Abstract
Contact investigations following the diagnosis of active tuberculosis (TB) are paramount for the control of the disease. Epidemiological data are very powerful for contact tracing but might be delayed and/or difficult to integrate, especially in the setting of multiple contact-tracing investigations. The aim of this study was to address the added-value of whole-genome sequencing (WGS) to routine local TB surveillance systems. From November 2016 to July 2017, the local TB surveillance system identified three clusters that could constitute a unique larger outbreak. Epidemiological and clinical information were integrated with WGS genotyping data of Mycobacterium tuberculosis strains obtained using a simple DNA extraction method coupled with sequencing using an Illumina MiSeq platform and an in-house bioinformatics pipeline for single nucleotide polymorphism (SNP) analysis. Epidemiological investigations identified three putative TB clusters potentially interrelated including eight patients with active TB. Seven M. tuberculosis isolates were available and analysed by WGS. Using a 5-SNP threshold to define recent transmission, WGS-based genotyping supported the occurrence of the three clusters as well as a link between clusters 1 and 2 (SNP ≤1), constituting a larger outbreak. This outbreak was clearly delineated by refuting a potential link with the third cluster (SNP >500). Genotyping data did not support the belonging of patient 7 to any studied cluster. This study illustrates the usefulness of WGS genotyping for routine TB surveillance systems in local communities to rapidly confirm or disprove epidemiological hypotheses and delineate TB clusters, especially in the context of multiple contact-tracing investigations.
Keywords: Contact investigation, genotyping outbreak, molecular epidemiology, Mycobacterium tuberculosis, whole-genome sequencing
Introduction
Tuberculosis (TB) has become one of the worst global infectious epidemics. According to the latest report of the WHO, TB is killing more people than AIDS and malaria combined [1]. In 2016, 10.4 million people were diagnosed with TB, leading to 1.7 million deaths. Although molecular methods such as rapid PCR-based tests have dramatically improved TB diagnosis [2], [3], [4], [5], [6], [7], [8], it is paramount to limit disease transmission following a new diagnosis. Contact tracing is performed by TB surveillance systems in high-income/low-prevalence countries for people exposed to a patient with contagious TB, to identify contacts who should receive preventive therapy and to reduce further transmission [7], [9]. Epidemiological investigations are useful to identify contacts and define patient clusters, but often cannot distinguish co-occurring transmissions of different strains at a local or global scale. Molecular epidemiology comprises integrating molecular genotyping results with existing epidemiological data, adding an important value to study the chain of transmission and spread of TB. To corroborate or disprove epidemiological links, several genotyping methods have been implemented such as restriction fragment length polymorphism, spoligotyping and the Mycobacterial Interspersed Repetitive Unit (MIRU 12, 15 and 24) [10]. Nevertheless, the resolution of conventional genotyping methods, such as restriction fragment length polymorphism or Mycobacterial Interspersed Repetitive Unit, is not sufficient and may overestimate transmission [11]. Strain genotyping based on the identification of single nucleotide polymorphisms (SNPs) using Mycobacterium tuberculosis whole-genome sequencing (WGS) has a higher discriminatory power than all other molecular typing methods in that it enables the identification of single mutations in the entire genome [10].
A growing number of publications demonstrated the utility of M. tuberculosis WGS in critical situations, such as international outbreaks [12]. For instance, a recent study demonstrated the presence of a circulating cluster of multidrug-resistant TB (MDR-TB) strains in European countries involving patients from countries of the African Horn and Sudan [13]. The sequencing of M. tuberculosis strains facilitated and accelerated the reconstitution of the clusters [14]. On a local scale, it is sometimes difficult to delineate clusters based on epidemiological data alone and some doubts may persist about the link between two patients. This may occur in the setting of two distinct contact-tracing investigations. Next-generation sequencing technologies have facilitated the use of WGS-based molecular genotyping, which is currently the method of choice to support or disprove suspicions of transmission [15].
In this study, we describe the implementation and evaluation of a simple, rapid and reliable M. tuberculosis genotyping method using next-generation sequencing technologies. To address the added value of such an approach to support local health-care authorities in routine TB surveillance, we applied this method to the investigation of three local TB clusters suspected on the basis of clinical and epidemiological data.
Methods
Patients, criteria for contact tracing, definitions for case assessment
Vaud is one of the largest cantons of Switzerland with 794 000 inhabitants and around 65 new TB diagnoses per year. The TB surveillance system implies systematic new TB patient notification to the Medical Officer of Public Health and to the Swiss Federal Office of Public Health. Contact-tracing investigation is systematically performed for every new case of pulmonary contagious TB according to the WHO and national Swiss guidelines published by the Swiss Lung Association guidelines [16], [17]. Epidemiological investigations including sociodemographic information and clinical data were performed by TB nurses and lung specialists at the Outpatient TB Clinic of the Medical Policlinic of Lausanne and at the Lung Association of Vaud County [18]. According to National Swiss Guidelines published by the Swiss Lung Association, contact tracing should be performed for the identification of possible infections with M. tuberculosis in individuals recently exposed to an index TB patient with transmissible TB (smear-positive pulmonary TB) [16]. In this situation, individuals in direct and close contact with the patient including household members, close contacts, workmates and schoolmates are screened for latent TB infection. A limited contact investigation (only for household members or close contacts) is performed for patients with pulmonary TB with negative acid-fast bacilli smear but positive PCR and/or culture. Groups at high risk of TB infection are identified and tested, prioritizing immunocompromised patients and young infants [9].
Microbiological and molecular procedures
For microbiological analysis each specimen was split for the purposes of acid-fast bacilli staining, Xpert MTB/RIF (Cepheid, Sunnyvale, CA, USA), DNA extraction for home-made specific M. tuberculosis real-time PCR and mycobacterial culture as previously described [2], [7]. Acid-fast bacilli detection was accomplished by fluorescent auramine staining and fluorescence microscopy examination. Respiratory specimens were liquefied and decontaminated by standard procedure using NaOH and N-acetyl-l-cysteine. Mycobacterial culture and first-line drug susceptibility testing were carried out using a Mycobacteria Growth Indicator Tube (MGIT 960, Becton Dickinson, Baltimore, MD, USA). The Xpert MTB/RIF was performed for expectorated pulmonary samples (and for one bronchial aspirate that was received from another hospital). The TaqMan real-time PCR based on IS6110 amplification was achieved on the fully automated molecular diagnostic platform of our institute [19].
For next-generation sequencing, M. tuberculosis DNA was extracted from 1 mL of heat-inactivated (30 minutes at 56°C in proteinase K and ATL buffer, Qiagen, Hilden, Germany) positive MGIT using the QIAmp mini Kit (Qiagen, Hilden, Germany). Quantitative and qualitative measurements of the DNA were performed using Qubit Fluorometry 0.2 (Thermo Fisher Scientific) and real-time PCR [19]. DNA libraries were prepared using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA) and checked for quality using a fragment analyser (Advanced Analytical Technologies, Inc. (AATI), Santa Clara, CA, USA) before sequencing using a MiSeq sequencer (Illumina). Raw sequencing data were submitted under NCBI BioProject accession number PRJNA526295.
Bioinformatics analyses and data
Samples were genotyped by mapping the sequence data against the reference genome of H37Rv (accession number NC_000962.3) and calling variants with GATK’s HaplotypeCaller. Genotype calls with coverage <10 reads or supported by <75% of the observations were masked and discarded when enumerating differences between isolates. Complex and indels mutations were also discarded and distances in SNPs were calculated over the full-length of the H37Rv genome, as well as over the genomic regions of the core genome multilocus sequence typing schema for the M. tuberculosis complex defined by www.cgMLST.org. Phylogenetic trees were reconstructed using RAxML and the GTRCAT model over the full alignment including the called SNPs. The sequence of the reference H37Rv (lineage 4) as well as Sequence Read Archive data from isolates of the six other lineages of M. tuberculosis were included in the alignment to determine the lineages of the samples (see Supplementary material, Table S2).
The data have been made available under NCBI BioProject PRJNA526295: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA526295.
Ethical approval
The Swiss Ethics Committee of the canton of Vaud has strictly reviewed and approved this study and the project is referenced as 2018-00023.
Results
Proposed transmission network based on clinical and epidemiological investigation
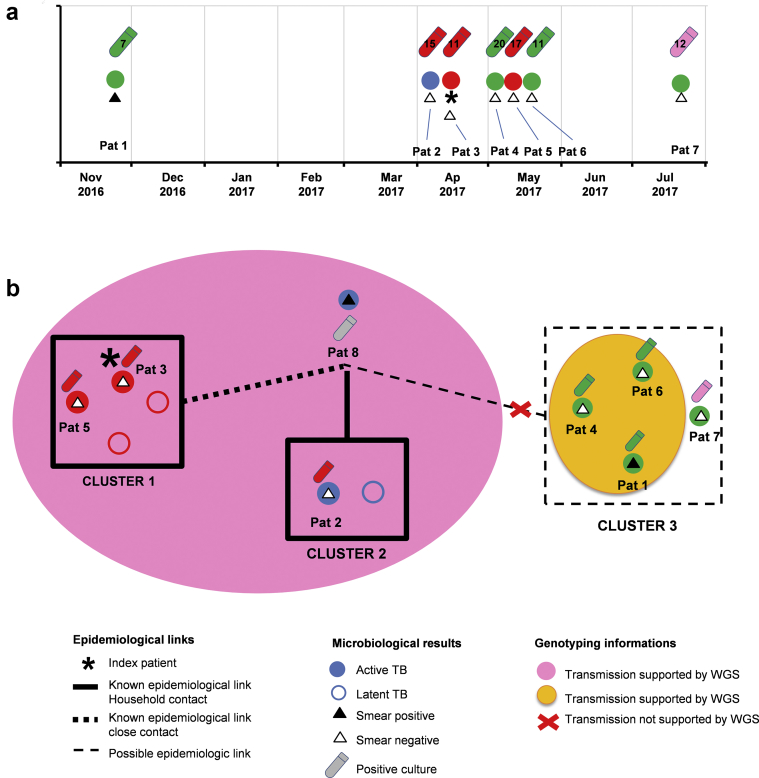
Between November 2016 and July 2017, we focused on seven individuals who were diagnosed with active pulmonary TB among all the patients notified in that period (Fig. 1).
Fig. 1.
Proposed transmission network based on epidemiological investigation and whole-genome sequencing. (a) Date of tuberculosis diagnosis and microbiological findings; (b) proposed transmission network based on epidemiological, clinical, microbiological and genomic data.
The first contact investigation was performed in April 2017 around patient 3, who had undergone a lobectomy because of a suspected lung tumour (Fig. 1). Lung aspirates were negative both for AFB detection and for M. tuberculosis complex PCR. The culture became positive for M. tuberculosis complex within 11 days. Household contact investigation identified a patient with positive interferon-γ release assay. Because of a slightly abnormal chest X-ray, a CT scan was performed on this patient and did not show any abnormality. Initial microbiological analyses (microscopy and Xpert MTB/RIF) and later culture for M. tuberculosis complex were negative. A second household contact, a young boy, had a 10-mm positive Mantoux test with a normal X-ray and normal clinical examination; a preventive treatment was initiated for these two individuals (Fig. 2, Table 1). A third household contact (patient 5) showed an abnormal X-ray and CT scan with minimal tree in bud-branching infiltrates and micro-nodules in the right lung, indicating recent acquisition of active pulmonary TB (Fig. 1). Respiratory samples were smear and Xpert MTB/RIF negative but the culture grew M. tuberculosis after 17 days. The patient received a standard anti-TB treatment regimen. The lung specialists actively continued to search for the index patient, because they assumed that all of these patients suffered from early stages of a smear-negative TB, suggesting that none of them were the index patient.
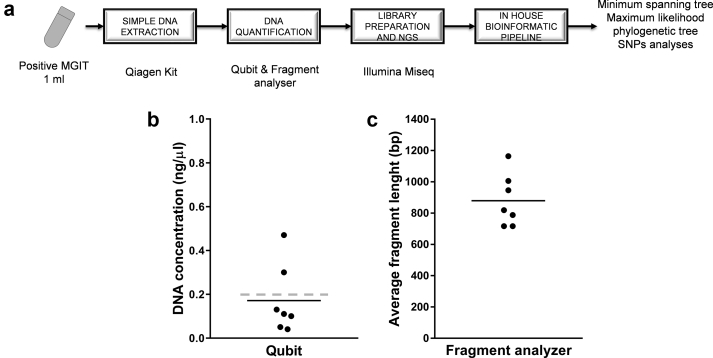
Fig. 2.
Microbiology workflow and DNA extraction results. (a) Analytic workflow from positive Mycobacterium tuberculosis culture (MGIT) to phylogenetic trees. (b) DNA yield obtained from 1 mL of positive MGIT, concentrations of DNA measured by Qubit; the grey dashed line represents the requested concentration for library preparation according to Illumina standard protocols. (c) Average fragment length of extracted DNA for each isolate. The line represents mean values.
Table 1.
Clinical and microbiological data of patients
| Patient | Date of TB diagnosisa | Type of sample | Smear microscopy | Culture time to positivity | Xpert MTB/RIF | rpoB mutation Xpert | MTBC TAQMAN (Cp/mL) | Clinical manifestations |
|---|---|---|---|---|---|---|---|---|
| 1 | 2016.11.27 | Sputum | Positive ++++ | 7 | Positive/medium | Negative | ND | Extensive bilateral lung infiltrates |
| 2 | 2017.04.02 | Sputum | Negative | 15 | Negative | NA | ND | Pleural effusion |
| 3 | 2017.04.11 | Lung abscess | Negative | 11 | ND | ND | ND (Q INS/cf) | Pulmonary TB diagnosed after lobectomy |
| 4 | 2017.05.01 | Bronchial aspirate | Negative | 20 | Positive/very low | Negative | ND | Miliary TB |
| 5 | 2017.05.05 | Induced sputum | Negative | 17 | Negative | NA | ND | Abnormal X-ray and CT scan with tree in bud branching infiltrates and micro-nodules in the right lung |
| 6 | 2017.06.17 | Sputum | Negative | 11 | Positive/very low | Negative | 150 | Pulmonary TB |
| 7 | 2017.07.27 | Bronchial aspirate | Negative | 12 | ND | ND | ND | Poor health condition, lung infiltrates, cough |
NA, not applicable; ND, not done; UN, unknown.
Consultation and sampling date.
One month before TB was diagnosed in patient 3, patient 2 was diagnosed with pleural TB (Fig. 1, Patient 2). The Xpert MTB/RIF results were negative in pleural fluid but culture became positive for M. tuberculosis after 15 days. One household contact, a young girl, exhibited a 10-mm positive Mantoux test but the chest X-ray and clinical examination were normal and microbiological analysis remained negative. She also received preventive treatment. The interrogatory performed by the TB nurses revealed that patient 8, a contact of patient 2 who lived abroad, had symptoms compatible with TB (illness and cough) and had been in contact with patients 3 and 5. The patient was then tested abroad, which confirmed the diagnosis of a bilateral cavitary TB, with a sputum smear positive for M. tuberculosis (Fig. 1, Table 1). The suspected source patient was treated abroad for active TB.
During the same period, patients 1, 4, 6 and 7 were diagnosed with active pulmonary TB within the city with possible contacts and transmission (Fig. 1, Table 1). The first patient (patient 1), was diagnosed in November 2016 with an extensive bilateral pulmonary TB and a positive smear microscopy examination, also positive by Xpert MTB/RIF PCR. He was later confirmed positive for M. tuberculosis by culture. The second patient (patient 4), presented a miliary pulmonary TB with negative smear microscopy but positive at Xpert MTB/RIF PCR with very low bacterial load in tested respiratory samples (bronchoalveolar lavage and bronchial aspirate), and positive at M. tuberculosis culture, which grew within 20 days. The third patient (patient 6) presented with a pulmonary TB with negative smear microscopy but positive Xpert MTB/RIF with very low quantity and a culture that grew M. tuberculosis within 11 days. The fourth patient (patient 7) had a pulmonary TB with negative smear examination and the culture grew M. tuberculosis within 12 days. A possible epidemiological link was established between these four patients and patient 8 (Fig. 1, Table 1). The epidemiological and clinical information together identified three putative, potentially interrelated clusters.
DNA extraction from positive MGIT, DNA yield and MiSeq sequencing
DNA extraction, including heat inactivation of 1 mL of positive MGIT culture followed by the use of a QIAmp minikit (Qiagen) provided from 0.043 to 0.3 ng/μL of DNA quantified using a Qubit fluorometer (Fig. 1, and see Supplementary material, Table S1). This extraction method is easily applicable in the routine conditions of BSL3. Libraries were prepared using the Nextera XT procedure despite the fact that the amount of DNA was often lower than the recommended amount of 0.2 ng/μL (1 ng in total). The Fragment Analyser showed a molarity >4 nM for five isolates and <4 nM for two isolates (3 and 3.3) (Fig. 2, and see Supplementary material, Table S1). The average length of DNA fragments in the libraries always exceeded 717 bp and DNA was not excessively fragmented. When mapping against the H37Rv genome, the depth of coverage ranged from 81.0 × to 197.0 × in the various samples and 94.4% to 97.6% of the nucleotides were covered at more than 30 × depth. This shows that concentrations of DNA lower that those recommended by Illumina can be successfully used to perform WGS using Nextera XT protocol and a MiSeq sequencer.
Genotyping of M. tuberculosis isolates and proposed transmission network based on both epidemiological investigation and WGS
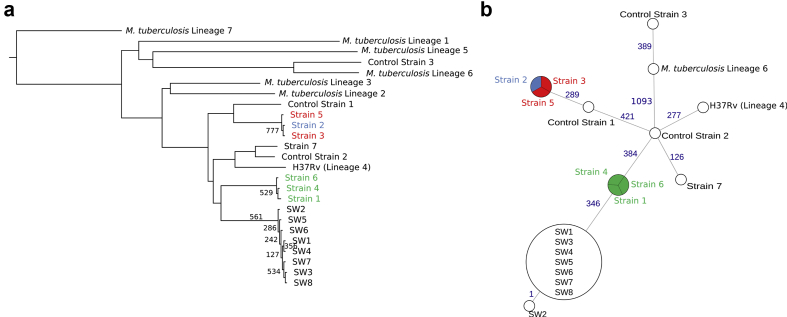
The phylogeny indicated that all isolates belonged to lineage 4 of M. tuberculosis (Fig. 3a). The isolates from patients 1, 4 and 6 presented no SNP differences over the core genome (Fig. 3b) nor over the full H37Rv genome (see Supplementary material, Fig. S1). Isolates from patients 2, 3 and 5 presented no SNP difference over the core genome (Fig. 3b) and only one SNP difference between each pair of isolates over the full H37Rv genome (see Supplementary material, Fig. S1). Finally, the isolate from patient 7 was not closely related to any of the other isolates, i.e. with more than 500 SNPs differences to the closest isolate (on the core genome). According to Walker et al. [13], in this study, we defined a genomic cluster as two or more strains that differed by no more than five SNPs. Altogether, the WGS-based genotyping supported the occurrence of three different clusters (1, 2 and 3) as well as the strong link between clusters 1 and 2. Finally, genotyping data did not support the belonging of patient 7 to cluster 3 or to any other studied cluster.
Fig. 3.
Phylogeny and relatedness of Mycobacterium tuberculosis isolates. (a) Maximum likelihood phylogenetic tree of the seven isolates and references for each lineage of M. tuberculosis three control patients, eight Swiss isolates from a previously published pan-European epidemic (SW1–8) [24] and references for each lineage over the core genome of M. tuberculosis. All bootstrap values are equal to 1000 except when mentioned. The list of SRA entries used for each lineage of M. tuberculosis and for SW1–8 is available in the Supplementary material (Table S2). (b) Minimum spanning tree of distances in number of single nucleotide polymorphisms between isolates and relevant lineages over the core genome of M. tuberculosis. Blue numbers represent the distances between isolates.
Discussion
Epidemiological investigations are useful to identify contacts of a patient with active TB. The canton of Vaud (Switzerland) is a highly populated region in the French-speaking part of Switzerland and an important crossway between different Swiss cities and other European countries. According to the WHO and European Respiratory Society recommendations for TB prevention programmes in industrialized countries with low TB incidence, contact investigation should be performed for all contagious pulmonary TB to reduce the transmission of the disease [20]. The Vaud Pulmonary League organizes and implements community-based surveys following a new TB diagnostic using, when necessary, an active detection strategy that allows the detection of latent TB or the discovery of individuals with secondary active TB [18]. However, epidemiological investigations are sometime unable to discriminate between multiple circulating clusters and it is sometime necessary to rely on available M. tuberculosis strains genotyping to support or to disprove the hypothesis of recent transmission. Strain genotyping based on WGS displays the highest discriminatory power among all molecular typing methods and does not represent a technological issue thanks to the last generation of sequencer coupled with simple and friendly bioinformatics pipelines [10]. WGS is now largely applied to critical situations such as suspicion of international outbreaks or MDR-TB outbreaks [13].
In this study, we describe the use of a rapid and reliable genotyping method based on WGS for an epidemiological approach to assist the local health-care authorities in the routine TB surveillance system. From November 2016 to July 2017 epidemiological investigations suspected three clusters of TB patients involving eight individuals with active TB. WGS suggested that the putative first and second clusters were due to recent transmission events (SNPs 0 or 1), hence constituting a single larger outbreak encompassing clusters 1 and 2 and clearly delineated this outbreak by infirming a potential link with the third cluster. Finally, within the suspected third cluster, one strain was distantly related to all other strains, and hence did not belong to any of the three investigated clusters. The identification of the link between cluster 1 and cluster 2 through patient 8 supported the suspected transmission between the two families identified by contact tracing and prevented the patient catching a plane, so avoiding potential further transmission.
We also report that a simple DNA extraction method easily applicable in BSL3 conditions, based on the QIAmp DNA extraction mini kit, can provide a sufficient amount of DNA for Illumina-based whole genome analysis of M. tuberculosis. For most of the mycobacterial strains the DNA yield obtained with this extraction method from 1 mL of positive MGIT, was lower than the requested threshold (0.2 ng/μL). Nevertheless, fragmented DNA libraries reached sufficient quality and quantity for Illumina sequencing. Read mapping against the H37Rv genome showed a sufficient depth of coverage for reliable SNPs calling in all strains. This supports previous results showing that successful whole genome analysis may be achieved with a very small amount of DNA obtained using simple DNA extraction methods [21]. Indeed despite the lower DNA yield, simple DNA extraction is more suitable for BSL3 conditions than complex methods based on ethanol precipitation or acetyl methyl bromide [21].
This study illustrates the usefulness of genotyping methods based on complete genome sequencing using a molecular epidemiology approach. This approach combines epidemiological information (spatial–temporal, clinical and societal) and genotyping information provided by WGS that currently represents the most discriminatory method and outperforms current reference methods. Furthermore, WGS also enables the investigation of microevolution events within patients and between different patients along transmission chains [22]. It can also help decipher community TB outbreaks by inferring chains of transmission between patients [23]. Caution should be taken, as strain relatedness is not sufficient to determine a direct transmission or an outbreak. Indeed similar strains can also be the result of a dominant circulating strain [11]. Since 2011, we have in our hospital, weekly and monthly meetings with specialists from different backgrounds working in the field of TB including: (i) health-care providers in contact with TB patients—TB nurses and lung specialists at the anti-TB dispensary, infectious disease specialists and co-workers of the hospital preventive medicine; (ii) health-care providers in contact with patient’s families in the community; and (iii) clinical microbiologists specialized in M. tuberculosis diagnosis and genomics. The different views and perspectives of the patients and their families, shared during these meetings, helps to build connections and to coordinate the different procedures, facilitate treatment supervision and contact investigations, and eventually permits identification of some gaps. This article also shows that these providers are not only delivering care to an individual patient with TB, but they are assuming an important public health function that entails a high level of responsibility to the community, as well as to the individual patient [18]. Hence, molecular epidemiological investigations could add value to contact-tracing investigations but do not replace clinic epidemiological efforts. This study clearly demonstrates that the public health authorities at the local level can rely on WGS for contact investigation and TB surveillance, in particular when multiple contact-tracing investigations might be linked. Efforts should now aim at implementing national and international M. tuberculosis genome databases, allowing easier sharing of data and better surveillance. The routine application of this approach will also facilitate collaborations and exchanges of information between the laboratories and the various public health actors at a national and even international level.
Conflict of interest
The authors disclose no conflict of interest.
Acknowledgements
This work has been carried out within the framework of a Grant of the Pulmonary League of the Vaud canton and a Swiss government excellence scholarships postdoctoral fellowship (Grant number 2017.0763/Jemen/OP). The authors would like to thank the member staff of the Laboratory of Molecular Diagnostic and Tuberculosis and of the Laboratory of Genomics and Metagenomics of the Institute of Microbiology of the University of Lausanne; in particular, we are grateful to René Brouillet, Grégory Gonzalez and Sébastien Aeby. We also thanks Dr Peter Helbing as well as all the member of the Pulmonary League of the Vaud canton (LPV) and of the Dispensaire anti-tuberculeux.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2019.100582.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.WHO . World Health Organization; Geneva: 2017. Global tuberculosis report. [Google Scholar]
- 2.Opota O., Zakham F., Mazza-Stalder J., Nicod L., Greub G., Jaton K. Added value of Xpert MTB/RIF Ultra for diagnosis of pulmonary tuberculosis in a low-prevalence setting. J Clin Microbiol. 2019;57(2) doi: 10.1128/JCM.01717-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opota O., Mazza-Stalder J., Greub G., Jaton K. The rapid molecular test Xpert MTB/RIF ultra: towards improved tuberculosis diagnosis and rifampicin resistance detection. Clin Microbiol Infect. 2019 doi: 10.1016/j.cmi.2019.03.021. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.Boehme C.C., Nabeta P., Hillemann D., Nicol M.P., Shenai S., Krapp F. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravorty S., Simmons A.M., Rowneki M., Parmar H., Cao Y., Ryan J. The New Xpert MTB/RIF Ultra: improving detection of mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio. 2017;8(4) doi: 10.1128/mBio.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorman S.E., Schumacher S.G., Alland D., Nabeta P., Armstrong D.T., King B. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: a prospective multicentre diagnostic accuracy study. Lancet Infect Dis. 2018;18:76–84. doi: 10.1016/S1473-3099(17)30691-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opota O., Senn L., Prod'hom G., Mazza-Stalder J., Tissot F., Greub G. Added value of molecular assay Xpert MTB/RIF compared to sputum smear microscopy to assess the risk of tuberculosis transmission in a low-prevalence country. Clin Microbiol Infect. 2016;22:613–619. doi: 10.1016/j.cmi.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Drancourt M., Michel-Lepage A., Boyer S., Raoult D. The point-of-care laboratory in clinical microbiology. Clin Microbiol Rev. 2016;29:429–447. doi: 10.1128/CMR.00090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox G.J., Barry S.E., Britton W.J., Marks G.B. Contact investigation for tuberculosis: a systematic review and meta-analysis. Eur Resp J. 2013;41:140–156. doi: 10.1183/09031936.00070812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannas A., Mazzarelli A., Di Caro A., Delogu G., Girardi E. Molecular typing of Mycobacterium Tuberculosis strains: a fundamental tool for tuberculosis control and elimination. Infect Dis Rep. 2016;8:6567. doi: 10.4081/idr.2016.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stucki D., Ballif M., Egger M., Furrer H., Altpeter E., Battegay M. Standard genotyping overestimates transmission of Mycobacterium tuberculosis among immigrants in a low-incidence country. J Clin Microbiol. 2016;54:1862–1870. doi: 10.1128/JCM.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Werf M.J., Ködmön C. Whole-genome sequencing as tool for investigating international tuberculosis outbreaks: a systematic review. Front Public Health. 2019;7(87) doi: 10.3389/fpubh.2019.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker T.M., Merker M., Knoblauch A.M., Helbling P., Schoch O.D., van der Werf M.J. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis. 2018;18:431–440. [Google Scholar]
- 14.Walker T.M., Ip C.L., Harrell R.H., Evans J.T., Kapatai G., Dedicoat M.J. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardy J.L., Johnston J.C., Sui S.J.H., Cook V.J., Shah L., Brodkin E. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 16.Lung Association Swiss. Swiss Lung Association; Wald: 2014. Tuberculosis in Switzerland, Guidance for healthcare professionals 2014. [Google Scholar]
- 17.World Health Organization . WHO; Geneva: 2017. Global tuberculosis report. [Google Scholar]
- 18.Mazza-Stalder J., Chevallier E., Opota O., Carreira A., Jaton K., Masserey E. Improvement in tuberculosis outcomes with a combined medical and social approach. Front Med. 2019 doi: 10.3389/fmed.2019.00135. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greub G., Sahli R., Brouillet R., Jaton K. Ten years of R&D and full automation in molecular diagnosis. Future Microbiol. 2016;11:403–425. doi: 10.2217/fmb.15.152. [DOI] [PubMed] [Google Scholar]
- 20.Lönnroth K., Migliori G.B., Abubakar I., D'Ambrosio L., de Vries G., Diel R. Towards tuberculosis elimination: an action framework for low-incidence countries. Eur Resp J. 2015;45:928–952. doi: 10.1183/09031936.00214014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Votintseva A.A., Pankhurst L.J., Anson L.W., Morgan M.R., Gascoyne-Binzi D., Walker T.M. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53:1137–1143. doi: 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merker M., Kohl T.A., Roetzer A., Truebe L., Richter E., Rüsch-Gerdes S. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker T.M., Monk P., Grace Smith E., Peto T.E.A. Contact investigations for outbreaks of Mycobacterium tuberculosis: advances through whole genome sequencing. Clin Microbiol Infect. 2013;19:796–802. doi: 10.1111/1469-0691.12183. [DOI] [PubMed] [Google Scholar]
- 24.Walker T.M., Merker M., Knoblauch A.M., Helbling P., Schoch O.D., van der Werf M.J. A cluster of multidrug-resistant Mycobacterium tuberculosis among patients arriving in Europe from the Horn of Africa: a molecular epidemiological study. Lancet Infect Dis. 2018;18:431–440. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.