Abstract
Mycousfurans (1 and 2), two new usnic acid congeners, along with (−)-mycousnine (3), (−)-placodiolic acid (4), and (+)-usnic acid (5), were isolated using high-performance liquid chromatography-ultraviolet (HPLC-UV)-guided fractionation of extracts of Mycosphaerella sp. isolated from a marine sediment. The planar structures of 1 and 2 were elucidated using 1D and 2D NMR spectra. The relative configurations of the stereogenic carbons of 1 and 2 were established via analysis of their nuclear Overhauser spectroscopy (NOESY) spectra, and their absolute configurations were determined using a comparison of experimental and calculated electronic circular dichroism (ECD) spectra. Compounds 1 and 2 were found to have antibacterial activity, showing moderate activity against Kocuria rhizophila and Staphylococcus aureus.
Keywords: usnic acid, mycousfurans, mycousnine, placodiolic acid, Mycosphaerella sp., antibacterial activity
1. Introduction
Dibenzofurans have been isolated from plants, mushrooms, and marine organisms. Although lichens were their first reported natural source, isolation of dibenzofurans from filamentous fungi has been increasingly reported [1]. Usnic acid (UA) is the most representative dibenzofuran natural product and has interesting chemical and pharmacological properties with a broad spectrum of biological activities such as antibacterial, antiviral, anti-inflammatory, antiprotozoal, antifungal, anti-proliferative, phytotoxic, UV filter, and anti-osteoclastogenic activities [2,3,4,5,6]. UA is generally distributed in lichen genera such as Usnea (Usneaceae), Cladonia (Cladoniaceae), Hypotrachyna (Parmeliaceae), Lecanora (Lecanoraceae), Ramalina (Ramalinaceae), Evernia, Parmelia (Parmeliaceae), and Alectoria (Alectoriaceae) [7]. There are also a few reports on the isolation of UA or its derivatives from non-lichen sources [4].
Mycosphaerella is the largest genus of Ascomycota, with more than 10,000 species. Mycosphaerella species produce secondary metabolites including rosigenin [8], rubellins A and B [9], (−)-mycousnine, and (+)-isomycousnine [10]. Mycousfuranine and mycousnicdiol have also been recently reported to possess antifungal activity [11]. Mycosphaerella species are generally known as foliicolous plant pathogens, isolated from the leaves of plants; however, some species are also found in marine environments. M. ascopliylli and M. pelvetiae are endophytes of the brown algae Ascophyllum nodusum and Pelvetia canaliculate, respectively, while M. apophlaeae is the symbiont of the rhodophyte, Apophlaea lyallii [12,13,14]. In addition, there are recent taxonomy studies demonstrating that Mycosphaerella is not just a terrestrial genus but is spread across marine environments as well. For example, Mycosphaerella sp. was one of the two dominant fungal communities in samples collected from salt marshes in California Bay and the Atlantic east coast of USA [15,16].
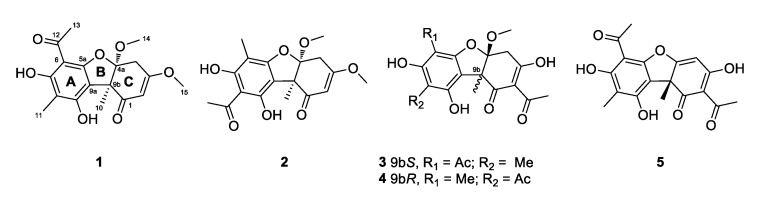
During the course of the chemical analysis of cultured fungal strains, isolated from marine sediments, we isolated two new usnic acid congeners, mycousfurans A and B (1 and 2), along with the previously reported compounds, (−)-mycousnine (3), (−)-placodiolic acid (4), and (+)-usnic acid (5), from extracts of Mycosphaerella sp. (Figure 1). Herein, we describe the isolation, structural elucidation, and bioactivities of mycousfurans A and B (1 and 2).
Figure 1.
Chemical structures of mycousfurans A and B (1 and 2), (−)-mycousnine (3), (−)-placodiolic acid (4), and (+)-usnic acid (5), isolated from Mycosphaerella sp.
2. Results
2.1. Isolation and Structure Elucidation
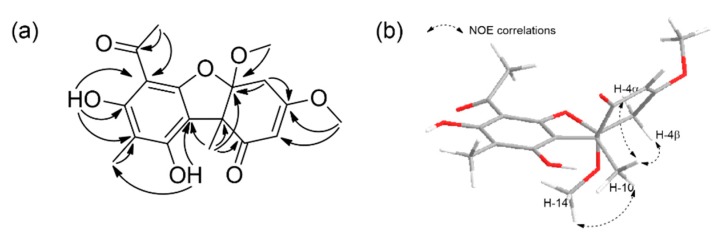
Compound 1 was obtained as an amorphous yellowish powder, and its molecular formula was determined to be C18H20O7 based on a (+)-high-resolution electrospray ionization mass spectrometry (HRESIMS) m/z 349.1302 [M + H]+, indicating 9 degrees of unsaturation. The IR spectrum of 1 indicated the presence of hydroxyl (3387, 3232 cm−1) and ketone functionalities (1616 cm−1), and the UV spectrum showed similar absorption patterns to those of dibenzofuran derivatives. The 1H NMR spectrum of 1 showed five methyl singlets (δH 1.62, 2.04, 2.61, 3.49, 3.81), two doublets at δH 2.96, J = 17.5 Hz (1H) and δH 3.15, J = 17.5 Hz (1H), a singlet of an olefinic proton (δH 5.55, s), and two singlets of phenolic hydroxyl protons (δH 9.34, 13.34). The 13C NMR, in combination with the heteronuclear single quantum correlation (HSQC) spectrum, showed two ketone carbonyls (δC 200.5, 201.2), six non-protonated aromatic carbons (δC 102.0, 106.5, 107.5, 157.1, 159.6, 163.3), five methyl carbons (δC 7.4, 16.6, 31.3, 50.9, 56.9), two bridgehead quaternary carbons (δC 57.9, 111.6), a methine sp2 carbon (δC 100.6), and one methylene sp3 carbon (δC 34.3) (Table 1). The heteronuclear multiple bond correlation (HMBC) correlations from the phenolic proton 7-OH (δH 9.34) to C-6 (δC 102.0), C-7 (δC 163.3), and C-8 (δC 107.5); from the phenolic proton 9-OH (δH 13.34) to C-8 (δC 107.5) and C-9a (δC 106.5); from the methyl protons H3-13 (δH 2.61, s) to C-6 (δC 102.0) and C-12 (δC 201.2); and from H3-11 (δH 2.04, s) to C-8 (δC 107.5), established the substitution pattern of the A ring. The HMBC correlations from H2-4 (δH 2.96, d, J = 17.5 and 3.15, d, J = 17.5) to C-2 (δC 106.6) and C-4a (δC 34.3); from H3-15 (δH 3.81, s) to C-2 (δC 106.6) and C-3 (δC 175.4), and from the angular methyl protons H3-10 (δH 1.62, s) to C-1 (δC 200.5), C-4a (δC 34.3), and C-9b (δC 57.9) established the substitution pattern of ring C. Finally, the linkage between C-9a and C-9b was corroborated by the observation of the cross peak from H3-10 to C-9a in the HMBC spectrum. Combining the 1D and 2D NMR data and the molecular formula, the presence of an ether linkage between C-4a (δC 111.6) and C-5a (δC 157.1) was proposed. Moreover, the HMBC correlation from the methoxyl protons H3-14 (δH 3.49, s) to C-4a permitted the placement of the methoxy group at C-4a, thus completing the structural assignment of 1, as shown in Figure 2. The NOESY correlations from H3-14/H3-10 and H2-4/H3-10 indicated that the B/C ring junction was cis-orientated (Figure S5) [10].
Table 1.
1H and 13C NMR spectroscopic data (700 MHz and 175 MHz in CDCl3) for mycousfurans (1–2).
| Position | 1 | HMBC | 2 | ||
|---|---|---|---|---|---|
| δC, Type | δH, mult. (J in Hz) | δC, Type | δH, mult. (J in Hz) | ||
| 1 | 200.5, C | 201.0, C | |||
| 2 | 100.6, CH | 5.55, s | 100.2, CH | 5.55, s | |
| 3 | 175.4, C | 175.9, C | |||
| 4α 4β |
34.3, CH2 | 3.15, d (17.5), 2.96, d (17.5) |
2, 4a 2, 4a |
34.2, CH2 | 3.20, d (17.5) 2.94, d (17.5) |
| 4a | 111.6, C | 110.6, C | |||
| 5a | 157.1, C | 160.7, C | |||
| 6 | 102.0, C | 100.4, C | |||
| 7 | 163.3, C | 165.5, C | |||
| 8 | 107.5, C | 107.1, C | |||
| 9 | 159.6, C | 156.4, C | |||
| 9a | 106.5, C | 106.1, C | |||
| 9b | 57.9, C | 58.9, C | |||
| 10 | 16.6, CH3 | 1.62, s | 1, 4a, 9a, 9b | 16.2, CH3 | 1.66, s |
| 11 | 7.4, CH3 | 2.04, s | 8 | 7.5, CH3 | 2.02, s |
| 12 | 201.2, C | 203.8, C | |||
| 13 | 31.3, CH3 | 2.61, s | 6, 12 | 32.9, CH3 | 2.73, s |
| 14 | 50.9, CH3 | 3.49, s | 4a | 50.3, CH3 | 3.47, s |
| 15 | 56.9, CH3 | 3.81, s | 2, 3 | 56.8, CH3 | 3.84, s |
| 7-OH | 13.34, s | 6, 7, 8 | 14.32, s | ||
| 9-OH | 9.34, s | 8, 9a | 9.61, s | ||
Figure 2.
Key HMBC (a) and NOESY (b) correlations of mycousfuran A (1).
Compound 2 was obtained as a yellow amorphous powder. Its molecular formula was determined to be C18H20O7 based on a (+)-HRESIMS m/z 349.1305 [M + H]+ and 13C NMR data. Interpretation of the NMR data revealed that the structure of 2 was almost identical to that of 1, except that 2 possessed a methyl group at C-6 and an acetyl group at C-8 (Figure 1). The HMBC correlations from H3-10 (δH 1.66, s) to C-9a (δC 106.1), 9-OH (δH 9.61, s) to C-8 (δC 107.1) and C-9a (δC 106.1), H3-13 (δH 2.73, s) to C-8, and H3-11 (δH 2.02, s) to C-5a (δC 160.7) supported the positions of the acetyl group at C-8, and consequently placed a methyl group at C-6 (δC 100.4) (Figures S9 and S10). Thus, 2 was defined as a regioisomer of 1.
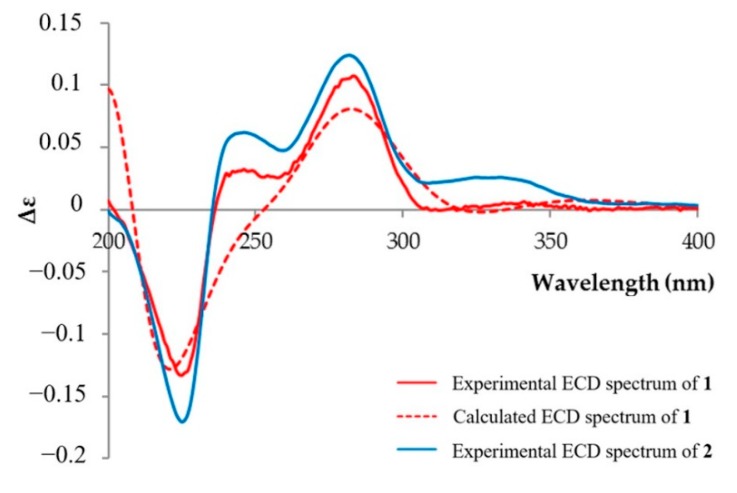
The absolute configurations of the stereogenic carbons of 1 were established via the comparison of the experimental electronic circular dichroism (ECD) spectrum with that generated by the computer-assisted ECD calculation. The ECD spectrum of 1 fits well with that of the calculated ECD spectrum of 4aS and 9bR stereoisomers (Figure 3). Both experimental and calculated ECD spectra of 1 showed the negative absorption in the range from 210 to 235 nm and the positive absorption from 250 to 310 nm (Figure 3). Therefore, the absolute configurations of C-4a and C-9a in 1 was established as 4aS and 9bR. The experimental ECD spectrum of 2 was almost identical to that of 1, leading to the conclusion that the absolute configurations of C-4a and C-9a in 2 were 4aS and 9bR.
Figure 3.
Experimental and calculated ECD spectra of 1 and 2.
Three known UA derivatives were also isolated together with 1 and 2 and they were identified as (−)-mycousnine (3) [10], (−)-placodiolic acid (4) [17], and (+)-usnic acid (5) [18] via comparison of their NMR and MS data with those reported in the literatures.
2.2. Bioactivity
Compounds 1–5 were tested for their antibacterial activity using three Gram-positive bacteria (Bacillus substilis ATCC 6633, Kocuria rhizophila ATCC 9341, Staphylococcus aureus ATCC 6538) and three Gram-negative bacteria (Escherichia coli ATCC 11775, Salmonella typhimurium ATCC 14208, Klebsiella pneumonia ATCC 4352). Compound 1 exhibited the minimal inhibitory concentration (MIC) values of 8 μg/mL and 32 μg/mL, while 2 exhibited MIC values of 16 μg/mL and 32 μg/mL, against K. rhizophila and S. aureus, respectively (Table 2). Compounds 1 and 2 showed no antibacterial activity against B. substilis and Gram-negative bacteria. Since the MIC values of 3–5 indicated stronger antibacterial activity than that of 1 and 2 against Gram-positive bacteria, it was suggested that the substituents in ring C could play a role in the antibacterial activity.
Table 2.
The MIC values (g/mL)1 of 1–5 against Gram-positive and Gram-negative bacteria.
| Compound | Gram (+) Bacteria | Gram (−) Bacteria | ||||
|---|---|---|---|---|---|---|
| B. subtilis ATCC 6633 |
K. rhizophila ATCC 9341 |
S. aureus ATCC 6538 |
E. coli ATCC 11775 |
S. typhimurium ATCC 14208 |
K. pneumonia ATCC 4352 |
|
| 1 | >128 | 8 | 32 | >128 | >128 | >128 |
| 2 | >128 | 16 | 32 | >128 | >128 | >128 |
| 3 | 4 | 8 | 4 | >128 | >128 | >128 |
| 4 | 4 | 8 | 4 | >128 | >128 | >128 |
| 5 | 2 | 8 | 16 | >128 | >128 | >128 |
| Vancomycin | 0.25 | 0.25 | 0.5 | >128 | >128 | >128 |
| Ampicillin | 0.5 | 0.25 | 2 | 16 | 8 | >128 |
1 Each sample was tested in triplicate and repeated three times.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were measured using an Autopol III (Rudolph Research Analytical, Hackettstown, NJ, USA) polarimeter with a 5-cm cell. ECD spectra were recorded using a Chirascan™-plus CD Spectrometer (Applied Photophysics Ltd., Surrey, UK) and the UV spectra were recorded on a Scinco UVS-2100 spectrophotometer (Sinco, Daejeon, Korea). IR spectra were obtained using a Scimitar 800 FT-IR spectrometer (Varian Inc., Palo Alto, CA, USA). NMR spectra were recorded on a Bruker Avance 700 MHz spectrometer (Bruker Biospin Group, Karlsruhe, Germany); The residual solvent signals of CDCl3 (δH 7.26, δC 77.0) were referenced for the 1H and 13C chemical shift values. HRESIMS spectra were obtained using a JEOL JMS-AX505WA mass spectrometer (JEOL Ltd., Tokyo, Japan). Low-resolution LC-MS data were obtained using an Agilent Technologies 6120 quadrupole LC/MS system (Agilent Technologies, Santa Clara, CA, USA) with a reversed-phase C18 column (Phenomenex Luna C18 (2), 50 mm × 4.6 mm, 5 μm) at a flow rate of 1.0 mL/min. Column chromatography separation was performed using a C18 column (40–63 m, ZEO prep 90), eluting with a gradient of methanol and water. The fractions were purified using a WATERSTM (Milford, MA, USA) 1525 binary HPLC (high-performance liquid chromatography) pump, equipped with a WATERS 2489 UV visible detector using a WATERS reversed-phase HPLC Watchers 120 ODS-BP (250 mm × 10 mm, 5 μm) column, eluting with 80% CH3CN in H2O at flow rate of 2.5 mL/min.
3.2. Fungal Material
The strain F8015-2B was isolated from a marine sediment at a 5-m depth in Donghae-si, Gangwon-do, South Korea. The collected sediment was dried on a clean bench for 24 h and then crushed using a sterile spoon. The powder was stamped onto 1/3 marine agar medium and incubated at 27 °C. After two weeks, fungal spores were observed. The spores were cultured via repeated inoculation on potato dextrose agarplates. F8015-2B was identified as Mycosphaerella sp. based on a 99.6% (496/498) similarity of 18S rRNA genes to the Mycosphaerella nawae strain MY3.
3.3. Fermentation, Extraction, and Purifircation
The strain F8015-2B was cultured in 6 × 2.5-L Ultra Yield Flasks (Thomson Instrument Company, Oceanside, CA, USA), each containing 1 L of potato dextrose broth (PDB) dissolved in seawater. The fungus was cultivated on seed agar blocks in 6 × 2.5-L Ultra Yield Flasks, each containing 1 L of PDB dissolved in seawater at 27 °C and 140 rpm in a shaking incubator. After seven days, the mycelia were filtered from the broth using gauze filtration and extracted with acetone and methanol. The broth was extracted with EtOAc and evaporated to obtain the crude extract (4.01 g).
The crude extract was fractionated into eight fractions with a silica gel open column chromatography using a step-gradient with a mixture of CH2Cl2 and MeOH as an eluent. Fractions 1 (974.9 mg), 2 (280.1 mg), and 3 (420.1 mg) were subjected to a reversed-phase HPLC (Phenomenex luna C18 column, 250 mm × 10 mm, 5 μm, flow rate = 2.0 mL/min) and eluted with 65% CH3CN in distilled water to yield 1 (7.5 mg), 2 (3.3 mg), 3 (56.3 mg), 4 (28.7 mg), and 5 (6.8 mg).
Mycousfuran A (1): amorphous powder, [α]D25 + 11 (c 1.00, CHCl3); UV (MeOH) λmax (log ε) 200 (2.15), 281 (2.09), and 349 (1.24) nm; IR (KBr) νmax 3387, 3232, and 1616 cm−1; CD λext (MeOH) nm (Δε): 282 (+0.11), 246 (+0.03) [236(0)]; 1H and 13C NMR data, see Table 1; (+)-HRESIMS, m/z 349.1302 [M + H]+ (calcd for C18H20O7, 349.1287).
Mycousfuran B (2): amorphous yellowish powder, [α]D25 + 15 (c 1.00, CHCl3); UV (MeOH) λmax (log ε) 200 (2.15), 281 (2.09), and 349 (1.24) nm; IR (KBr) νmax 3325, 3198, and 1625 cm−1; CD λext (MeOH) nm (Δε): 282 (+0.12), 246 (+0.06) [236(0)]; 1H and 13C NMR data, see Table 1; (+)-HRESIMS, m/z 349.1305 [M + H]+ (calcd for C18H20O7, 349.1287).
3.4. Computer-Assisted Conformational Analyses and ECD Calculations
Preliminary conformational analyses of 1 and 2 were performed with Merck Molecular Force Field (MMFF) by Spartan 10 (Wavefunction, Irvine, CA, USA). The two lowest energy conformers of 1 and 2 were geometrically optimized with the B3LYP/6-31G(d,p) level of density functional theory (DFT) in methanol using Gaussian 16 (Expanding the limits of computational chemistry, Wallingford, CT, USA). The computer-assisted ECD calculation was carried out with the B3LYP/6-31G(d,p) level of time-dependent density functional theory (TDDFT). The calculated ECD spectra of 1 and 2 were obtained via visualization of SpecDis version 1.71 (SpecDis, Berlin, Germany) in combination with the calculated ECD spectra of each conformer on the basis of Boltzmann distribution theory and their relative Gibbs free energy.
3.5. Antibacterial Activity
Three Gram-positive (Bacillus substilis ATCC 6633, Kocuria rhizophila ATCC 9341, Staphylococcus aureus ATCC 6538) and three Gram-negative (Escherichia coli ATCC 11775, Salmonella typhimurium ATCC 14208, Klebsiella pneumonia ATCC 4352) strains were used. These bacteria were inoculated onto a Mueller–Hinton agar medium and allowed to grow for 24 h at 37 °C. The bacterial colonies were cultivated in 15-mL round-bottom tubes containing 5 mL of Mueller–Hinton broth (MHB) at 37 °C and 220 rpm for 24 h. One hundred microliter aliquots of test compounds and positive controls (vancomycin and ampicillin) at a concentration of 256 µg/mL in DMSO were added to different wells of a 96-well microtiter plate containing 50 µL of MHB. The samples were serially diluted and 50 µL of bacterial MHB medium was adjusted to a concentration of 1/100 dilution. McFarland 0.5% standard was added to the wells. The 96-well microtiter plate was incubated for 24 h at 37 °C. Subsequently, the minimum inhibitory concentration was determined as the concentration of compounds inhibiting bacterial growth [19].
4. Conclusions
In conclusion, mycousfurans A and B (1 and 2) and other usnic acid congeners, were isolated from a marine sediment-derived fungus Mycosphaerella sp. The structures of 1 and 2 were established using 1D and 2D NMR spectra. The absolute configurations of the stereogenic carbons of 1 and 2 were determined using NOESY experiments and a comparison between the experimental and calculated ECD spectra. Compounds 1 and 2 exhibited antibacterial activity against K. rhizophila, S. aureus, and E. coli. The present study is the first report of the antibacterial compounds, produced by Mycosphaerella sp., which was isolated from the marine environment.
Supplementary Materials
The following are available online at https://www.mdpi.com/1660-3397/17/7/422/s1, Figure S1: 1H NMR spectrum (700 MHz, CDCl3) of 1, Figure S2: 13C NMR spectrum (175 MHz, CDCl3) of 1, Figure S3: HSQC spectrum (700 MHz, CDCl3) of 1, Figure S4: HMBC spectrum (700 MHz, CDCl3) of 1, Figure S5: NOESY spectrum (700 MHz, CDCl3) of 1, Figure S6: 1H NMR spectrum (700 MHz, CDCl3) of 2, Figure S7: 13C NMR spectrum (175 MHz, CDCl3) of 2, Figure S8: HSQC spectrum (700 MHz, CDCl3) of 2, Figure S9: HMBC spectrum (700 MHz, CDCl3) of 2, Figure S10: Expanded HMBC of 2.
Author Contributions
J.L. (Jihye Lee) isolated the compounds, elucidated the chemical structure, and performed bioassays. J.L. (Jusung Lee) performed the large-scale culture of fungal strains and elucidated the chemical structure. G.J.K., J.-W.N., and H.C calculated the ECD spectra and wrote the manuscript. W.W. performed sampling and isolated a fungal strain from a sediment. I.Y. contributed NMR analysis and wrote the manuscript. S.-J.N. was the project leader guiding the chemical analysis experiments. H.K. headed the project and chemical analysis, provided the microbial strains, and wrote the manuscript.
Funding
This research was funded by Basic Science Research Program through the National Research Foundation of Korea (NRF) of the Ministry of Science, ICT & Future Planning, grant number NRF-2017R1D1A1B03028172 (to S.-J.N.) and NRF-2019R1A2C2005492 (to H.K.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Millot M., Dieua A., Tomasib S. Dibenzofurans and derivatives from lichens and ascomycetes. Nat. Prod. Rep. 2016;33:801–811. doi: 10.1039/C5NP00134J. [DOI] [PubMed] [Google Scholar]
- 2.Ingólfsdóttir K. Molecules of interest usnic acid. Phytochemistry. 2002;61:729–736. doi: 10.1016/S0031-9422(02)00383-7. [DOI] [PubMed] [Google Scholar]
- 3.Luzina O.A., Salakhutdinov N.F. Usnic Aacid and its derivatives for pharmaceutical use: A patent review (2000-2017) Expert Opin. Ther. Pat. 2018;28:477–491. doi: 10.1080/13543776.2018.1472239. [DOI] [PubMed] [Google Scholar]
- 4.Cocchietto M., Skert N., Nimis P.L., Sava G. A review on usnic acid, an interesting natural compound. Naturwissenschaften. 2002;89:137–146. doi: 10.1007/s00114-002-0305-3. [DOI] [PubMed] [Google Scholar]
- 5.Alahmadi A.A. Usnic acid biological activity: History, evaluation and usage. Int. J. Basic Clin. Pharmacol. 2017;6:2752–2759. doi: 10.18203/2319-2003.ijbcp20175072. [DOI] [Google Scholar]
- 6.Kim K.-J., Jeong M.-H., Lee Y., Hwang S.-J., Shin H.-B., Hur J.-S., Son Y.-J. Effect of usnic acid on osteoclastogenic activity. J. Clin. Med. 2018;7:345. doi: 10.3390/jcm7100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzina O.A., Salakhutdinov N.F. Biological activity of usnic acid and tts derivatives: Part 1. Activity against unicellular organisms. Russ. J. Bioorg. Chem. 2016;42:115–132. doi: 10.1134/S1068162016020084. [DOI] [Google Scholar]
- 8.Albinati A., Bruckner S., Camarda L., Nasini G. Rosigenin, an unusual metabolite from Mycosphaerella rosigena. Tetrahedron. 1980;36:117–121. doi: 10.1016/0040-4020(80)85033-2. [DOI] [Google Scholar]
- 9.Arnone A., Camarda L., Nasini G., Assante G. Secondary metabolites. Part 15. Structure elucidation of rubellins A and B, two novel anthraquinone metabolites from Mycosphaerella rubella. J. Chem. Soc. 1986;1:255–260. doi: 10.1039/p19860000255. [DOI] [Google Scholar]
- 10.Sassa T., Igarashi M. Structures of (−)-mycousnine, (+)-isomycousnine and (+)-oxymycousnine. Agric. Biol. Chem. 1990;54:2231–2237. doi: 10.1271/bbb1961.54.2231. [DOI] [Google Scholar]
- 11.De Oliveira D.M., Pereira C.B., Mendes G., Junker J., Kolloff M., Rosa L.H., Rosa C.A., Alves T.M.A., Zani C.L., Johann S., et al. Two new usnic acid derivatives from the endophytic fungus Mycosphaerella sp. Naturforsch C. 2018;73:449–456. doi: 10.1515/znc-2017-0162. [DOI] [PubMed] [Google Scholar]
- 12.Webber F.C. Observations on the structure, life history and biology of Mycosphaerella ascophylli. Trans. Br. Mycol. Soc. 1967;50:583–601. doi: 10.1016/S0007-1536(67)80090-1. [DOI] [Google Scholar]
- 13.Kohlmeyer J. Geography of marine fungi. Aust. J. Bot. Suppl. Ser. 1983;10:67–76. doi: 10.1071/BT8310067. [DOI] [Google Scholar]
- 14.Fries N. Physiological characteristics of Mycosphaerella ascophylli, a fungal endophyte of the marine brown alga Ascophyllum nodosum. Physiol. Plant. 1979;45:117–121. doi: 10.1111/j.1399-3054.1979.tb01674.x. [DOI] [Google Scholar]
- 15.Lyons J.I., Alber M., Hollibaugh J.T. Ascomycete fungal communities associated with early decaying leaves of Spartina spp. from central California estuaries. Oecologia. 2010;162:435–442. doi: 10.1007/s00442-009-1460-4. [DOI] [PubMed] [Google Scholar]
- 16.Newell S.Y., Blum L.K., Crawford R.E., Dai T., Dionne M. Autumnal biomass and potential productivity of salt marsh fungi from 29° to 43° north latitude along the United States Atlantic coast. Appl. Environ. Microbiol. 2000;66:180–185. doi: 10.1128/AEM.66.1.180-185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connolly J.D., Freer A.A., Huneck S. Crystal Structure of (-)-placodiolic acid, a dibenzofuran derivative from the lichen Rhizoplaca chrysoleuca. Phytochem. 1984;23:702. doi: 10.1016/S0031-9422(00)80419-7. [DOI] [Google Scholar]
- 18.Seo C., Sohn J.H., Park S.M., Yim J.H., Lee H.K., Oh H. Usimines A-C, bioactive usnic acid derivatives from the Antarctic lichen Stereocaulon alpinum. J. Nat. Prod. 2008;71:710–712. doi: 10.1021/np070464b. [DOI] [PubMed] [Google Scholar]
- 19.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.