at day 7.5 and reduced the number of CDX2+ nuclei and YAP1+ nuclei in blastocysts at day 8.5 of development. Inhibition of MAPK1/2 with PD0325901 did not affect blastocyst development but increased the number of epiblast cells. Results indicate a role for YAP1 and AMOT in function of TE in the bovine blastocyst. YAP1 can also affect function of the epiblast and hypoblast, and MAPK signaling is important for inner cell mass differentiation by reducing epiblast numbers.
Keywords: preimplantation embryo, epiblast, blastocyst, trophoblast, hypoblast
The cell differentiation regulators, AMOT, MAP2K1/2, and YAP1, are involved in differentiation of the bovine blastocyst.
Introduction
The first differentiation event in the mammalian embryo occurs when the totipotent morula goes through processes of compaction and cell polarization that lead to formation of the blastocyst containing a pluripotent inner cell mass (ICM), a differentiated trophectoderm (TE), and a blastocoel [1, 2]. The second differentiation event results in the partition of cells in the ICM into pluripotent epiblast and differentiated hypoblast (also known as primitive endoderm). Together, the first three cell types of the blastocyst are precursors of fetal tissues (epiblast), yolk sac and extraembryonic endoderm (hypoblast), and placental tissues (TE) [2, 3]. In the bovine embryo, these events occur between days 6 and 7 (first differentiation) and days 7 and 9 (second differentiation).
The mouse is the best studied model for understanding the mechanisms by which blastocyst differentiation is achieved. In this species, differential activation of the Hippo signaling pathway is an important determinant of cell fate with respect to TE or ICM [4–6]. Activation of the Hippo pathway in the inner cells of the embryo leads to inactivation of the downstream regulator, yes-associated protein 1 (YAP1) via phosphorylation. As a result, YAP1 does not accumulate in the nucleus, and pluripotency is favored by transcription of genes such as POU class 5 homeobox 1 (Pou5f1) and SRY box 2 (Sox2) [7, 8]. In the outer cells of the embryo, the Hippo pathway is inactivated so that YAP1 remains nonphosphorylated and is translocated into the nucleus where it associates with TEA domain transcription factor 4 (TEAD4) and activates transcription of genes promoting differentiation of TE including caudal type homeobox 2 (Cdx2) and GATA binding protein 3 (Gata3) [9–11].
Activation of Hippo signaling in cells destined to become ICM is achieved by gap junctions that allow cell to cell communication to activate WW and C2 domain containing 1 (WWC1, also known as KIBRA) and neurofibromin (NF2) which synergistically act to phosphorylate large tumor suppressor kinase 1/2 (LATS1/2). Simultaneously, serine/threonine kinase 4/3 (STK4/3, also known as MST1/2) are activated by an unknown signal and further reinforce phosphorylation of LATS1/2 [12–14]. Phosphorylated LATS1/2 is functionally active and phosphorylates YAP1. Angiomotin (AMOT) is another protein that participates in inactivation of YAP1. AMOT, which is localized at the basolateral membrane of inner embryonic cells through interactions with cadherin 1 (CDH1), can activate LATS1/2 [5, 6, 15], directly phosphorylate YAP1 [5, 6], and bind to and retain YAP1 at the plasma membrane so that it cannot enter the nucleus [16]. Inactivation of YAP1 does not occur in the outer cells of the compact morula destined to become TE cells. These cells become polarized and connected through tight junctions that inhibit cell to cell communication. Lack of gap junctions prevents MST1/2 activation. In addition, AMOT is phosphorylated in inner cells to interact with LATS1/2 or YAP1 to reinforce inactivation of YAP1. In the outer cells, AMOT is unphosphorylated [15, 17] and inactivation of YAP1 is relieved.
In the mouse, formation of hypoblast cells involves actions of fibroblast growth factor 4 (FGF4) from future epiblast cells acting on future hypoblast cells via fibroblast growth factor receptor 2 (FGFR2) to activate mitogen-activated protein kinase (MAPK) signaling, transcription of GATA binding protein 6 (Gata6) and differentiation into hypoblast cells [18–20]. Cells that secrete FGF4 have low numbers of FGF2R so that Gata6 expression is low, expression of the pluripotency factor NANOG homeobox (Nanog) is high, and cells become epiblast [21].
Involvement of the Hippo signaling pathway in formation of the blastocyst in the cow is largely unknown, but the role of two downstream effectors, CDX2 and TEAD4, have been evaluated. Embryos that were deficient for CDX2 could develop into blastocysts [22, 23], but there was disruption in regulation of GATA3 [24] and TE tight junctions [23]. After transfer of CDX2-knockout embryos to recipient females, the embryo was not able to elongate at day 14, although expression of interferon-τ (IFNT) was the same in control and CDX2-mutant embryos [22]. These findings suggest that CDX2 is not required for blastocyst formation in the bovine but is necessary for its proper function including subsequent elongation. Similarly, bovine embryos were able to develop to the blastocyst stage when treated with an RNA interference molecule to downregulate TEAD4 from the zygote to the blastocyst stage [25]. Even though treatment decreased TEAD4 mRNA abundance, there was no effect on number of ICM or TE cells in blastocysts at day 7 [25].
There is also little information about processes controlling hypoblast differentiation in the bovine embryo. Embryos that were treated with FGF4 and heparin developed an ICM that was rich in hypoblast cells [20]. Treatment of bovine blastocysts with FGF2 also increased the outgrowths of hypoblast-like cells and expression of GATA4 and GATA6 [26]. However, inhibition of FGF4 did not result in a decrease in hypoblast cells in bovine blastocysts [20]. Thus, it may be that the fibroblast growth factor signaling pathway is involved in differentiation of the hypoblast but that FGF4 is not essential for that process.
In the current series of experiments, we evaluated the role of Hippo signaling regulators, YAP1 and AMOT, as well as the MAPK pathway in formation and function of TE, hypoblast, and epiblast in the cow. The first objective was to characterize developmental patterns in localization of YAP1 and CDX2 during early preimplantation stages of development by immunolocalization and fluorescent imaging. The second objective was to analyze the consequences of disruption of YAP1 activity by either chemical interference of the interaction of YAP1 with TEAD4 using the inhibitor verteporfin [27] or knockdown of the mRNA for YAP1. It was hypothesized that these treatments would prevent CDX2 transcription, formation of TE, and a blastocyst with a normal blastocoele and capability of hatching. A third objective was to determine whether interference with AMOT biosynthesis through mRNA knockdown of AMOT would increase TE formation. For experiments with both YAP1 and AMOT, effects on hypoblast were also examined because, like the TE, the hypoblast is an epithelium, and it is possible that the two cell types share some aspects of control of differentiation. A fourth objective was to study the effects of disruption of the MAPK pathway by inhibiting mitogen-activated protein kinase kinase (MAP2K1/2, also known as MEK) with PD0325901. It was hypothesized that the ratio of epiblast cells would increase and hypoblast cells would decrease because of an essential role for the MAPK pathway in formation of epiblast and hypoblast.
Materials and methods
In vitro production of bovine embryos
All experiments were performed with embryos produced in vitro using the protocol previously described [28, 29]. Sperm and oocytes were from a mixture of animals of various Bos taurus breeds as well as cattle containing an admixture of B. taurus and B. indicus. The procedures were done following the protocol previously described [28, 29]. Presumptive zygotes were collected and exposed to hyaluronidase (1000 U/ml in approximately 0.5 ml HEPES-Tyrode albumin lactate pyruvate [TALP]) to remove the cumulus cells, washed three times in HEPES-TALP, and placed in groups of 25–30 zygotes per 50 μl drop of synthetic oviductal fluid-bovine embryo 2 (SOF-BE2) [30] covered with mineral oil in a humidified gas atmosphere of 5% (v/v) CO2, 5% (v/v) O2, and the balance nitrogen, at 38.5°C. The proportion of embryos that cleaved was assessed at day 3 after fertilization and the proportion of embryos that became blastocysts was verified at day 7.5 (182 ± 2 h) postfertilization.
A replicate was defined as a single in vitro fertilization procedure involving 200–300 cumulus-oocyte complexes and sperm for fertilization from a pool of three bulls. A total of 19 bulls were used for the different replicates throughout all the experiments. Only replicates with a cleavage rate ≥65% in control embryos were used.
Immunofluorescent analysis of embryos
Antibodies used are described in Supplemental Table S1. Procedures for immunolabeling were performed at room temperature unless otherwise stated. For dual labeling of immunoreactive YAP1 and CDX2, oocytes and embryos were incubated in permeabilization solution [Dulbecco phosphate-buffered saline (DPBS) + polyvinylpyrrolidone (PVP) containing 0.25% (v/v) Triton X-100] for 30 min and then in blocking buffer [5% (w/v) bovine serum albumin (BSA) in DPBS] for 1 h. Oocytes and embryos were then transferred to rabbit monoclonal antibody against endogenous YAP1 (1 μg/ml; Cell Signaling Technology, Danvers, MA, USA), and incubated overnight at 4°C. Afterwards, the embryos were washed three times in washing buffer [DPBS + 0.1% BSA (w/v) and 0.1% (v/v) Tween-20] and incubated for 1 h in the first secondary antibody: Alexa Fluor 555 conjugated goat polyclonal anti-rabbit IgG (1 μg/ml; ThermoFisher Scientific, Waltham, MA, USA). The oocytes and embryos were washed another three times in washing buffer and incubated for 1 h in the second primary antibody, which was mouse monoclonal antibody against CDX2 (Biogenex, Fremont, CA, USA) at the 1 μg/ml working concentration provided. Oocytes and embryos were washed again and incubated for 1 h in the second secondary antibody [1 μg/ml fluorescein isothiocyanate (FITC) conjugated goat polyclonal anti-mouse IgG; Abcam, Cambridge, MA, USA]. Then, embryos were washed three times and counterstained with 1 μg/ml Hoechst 33342 in DPBS-PVP for 15 min, washed once in DPBS-PVP, and transferred to a 10 μl drop of SlowFade Gold antifade reagent (ThermoFisher Scientific) on a glass microscope slide and covered with a coverslip. To determine nonspecific labeling, primary antibodies were replaced with rabbit or mouse IgG (1 μg/ml).
The same procedures were used for dual labeling of NANOG (epiblast marker) and GATA6 (hypoblast marker) except that the primary antibodies were substituted with rabbit polyclonal antibody against human GATA6 (Santa Cruz Biotechnology, Dallas, TX, USA) and mouse polyclonal antibody against human NANOG (eBioscience, San Diego, CA, USA); both were used at 1 μg/ml.
Images were observed with a 40X objective using a Zeiss Axioplan 2 epifluorescence microscope (Zeiss, Göttingen, Germany) and Zeiss filter sets 02 [4,6-diamidino-2-phenylindole], 03 (FITC), and 04 (rhodamine). Digital images were acquired using AxioVision software (Zeiss) and a high-resolution black and white Zeiss AxioCam MRm digital camera. Image J V. 1.48 (National Institutes of Health, Bethesda, MD, USA) was used to visualize images, count the number of cells using the cell counter tool, and measure protein labeling intensity using the selection of area and measure options. The protein labeling intensity was adjusted by selecting a region in the background, measuring the intensity, and subtracting this number from the measured protein labeling intensity. Depending on the experiment, immunofluorescent intensity for a specific marker was determined for the entire area of the embryo, all nuclei in the embryo, or all nuclei in a subset of nuclei that was positive for a second specific marker (for example, CDX2 or YAP1). Thus, labeling of nuclear YAP1 in TE cells was assessed for cells positive for CDX2, labeling of nuclear YAP1 cells in epiblast was assessed for cells positive for NANOG and labeling of nuclear YAP1 cells in hypoblast was assessed for cells positive for GATA6 and that could be identified spatially as being located in the ICM.
RNA isolation
Embryos for analysis of reverse transcription (RT)-PCR were washed three times in DPBS-PVP, incubated in 0.1% (w/v) protease from Streptococcus griseus (Sigma-Aldrich) in DPBS for ∼3 min or until the zonae dissolved, washed three times in fresh DPBS-PVP, snap frozen in liquid nitrogen and stored at –80°C until RNA isolation.
For RNA extraction, the PicoPure RNA isolation kit (Applied Biosystems, Carlsbad, CA, USA) was used following the manufacturer's instructions. Isolated RNA (15 μl) was treated with 1 μl (2 U) of DNaseI (New England Biolabs, Ipswich, MA, USA) for removal of DNA contamination prior to reverse transcription. The High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used for reverse transcription following the manufacturer's instructions. For each sample, there was a negative control in which reverse transcriptase was omitted. The cDNA was stored at –20°C until further gene expression analysis.
Gene expression analysis using high-throughput RT-PCR
The Fluidigm qPCR microfluidic device Biomark HD system was used to analyze the effect of YAP1 and AMOT knockdown on 96 selected genes. Primers for 96 genes (Supplemental File S1) were designed by Fluidigm Delta Gene assays (Fluidigm Co., San Francisco, CA, USA) and optimized by Miami Center for AIDS Research at the University of Miami Miller School of Medicine. Amongst the genes analyzed were 3 housekeeping genes, 9 epiblast potential specific markers, 9 markers of trophoblast in one or more species, 11 hypoblast markers, 16 chemokine signaling pathway genes, 9 Hippo signaling pathway genes, 13 epigenetic modification gene markers, 14 tight junctions, cell polarity and axon guidance, and another 12 genes of interest. All 96 primer pairs were validated except for the primer set for Krüppel-like factor 2. Thus, results for this gene were not used for further analysis. The cDNA of all the pools of embryos was preamplified following the guidelines for the Ambion Single Cell-to-CT kit (ThermoFisher Scientific) and diluted in 2 fold down to a single cell equivalent.
The procedures for gene amplification were the same as previously described [31]. A total of 40 PCR cycles were performed using the 96.96 dynamic array integrated fluidic circuit developed by the manufacturer. The cutoff for detectable genes was those with Ct > 27. The geometric mean of the three housekeeping genes, hypoxanthine guanine phosphoribosyl transferase (HPRT1), H2A histone family member Z (H2AFZ), and succinate dehydrogenase complex subunit A flavoprotein (SDHA), was calculated and used to obtain the delta Ct (dCt) values of the other genes. Fold changes were then calculated as 2−dCt relative to the geometric mean of the housekeeping genes. The dCt was used for statistical analysis, and the fold change was used to display the data.
Experiment 1: developmental changes in immunoreactive YAP1 and CDX2
All oocytes and embryos were produced in vitro. MII oocytes (n = 5) were harvested at 22–24 h after maturation and denuded of cumulus cells using hyaluronidase as described elsewhere [28, 29]. Embryos were collected from culture drops at specific hours postinsemination (hpi): 28–32 hpi (2-cell, n = 2), 44–48 hpi (3–4 cell, n = 5), 50–55 hpi (5–8 cell, n = 3), 72 hpi (9–16 cell, n = 7), 120 hpi (day 5 morula, n = 8), 144 hpi (day 6 morula, n = 11), 168 hpi (day 7 blastocyst, n = 6), 192 hpi (day 8 blastocyst, n = 7), and 216 hpi (day 9 blastocyst, n = 10). Oocytes and embryos were washed three times in DBPS containing 0.1% (w/w) PVP (Kodak, Rochester, NY, USA), fixed in 4% (w/v) paraformaldehyde diluted in DPBS-PVP for 15 min, washed another three times in DPBS-PVP, and stored at 4°C until protein immunolocalization procedures for YAP1 and CDX2 as described earlier.
Experiment 2: inhibition of interactions between YAP1 and TEAD4
Verteporfin (VP) is a molecule that inhibits interactions between YAP1 and TEAD4 [27]. For Experiment 2, it was tested whether VP would alter the proportion of embryos becoming blastocysts, block blastocoel formation or expansion, affect cell allocation to various lineages, and alter blastocyst hatching.
Bovine embryos were randomly divided into groups and cultured in 45 μl SOF-BE2. Embryos were treated at day 5 after insemination with 5 μl VP (Sigma-Aldrich, St. Louis, MO, USA) or vehicle [SOF-BE2 containing 1% (v/v) dimethyl sulfoxide (DMSO)] to produce a final concentration of 10 μM VP or equivalent amount of vehicle [0.1% (v/v) DMSO].
Embryos were evaluated for formation of the blastocoel at day 7.5 postinsemination. Some blastocysts were harvested at this point. The remaining blastocysts were placed in one of the drops containing the same treatments as originally, and cultured until 8.5 or day 9.5. Harvested blastocysts were analyzed for immunolocalization of YAP1 and CDX2 at days 7.5, 8.5, and 9.5 postinsemination or for immunolocalization of NANOG and GATA6 at day 9.5.
A total of four replicates were used to produce embryos for the experiment with the total number of blastocysts examined being the following: n = 18 (day 7.5 for CDX2 and YAP1), n = 18 (day 8.5 for CDX2 and YAP1), n = 11 (day 9.5 for CDX2 and YAP1), and n = 9 (day 9.5 for GATA6 and NANOG) for embryos treated with VP and n = 28 (day 7.5 for CDX2 and YAP1), n = 23 (day 8.5 for CDX2 and YAP1), n = 11 (day 9.5 for CDX2 and YAP1), and n = 7 (day 9.5 for GATA6 and NANOG) for embryos treated with vehicle.
Experiment 3: knockdown of YAP1
After fertilization, putative zygotes were randomly divided in groups and placed in culture medium containing either 3 μM antisense GapmeR (LNA GapmeRs in vivo Ready, Exiqon, Inc., Woburn, MA, USA) directed against YAP1 (5΄-GAGCACTTTGACTGAT-3΄), 3 μM of a standard negative control GapmeR designed by the company (5΄-AACACGTCTATACGC-3΄), or vehicle [1.6% (v/v) diethyl pyrocarbonate treated double-distilled water]. Embryos were evaluated for blastocyst formation at day 7.5 after insemination and were collected at either day 7.5, 8.5, or 9.5. For five replicates, blastocysts were processed for immunolocalization of YAP1 and CDX2 on days 7.5 and 8.5 and for immunolocalization of GATA6 and NANOG on day 9.5. The number of blastocysts used for the immunolocalization analysis was n = 6 (day 7.5 for CDX2 and YAP1), n = 10 (day 8.5 for CDX2 and YAP1), and n = 3 (day 9.5 for GATA6 and NANOG) for the YAP1 targeting GapmeR; n = 23 (day 7.5 for CDX2 and YAP1), n = 15 (day 8.5 for CDX2 and YAP1), and n = 3 (day 9.5 for GATA6 and NANOG) for the standard negative control GapmeR; and n = 11 (day 7.5 for CDX2 and YAP1), n = 19 (day 8.5 for CDX2 and YAP1), and n = 6 (day 9.5 for GATA6 and NANOG) for embryos treated with vehicle. For five replicates, pools of day 8.5 blastocysts, representing n = 6, 9, 7, 10, and 12 YAP1 targeting GapmeR-treated blastocysts, n = 19, 12, 9, 16, and 13 standard negative control GapmeR-treated blastocysts, and n = 17, 15, 11, 10, and 14 vehicle-treated blastocysts, were used for analysis of mRNA for 96 genes as described above.
Experiment 4: knockdown of AMOT
This experiment was conducted as for Experiment 3 except that the GapmeR against AMOT was 5΄-GAACGCTGCTGGAGTA-3΄. For five replicates, blastocysts were collected at day 7.5 and 8.5 and used for immunolabeling for YAP1 and CDX2 or GATA6 and NANOG (day 8.5 only). The number of blastocysts was n = 23 (day 7.5 for CDX2 and YAP1), n = 18 (day 8.5 for CDX2 and YAP1), and n = 19 (day 8.5 for GATA6 and NANOG) for embryos treated with AMOT targeting GapmeR; n = 21 (day 7.5 CDX2 and YAP1), n = 24 (day 8.5 for CDX2 and YAP1), and n = 22 (day 8.5 for GATA6 and NANOG) for embryos treated with standard negative control GapmeR; and n = 14 (day 7.5 for CDX2 and YAP1), n = 20 (day 8.5 for CDX2 and YAP1), and n = 15 (day 8.5 for GATA6 and NANOG) for embryos treated with vehicle. For three replicates, blastocysts were collected at day 7.5 to produce three pools each of 14, 30, 31 AMOT targeting GapmeR-treated blastocysts; n = 26, 23, and 37 standard negative control GapmeR-treated blastocysts; and n = 28, 29 and 27 vehicle-treated blastocysts for RNA analysis.
Experiment 5: inhibition of MAP2K1/2
PD0325901 is a molecule that inhibits MAP2K1/2 and which has been reported to increase number of epiblast cells and decrease number of hypoblast cells in bovine blastocysts [27]. To verify this observation, embryos were randomly divided into groups and cultured in 45 μl SOF-BE2. Embryos were treated at day 6 after insemination with 5 μl MAP2K1/2 inhibitor (PD0325901; Sigma-Aldrich) or vehicle [SOF-BE2 containing 1% (v/v) DMSO] to produce a final concentration of 0.5 μM inhibitor or equivalent amount of vehicle [0.1% (v/v) DMSO]. Embryos were evaluated for formation of the blastocoel at day 7.5 and 8.5 postinsemination. Blastocysts were collected at day 8.5 postinsemination for immunolocalization of NANOG and GATA6.
A total of seven replicates were used to produce embryos for the experiment with the total number of blastocysts examined for NANOG and GATA6 labeling being n = 26 (day 8.5 for GATA6 and NANOG) for MAP2K1/2 inhibitor and n = 25 (day 8.5 for GATA6 and NANOG) for vehicle.
Statistical analysis
The SAS v 9.4 software package (SAS Institute Inc, Cary, NC, USA) was used for statistical analysis. The generalized linear mixed models procedures (Proc GLIMMIX) was used to evaluate the effects of treatment on the percent of putative zygotes to become blastocysts and the percent of blastocysts that were hatching or hatched from the zona pellucida. Each embryo was considered as an individual observation, and development and hatching considered as a binary variable (0 = did not occur; 1 = occurred). Treatment was considered as a fixed effect and replicate was considered random.
Treatment effects on other variables were analyzed by analysis of variance using the generalized linear models procedure (Proc GLM) of SAS. Main effects of treatment and replicate were considered fixed. For knockdown experiments, differences between treatments were separated into two individual orthogonal contrasts as follows: YAP1 targeting GapmeR or AMOT targeting GapmeR vs two controls (standard negative GapmeR and vehicle control) and standard negative control vs vehicle. For analysis of gene expression in the knockdown studies, treatment effects were determined by analysis of dCt data but results are shown as fold change relative to housekeeping genes.
Unless otherwise stated, data shown are least-squares means ± SEM.
Results
Experiment 1: developmental changes in YAP1 and CDX2
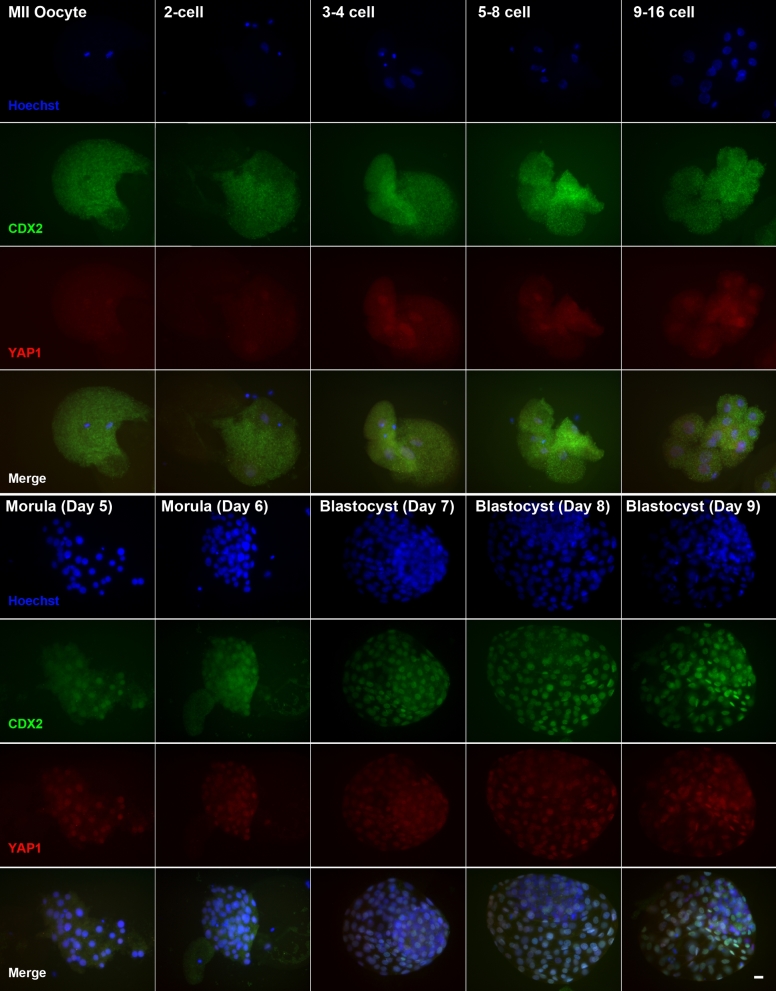
The presence of YAP1 and CDX2 in the nucleus and cytoplasm of MII oocytes and embryos throughout development to the blastocyst stage was evaluated by dual immunofluorescence labeling. Representative images are shown in Figure 1. YAP1 was localized exclusively in the cytoplasm of the MII oocyte. For embryos from the 2-cell stage to the 9–16-cell stage, YAP1 continued to be present in the cytoplasm but also was present in nuclei. From the day 5 morula onwards, YAP1 was present in most cells and primarily in the nuclear compartment.
Figure 1.
Immunolocalization of CDX2 and YAP1in the bovine oocyte and early embryo. MII oocytes and embryos were labeled with antibody against CDX2 (green) and YAP1 (red) or with Hoechst 33342 to label nuclei (blue). Merged panels contain all three fluorochrome channels. Scale bar = 20 μM.
To determine whether YAP1 was limited to cells of the TE or also occurs in epiblast cells of the ICM, some blastocysts were labeled with antibodies against YAP1 and NANOG. Expression of NANOG is limited to nuclei of the epiblast of the ICM [32]. As shown in Supplemental Figure S1, cells positive for NANOG did not label with YAP1.
In early stages of development (MII oocytes and embryos through the 9–16 cell stage), CDX2 was localized exclusively in the cytoplasm. By the morula stage, the pattern of localization changed so that CDX2 became located primarily in nuclei. At the blastocyst stages (days 7–9), intensity of nuclear CDX2 was greater than at earlier stages.
Experiment 2: inhibition of YAP1-TEAD interactions by treatment with verteporfin
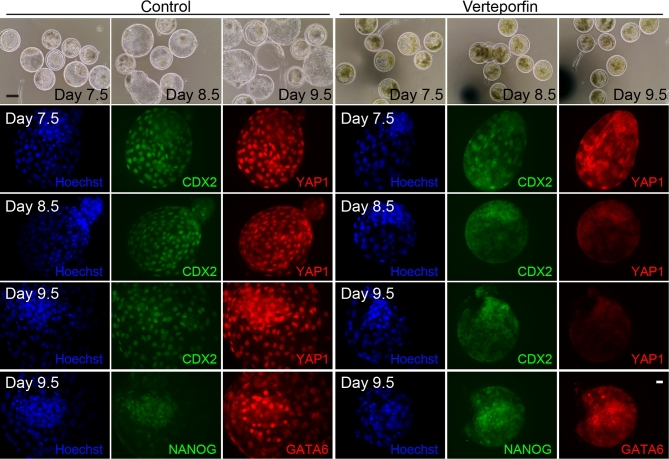
Representative images for labeling of blastocysts with antibodies against YAP1, CDX2, NANOG, and GATA6 are shown in Figure 2, while results from quantitative analyses are summarized in Table 1. In the late bovine blastocyst, some nuclei in the ICM considered to be hypoblast label brightly with GATA6, while cells in the TE also label for GATA6 but less intensely [32]. Accordingly, cells positive for GATA6 in the ICM were considered hypoblast. Treatment with VP decreased (P < 0.05) the proportion of putative zygotes that became blastocysts at day 7.5 postinsemination. Verteporfin also decreased the total number of nuclei in blastocysts at days 7.5 and 8.5 (P < 0.001) but not at day 9.5 (Table 1). Verteporfin also decreased (P < 0.001) the number of nuclei that were CDX2+ at all three points examined (days 7.5, 8.5, and 9.5) and decreased (P < 0.05 or less) the number of YAP1+ cells at the three time points examined (days 7.5, 8.5, and 9.5). Verteporfin also caused a significant decrease in numbers of NANOG+ and GATA6+ nuclei at day 9.5 (Table 1). Note that the reduction in percent of nuclei positive for CDX2, YAP1, GATA6, and NANOG does not reflect absence of the proteins; cytoplasmic labeling was noticeable for all four proteins (Figure 2).
Figure 2.
Representative images of blastocysts derived from embryos cultured in absence or presence of verteporfin (VP; YAP1-TEAD4 inhibitor) from days 5–9.5 of development. The top row represents bright field images that are shown to indicate the effect of VP on morphology and hatching. While the control group (left panels) underwent blastocyst formation, expansion, and hatching, the treated group (right panels) experienced blastocyst formation (indicated by presence of blastocoel) but blastocyst expansion and hatching from the zona pellucida was interrupted. Color panels show that VP led to loss of YAP1 (red) labeling, absence of CDX2 (green) nuclear labeling (but not cytoplasmic labeling), and reduced labeling of GATA6+ (red) and NANOG+ (green) in ICM nuclei (but not cytoplasm). In each image, nuclei were labeled with Hoechst 33342 (blue). Scale bar on bright field images = 50 μM; scale bar on fluorescent images = 20 μM.
Table 1.
Effects of verteporfin on characteristics of blastocyst development in the bovine embryo.a
| Vehicle | Verteporfinb | |
|---|---|---|
| Proportion of putative zygotes that reached the blastocyst stage, day 7.5 (N = 8)a | 22.2 ± 1.5% (n = 923) | 16.7 ± 1.5% (n = 887)* |
| Proportion of blastocysts that were undergoing hatching from the zona pellucida, day 8.5 (N = 5)a,c | 12.2 ± 1.5% (n = 82) | 1.7 ± 1.5% (n = 58)* |
| Proportion of blastocysts that hatched from the zona pellucida, day 9.5 (N = 5)a,c | 21.0 ± 1.5% (n = 38) | 0.0 ± 1.5% (n = 31)* |
| Total number of nuclei (Hoechst+), day 7.5 (N = 4)a | 131.8 ± 6.3 (n = 28) | 103.2 ± 7.5 (n = 18)*** |
| Total number of nuclei (Hoechst+), day 8.5 (N = 4)a | 150.6 ± 10.2 (n = 23) | 107.1 ± 9.5 (n = 18)*** |
| Total number of nuclei (Hoechst+), day 9.5 (N = 4)a | 103.7 ± 6.9 (n = 11) | 108.9 ± 6.6 (n = 11) |
| Number of CDX2− nuclei (ICM), day 7.5 (N = 4)a | 53.5 ± 3.9 (n = 28) | 65.5 ± 4.7 (n = 18)* |
| Number of CDX2− nuclei (ICM), day 8.5 (N = 4)a | 30.3 ± 6.2 (n = 23) | 108 ± 5.8 (n = 18)*** |
| Number of CDX2− nuclei (ICM), day 9.5 (N = 4)a | 30.3 ± 8.2 (n = 11) | 106.8 ± 8.6 (n = 11)*** |
| Number of CDX2+ nuclei (TE), day 7.5 (N = 4)a | 78.3 ± 5.8 (n = 28) | 37.7 ± 6.9 (n = 18)*** |
| Number of CDX2+ nuclei (TE), day 8.5 (N = 4)a | 120.3 ± 7.3 (n = 23) | −0.9 ± 6.8 (n = 18)*** |
| Number of CDX2+ nuclei (TE), day 9.5 (N = 4)a | 81.3 ± 5.3 (n = 11) | 6.7 ± 5.5 (n = 11)*** |
| Number of YAP1+ nuclei, day 7.5 (N = 4)a | 64.0 ± 6.4 (n = 28) | 35.4 ± 8.6 (n = 18)* |
| Number of YAP1+ nuclei, day 8.5 (N = 4)a | 59.8 ± 7.6 (n = 23) | 4.8 ± 4.9 (n = 18)*** |
| Number of YAP1+ nuclei, day 9.5 (N = 4)a | 85.0 ± 6.9 (n = 11) | 11.8 ± 8.3 (n = 11)*** |
| Number of GATA6+ nuclei (hypoblast), day 9.5 (N = 4)a | 39.7 ± 6 (n = 7) | −3.3 ± 5.4 (n = 9)*** |
| Number of NANOG+ nuclei (epiblast), day 9.5 (N = 4)a | 20.0 ± 4.2 (n = 7) | −2.0 ± 3.8 (n = 9)** |
aN = Number of replicates; n = total number of embryos per treatment.
b*P < 0.05 (except for percent hatching at day 8.5 where P = 0.05); **P < 0.01; ***P < 0.001.
cA blastocyst was considered to be undergoing hatching; it was hatching from the zona pellucida or had completed hatching.
A subset of blastocysts that formed at day 7.5 was cultured for further 2 days to evaluate whether VP would affect hatching from the zona pellucida. Representative images of hatching are shown in Figure 2. The percentage of blastocysts that either hatched or were undergoing hatching at day 8.5 was 12.2 ± 1.5% for vehicle vs 1.7 ± 1.5% for VP (P = 0.05). For a separate set of blastocysts, the percent that hatched or were hatching at day 9.5 was 21 ± 1.5% for vehicle vs 0 ± 1.5% for VP (P < 0.05).
Experiment 3: knockdown of YAP1
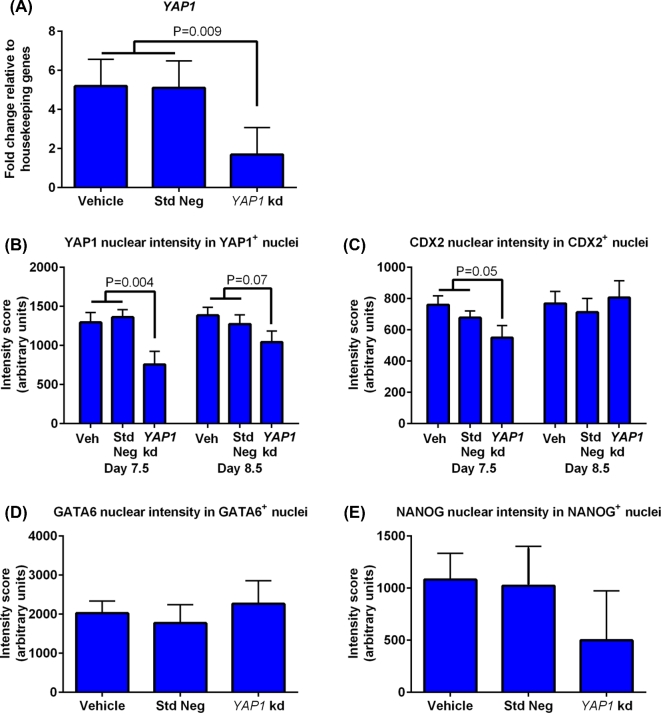
The effectiveness of knockdown was confirmed with qPCR (Figure 3A) and immunoreactive YAP1 labeling at day 8.5 (Figure 3B). Addition of the YAP1 targeting GapmeR did not affect cleavage rate (74.2 ± 2.7% for vehicle, 72.3 ± 3.5% for standard negative control GapmeR, and 71.5 ± 3.1% for YAP1 targeting GapmeR) but significantly reduced the percent of putative zygotes becoming a blastocyst at days 7.5 and 8.5 and the percent of blastocysts that underwent hatching from the zona pellucida at days 7.5 and 8.5 (Table 2).
Figure 3.
Amounts of YAP1 mRNA and labeling intensity for CDX2, YAP1, GATA6, and NANOG as affected by treatment with YAP1 targeting GapmeR. (A) Analysis of YAP1 mRNA by high-throughput RT-PCR (Fluidigm) for day 8.5 blastocysts (5 pools per treatment). Data are presented as fold change relative to the geometric mean of housekeeping genes (HPRT1, H2AFZ, and SHDA). (B–E) Intensity of immunoreactive YAP1 (B), CDX2 (C), GATA6 (D), and NANOG (E) in nuclei positive for the same protein. Immunoreactivity of GATA6 and NANOG were measured at day 9.5 only. The number of embryos per group was n = 6 (day 7.5), n = 10 (day 8.5), and n = 3 (day 9.5) for those treated with YAP1 targeting GapmeR; n = 23 (day 7.5), n = 15 (day 8.5), and n = 3 (day 9.5) for those treated with standard negative control GapmeR; and n = 11 (day 7.5), n = 19 (day 8.5), and n = 6 (day 9.5) for those treated with vehicle. Data are least-squares means ± SEM results. The P-values for effects of treatment are indicated by the value above the bars. Std Neg, standard negative control GapmeR; YAP1 kd, YAP1 targeting GapmeR.
Table 2.
Consequences of YAP1 knockdown in the bovine embryo.
| Vehicle | Negative control GapmeRb | YAP1 targeting GapmeRc | |
|---|---|---|---|
| Proportion of putative zygotes that reached the blastocyst stage, day 7.5 (N = 13)a | 15 ± 1.6% (n = 724) | 16.2 ± 2% (n = 751) | 9.1 ± 1.2% (n = 746)*** |
| Proportion of blastocysts that were undergoing hatching from the zona pellucida, day 7.5 (N = 5)a,d | 21 ± 6.6% (n = 54) | 20.3 ± 7% (n = 52) | 2.6 ± 2.6% (n = 37)* |
| Proportion of blastocysts that were undergoing hatching from the zona pellucida, day 8.5 (N = 5)a,d | 37 ± 7.2% (n = 68) | 25.7 ± 6% (n = 65) | 11.2 ± 5% (n = 43)** |
| Total number of nuclei (Hoechst+), day 7.5 (N = 6)a | 94.0 ± 8.6 (n = 11) | 103.7 ± 6.6 (n = 23) | 99.9 ± 11.7 (n = 6) |
| Total number of nuclei (Hoechst+), day 8.5 (N = 6)a | 112.0 ± 7.4 (n = 19) | 95.2 ± 8.6 (n = 15) | 101.3 ± 5.1 (n = 10) |
| Number of CDX2− nuclei (ICM), day 7.5 (N = 6)a | 55.4 ± 6.5 (n = 11) | 70.6 ± 5.1 (n = 23) | 87.5 ± 7.1 (n = 6)* |
| Number of CDX2− nuclei (ICM), day 8.5 (N = 6)a | 57.0 ± 5.1 (n = 19) | 56.5 ± 5.8 (n = 15) | 89.3 ± 7.1 (n = 10)*** |
| Number of CDX2+ nuclei (TE), day 7.5 (N = 6)a | 38.7 ± 5.2 (n = 11) | 33.1 ± 4 (n = 23)* | 12.5 ± 7.1 (n = 6)** |
| Number of CDX2+ nuclei (TE), day 8.5 (N = 6)a | 54.8 ± 5.1 (n = 19) | 38.7 ± 5.8 (n = 15) | 12.0 ± 7.1 (n = 10)*** |
| Number of YAP1+ nuclei, day 7.5 (N = 6)a | 44.7 ± 5.2 (n = 11) | 48.0 ± 4.0 (n = 23) | 41.1 ± 7.1 (n = 6) |
| Number of YAP1+ nuclei, day 8.5 (N = 6)a | 56.8 ± 5.8 (n = 19) | 43.3 ± 6.6 (n = 15) | 27.8 ± 7.1 (n = 10) * |
| Number of GATA6+ nuclei (hypoblast), day 9.5 (N = 4)a | 28.2 ± 5.5 (n = 6) | 16.0 ± 8.1 (n = 3) | 2.5 ± 10.4 (n = 3) |
| Number of NANOG+ nuclei (epiblast), day 9.5 (N = 4)a | 15.8 ± 5.1 (n = 6) | 2.1 ± 7.5 (n = 3) | 2.8 ± 9.6 (n = 3) |
aN = Number of replicates; n = total number of embryos per treatment.
bVehicle vs negative control GapmeR orthogonal contrast. *P < 0.05.
cControls vs treatment orthogonal contrast. *P < 0.05; **P < 0.01; ***P < 0.001.
dA blastocyst was considered to be undergoing hatching if it was hatching from the zona pellucida or had completed hatching.
Treatment with YAP1 targeting GapmeR did not affect total cell number of blastocysts at either days 7.5 or 8.5, but the number of cells that were CDX2+ cells was decreased at days 7.5 (P < 0.01) and 8.5 (P < 0.0001) when compared to control groups (Table 2). YAP1 targeting GapmeR did not reduce the number of cells that had YAP1+ nuclei at day 7.5 but did reduce these measurements at day 8.5 (P < 0.05) (see Supplemental Figure S2 for representative images of day 7.5 blastocysts labeled with antibodies against CDX2 and YAP1). The majority of the day 9.5 embryos showed poor labeling for GATA6 and NANOG. As a result, there were few GATA6+ or NANOG+ nuclei in the YAP1 knockdown group at day 9.5 but the difference was not significant.
The fluorescent intensity of each immunoreactive protein was measured in nuclei that were positive for this protein (Figure 3). Intensity for CDX2+ in CDX2+ nuclei was lower for the YAP1 targeting GapmeR group than for controls at day 7.5 (P ≤ 0.05) but not at day 8.5 (Figure 3C; Supplemental Figure S2). The intensity for YAP1 in YAP1+ nuclei (Figure 3B; Supplemental Figure S2) was also reduced by YAP1 targeting GapmeR at day 7.5 (P < 0.01) and day 8.5 (P = 0.07). Treatment with YAP1 targeting GapmeR did not affect intensity of labeling for GATA6+ (Figure 3D) or NANOG+ nuclei (Figure 3E).
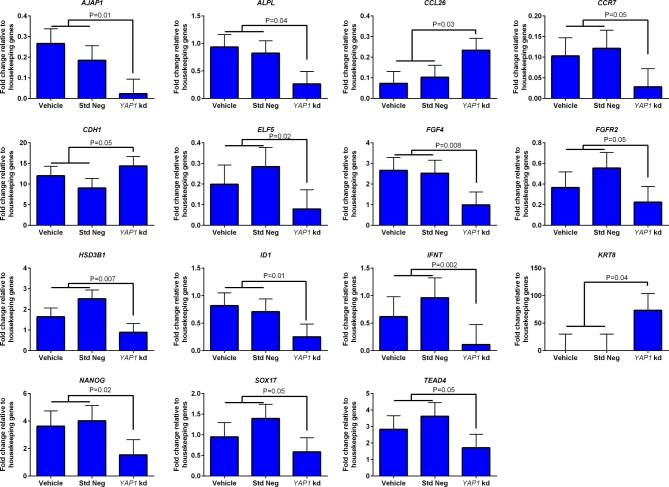
Effects of treatment on transcript abundance for 92 genes at day 8.5 was also examined (Figure 4). Treatment of embryos with YAP1 targeting GapmeR significantly reduced transcript abundance for 12 genes [adherens junction associated protein 1 (AJAP1), alkaline phosphatase liver/bone/kidney (ALPL), chemokine (C-C motif) receptor 7 (CCR7), E74-like factor 5 (ELF5), FGF4, FGFR2, hydroxy-delta-5-steroid dehydrogenase 3 beta- and steroid delta-isomerase 1 (HSD3B1), inhibitor of DNA binding 1 (ID1), IFNT, NANOG, SRY (sex-determining region Y)-box 17 (SOX17) and TEAD4]. In addition, YAP1 targeting GapmeR increased transcript abundance for three genes [chemokine (C-C motif) ligand 26 (CCL26), CDH1, and keratin 8 (KRT8)]. While not significant, there was also a tendency (P = 0.09) for CDX2 expression to be lower for blastocysts treated with YAP1 targeting GapmeR. Abundance of CDX2, expressed as a fold change relative to the geometric means of housekeeping genes was 4.57 for vehicle, 4.63 for standard negative GapmeR, and 2.35 for the YAP1 targeting GapmeR (SEM = 1.1).
Figure 4.
Genes whose expression was affected by knockdown of YAP1 in blastocysts at day 8.5 of development as determined by high-throughput RT-PCR (Fluidigm) (n = 5 pools of blastocysts per treatment). Data are represented as fold change relative to the geometric mean of housekeeping genes (HPRT1, H2AFZ, and SDHA) and are least-squares means ± SEM. The P-values for effects of treatment are indicated by the value above the bars. Std Neg, standard negative control GapmeR; YAP1 kd, YAP1 targeting GapmeR.
Experiment 4: AMOT knockdown
As shown in Figure 5, AMOT targeting GapmeR knocked down AMOT mRNA in blastocysts at day 7.5 postinsemination (P < 0.001). Treatment with AMOT targeting GapmeR had no negative effects on the proportion of putative zygotes that cleaved (64.7 ± 4.8% for vehicle, 70.8 ± 4.3% for standard negative control GapmeR, and 70.7 ± 4.3% for AMOT targeting GapmeR). The treatment decreased slightly the percent of putative zygotes that developed to the blastocyst stage at day 7.5 but had no effect on development to the blastocyst stage at day 8.5 (Table 3). There was no effect of AMOT targeting GapmeR on the percent of blastocysts that underwent hatching (Table 3).
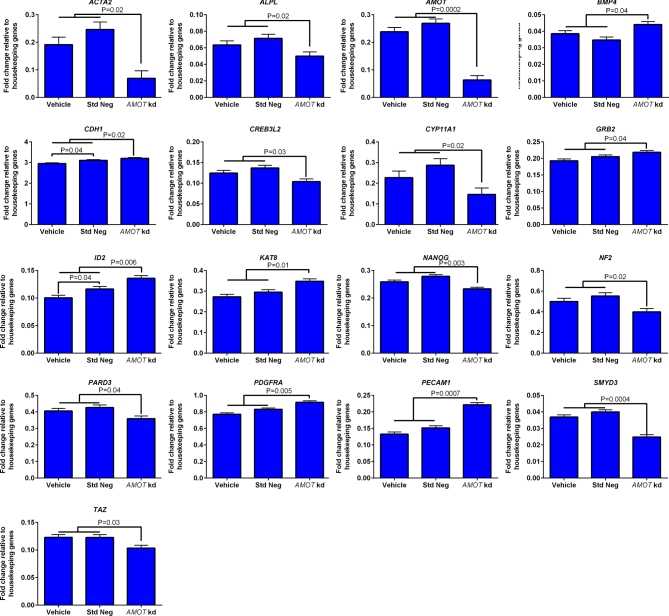
Figure 5.
Genes whose expression was affected by knockdown of AMOT in blastocysts at day 7.5 of development as determined by high-throughput RT-PCR (Fluidigm) (n = 3 pools of blastocysts per group). Data are represented as fold change relative to the geometric mean of housekeeping genes (HPRT1, H2AFZ, and SDHA) and are least-squares means ± SEM. The P-values for effects of treatment are indicated by the value above the bars. Std Neg, standard negative control GapmeR; AMOT kd, AMOT targeting GapmeR.
Table 3.
Effects of AMOT knockdown on the bovine embryo.
| Vehicle | Negative control GapmeRb | AMOT targeting GapmeRc | |
|---|---|---|---|
| Proportion of putative zygotes that reached the blastocyst stage, day 7.5 (N = 8)a | 26 ± 2.5% (n = 562) | 31 ± 2.6% (n = 590) | 24 ± 2.3% (n = 613)* |
| Proportion of putative zygotes that reached the blastocyst stage, day 8.5 (N = 5)a | 28.1 ± 3% (n = 344) | 30.8 ± 3% (n = 380) | 26 ± 2.8% (n = 368) |
| Proportion of blastocysts that were undergoing hatching from the zona pellucida, day 7.5 (N = 8)a | 12 ± 2.6% (n = 153) | 13 ± 2.5% (n = 187) | 8.5 ± 2.3% (n = 153) |
| Proportion of blastocysts that were undergoing hatching from the zona pellucida, day 8.5 (N = 5)a | 29 ± 6.4% (n = 51) | 34 ± 5.9% (n = 64) | 19 ± 5.7% (n = 49) |
| Total number of nuclei (Hoechst+), day 7.5 (N = 5)a | 104.0 ± 8.5 (n = 14) | 108.4 ± 7.2 (n = 21) | 124.4 ± 6.9 (n = 23) |
| Total number of nuclei (Hoechst+), day 8.5 (N = 5)a | 125.0 ± 6.2 (n = 35) | 115.6 ± 5.5 (n = 46) | 107.0 ± 5.2 (n = 37) |
| Number of CDX2− nuclei (ICM), day 7.5 (N = 5)a | 43.8 ± 6.3 (n = 14) | 52.8 ± 5.3 (n = 21) | 62.8 ± 5.1 (n = 23) |
| Number of CDX2− nuclei (ICM), day 8.5 (N = 5)a | 44.1 ± 4.3 (n = 20) | 47.7 ± 3.8 (n = 24) | 50.0 ± 3.8 (n = 18) |
| Number of CDX2+ nuclei (TE), day 7.5 (N = 5)a | 59.7 ± 4.9 (n = 14) | 55.6 ± 4.1 (n = 21) | 61.6 ± 4.0 (n = 23) |
| Number of CDX2+ nuclei (TE), day 8.5 (N = 5)a | 79.0 ± 5.9 (n = 20) | 78.1 ± 5.2 (n = 24) | 61.5 ± 5.1 (n = 18)** |
| Number of YAP1+ nuclei, day 7.5 (N = 5)a | 56.0 ± 5.8 (n = 14) | 54.1 ± 4.9 (n = 21) | 60.2 ± 4.7 (n = 23) |
| Number of YAP1+ nuclei, day 8.5 (N = 5)a | 78.4 ± 5.9 (n = 20) | 79.2 ± 5.2 (n = 24) | 56.4 ± 5.1 (n = 18) *** |
| Number of GATA6+ nuclei (hypoblast), day 8.5 (N = 5)a | 25.0 ± 2.1 (n = 15) | 18.7 ± 1.9 (n = 22) | 24.7 ± 1.7 (n = 19)* |
| Number of NANOG+ nuclei (epiblast), day 8.5 (N = 5)a | 12.4 ± 1.5 (n = 15) | 8.4 ± 1.4 (n = 22) | 8.2 ± 1.2 (n = 19) |
aN = Number of replicates; n = total number of embryos per treatment.
bThere were no significant differences between vehicle negative control GapmeR.
cControls vs treatment orthogonal contrast. *P < 0.05; **P < 0.01; ***P < 0.001.
Results for cell number on blastocysts are shown in Table 3. Total cell number of blastocysts at day 7.5 or day 8.5 was not different between treatments. Similarly, there was no effect of treatment on the number of cells at day 7.5 that were CDX2+ or YAP1+ cells. At day 8.5, in contrast, blastocysts produced with the AMOT targeting GapmeR had significantly lower number (P < 0.01) of cells that were CDX2+ or YAP1+. Treatment with the AMOT targeting GapmeR increased slightly the number of ICM nuclei that were GATA6+ as compared to the two controls (P < 0.05). Numerically, the difference only existed for the comparison of the two GapmeR treatments. There was no effect of treatment on the number of ICM nuclei that were NANOG+.
Effects of treatment on transcript abundance for 92 genes at day 7.5 was also examined (Figure 5). Treatment of embryos with AMOT targeting GapmeR significantly reduced transcript abundance for 10 genes [actin alpha 2 smooth muscle/aorta (ACTA2), ALPL, AMOT (mentioned earlier), cAMP responsive element binding protein 3 like 2 (CREB3L2), cytochrome P450, family 11, subfamily A, polypeptide 1 (CYP11A1), NANOG, neurofibromin 2 (NF2), par-3 family cell polarity regulator (PARD3), SET and MYND domain containing 3 (SMYD3), and tafazzin (TAZ)]. In addition, AMOT targeting GapmeR increased transcript abundance for seven genes [bone morphogenetic protein 4 (BMP4), CDH1, growth factor receptor bound protein 2 (GRB2), inhibitor of DNA binding 2/HLH protein (ID2), KAT8, platelet-derived growth factor receptor alpha (PDGFRA), and platelet and endothelial cell adhesion molecule 1 (PECAM1)].
As compared to the previous experiment with the AMOT targeting GapmeR (in which the same vehicle and standard negative GapmeR were used), the standard negative GapmeR upregulated slightly expression of five genes when compared to the vehicle [ASH1L (P = 0.01), CCR2 (P = 0.005), CDH1 (P = 0.04), ID2 (P = 0.04), LATS1 (P = 0.05) (Figure 5 for CDH1 and ID2)].
Experiment 5: inhibition of MAP2K1/2
Results are shown in Table 4. Treatment with 0.5 μM MAP2K1/2 inhibitor did not affect the proportion of putative zygotes becoming blastocysts. However, blastocysts in the MAP2K1/2 inhibitor group had more cells than control blastocysts (P < 0.01), and this increase resulted from an increase in number of CDX2− cells (i.e., cells in the ICM) rather than from an increase in number of cells that were CDX2+. Moreover, the increase in number of cells in the ICM was ascribed to an increased number of NANOG+ cells (i.e., epiblast) (P < 0.001) rather than GATA6+ cells (i.e., hypoblast).
Table 4.
Effects of treatment with MAP2K1/2 inhibitor on development of bovine embryos to the blastocyst stage.
| Vehicle | PD0325901b | |
|---|---|---|
| Proportion of putative zygotes that reached the blastocyst stage, day 7.5 (N = 7)a | 23.1 ± 1.6% (n = 471) | 25.3 ± 1.6% (n = 503) |
| Proportion of putative zygotes that reached the blastocyst stage, day 8.5 (N = 7)a | 22.8 ± 1.5% (n = 395) | 23.6 ± 1.5% (n = 415) |
| Total number of nuclei, day 8.5 (N = 7)a | 104.0 ± 4.4 (n = 25) | 119.9 ± 4.2 (n = 26)** |
| Number of CDX2− nuclei (ICM), day 8.5 (N = 7)a | 48.2 ± 3.5 (n = 25) | 62.5 ± 3.3 (n = 26)** |
| Number of CDX2+ nuclei (TE), | ||
| day 8.5 (N = 7)a | 56.7 ± 3.6 (n = 25) | 57.4 ± 3.5 (n = 26) |
| Number of GATA6+ nuclei (hypoblast), | ||
| day 8.5 (N = 7)a | 36.0 ± 2.4 (n = 25) | 37.2 ± 2.3 (n = 26) |
| Number of NANOG+ nuclei (epiblast), | ||
| day 8.5 (N = 7)a | 12.2 ± 2.0 (n = 25) | 25.3 ± 1.9 (n = 26)*** |
aN = Number of replicates; n = total number of embryos per treatment.
b**P < 0.01; ***P < 0.001.
Discussion
Results reported here are indicative of a role of the Hippo pathway members, YAP1 and AMOT, in the formation and function of TE in the bovine blastocyst that developed in vitro. Moreover, results indicate that YAP1 can also affect function of the epiblast and hypoblast and that, as seen earlier [20], MAPK signaling is important for ICM differentiation by reducing epiblast numbers.
A central regulator of differentiation of the TE is the transcription factor CDX2. Transcription of Cdx2 in the mouse is dependent upon interactions of YAP1 and TEAD4. Embryonic transcription of Cdx2 is not required for blastocyst formation in mice [33], but a recent study showed that the Cdx2 derived from the oocyte is important for formation of the blastocyst [34]. In the cow, too, formation of the TE is not dependent on embryonic-derived CDX2 because embryos develop to the blastocyst stage when the gene is deleted [22] or knocked down using siRNA technology [23, 24]. Similarly, blastocyst formation is not compromised in embryos with downregulated TEAD4, which is required for transcription of CDX2 [25]. However, blastocysts formed when CDX2 is low or absent experience abnormalities in GATA3 expression [24], maintenance of tight junctions [23] and trophoblast elongation after transfer to recipient females [22].
Present results suggest that, like in the mouse, accumulation of nuclear CDX2 is dependent upon YAP1. In particular, both VP, which blocks the interaction of YAP1 and TEAD4 [27], and a YAP1 targeting GapmeR, which greatly reduced amount of YAP1 mRNA and protein, decreased the percent of embryos that became blastocysts as well as the percent of blastomeres in the blastocyst that had nuclei that were positive for CDX2 or that positive for YAP1. Like for blastocysts formed in the presence of inadequate CDX2, blastocysts produced in the presence of both VP and the YAP1 targeting GapmeR were dysfunctional as indicated by a reduced capacity for undergoing hatching from the zona pellucida. Effects of VP were more exaggerated than for the YAP1 targeting GapmeR, possibly because VP can also affect autophagy [35, 36] and RAS signaling [37]. However, most effects of VP paralleled those of the YAP1 targeting GapmeR.
It is likely that interactions between YAP1 and CDX2 to regulate differentiation of the TE involve more than regulation of CDX2 transcription by YAP1-TEAD4. Indeed, immunoreactive CDX2 was present in embryos after treatment with VP and the YAP1 targeting GapmeR. A similar phenomenon was observed when examining developmental changes in immunoreactive YAP1 and CDX2. In the early stages of development, CDX2 was localized primarily to the cytoplasm and did not become primarily nuclear in location until the morula stage of development. Thus, it is likely that development of the TE involves regulation of CDX2 accumulation in the nucleus [33, 38] and that YAP1 is involved in this process [11]. This pattern of protein expression is not surprising as it has been observed in the mouse embryo [39]. Like the mouse [34], transcription factors of maternal origin may be important for differentiation of the embryo. In bovine embryos, transcript for CDX2 is low or absent prior to the morula stage and then increases up to the blastocyst stage [40, 41]. In contrast to transcript abundance, it was observed here that CDX2 protein was present in all stages of development examined including the MII oocyte.
The YAP1 targeting GapmeR also disrupted gene expression in the blastocyst in a way consistent with altered function of TE. Thus, treatment with YAP1 targeting GapmeR significantly reduced other genes characteristically expressed in TE in cattle or other species including ELF5, HSD3B1, ID1, IFNT, and TEAD4. There was also a tendency for reduced transcript abundance of CDX2. Expression of other genes associated with TE (EOMES, GATA3, MBNL3, and PECAM1) was not significantly affected by treatment. Moreover, the TE marker KRT8 was upregulated by treatment with YAP1 targeting GapmeR. Data from our laboratory [42] indicate that there is heterogeneity of gene expression among TE cells, and it is possible that YAP1 affects gene expression in some TE subpopulations differently than others.
The YAP1 targeting GapmeR did not have a clear effect on numbers of epiblast and hypoblast cells, but it did disrupt expression of genes characteristically expressed by those cell types. In particular, treatment decreased expression of three genes that are markers of hypoblast in cattle (ALPL, FGFR2, and SOX17) and three markers of epiblast (AJAP1, FGF4, and NANOG) [42]. It can be inferred, therefore, that YAP1 is important for function of hypoblast and epiblast, either directly, because of actions on cells of the ICM, or indirectly, because TE function is compromised.
Although results from experiments in which actions of YAP1 were disrupted were mostly similar to what would be expected based on results in the mouse, this was not the case for AMOT. Based on experiments in mice [5], it was hypothesized that the AMOT targeting GapmeR would alleviate inhibition of YAP1 by AMOT and increase the number of cells designated as TE (i.e., nuclei labeled with CDX2 or YAP1). Instead, treatment with the AMOT targeting GapmeR decreased the number of cells in the day 8.5 blastocyst that were positive for CDX2 and YAP1. There was no effect of the AMOT targeting GapmeR on number of cells positive for CDX2 or YAP1 at day 7.5 of development but treatment did reduce expression of markers for TE (ACTA2, CYP11A1, and TAZ) as well as markers of hypoblast (ALPL) and epiblast (NANOG). The AMOT targeting GapmeR also increased expression of PDGFRA (hypoblast). A role for AMOT in development of the TE is also indirectly supported by the observation that expression of AMOT is greater in TE than ICM at days 7.5 and 8 [43, 44]. Further work is needed to define how AMOT participates in function of the TE.
It is not clear whether the effect of AMOT targeting GapmeR on expression of ALPL and NANOG represents a direct role in differentiation of the ICM into epiblast and hypoblast or indirect effects caused by disruption of TE function. While the AMOT targeting GapmeR had no effect on the number of NANOG+ cells, numbers of CDX2+ cells were decreased and numbers of GATA6+ cells were increased.
Treatment of embryos with the MAP2K1/2 inhibitor did not affect competence of embryos to become blastocysts but the number of ICM cells increased as a result of an increase in NANOG+ epiblast cells. In mice, MAPK is activated when FGF4 from epiblast cells binds to FGFR2 in the hypoblast to trigger Gata6 expression and subsequent downregulation of Nanog transcription. In cattle, knockdown of FGFR2 does not affect expression of epiblast and hypoblast markers NANOG and GATA6 but decreases the hypoblast marker HNF4A [45]. Also, addition of FGF4 [20] and FGF2 [26] can promote hypoblast formation. It is not clear whether the endogenous ligand for MAPK signaling in the bovine blastocyst is FGF4 because inhibition of activity of this molecule had no effect on hypoblast numbers [20]. That the MAP2K1/2 inhibitor affected epiblast numbers and not numbers of hypoblast suggests that the main role of the MAPK pathway in cattle is to suppress epiblast formation.
Altogether, these results demonstrate for the first time that YAP1 and AMOT play a role in the regulation of TE differentiation and function in the bovine embryo. Results also confirm the importance of MAPK signaling for differentiation of the ICM through inhibition of epiblast numbers. Experiments are consistent with the idea that YAP1 is also involved in the function of epiblast and hypoblast. More studies are required to expand this latter finding and to confirm whether this role is restricted to in vitro developed embryos or also occurs in vivo.
Supplementary Material
Acknowledgments
The authors thank Luana Teixeira Rodrigues (UNESP-Universidade Estadual Paulista, Araçatuba, São Paulo, Brazil) for aid in the process of sample collection and processing; owners and employees of Central Beef Packing Co. (Center Hill, FL), Adena Meat Products L.P. (Fort McCoy, FL), and Florida Beef Inc. (Zolfo Springs, FL) for providing ovaries; William Rembert and Eddie Cummings for ovary collection; and the Miami Center for AIDS Research (CFAR) at the University of Miami Miller School of Medicine, which is funded by a grant (P30AI073961) from the National Institutes of Health, and which performed qPCR.
Notes
Conference Presentation: Presented in part at the 49th Annual Meeting of the Society for the Study of Reproduction, July 16–20 2016, San Diego, California.
Edited by Dr. Myriam Hemberger, PhD, Babraham Institute.
Footnotes
Grant Support: This research was supported by Agriculture and Food Research Initiative Competitive Grant No. 2011-67015-30688 from the USDA National Institute of Food and Agriculture and from funds from the L.E. “Red” Larson Endowment. Verónica Negrón-Pérez was supported by a McKnight Doctoral Fellowship from the Florida Education Fund, Inc.
Supplementary data
Supplementary data are available at BIOLRE online.
https://academic.oup.com/biolreprod
Supplemental Table S1. Details of antibodies used.
Supplemental File S1. Information about genes for transcript abundance analysis. Data shown are the gene symbol, forward and reverse primers used for RT-PCR, the reference sequence identification number, the full gene name and reason for selection/previously reported effects. Note that the primers for KLF2 were not validated (highlighted in yellow).
Supplemental Figure S1. Representative image showing colocalization of YAP1 and NANOG in a day 9.5 bovine blastocyst. Left panel staining represents labeling for NANOG (i.e., epiblast cells; green), middle panel labeling for YAP1 (red), and the right panel the merged image. The dashed circle delineates the inner cell mass of the blastocyst. Scale bar = 20 μM.
Supplemental Figure S2. Representative images of day 7.5 blastocysts treated with vehicle, standard negative GapmeR (Std Negative), or YAP1 targeting GapmeR (YAP1 kd). The total number of nuclei (blue) was not affected by treatment but the nuclear intensity for CDX2+ (green) and YAP1+ (red) nuclei was decreased in the embryos treated with the YAP1 targeting GapmeR. Merged images are a composite of all three fluorochrome channels. Scale bar = 20 μM.
References
- 1. Pfeffer PL. Lineage commitment in the mammalian preimplantation. In: Juengel J, Miyamoto A, Price C, Reynolds LP, Smith MF, Webb R (eds.), Reproduction in Domestic Ruminants VIII. Context Products, Ashby-de-la-Zouch, England; 2014:89–103. [Google Scholar]
- 2. Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 2009; 10:91–103. [DOI] [PubMed] [Google Scholar]
- 3. Morris SA, Zernicka-Goetz M. Formation of distinct cell types in the mouse blastocyst. In: Kubiak JZ. (ed.), Mouse Development. 55 Berlin Heidelberg: Springer-Verlag; 2012:203–217. [DOI] [PubMed] [Google Scholar]
- 4. Lorthongpanich C, Issaragrisil S. Emerging role of the hippo signaling pathway in position sensing and lineage specification in mammalian preimplantation embryos. Biol Reprod 2015; 92:1–10. [DOI] [PubMed] [Google Scholar]
- 5. Hirate Y, Hirahara S, Inoue KI, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, Ohno S, Marikawa Y et al. Polarity-dependent distribution of angiomotin localizes hippo signaling in preimplantation embryos. Curr Biol 2013; 23:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paramasivam M, Sarkeshik A, Yates JR, Fernandes MJG, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell 2011; 22:3725–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, Ding S, Guan KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 2010; 24:1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorthongpanich C, Doris TPY, Limviphuvadh V, Knowles BB, Solter D. Developmental fate and lineage commitment of singled mouse blastomeres. Development 2012; 139:3722–3731. [DOI] [PubMed] [Google Scholar]
- 9. Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev 2008; 125:270–283. [DOI] [PubMed] [Google Scholar]
- 10. Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010; 137:395–403. [DOI] [PubMed] [Google Scholar]
- 11. Nishioka N, Inoue KI, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell 2009; 16:398–410. [DOI] [PubMed] [Google Scholar]
- 12. Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 1995; 9:534–546. [DOI] [PubMed] [Google Scholar]
- 13. Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 1995; 121:1053–1063. [DOI] [PubMed] [Google Scholar]
- 14. Li P, Chen Y, Mak KK, Wong CK, Wang CC, Yuan P. Functional role of Mst1/Mst2 in embryonic stem cell differentiation. PLoS One 2013; 8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hirate Y, Sasaki H. The role of angiomotin phosphorylation in the Hippo pathway during preimplantation mouse development. Tissue Barriers 2014; 2:e28127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Comms 2013; 4:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kono K, Tamashiro DAA, Alarcon VB. Inhibition of RHO–ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Bio 2014; 394:142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morris SA, Graham SJL, Jedrusik A, Zernicka-Goetz M. The differential response to Fgf signalling in cells internalized at different times influences lineage segregation in preimplantation mouse embryos. Open Biol 2013; 3:130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamanaka Y, Lanner F, Rossant J. FGF signal-dependent segregation of primitive endoderm and epiblast in the mouse blastocyst. Development 2010; 137:715–724. [DOI] [PubMed] [Google Scholar]
- 20. Kuijk EW, van Tol LTA, Van de Velde H, Wubbolts R, Welling M, Geijsen N, Roelen BA. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development 2012; 139:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morris SA, Teo RTY, Li H, Robson P, Glover DM, Zernicka-Goetz M. Origin and formation of the first two distinct cell types of the inner cell mass in the mouse embryo. Proc Natl Acad Sci USA 2010; 107:6364–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Dev Cell 2011; 20:244–255. [DOI] [PubMed] [Google Scholar]
- 23. Goissis MD, Cibelli JB. Functional characterization of CDX2 during bovine preimplantation development in vitro. Mol Reprod Dev 2014; 81:962–970. [DOI] [PubMed] [Google Scholar]
- 24. Sakurai N, Takahashi K, Fujii T, Hirayama H, Kageyama S, Hashizume T, Sawai K. The necessity of OCT-4 and CDX2 for early development and gene expression involved in differentiation of inner cell mass and trophectoderm lineages in bovine embryos. Cellular Reprogr 2016; 18:309–318. [DOI] [PubMed] [Google Scholar]
- 25. Sakurai N, Takahashi K, Emura N, Hashizume T, Sawai K. Effects of downregulating TEAD4 transcripts by RNA interference on early development of bovine embryos. J Repduction Dev 2017; 63:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang QE, Fields SD, Zhang K, Ozawa M, Johnson SE, Ealy AD. Fibroblast growth factor 2 promotes primitive endoderm development in bovine blastocyst outgrowths. Biol Reprod 2011; 85:946–953. [DOI] [PubMed] [Google Scholar]
- 27. Liu-chittenden Y, Huang B, Shim JS, Dev G, Chen Q, Lee S, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 2012; 26:1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ortega MS, Wohlgemuth S, Tribulo P, Siqueira LGB, Null DJ, Cole JB, Da Silva MV, Hansen PJ. A single nucleotide polymorphism in COQ9 affects mitochondrial and ovarian function and fertility in Holstein cows. Biol Reprod 2017; 96:652–663. [DOI] [PubMed] [Google Scholar]
- 29. Ortega MS, Rocha-Frigoni NAS, Mingoti GZ, Roth Z, Hansen PJ. Modification of embryonic resistance to heat shock in cattle by melatonin and genetic variation in HSPA1L. J Dairy Sci 2016; 99:9152–9164. [DOI] [PubMed] [Google Scholar]
- 30. Kannampuzha Francis J, Tribulo P, Hansen PJ. Actions of activin A, connective tissue growth factor, hepatocyte growth factor and teratocarcinoma derived growth factor 1 on the development of the bovine preimplantation embryo. Reprod Fertil Dev 2017; 29:1329–1339. [DOI] [PubMed] [Google Scholar]
- 31. Siqueira LGB, Hansen PJ. Sex differences in response of the bovine embryo to colony-stimulating factor 2. Reproduction 2016; 152:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Denicol AC, Block J, Kelley DE, Pohler KG, Dobbs KB, Mortensen CJ, Ortega MS, Hansen PJ. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J 2014; 28:3975–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005; 132:2093–2102. [DOI] [PubMed] [Google Scholar]
- 34. Jedrusik A, Cox A, Wicher K, Glover DM, Zernicka-Goetz M. Maternal-zygotic knockout reveals a critical role of Cdx2 in the morula to blastocyst transition. Dev Biol 2015; 398:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Donohue E, Tovey A, Vogl AW, Arns S, Sternberg E, Young RN, Roberge M. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem 2011; 286:7290–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liang N, Zhang C, Dill P, Panasyuk G, Pion D, Koka V, Gallazzini M, Olson EN, Lam H, Henske EP, Dong Z, Apte U et al. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J Exp Med 2014; 211:2249–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Garcia-Rendueles MER, Ricarte-Filho JC, Untch BR, Landa I, Knauf JA, Voza F, Smith VE, Ganly I, Taylor BS, Persaud Y, Oler G, Fang Y et al. NF2 loss promotes oncogenic RAS-induced thyroid cancers via YAP-dependent transactivation of RAS proteins and sensitizes them to MEK inhibition. Cancer Discov 2015; 5:1178–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jedrusik A, Parfitt DE, Guo G, Skamagki M, Grabarek JB, Johnson MH, Robson P, Zernicka-Goetz M. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev 2008; 22:2692–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deb K, Sivaguru M, Yong HY, Michael Roberts R. Cdx2 Gene expression and trophectoderm lineage specification in mouse embryos. Science 2006; 311:992–996. [DOI] [PubMed] [Google Scholar]
- 40. Jiang Z, Sun J, Dong H, Luo O, Zheng X, Obergfell C, Tang Y, Bi J, O’Neill R, Ruan Y, Chen J, Tian X et al. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics 2014; 15:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Denicol AC, Leão BCS, Dobbs KB, Mingoti GZ, Hansen PJ. Influence of sex on basal and dickkopf-1 regulated gene expression in the bovine morula. PLoS One 2015; 10:e0133587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Negrón-Pérez VM, Zhang Y, Hansen PJ. Single-cell gene expression of the bovine blastocyst. Reproduction 2017; 154:627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hosseini SM, Dufort I, Caballero J, Moulavi F, Ghanaei HR, Sirard MA. Transcriptome profiling of bovine inner cell mass and trophectoderm derived from in vivo generated blastocysts. BMC Dev Biol 2015; 15:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ozawa M, Sakatani M, Yao J, Shanker S, Yu F, Yamashita R, Wakabayashi S, Nakai K, Dobbs KB, Sudano MJ, Farmerie WG, Hansen PJ. Global gene expression of the inner cell mass and trophectoderm of the bovine blastocyst. BMC Dev Biol 2012; 12:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Akizawa H, Nagatomo H, Odagiri H, Kohri N, Yamauchi N, Yanagawa Y, Nagano M, Takahashi M, Kawahara M. Conserved roles of fibroblast growth factor receptor 2 signaling in the regulation of inner cell mass development in bovine blastocysts. Mol Reprod Dev 2016; 83:516–525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.