Abstract
Lactobacillus plantarum is a bacterium with probiotic properties and promising applications in the food industry and agriculture. So far, bacteriophages of this bacterium have been moderately addressed. We examined the diversity of five new L. plantarum phages via whole genome shotgun sequencing and in silico protein predictions. Moreover, we looked into their phylogeny and their potential genomic similarities to other complete phage genome records through extensive nucleotide and protein comparisons. These analyses revealed a high degree of similarity among the five phages, which extended to the vast majority of predicted virion-associated proteins. Based on these, we selected one of the phages as a representative and performed transmission electron microscopy and structural protein sequencing tests. Overall, the results suggested that the five phages belong to the family Myoviridae, they have a long genome of 137,973–141,344 bp, a G/C content of 36.3–36.6% that is quite distinct from their host’s, and surprisingly, 7 to 15 tRNAs. Only an average 41/174 of their predicted genes were assigned a function. The comparative analyses unraveled considerable genetic diversity for the five L. plantarum phages in this study. Hence, the new genus “Semelevirus” was proposed, comprising exclusively of the five phages. This novel lineage of Lactobacillus phages provides further insight into the genetic heterogeneity of phages infecting Lactobacillus sp. The five new Lactobacillus phages have potential value for the development of more robust starters through, for example, the selection of mutants insensitive to phage infections. The five phages could also form part of phage cocktails, which producers would apply in different stages of L. plantarum fermentations in order to create a range of organoleptic outputs.
Keywords: Lactobacillus plantarum, phage, new genus, annotation, comparative genomics, phylogenetics, isolation, diversity
1. Introduction
Lactobacillus plantarum is a gram-positive, non-sporeforming, lactic acid bacterium with probiotic qualities. It is commonly encountered in a variety of environments ranging from dairy products, meat, grape must and vegetable fermentations to the human mouth, gastrointestinal tract and stool, as well as sewage and cow dung [1]. Lately, the species has received attention as a promising biocontrol agent in agriculture due to its favourable stimulating effects on crop growth and yield, and its activity against phytopathogenic microbes [2]. From a winemaking perspective, L. plantarum has been proposed and utilised as an alternative malolactic fermentation (MLF) starter [3,4,5]. This is thanks to its wide variety of enzymes linked with aroma compounds, and its production of bacteriocins against spoilage bacteria [3]. Other characteristics of L. plantarum that render it an attractive MLF starter are its ability to endure conditions of low pH, high ethanol, sulfur dioxide, low temperatures, fatty acids, tannins and lysozyme [6]. Lastly, like other lactic acid bacteria (LAB), L. plantarum can both be a traditional preservative of fermented food products [1] and a food spoiler [7].
While fermentation starter strains of L. plantarum and other LAB are of great industrial value, their use is not challenge-free. Bacteriophage (phage) attacks often hamper their activity and by extension cause economic losses because of fermentation failure or poor product quality [8]. Owing to the acute threat they pose to industrially-relevant starters, Lactococcus and Streptococcus phages have been long and extensively studied [9,10,11]. On the other hand, the knowledge gaps on the biology of Lactobacillus phages and their genetic heterogeneity have negatively impacted taxonomic efforts [12,13]. More than 200 published Lactobacillus phages have never been sequenced [13], and of the 56 complete genome sequences just eight are formally classified (NCBI search). Nevertheless, it is clear that research on Lactobacillus phages is being revitalised. In their review in 2017, Murphy et al. [8] documented 27 complete genomes of Lactobacillus phages; a year on, this number has been doubled.
As a subset of Lactobacillus phages, L. plantarum phages have also been affected by the paucity of information and the moderate earlier interest in their host. The first reference on virulent phages infecting L. plantarum dates back to 1969 [14]. Since then, several virulent and temperate phages of L. plantarum have been discovered. A few of these phages were reported to have a high level of strain specificity [15]. Transmission electron microscopy (TEM) images of L. plantarum phages have principally placed them in the family Siphoviridae. The phages fri [16], phiPY1 and phiPY2 [17], phi22-D10 [18], and LP65 [19] are all virulent and are currently the sole representatives of the family Myoviridae, and a single account of a Podoviridae phage [20] is probably inaccurate, according to Villion and Moineau [13]. Confirmed sources of phages against L. plantarum are sewage, feces, silage, kimchi, sauerkraut, fermented coffee, chicha, whey, pear, cucumber, plant materials, salami and meat [13,19,21]. Noteworthy is the latest publication on the temperate phage PM411, as this phage represents the first to be associated with an agriculturally-relevant strain of L. plantarum [21]. In this study, we describe new L. plantarum phages isolated from organic household waste, and we provide further insight into the diversity of phages of Lactobacillus. On a global scale, our aim is to broaden current knowledge on the viral tree of life through a novel group of phages that infect Lactobacillus, a genus of probiotic bacteria with emerging roles in agriculture, as well as in the dairy and wine industries.
2. Materials and Methods
2.1. Bacterial Strains and Culture Media
Two bacterial strains of L. plantarum, strains L1 and MW-1, were used as indicator strains for phage isolation, propagation and characterisation. Both L1 and MW-1 were obtained from private collections and had initially been isolated from a wine fermentation sample and grapes, respectively. Bacterial cultures and phage propagations were performed in De Man, Rogosa and Sharpe (MRS) broth and agar (Difco Laboratories, Detroit, MI, USA). All cultures and propagations were grown overnight at 37 °C without shaking to ensure minimum aeration, unless otherwise stated.
2.2. Environmental Sampling, Isolation, Purification, and Enrichment of Phages
Organic household waste samples (mainly consisting of food-related waste) were collected from two different organic household waste treatment plants (hereafter named treatment plants A and B) in Denmark in February and May 2017 and split into two subsamples. The first part of the samples was centrifuged (5000× g, 15 min, 25 °C) and supernatants were passed through 0.45-μm pore size PVDF syringe filters (Merck Millipore, Darmstadt, Germany). The filtrates were stored at 4 °C until use. The other part was centrifuged (5000× g, 15 min, 25 °C) and phages were precipitated with polyethylene glycol (PEG). Specifically, PEG 8000 (10% w/v) was dissolved in 250 mL of supernatant, stored for 1 h at 4 °C and then centrifuged (10000× g, 10 min, 4 °C). Formed pellets were dried for 15 min, carefully resuspended in 5 mL of SM buffer (0.05 M TRIS, 0.1 M NaCl, 0.008 M MgSO4, pH 7.5) and stored overnight at 4 °C. Following that, they were treated as explained for the first part of samples, which resulted in concentrated filtrates. The presence of bacteriophages in both plain and concentrated filtrates was assessed using a double agar overlay assay [22]. Briefly, 0.1 mL of one indicator strain (106–107 colony-forming units (CFU)/mL) and 0.1 mL of one filtrate at a time were added in MRSφ, i.e., MRS broth supplemented with 0.4% w/v agarose and 10 mM CaCl2. Mixtures were poured on top of MRS agar and incubated overnight at 25 °C. To select for phages specific to L. plantarum, 16 mL of the plain filtrates were blended with 20 mL of 2× MRSφ and supplemented with 4 mL of an overnight culture of either strain L1 or strain MW-1. The mixtures were incubated overnight, and their filtrates tested for phages using the aforementioned overlay assay.
For the purification of phages, all plates deriving from the overlay assay were examined and single plaques were picked. Each plaque was then transferred into 0.7 mL of SM buffer, filtered through 0.45-μm pore size PVDF spin filters (Ciro, FL, USA) and propagated on the same strain. Occasionally, 100 mM glycine were also added to promote plaque formation and visibility [23]. This procedure was repeated twice to ensure purity and phage stocks were stored at 4 °C. Generally, high titers were produced by infecting 0.1 mL of the indicator strains (106–107 CFU/mL) with 0.5 mL of each phage isolate (105–106 plaque-forming units (PFUs)/mL) in 10 mL of MRSφ. In some cases though, phage lysates of high titer were produced as elaborated elsewhere [24] and kept at 4 °C until needed. The filtered phage lysates were further purified by a caesium chloride gradient according to the protocol of Sambrook [25].
2.3. Phage DNA Extraction, Library Preparation, and Sequencing
For the DNA extraction, 0.3 mL of filtered phage lysates ranging from 108–1010 PFUs/mL were used and the extraction was performed following a standard phenol/chloroform protocol [26]. DNA pellets were resuspended in 2 µL of TE buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH 7.5). Libraries were built with the Nextera® XT DNA kit (Illumina Inc., San Diego, CA, USA) and later sequenced on an Illumina MiSeq as a part of the flowcell using the MiSeq v2, 2 × 250 cycle chemistry. The library normalisation, pooling, and sequencing were carried out as described elsewhere [27].
2.4. Assemblies and Annotations
Illumina reads from the DNA sequencing were cleaned using VecScreen (NCBI) and Cutadapt (v. 1.8.3) [28] and assembled with SPAdes (v. 3.5.0) [29] as detailed in [30]. SPAdes assemblies were cross-verified by Unicycler (v. 0.4.3) [31]. Furthermore, assemblies were compared against those generated by CLC Genomic Workbench (v. 9.5.3; CLC bio, Aarhus, Denmark), by first applying the “merge overlapping pairs” tool for overlapping reads and subsequently the “trim sequences” and the “de novo assembly” tools. Phage genomes were auto-annotated by the RAST annotation server v. 2.0 [32] using the RASTtk annotation scheme and GeneMark [33] as a gene caller. Protein functions were ascribed only after manually corroborating RASTtk predictions with BLASTp [34] and HHpred [35], and occasionally with Pfam [36], TMHMM [37] and DELTA-BLAST [38]. The tool tRNAscan-SE (v. 2.0) [39] was used to search for existing tRNA genes. All phage genomes were scanned against ISFinder to identify insertion sequences using BLASTn and the default pipeline of ISFinder [40].
2.5. Transmission Electron Microscopy
The caesium chloride-treated phage lysate was adsorbed to freshly prepared ultra-thin carbon film and treated with 1% (v/v) EM-grade glutaraldehyde (20 min) for fixation. Subsequently, negative staining was done with 2% (w/v) uranyl acetate. The specimen was picked up with 400-mesh copper grids (Plano, Wetzlar, Germany) and analysed using a Tecnai 10 transmission electron microscope (Thermo Fisher, Eindhoven, the Netherlands) at an acceleration voltage of 80 kV. Micrographs were taken with a MegaView G2 CCD-camera (EMSIS, Muenster, Germany).
2.6. Identification of Structural Proteins
To identify the proteins, we followed a procedure described elsewhere [41] with minor modifications. In short, 100 µL of the caesium chloride-treated phage lysate were transferred to Amicon Ultra filter unit (MWCO 30k Da) and centrifuged at 14,000× g for 20 min and further desalted four times with 450 µL water. The filtrate containing the phage particles (10 µL) was denaturised in 25 µL buffer consisting of 6 M urea, 5 mM dithiothreitol and 50 mM Tris-HCl (pH 8). The phage particles were destabilised by five successive freeze-thawing cycles followed by an incubation at 60 °C for an hour to reduce the phage proteins. The proteins were alkylated by adding 25 µL 100 mM iodoacetamide and 150 µL 50 mM ammonium bicarbonate, then incubated for 45 min at room temperature. Phage proteins were digested with 0.8 µg trypsin dissolved in 40 µL 50 mM ammonia bicarbonate and incubated for a day at 37 °C. The protein digest was diluted 1:1 with 0.05% trifluoroacetic acid. The phage peptides were analysed with an Ultimate 3000 RSLCnano UHPLC system hyphenated with a Q Exactive HF mass spectrometer (ThermoFisher Scientific, Denmark). An amount of 21.4 µL of sample was loaded on a pre-concentration trap (a C18 100 µm × 5 mm cartridge) and eluted onto an analytical column (75 µm × 250 mm, 2 µm C18) with a chromatographic triple-phasic 53 min gradient ranging from 1% to 64% mobile phase B (98% acetonitrile and 0.1% formic acid) at 300 nL per minute. The total analysis time was 65 min, and mobile phase A consisted of 2% acetonitrile and 0.1% formic acid. The high-resolution mass spectrometer was operated with positive electrospray ionisation in data-dependent mode by automatically switching between MS and MS/MS fragmentation. Based on a survey MS scan in the Orbitrap, operated at a mass resolution of 120,000 at m/z 200 with a target of 3e6 ions and a maximum injection time at 50 ms, the twelve most intense peptide ions were selected for MS/MS fragmentation in subsequent scans. The selected ions were isolated (in a m/z 1.4 window) and higher-energy collision dissociation was done at a normalised collision energy (28), and fragments recorded in centroid mode at a resolution of 60,000 (m/z 200) with a 250 ms max filling time and target of 1e5 ions. The generated high-resolution data was analysed in a Proteome Discoverer 2.2 (ThermoFisher Scientific) and searched against predicted phage proteins by the Sequest HT algorithm in an iterative processing pipeline. The search criteria were: enzyme, trypsin (full); dynamic modifications, methionine oxidation and acetyl (N-terminus); precursor mass tolerance, 5 ppm; and, fragment mass tolerance, 20 mDa. The processed data were filtered in a Proteome Discoverer consensus workflow with the Peptide Validator algorithm (q-value < 0.01) to ensure the peptide-spectrum match had a false discovery rate under 1%.
2.7. Comparative Genomics and Phylogenetics to Distant Relatives
In accordance with the International Committee on Taxonomy of Viruses (ICTV) [42], each phage sequence was initially compared to the viral nucleotide collection database (taxid:10239; nt/nr) using the BLASTn algorithm. Phage records that showed some degree of similarity with the new phage isolates of this study were selected and included in an all-against-all, quantitative DNA comparison with Gegenees (v. 2.1) [43]. The Gegenees analysis generally relies on the division of a full genome (query) into fragments and the search for BLAST “seeds” of each fragment against another genome (reference) [43]. The final phylogenomic distance of a query from a reference is the average value of all fragments’ BLASTn scores but expressed as a percentage of the score each fragment would yield towards itself (at 100% identity). In this study, the Gegenees analysis was performed with a customised setting of 50/25, fragment size/sliding step size, and an “accurate” threshold of 0%. In addition, two phylogenetic trees, one for the major capsid protein and one for the large subunit terminase, were constructed using the default pipeline of “One Click mode” (http://phylogeny.Lirmm.fr/) [44]. The tree leaves comprised the phages of this study and selected phage records from the Gegenees comparisons, which had earlier yielded average nucleotide similarity of at least 0.05 or higher with Gegenees. Homology between and within proteomes was assessed via a BLASTp comparison of the new phages and their closest phage relative using the CMG-biotools system [45]. In this type of analysis, a pair of amino acid sequences is considered homologous (paralogous or orthologous) if the length of the alignment is at least 50% of the longest sequence and the identity of the alignment is at least 50%. Paralogue hits were determined using MAFFT [46] (v. 7.388) with the following arguments: algorithm FFT-NS-i x1000; scoring matrix 200PAM/k = 2; gap open penalty 1.53; offset value 0.123, and then BLASTp. Finally, the genomes of the new phages and the two most related phage genomes were examined for common synteny with Easyfig by tandemly comparing phage pairs with BLASTn [47].
2.8. Phage Genomic Data Availability
Assembled and annotated genomes of Semele, Bacchae, Iacchus, Dionysus and Bromius have been uploaded to GenBank under accession numbers MG765279, MG765277, MH809529, MH809530 and MH809531, respectively.
3. Results and Discussion
3.1. Isolation of Phages and Basic Features
Each one of the phages Semele, Bacchae, Iacchus, Dionysus and Bromius was isolated from a different in vitro treatment of the original organic household waste samples (treatment details in Section 2.2.). Phage Bacchae was isolated from the sample of treatment plant A, and Semele, Iacchus, Dionysus and Bromius came from the sample collected from treatment plant B. All five isolates formed clear plaques with a diameter of approximately 0.5 mm on their respective host strains, L1 and MW-1 (see Figure S6 for an example of a plate with plaques). In particular, phages Semele and Bacchae were isolated from an infected lawn of strain L1, while phages Iacchus, Dionysus and Bromius were isolated from an infected lawn of strain MW-1.
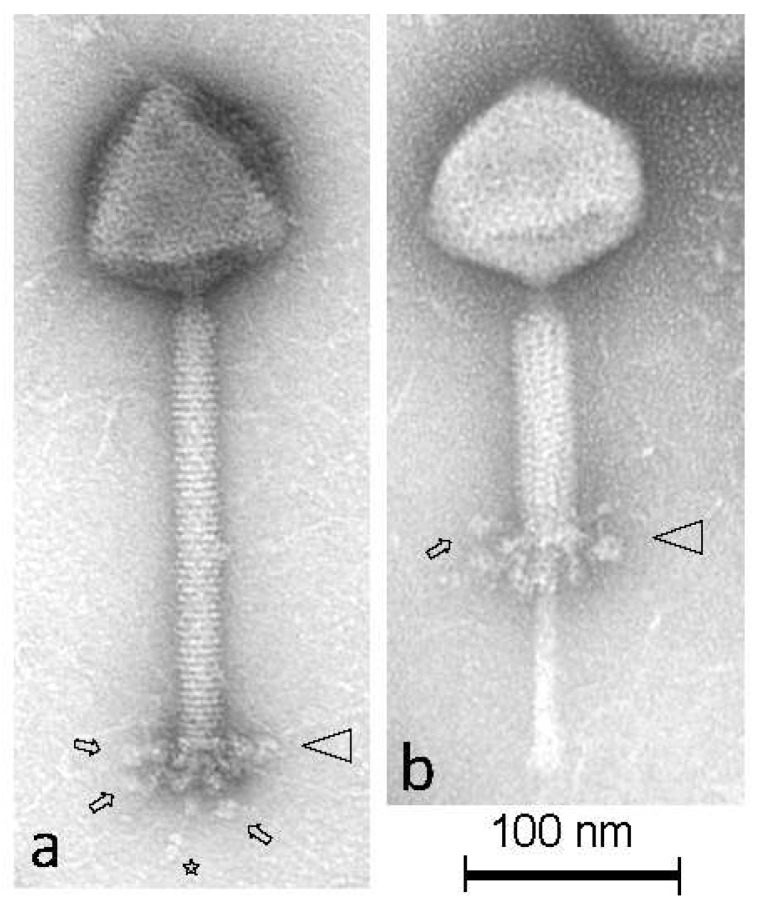
Transmission electron microscopy imaging analysis was carried out with a representative phage, i.e., phage Dionysus (Figure 1, Table 1). A representative phage was chosen due to the five phages being quite unstable and difficult to propagate at high titers. Nevertheless, Dionysus can sufficiently represent the studied phages because the vast majority of morphogenesis genes of the five phages were proven to be highly homologous (please read Section 3.3. and Spreadsheet S1 for comparisons with the virions of phages Semele and Bacchae). The only obvious exception was one gene predicted to encode a tail protein in all five phages (AUV60228 of Semele, AYH92260 of Bromius, AUV59956 of Bacchae, AYH92087 of Dionysus and AYH91916 of Iacchus). Dionysus revealed a remarkably high stability, as only a low number of phage particles with contracted tail sheaths were detected (Figure 1b). The phage has a large isometric capsid (diameter: 93 nm) with characteristic (“blackberry-like”) surface decorations. At the distal end of the (contractible) tail (length 222 nm), a complex baseplate structure is visible, and unique flexible appendages (length ca. 22 nm) with terminal globular structures (ca. 9 nm in diameter) are attached under the baseplate. A flexible thin tail fiber (length: 30 nm) terminating with approximately three distinct knob-like structures protrudes under the baseplate. These morphological characteristics classify phage Dionysus into the order Caudovirales and the family Myoviridae. This is in agreement with existing records of phages infecting L. plantarum, all of which fall into the order Caudovirales, while five of those belong to the family Myoviridae [13].
Figure 1.
Transmission electron micrographs of L. plantarum phage Dionysus negatively stained with 2% (w/v) uranyl acetate. Triangles and arrows indicate the terminal baseplate structure and representative flexible appendages with terminal globular structures, respectively, attached underneath them. The single distal tail fibers terminating with three distinct knob-like structures are indicated by the star symbol. Phage Dionysus is shown with extended tail sheath (a) and contracted tail sheath (b), substantiating that it is a Myoviridae phage.
Table 1.
Dimensions of phage Dionysus as measured with TEM.
| Structural Components | Dimensions (nm) | Counted Phage Particles |
|---|---|---|
| Head Diameter | 92.7 ± 3.5 | 17 |
| Tail Length (with Baseplate) | 222.4 ± 9.0 | 17 |
| Tail Width | 20.9 ± 0.6 | 17 |
| Baseplate Width | 44.1 ± 3.9 | 16 |
| Baseplate Length | 25.0 ± 2.4 | 16 |
| Baseplate Appendages Length | 21.8 ± 1.8 | 10 |
| Globular Structure Diameter | 9.2 ± 1.3 | 12 |
| Tail Fiber Length | 30.4 ± 2.2 | 13 |
All five genomes of this study were assembled into a single major contig of size range 137,973–141,344 bp, G/C content range 36.3–36.6%, and with average coverage range 77.1–600.6× (Table 2). The noted genome size is close to the longest observed genome for L. plantarum phages (145,162 bp, phage LpeD, accession number MF787246.1).
Table 2.
Basic genomic characteristics of the five Lactobacillus phage isolates.
| Phage Isolate | Open Reading Frames with Assigned Function | Genome Size (bp) | G/C Content (%) | tRNA Genes |
|---|---|---|---|---|
| Semele | 40/177 | 139,450 | 36.3 | 11 |
| Bacchae | 38/180 | 141,124 | 36.3 | 15 |
| Iacchus | 44/170 | 137,973 | 36.5 | 7 |
| Dionysus | 42/172 | 141,344 | 36.6 | 7 |
| Bromius | 39/172 | 140,527 | 36.5 | 7 |
3.2. Analyses of DNA Sequences and Protein Predictions
Annotations of Semele, Bacchae, Iacchus, Dionysus and Bromius genomes assigned functions to 40 of the 177, 38 of the 180, 44 of the 170, 42 of the 172, and 39 of the 172 open reading frames (ORFs), respectively. Of the total number of ORFs, one-third is harboured by the antisense strand. These genomes were also shown to encode clusters of 11, 15, 7, 7, and 7 tRNA genes, accordingly. Additional analyses presented in Section 3.3 suggest that these phage isolates are new species and could be members of a new Lactobacillus phage genus. Below, we discuss the different categories of proteins produced by these five phage genomes.
3.2.1. Transcription and Translation Takeover
A module of 7–15 tRNA-encoding genes, often accompanied by between one and three repeat regions, points to a potential mechanism of host transcription and translation takeover in all five Lactobacillus phage genomes. These tRNAs are predicted to carry 7–11 different amino acids, with a common core of seven amino acids, with the first two (Gly, Thr) and the final (Arg) maintaining their position across modules (please see Figures S1–S5 for details on the order of tRNAs and their frequency). Specifically, Semele’s tRNAs carry the amino acids Gly, Thr, Trp, Met, Phe, Leu, Asn, Ile, Tyr, and Arg; Bacchae’s tRNAs deliver the amino acids Gly, Thr, Trp, Met, Phe, Leu, Gln, Ser, Asn, Ile, and Arg; Iacchus’, Dionysus’ and Bromius’ tRNAs deliver the amino acids Gly, Thr, Leu, Asn, Ile, Phe, and Arg. Interestingly, similar observations were made for the genome of the distantly related phage LP65 of L. plantarum, wherein 14 tRNAs were identified. In particular, the equivalent LP65 module has the same core of tRNA genes and shares the same first two and one last amino acids. Additionally, all five phages in this study possess two additional transcription-related elements, an RNA ligase and an RNA polymerase sigma factor. The RNA ligase is homologous to the T4 RNA ligase by the gene mlA (Pfam domain search). This enzyme has a double activity in T4. It participates in the repair of tRNAs to reverse the damage inflicted by host defense enzymes and it catalyses the attachment of tail fibers [48]. An RNA polymerase sigma factor has been described in phages before. In T4, the phage-encoded sigma factor serves the binding of the RNA polymerase to promoters different from the ones recognized by the host sigma factor [49]. In other words, the two sigma factors direct the transcription of the phage DNA independently and are complementary to each other. A hypothetical connection between the T4 sigma factor and the phage’s DNA packaging has also been described [50].
3.2.2. DNA Metabolism, Replication, Recombination, and Repair
The five phage genomes seem to have a substantial DNA replication, recombination, and repair system, which is fundamentally shared among them (Figures S1–S5). In confirmation of earlier observations, the genes of this system are arranged in modules [48] and are situated adjacent to DNA metabolism genes [48]. The replisome genes encode a DNA primase, a DNA polymerase, one or two DNA helicases, one or two DNA-binding proteins of unknown function and a recombinase A, not to mention putative recombination or repair-related proteins. Not least, a nucleotide precursor complex suggests that these phages may manipulate the nucleotide pool of their hosts and regulate DNA biosynthesis independently. Such a mechanism would be crucial given that their G/C content (Table 2) differs considerably from their host’s (~44.5%) [48]. Indeed, the predicted phosphatase enzymes phosphatase/phosphodiesterase and deoxynucleoside kinase could allow for the dephosphorylation [16] and phosphorylation [17] of nucleotides, accordingly. Moreover, a gene associated with the nucleoside deoxyribosyltransferase of Lactobacillus phage ATCC 8014-B2 could catalyse the transfer between purines and/or pyrimidines, just as found for many lactobacilli, and thus contribute to nucleotide recycling [51]. The array of genes for a nicotinamide riboside transporter, a nicotinamide-nucleotide adenylyltransferase and ADP-ribose pyrophosphatase may be involved in a potential pyridine nucleotide (NAD+) salvage pathway. In vitro tests to support an analogous observation have recently been performed for a Vibrio bacteriophage [52]. A gene producing a nicotinamide mononucleotide transporter, present in all the phages studied here, may be another unit of this pathway. At least 35 phages of different bacterial species have a nicotinamide mononucleotide transporter, including L. plantarum phages LpeD and LP65, but its exact functionality has not been verified [53]. The only information available suggests that this transporter can pass nicotinamide mononucleotide molecules across membranes and that it is similar to its bacterial counterpart produced by L. plantarum [53]. The latter hints at a recent gene transfer between host and phages [53]. The fact that the five phages and their host share a gene, and the striking difference in G/C content among the five phages and L. plantarum may mean that these phages have a host range beyond L. plantarum. Hosts other than their primary host have been demonstrated for L. plantarum phages before [17,18,19].
3.2.3. Self-Splicing/Selfish Genetic Elements
Self-splicing/selfish genetic elements are well-represented and interspersed throughout the genomes of the five phages (Figures S1–S5). One example is an unknown mobile genetic element protein, conserved in four of the five phages of this study and typically placed next to and upstream of the tRNA module. Another instance is that of transposases. Two transposases are observed in the genomes of Dionysus, Semele and Iacchus, and their neighbouring putative AAA ATPase probably controls their transposition to target DNAs [54]. None of these transposases returned any significant similarity to entries of inserted sequence elements in ISFinder [40]. Since lysogeny is widespread in lactobacilli [55], transposable genetic elements from the chromosome could partially drive the evolution of virulent Lactobacillus phages from temperate ones, as reported for Lactobacillus phage φFSW [9]. However, a BLASTn search against the bacterial database did not return any similarity of Dionysus, Semele or Iacchus to prophage records. In the genomes of Semele and Iacchus, a protein of unknown function splits the DNA polymerase gene into two fragments. This protein cannot be a genetic switch since it has the same direction as the DNA polymerase, but it may either be part of a self-splicing group I intron or a mini-intein that could regulate the expression of the DNA polymerase. Group I introns are ribozymes capable of self-splicing from primary transcripts [56]. They interrupt tRNA, rRNA, and protein-coding genes and sometimes contain their own homing endonucleases (intron-homing) [57]. Mini-inteins are protein splicing elements that can ligate the polypeptide produced by the gene they disrupt post-translationally [58]. Their main difference from large inteins is the absence of a homing endonuclease domain, which enables large inteins to catalyse their self-excision [59]. HNH homing endonuclease genes are common in phage genomes [60] and are sometimes expressed to exclude other competing phages [61]. In the genomes of all five phages some of the HNH homing endonucleases reside next to hypothetical proteins, which may hint at the existence of spliced genes. Group I introns and inteins have already been discovered in phages, with the closest examples those of Staphylococcus, Bacillus and Lactococcus phages, and cases of both splicing DNA polymerase genes have been described there [62,63]. Regarding Lactobacillus phages, JCL1032 of Lactobacillus delbrueckii is the only one proven to have group I introns so far [64]. On the other hand, many Lactobacillus phages have genes for HNH endonucleases, with phage P2 predicted to have as many as eight in its genome (accession number KY381600.1) and phage LP65 of L. plantarum predicted to have two (accession number NC_006565.1).
3.2.4. Morphogenesis, DNA Packaging, and Membrane Transport
Based on in silico analyses, the proteins that form the virion of the five Lactobacillus phages encompass a portal protein, a major capsid protein, a tail sheath protein, three unidentified tail proteins, and two baseplate proteins. The small and large subunit of a phage terminase are key enzymes of the DNA translocation and head filling [65]. A terminase DNA packaging subunit, characterised as the large subunit terminase, has been used to define the start of the five genomes here. These assigned functions were experimentally investigated by LC-MS/MS analysis on phage Dionysus and four virion-associated proteins were detected (Table 3). Moreover, twelve more non-virion-associated proteins of Dionysus were verified (Spreadsheet S2) due to the sensitivity of the proteomic analysis. Regarding the virion-associated proteins, in silico predictions were corroborated for the major capsid protein (peg. 170) and the portal protein (peg. 2). The other two proteins were a hypothetical protein in the vicinity of a baseplate protein (peg. 151) and a hydrolase (peg. 5) that could potentially be a virion-associated peptidoglycan hydrolase. However, the hypothetical protein and the hydrolase had low protein false discovery rate confidence (FDR < 1%), thus the uncertainty of these protein discoveries is high.
Table 3.
Mass spectrometry data for virion-associated proteins of phage Dionysus.
| Description | FDR | Coverage (%) | No of Peptides | Molecular Mass (kDa) | Gene |
|---|---|---|---|---|---|
| Major capsid protein | High | 2 | 1 | 53.5 | peg. 170 |
| Putative portal protein | High | 2 | 1 | 60.9 | peg. 2 |
| Hypothetical protein | Low | 3 | 1 | 101.9 | peg. 151 |
| Hydrolase | Low | 5 | 1 | 35.9 | peg. 5 |
3.2.5. Cell Wall and Membrane Degradation
A phage lytic cycle ends with the burst of the host cell and the release of progeny phages. The burst is accomplished due to the action of two groups of enzymes, holins and (endo)lysins. Holins are responsible for the formation of transmembrane holes, which allow lysins to reach and degrade the cell wall [66]. Out of the five different classes of lysins, the Lactobacillus phages studied thus far utilise only two, muramidases and amidases [67]. Holins usually have two or three transmembrane domains, although a few cases of holins with just one transmembrane domain have also been found [68]. For Lactobacillus phages there are examples of two- or three-transmembrane domain holins [69,70]. A TMHMM search coupled with DELTA-BLAST attributed traits typical of a holin to a hypothetical protein in all five phages (protein accession numbers: AYH92097, AUV60238, AUV59966, AYH91927, AYH92270). This hypothetical protein is right upstream of the lysin gene and was predicted to have a pair of transmembrane domains and low similarity to a holin of Pediococcus pentosaceus. All five phage genomes in this study possess at least one lysin. At the same time, the absence of lysogeny-related genes (integrases, excisionases and repressors of phage reproduction and lysis genes) indicates that these phages are most likely purely virulent [71]. However, since less than 24% of the genes in the five phages could be ascribed a function, and since some genes encoding hypothetical proteins were located at the antisense strand, it is possible that lysogeny-related genes remain undetected. New infections are initiated by the recognition and irreversible attachment of progeny phages to host bacterial receptors. The genomes of Bromius, Dionysus and Iacchus feature a glycerophosphodiester phosphodiesterase that has been described as a baseplate component of various bacteriophages against gram-positive bacteria, including L. delbrueckii phages [72]. Notably, this enzyme seems to facilitate the attachment of phage Ld17 of L. delbrueckii by degrading glycerophosphodiesters of the cell envelope [73]. Owing to glycerophosphodiester phosphodiesterases and other degrading proteins, receptor-binding proteins can access bacterial receptors masked by surface molecules and catalyse phage adsorption [74]. Peptidoglycan-degrading enzymes of the phage tail tip are indispensable for phage DNA ejection which occurs after adsorption [73]. We presume that an annotated tail protein occasionally predicted as putative peptidoglycan hydrolase, which lies near a tail protein in the genomes of the five phages (AYH91914 of Iacchus, AYH92085 of Dionysus, AUV59954 of Bacchae, AYH92258 of Bromius, AUV60225 of Semele), is participating in this process [75].
3.2.6. Other Predicted Proteins
In the region close to the DNA metabolism, replication, recombination, and repair genes, three coding sequences have been assigned a function but their exact purpose in the phage genome is unclear. One of them encodes a protein homologous with a DNA-binding ferritin-like protein from Lactococcus lactis, which provides protection against oxidative damage in bacteria [76]. The aerobic metabolism of L. plantarum leads to the production of H2O2 that the bacterium detoxifies via a manganese catalase (pseudo-catalase) [77]. Free Fe can interfere with the manganese complexes that are used by L. plantarum’s catalase to lower reactive oxygen levels [78]. Thus, the DNA-binding ferritin-like protein could help L. plantarum tolerate the hydrogen peroxide by binding to and storing excess free Fe that would otherwise damage the cell [79]. The second coding sequence encodes a protein that has homology with a low temperature requirement C protein of Listeria monocytogenes, which may be involved in lipid metabolism [80]. The protein of the third coding sequence is homologous with a YopX protein from Bacillus subtilis, an uncharacterised protein of bacteriophages and prophages of gram-positive bacteria [81]. This latter is solely produced by phages Dionysus and Iacchus.
3.3. The Diversity of the Five Phages Supports the Introduction of a New Lactobacillus Phage Genus
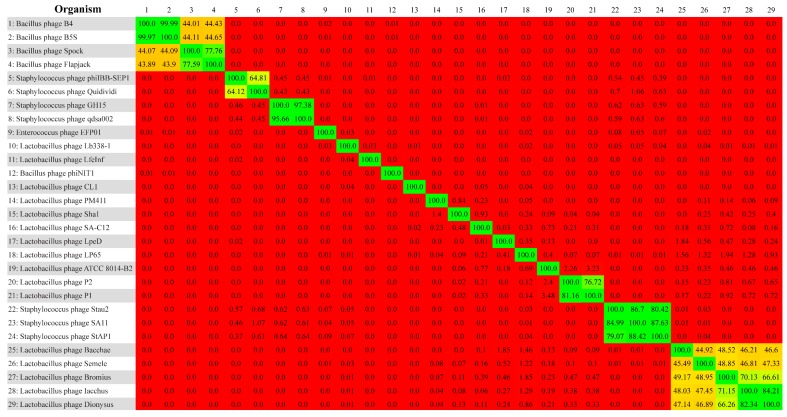
According to BLASTn, all five phages of this study show a low degree of similarity to Lactobacillus phage LpeD (70% identity over 51–55% query cover) and an even lower similarity to Lactobacillus phage LP65 (83–88% identity over 20–23% query cover). The criterion considered here as a first-hand indicator of high nucleotide similarity between phage genomes is the one recommended by ICTV. Namely, this criterion sets a threshold of >50% sequence similarity when multiplying the BLASTn query cover by identity [42]. Thereon, 24 phage records that returned some degree of similarity, as well as the five new phages of this study, were subjected to an all-against-all BLASTn comparison by Gegenees. In the resulting heatplot of phylogenomic data (Figure 2), the phages presented in this study fall into one group that is separate from the other 24 phages. The calculated score distances within this group, which are equal to the average normalised similarities of fragments by BLAStn, were 44.92% or higher at a nucleotide level.
Figure 2.
BLASTn heatplot generated using Gegenees. All-against-all comparisons were run with fragment length 50 bp, step size 25 bp, and threshold 0%. The studied phages (25–29) form a separate group.
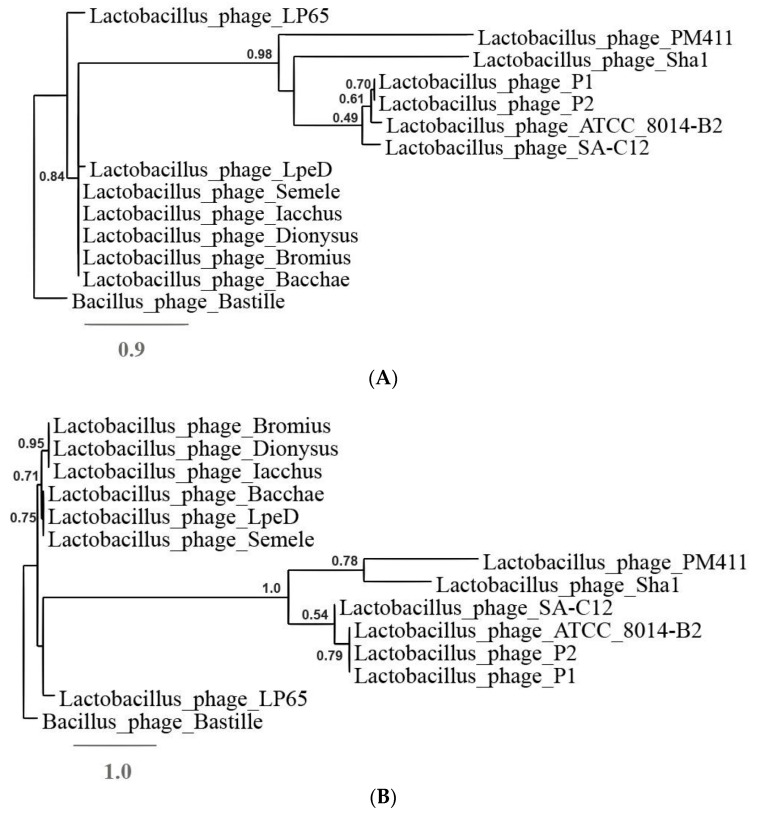
The Gegenees distinction is consistent with the outcome of the major capsid protein and large subunit terminase trees (Figure 3A,B). In both phylogenetic trees phages Semele, Bacchae, Iacchus, Dionysus and Bromius clearly cluster together and in the same clade with phage LpeD. In order to clarify if LpeD pertains to the same genus as the five new phages, we have chosen to include it in the remaining comparisons.
Figure 3.
Phylogenetic trees for phages Semele, Bacchae, Dionysus, Iacchus and Bromius and other Lactobacillus phages yielding average similarity of at least 0.05 or higher with Gegenees. (A) Tree constructed using the major capsid proteins, (B) tree constructed using the large subunit terminases. The amino acid sequences were compared with the “One Click mode” (http://phylogeny.Lirmm.fr/). Bacillus phage Bastille proteins were used as outliers.
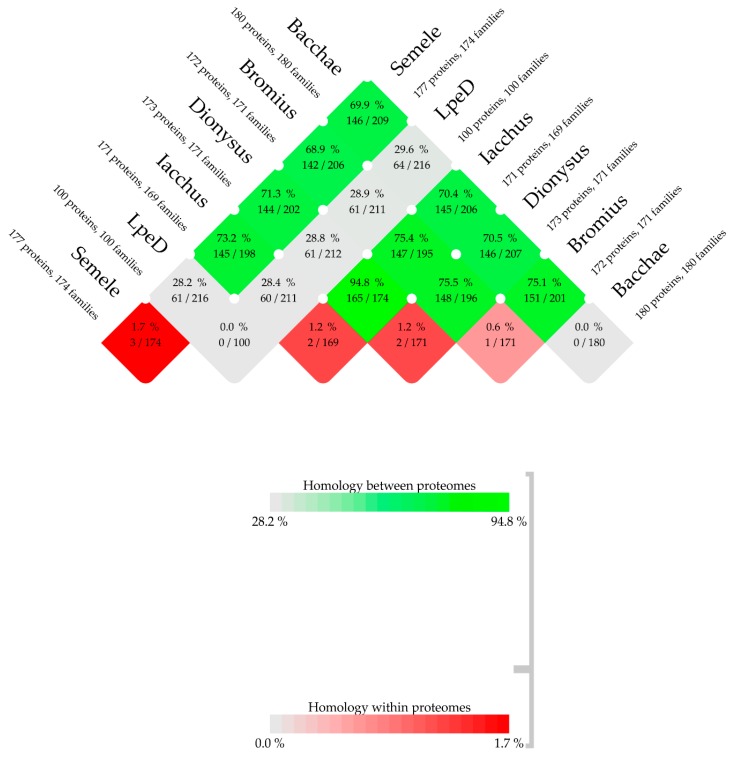
Total protein checks undertaken with CMG-biotools system revealed significant homology among the proteomes of the five phages, while the homology between each of the five phages and phage LpeD was consistently less than 30% (Figure 4). Furthermore, homology within proteomes of Semele, Iacchus, Dionysus and Bromius was traced to the family of transposases and one or two other hypothetical protein families using MAFFT and BLASTp (Spreadsheets S3–S6). This is surprising, given that duplicates (paralogues) in the same genome are rather rare for double-stranded DNA phages [82]. Transposases are motors of genome plasticity and adaptation [83]. Phages of L. plantarum are parasites of a versatile bacterium that can grow and survive in a raft of environmental niches. Hence, the abundance of transposases may assist phages Semele, Iacchus and Dionysus, which have two paralogous transposases, to adapt to novel niches and hosts by facilitating horizontal gene transfer with their hosts upon infection [84,85]. Moreover, the over-representation of transposases in the genomes of all five phages could reflect an abundance of transposases in the genome of host bacteria [86]. This scenario is consistent with the nature of the organic household waste samples, where a plethora of nutrients could translate to higher microbial densities, and thus higher rates of DNA exchange, as reported for bacteria from the Baltic Sea [60]. We investigated whether the transposases of the five new phages have a bacterial version by reviewing the BLASTp results. It turned out that all examined transposases showed significant homology with more than 20 different Lactobacillus sp., but unexpectedly not with their immediate host, L. plantarum.
Figure 4.
BLASTp comparison between and within proteomes of the five phages and their closest phage relative LpeD using CMG-biotools system. Phage pairs with >50% proteome homology are shown in green and those with <50% are depicted in grey. Red signifies the presence of one or more groups of paralogous proteins within a phage proteome.
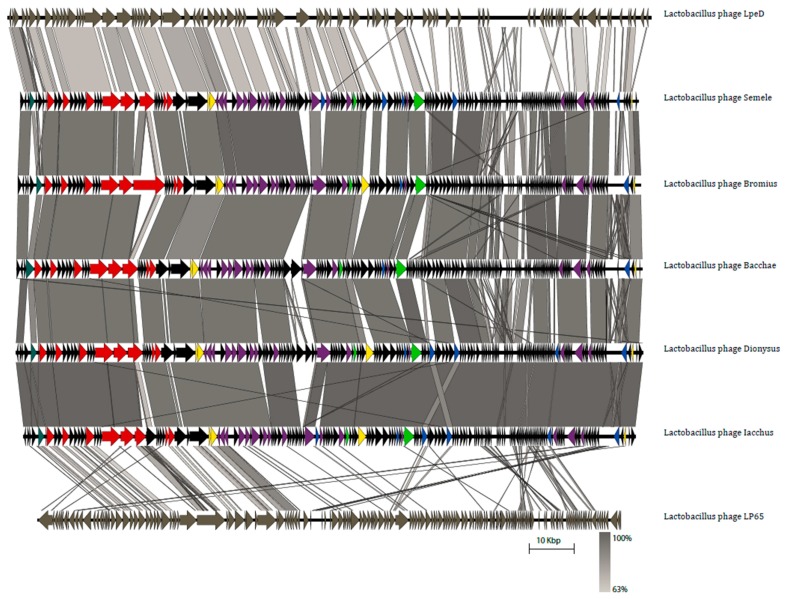
Finally, yet importantly, Easyfig comparisons revealed a striking conservation of gene order and nucleotide homology amongst the five new phages. When these were compared to phage LpeD, we could still observe gene order conservation to a large extent, but the nucleotide homology was generally low (top genome; Figure 5). Concerning LP65, the genome of this phage displayed relatively low synteny and nucleotide homology against the five phage genomes described here (bottom genome; Figure 5). Given the distinctive characteristics of these five phage isolates, we propose a new Lactobacillus phage genus, that we term “Semelevirus”. The genus “Semelevirus” has as members the phages Semele (founder), Bacchae, Iacchus, Dionysus and Bromius.
Figure 5.
BLASTn comparisons of the genomes of the five phages with distant relatives LpeD and LP65 using Easyfig. Arrows represent the locations of genes and lines represent the level of homology between each tandemly placed group of phages. In the genomes of the five phages, genes are coloured according to the assigned function of the protein each gene encodes. These are: turquoise for DNA packaging, yellow for cell wall and membrane degradation, green for transcription and translation takeover, and blue for selfish genetic elements. Genes in red are part of the morphogenesis module and genes in deep purple are part of the DNA metabolism, replication, recombination and repair module. Black colour denotes genes that encode either hypothetical proteins or other predicted proteins with unclear functions. Figures S1–S5 follow the same colour scheme.
4. Outlook
Lactobacillus phages have been only moderately addressed in comparison to other LAB phages. However, the doubling of publicly available, complete-genome sequences from 2017–2018 could imply a boom in this field of research. The many advantages displayed by their hosts, and especially by L. plantarum, may justify the fueled interest in these phages. There are quite some arguments to support the continuation of this trend in the coming years. Lactobacillus phages, as with other notorious LAB phages, can preclude obtaining high quality fermentation products on a regular basis. Hence, a systematic analysis of existing and newly-discovered Lactobacillus phage genomes would guarantee a more thorough understanding of their origin, evolution, and relationships with other phages [8]. Eventually, better knowledge on the phages of Lactobacillus could help to choose starter strains, and adjunct or aroma cultures with different phage sensitivity for strain rotation, and develop more efficient phage cocktails for inhibition of unwanted bacterial growth. At the same time, we believe that improved classification schemes are crucial to aid phage applications or to design efficient control strategies against phages, for example, by selecting mutants insensitive to infections of a range of phages. Should we expect an increasing number of Lactobacillus phages in the future, this need is even more urgent.
Looking at L. plantarum phages, we can discern some cases where steady accumulation of knowledge is essential. As mentioned, L. plantarum is an emerging biocontrol agent, as well as biostimulation agent of crops. Even if phages can impede successful application of this bacterium in the field they can still be sufficiently anticipated and tackled when we know enough about their biological profile. The same is true for L. plantarum starters used in the food industry. On the other hand, L. plantarum phages could be valuable biocontrol tools for the food industry [87]. First, they could be used to prevent growth of their host in those biotechnological processes in which L. plantarum is undesirable, such as in meat, beer, wine, or orange juice [7]. For example, phage cocktail preparations could minimize the addition of sulfur dioxide in those types of alcoholic beverages that should not undergo MLF. Studies on phages of beer spoiler strains of Lactobacillus brevis, Lactobacillus paraplantarum and Pediococcus damnosus have already introduced the concept of adding phages to inhibit these bacteria [88,89], [90]. Secondly, phage cocktails could offer a means to manipulate the MLF in wine, cider, and sour beer. Winemakers would have the chance to experiment by adding phages in aliquots of the same initial product but at different stages of the MLF and later evaluating the effects in each of the final wine products. Such an intervention may give rise to wines with novel characteristics, since through bacterial lysis phages can induce the release of enzymes for flavour development [91,92].
Acknowledgments
The authors would like to acknowledge all members of the MicroWine consortium, some for fruitful discussions and others for contributing to the collection of environmental samples. We would also like to thank the companies BioVækst (treatment plant A) and HCS (treatment plant B) for providing the organic waste samples, and Angela Back from the Max Rubner-Institut for technical support in the TEM analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/11/7/611/s1, Figures S1–S5, [93]: Genome Maps of Phages Semele, Iacchus, Dionysus, Bromius and Bacchae (respectively) and Visualization of All Encoded Proteins with Assigned Functions; Figure S6: Example of a Plate with Plaques Produced by Phage Dionysus on L. plantarum MW-1; Spreadsheet S1: TEM Analysis Results (Virion Micrographs And Dimensions) for Phages Semele and Bacchae; Spreadsheet S2: Mass Spectrometry Data for Non-Virion-Associated Proteins of Phage Dionysus; Spreadsheets S3–S6: MAFFT Results of Pairwise Protein Comparisons Within the Proteomes of Phages Semele, Dionysus, Iacchus and Bromius (respectively) to Determine Paralogous Proteins.
Author Contributions
Conceptualization, I.K., A.B.C. and L.H.H.; Methodology; I.K., A.B.C., M.H. and W.K.; Formal analysis, I.K., A.Z. and H.N.; Investigation, I.K., A.B.C., A.M.D. and H.N.; Validation, I.K., A.B.C., L.E.-J., W.K., A.Z., M.H., H.N. and L.H.H.; Writing-original draft preparation, I.K.; Review and editing, A.B.C., W.K., L.E.-J. and L.H.H.; Visualization, I.K., A.Z. and H.N.; Supervision and project administration, L.E.-J. and L.H.H.; Funding acquisition, L.H.H.
Funding
This research was funded by the Horizon 2020 Program of the European Commission within the Marie Skłodowska-Curie Innovative Training Network “MicroWine” (grant number 643063), the Human Frontier Science Program (Grant HFSP - RGP0024/2018), and the Danish Research Council for Technology and Production project “Phytoprotect” (grant number DFF – 4184-00070B).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.König H., Fröhlich J. Lactic Acid Bacteria. In: König H., Unden G., Fröhlich J., editors. Biology of Microorganisms on Grapes, in Must and in Wine. Springer International Publishing; Cham, Switzerland: 2017. pp. 3–41. [Google Scholar]
- 2.Lamont J.R., Wilkins O., Bywater-Ekegärd M., Smith D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 2017;111:1–9. doi: 10.1016/j.soilbio.2017.03.015. [DOI] [Google Scholar]
- 3.Du Toit M., Engelbrecht L., Lerm E., Krieger-Weber S. Lactobacillus: The Next Generation of Malolactic Fermentation Starter Cultures—An Overview. Food Bioprocess Technol. 2011;4:876–906. doi: 10.1007/s11947-010-0448-8. [DOI] [Google Scholar]
- 4.Lerm E., Engelbrecht L., Du Toit M. Selection and characterisation of Oenococcus oeni and Lactobacillus plantarum South African wine isolates for use as malolactic fermentation starter cultures. S. Afr. J. Enol. Vitic. 2011;32:280–295. doi: 10.21548/32-2-1388. [DOI] [Google Scholar]
- 5.López I., López R., Santamaría P., Torres C., Ruiz-Larrea F. Performance of malolactic fermentation by inoculation of selected Lactobacillus plantarum and Oenococcus oeni strains isolated from Rioja red wines. Vitis. 2008;47:123–129. [Google Scholar]
- 6.Du Toit M. Novel lactic acid bacteria for use as MLF starter cultures. Acenología Enoreports. 2012;130:1–6. [Google Scholar]
- 7.De Vries M.C., Vaughan E.E., Kleerebezem M., de Vos W.M. Lactobacillus plantarum—Survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 2006;16:1018–1028. doi: 10.1016/j.idairyj.2005.09.003. [DOI] [Google Scholar]
- 8.Samson J.E., Moineau S. Bacteriophages in Food Fermentations: New Frontiers in a Continuous Arms Race. Annu. Rev. Food Sci. Technol. 2013;4:347–368. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- 9.Sechaud L., Cluzel P.-J., Rousseau M., Baumgartner A., Accolas J.-P. Bacteriophages of lactobacilli. Biochimie. 1988;70:401–410. doi: 10.1016/0300-9084(88)90214-3. [DOI] [PubMed] [Google Scholar]
- 10.Martínez B., García P., Gonzalez A.R., Piuri M., Raya R.R. Bacteriophages of Lactic Acid Bacteria and Biotechnological Tools. In: Mozzi F., Raya R.R., Vignolo G.M., editors. Biotechnology of Lactic Acid Bacteria: Novel Applications. John Wiley & Sons, Ltd.; Chichester, UK: 2015. pp. 100–119. [Google Scholar]
- 11.Murphy J., Mahony J., Fitzgerald G.F., van Sinderen D. Bacteriophages infecting lactic acid bacteria. In: Fox P.F., McSweeney P.L.H., Cogan T.M., Guinee T.P., editors. Cheese. Academic Press; San Diego, CA, USA: 2017. pp. 249–272. [Google Scholar]
- 12.Mahony J., van Sinderen D. Current taxonomy of phages infecting lactic acid bacteria. Front. Microbiol. 2014;5:7. doi: 10.3389/fmicb.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villion M., Moineau S. Bacteriophages of Lactobacillus. Front. Biosci. 2009;14:1661–1683. doi: 10.2741/3332. [DOI] [PubMed] [Google Scholar]
- 14.Sakurai T., Takahashi T., Kamiyama K., Arai H. Isolation of bacteriophages parasitic on Lactobacillus casei and L. plantarum and their several properties. Virus. 1969;19:311–324. doi: 10.2222/jsv.19.311. [DOI] [Google Scholar]
- 15.Lu Z., Breidt F., Fleming H.P., Altermann E., Klaenhammer T.R. Isolation and characterization of a Lactobacillus plantarum bacteriophage, φJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003;84:225–235. doi: 10.1016/S0168-1605(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 16.Trevors K.E., Holley R.A., Kempton A.G. Isolation and characterization of a Lactobacillus plantarum bacteriophage isolated from a meat starter culture. J. Appl. Bacteriol. 1983;54:281–288. doi: 10.1111/j.1365-2672.1983.tb02618.x. [DOI] [Google Scholar]
- 17.Doi K., Zhang Y., Nishizaki Y., Umeda A., Ohmomo S., Ogata S. A comparative study and phage typing of silage-making Lactobacillus bacteriophages. J. Biosci. Bioeng. 2003;95:518–525. doi: 10.1016/S1389-1723(03)80054-3. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z., Breidt F., Plengvidhya V., Fleming H.P. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 2003;69:3192–3202. doi: 10.1128/AEM.69.6.3192-3202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chibani-Chennoufi S., Dillmann M.-L., Marvin-Guy L., Rami-Shojaei S., Brüssow H. Lactobacillus plantarum bacteriophage LP65: A new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 2004;186:7069–7083. doi: 10.1128/JB.186.21.7069-7083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon S.S., Barrangou-Poueys R., Breidt F., Klaenhammer T.R., Fleming H.P. Isolation and Characterization of Bacteriophages from Fermenting Sauerkraut. Appl. Environ. Microbiol. 2002;68:973–976. doi: 10.1128/AEM.68.2.973-976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daranas N., Bonaterra A., Francés J., Cabrefiga J., Montesinos E., Badosa E. Monitoring Viable Cells of the Biological Control Agent Lactobacillus plantarum PM411 in Aerial Plant Surfaces by Means of a Strain-Specific Viability Quantitative PCR Method. Appl. Environ. Microbiol. 2018;84:e00107-18. doi: 10.1128/AEM.00107-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kropinski A.M., Mazzocco A., Waddell T.E., Lingohr E., Johnson R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In: Clokie M.R.J., Kropinski A.M., editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Humana Press; Totowa, NJ, USA: 2009. pp. 69–76. [DOI] [PubMed] [Google Scholar]
- 23.Lillehaug D. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 1997;83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 24.Pires D., Sillankorva S., Faustino A., Azeredo J. Use of newly isolated phages for control of Pseudomonas aeruginosa PAO1 and ATCC 10145 biofilms. Res. Microbiol. 2011;162:798–806. doi: 10.1016/j.resmic.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J. In: Molecular Cloning: A Laboratory Manual. 2nd ed. Fritsch E.F., Maniatis T., editors. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1989. [Google Scholar]
- 26.Moineau S., Pandian S., Klaenhammer T.R. Evolution of a Lytic Bacteriophage via DNA Acquisition from the Lactococcus lactis Chromosome. Appl. Environ. Microbiol. 1994;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kot W., Vogensen F.K., Sørensen S.J., Hansen L.H. DPS—A rapid method for genome sequencing of DNA-containing bacteriophages directly from a single plaque. J. Virol. Methods. 2014;196:152–156. doi: 10.1016/j.jviromet.2013.10.040. [DOI] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 29.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen T.K., Carstens A.B., Browne P., Lametsch R., Neve H., Kot W., Hansen L.H. The first characterized phage against a member of the ecologically important sphingomonads reveals high dissimilarity against all other known phages. Sci. Rep. 2017;7:13566. doi: 10.1038/s41598-017-13911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Besemer J., Borodovsky M. GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 38.Boratyn G.M., Schäffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carstens A.B., Kot W., Lametsch R., Neve H., Hansen L.H. Characterisation of a novel enterobacteria phage, CAjan, isolated from rat faeces. Arch. Virol. 2016;161:2219–2226. doi: 10.1007/s00705-016-2901-0. [DOI] [PubMed] [Google Scholar]
- 42.Tolstoy I., Kropinski A.M., Brister J.R. Bacteriophage Taxonomy: An Evolving Discipline. In: Azeredo J., Sillankorva S., editors. Bacteriophage Therapy. Volume 1693. Humana Press; New York, NY, USA: 2018. pp. 57–71. [DOI] [PubMed] [Google Scholar]
- 43.Ågren J., Sundström A., Håfström T., Segerman B. Gegenees: Fragmented Alignment of Multiple Genomes for Determining Phylogenomic Distances and Genetic Signatures Unique for Specified Target Groups. PLoS ONE. 2012;7:e39107. doi: 10.1371/journal.pone.0039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vesth T., Lagesen K., Acar Ö., Ussery D. CMG-Biotools, a Free Workbench for Basic Comparative Microbial Genomics. PLoS ONE. 2013;8:e60120. doi: 10.1371/journal.pone.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller E.S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Rüger W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Travers A.A. Bacteriophage Sigma Factor for RNA Polymerase. Nature. 1969;223:1107–1110. doi: 10.1038/2231107a0. [DOI] [PubMed] [Google Scholar]
- 50.Malys N., Chang D.-Y., Baumann R.G., Xie D., Black L.W. A Bipartite Bacteriophage T4 SOC and HOC Randomized Peptide Display Library: Detection and Analysis of Phage T4 Terminase (gp17) and Late σ Factor (gp55) Interaction. J. Mol. Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 51.Mikhailopulo I. Biotechnology of Nucleic Acid Constituents—State of the Art and Perspectives. Curr. Org. Chem. 2007;11:317–335. doi: 10.2174/138527207780059330. [DOI] [Google Scholar]
- 52.Lee J.Y., Li Z., Miller E.S. Vibrio Phage KVP40 Encodes a Functional NAD+ Salvage Pathway. J. Bacteriol. 2017;199 doi: 10.1128/JB.00855-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greiner T., Moroni A., van Etten J., Thiel G. Genes for Membrane Transport Proteins: Not So Rare in Viruses. Viruses. 2018;10:456. doi: 10.3390/v10090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias-Palomo E., Berger J.M. An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment. Cell. 2015;162:860–871. doi: 10.1016/j.cell.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stetter K.O. Evidence for frequent lysogeny in lactobacilli: Temperate bacteriophages within the subgenus Streptobacterium. J. Virol. 1977;24:685–689. doi: 10.1128/jvi.24.2.685-689.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hausner G., Hafez M., Edgell D.R. Bacterial group I introns: Mobile RNA catalysts. Mob. DNA. 2014;5:8. doi: 10.1186/1759-8753-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrov V.M., Ratnayaka S., Nolan J.M., Miller E.S., Karam J.D. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 2010;7:292–311. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mills K.V. Self-Splicing Proteins. In: Rawlings N.D., Salvesen G., editors. Handbook of Proteolytic Enzymes. Academic Press; Cambridge, MA, USA: 2013. pp. 315–321. [Google Scholar]
- 59.Elleuche S., Pöggeler S. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl. Microbiol. Biotechnol. 2010;87:479–489. doi: 10.1007/s00253-010-2628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kot W., Hansen L.H., Neve H., Hammer K., Jacobsen S., Pedersen P.D., Sørensen S.J., Heller K.J., Vogensen F.K. Sequence and comparative analysis of Leuconostoc dairy bacteriophages. Int. J. Food Microbiol. 2014;176:29–37. doi: 10.1016/j.ijfoodmicro.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Goodrich-Blair H., Shub D.A. Beyond homing: Competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/S0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- 62.Perler F.B. InBase: The Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landthaler M., Shub D.A. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: Introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. USA. 1999;96:7005–7010. doi: 10.1073/pnas.96.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riipinen K.A., Alatossava T. Two self-splicing group I introns interrupt two late transcribed genes of prolate-headed Lactobacillus delbrueckii phage JCL1032. Arch. Virol. 2004;149:2013–2024. doi: 10.1007/s00705-004-0329-4. [DOI] [PubMed] [Google Scholar]
- 65.Mitchell M.S., Rao V.B. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J. Biol. Chem. 2006;281:518–527. doi: 10.1074/jbc.M507719200. [DOI] [PubMed] [Google Scholar]
- 66.Ugorčáková J., Bukovská G. Lysins and holins: Tools of phage-induced lysis. Biologia. 2003;58:327–334. [Google Scholar]
- 67.Josephsen J.Y., Neve H.O. Bacteriophage and antiphage mechanisms of lactic acid bacteria. In: Salminen S., von Wright A., Ouwehand A., editors. Lactic Acid Bacteria: Microbiological and Functional Aspects. CRC Press; New York, NY, USA: 2004. pp. 295–350. [Google Scholar]
- 68.Wang I.-N., Smith D.L., Young R. Holins: The Protein Clocks of Bacteriophage Infections. Annu. Rev. Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 69.Henrich B., Binishofer B., Bläsi U. Primary structure and functional analysis of the lysis genes of Lactobacillus gasseri bacteriophage phi adh. J. Bacteriol. 1995;177:723–732. doi: 10.1128/jb.177.3.723-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oki M., Kakikawa M., Nakamura S., Yamamura E.-T., Watanabe K., Sasamoto M., Taketo A., Kodaira K.-I. Functional and structural features of the holin HOL protein of the Lactobacillus plantarum phage φg1e: Analysis in Escherichia coli system. Gene. 1997;197:137–145. doi: 10.1016/S0378-1119(97)00252-7. [DOI] [PubMed] [Google Scholar]
- 71.Fogg P.C.M., Rigden D.J., Saunders J.R., McCarthy A.J., Allison H.E. Characterization of the relationship between integrase, excisionase and antirepressor activities associated with a superinfecting Shiga toxin encoding bacteriophage. Nucleic Acids Res. 2011;39:2116–2129. doi: 10.1093/nar/gkq923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casey E., Mahony J., O’Connell-Motherway M., Bottacini F., Cornelissen A., Neve H., Heller K.J., Noben J.-P., Dal Bello F., van Sinderen D. Molecular characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages. Appl. Environ. Microbiol. 2014;80:5623–5635. doi: 10.1128/AEM.01268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cornelissen A., Sadovskaya I., Vinogradov E. The baseplate of Lactobacillus delbrueckii bacteriophage Ld17 harbours a glycerophosphodiesterase. J. Biol. Chem. 2016;291:16816–16827. doi: 10.1074/jbc.M116.728279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Samson J.E., Magadán A.H., Sabri M., Moineau S. Revenge of the phages: Defeating bacterial defences. Nat. Rev. Microbiol. 2013;11:675–687. doi: 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 75.Rodríguez-Rubio L., Martínez B., Donovan D.M., Rodríguez A., García P. Bacteriophage virion-associated peptidoglycan hydrolases: Potential new enzybiotics. Crit. Rev. Microbiol. 2013;39:427–434. doi: 10.3109/1040841X.2012.723675. [DOI] [PubMed] [Google Scholar]
- 76.Pulliainen A.T., Kauko A., Haataja S., Papageorgiou A.C., Finne J. Dps/Dpr ferritin-like protein: Insights into the mechanism of iron incorporation and evidence for a central role in cellular iron homeostasis in Streptococcus suis. Mol. Microbiol. 2005;57:1086–1100. doi: 10.1111/j.1365-2958.2005.04756.x. [DOI] [PubMed] [Google Scholar]
- 77.Guidone A., Ianniello R.G., Ricciardi A., Zotta T., Parente E. Aerobic metabolism and oxidative stress tolerance in the Lactobacillus plantarum group. World J. Microbiol. Biotechnol. 2013;29:1713–1722. doi: 10.1007/s11274-013-1334-0. [DOI] [PubMed] [Google Scholar]
- 78.Archibald F.S., Duong M.N. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 1984;158:1–8. doi: 10.1128/jb.158.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dai G., Li R., Chen H., Jiang C., You X., Wu Y. A ferritin-like protein with antioxidant activity in Ureaplasma urealyticum. BMC Microbiol. 2015;15:145. doi: 10.1186/s12866-015-0485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumaran D., Bonanno J.B., Burley S.K., Swaminathan S. Crystal structure of phosphatidylglycerophosphatase (PGPase), a putative membrane-bound lipid phosphatase, reveals a novel binuclear metal binding site and two “proton wires”. Proteins. 2006;64:851–862. doi: 10.1002/prot.21039. [DOI] [PubMed] [Google Scholar]
- 81.Gutiérrez D., Martínez B., Rodríguez A., García P. Genomic characterization of two Staphylococcus epidermidis bacteriophages with anti-biofilm potential. BMC Genom. 2012;13:228. doi: 10.1186/1471-2164-13-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kristensen D.M., Cai X., Mushegian A. Evolutionarily conserved orthologous families in phages are relatively rare in their prokaryotic hosts. J. Bacteriol. 2011;193:1806–1814. doi: 10.1128/JB.01311-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schumann W. Dynamics of the Bacterial Chromosome: Structure and Function. Wiley-VCH; Weinheim, Germany: 2006. [Google Scholar]
- 84.Ignacio-Espinoza J.C., Sullivan M.B. Phylogenomics of T4 cyanophages: Lateral gene transfer in the ‘core’ and origins of host genes. Environ. Microbiol. 2012;14:2113–2126. doi: 10.1111/j.1462-2920.2012.02704.x. [DOI] [PubMed] [Google Scholar]
- 85.Millard A.D., Zwirglmaier K., Downey M.J., Mann N.H., Scanlan D.J. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: Implications for mechanisms of cyanophage evolution. Environ. Microbiol. 2009;11:2370–2387. doi: 10.1111/j.1462-2920.2009.01966.x. [DOI] [PubMed] [Google Scholar]
- 86.Vigil-Stenman T., Ininbergs K., Bergman B., Ekman M. High abundance and expression of transposases in bacteria from the Baltic Sea. ISME J. 2017;11:2611–2623. doi: 10.1038/ismej.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Melo A.G., Levesque S., Moineau S. Phages as friends and enemies in food processing. Curr. Opin. Biotechnol. 2018;49:185–190. doi: 10.1016/j.copbio.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 88.Kelly D., Neve H., McAuliffe O., Ross R.P., Arendt E.K., Coffey A. Isolation and Characterization of Bacteriophages That Inhibit Strains of Pediococcus Damnosus, Lactobacillus Brevis, and Lactobacillus paraplantarum That Cause Beer Spoilage. J. Am. Soc. Brew. Chem. 2011;69:8–12. doi: 10.1094/ASBCJ-2010-1119-01. [DOI] [Google Scholar]
- 89.Deasy T., Mahony J., Neve H., Heller K.J., van Sinderen D. Isolation of a Virulent Lactobacillus brevis Phage and Its Application in the Control of Beer Spoilage. J. Food Prot. 2011;74:2157–2161. doi: 10.4315/0362-028X.JFP-11-262. [DOI] [PubMed] [Google Scholar]
- 90.Kelly D., O’Sullivan O., Mills S., McAuliffe O., Ross R.P., Neve H., Coffey A. Genome sequence of the phage clP1, which infects the beer spoilage bacterium Pediococcus damnosus. Gene. 2012;504:53–63. doi: 10.1016/j.gene.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 91.Wang X., Kim Y., Ma Q., Hong S.H., Pokusaeva K., Sturino J.M., Wood T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lepeuple A.-S., Van Gemert E., Chapot-Chartier M.-P. Analysis of the bacteriolytic enzymes of the autolytic lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: Identification of a prophage-encoded enzyme. Appl. Environ. Microbiol. 1998;64:4142–4148. doi: 10.1128/aem.64.11.4142-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.SnapGene Software From GSL Biotech. [(accessed on 1 October 2018)]; Available online: https://www.snapgene.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.