Abstract
Status epilepticus (SE) is an abnormally prolonged seizure that results from either a failure of mechanisms that terminate seizures or from initiating mechanisms that inherently lead to prolonged seizures. Here we report that mice experiencing a 3 hours of SE caused by pilocarpine exhibit a rapid increase in expression of type 2 iodothyronine deiodinase gene (Dio2) and a decrease in the expression of type 3 iodothyronine deiodinase gene in hippocampus, amygdala and prefrontal cortex. Type 3 iodothyronine deiodinase in hippocampal sections was seen concentrated in the neuronal nuclei, typical of ischemic injury of the brain. An unbiased analysis of the hippocampal transcriptome of mice undergoing 3 hours of SE revealed a number of genes, including those involved with response to oxidative stress, cellular homeostasis, cell signaling, and mitochondrial structure. In contrast, in mice with targeted disruption of Dio2 in astrocytes (Astro D2KO mouse), the highly induced genes in the hippocampus were related to inflammation, apoptosis, and cell death. We propose that Dio2 induction caused by SE accelerates production of T3 in different areas of the central nervous system and modifies the hippocampal gene expression profile, affecting the balance between adaptive and maladaptive mechanisms.
Status epilepticus in mice increases Dio2 expression in the hippocampus, amygdala, and prefrontal cortex, affecting balance between adaptive and maladaptive mechanisms.
The thyroid gland secretes T4, which is subsequently activated via conversion to T3. T3 binds to nuclear thyroid hormone receptors (TR) and affects the expression of thyroid hormone responsive genes in virtually every tissue and organ including the brain, liver, muscles, adipose tissue, and skeleton (1).
Three different types of iodothyronine deiodinases metabolize T4 and T3 [i.e., type 1, type 2 (D2), and type 3 (D3) iodothyronine deiodinases, each exhibiting different catalytic properties and targeting different iodine residues in the T4 or T3 molecules] (2). D2 catalyzes the outer-ring deiodination of T4, and its expression is widespread in the central nervous system (CNS), primarily in the glial cells; D2-generated T3 enhances thyroid hormone signaling (3–5). Although type 2 iodothyronine deiodinase gene (Dio2) is induced by cAMP (6, 7) and mildly suppressed by T3 (8, 9), D2 activity is primarily regulated at a posttranslational level through T4-induced ubiquitination and proteasomal degradation (10, 11).
In contrast, D3 is predominantly expressed in neurons, where it terminates thyroid hormone signaling (12, 13) by catalyzing the inner-ring deiodination of T4 and T3 to rT3 and 3,3′-diiodothyronine, respectively. The CNS areas in which it is possible to detect D3 expression in adult rats include the II through IV layers of cerebral cortex, including pyramidal and granulated cells of dentate gyrus, and II layer of piriform cortex (14). Given the reciprocal regulation of Dio2 and type 3 iodothyronine deiodinase gene (Dio3) by thyroid hormone, the presence of these enzymes in the CNS provides a regional adaptation during iodine deficiency (15). In addition, the unique distribution of Dio2 and Dio3 expression in different cells of the CNS provides an example of how thyroid hormone signaling is modulated at a local level, relatively independent of circulating levels of T3 (2, 3). In the CNS, D2-generated T3 exits the glial cells and reaches neighboring neurons, where it modulates transcription of T3-responsive genes in a paracrine fashion (16); this mechanism is minimized/modulated by D3 expression in neurons.
Brain ischemia/hypoxia has been shown to acutely induce Dio3 expression and D3 redistribution to the nucleus, which in cell models reduced the paracrine effects of T3 (17). In contrast, other studies indicate that cerebral ischemia increases D2 activity in cerebral cortex and striatum, whereas the D3 activity remains stable (18). In another study, D2 activity was induced in a cell model by hypoxia without changes in Dio2 mRNA levels (19). In this case, hypoxia stabilized and prolonged D2’s half-life by decreasing its susceptibility to the ubiquitin-proteasome pathway, whereas D3 activity was not affected by hypoxia. Traumatic brain injury was also shown to increase Dio2 mRNA expression, although it is not clear how much of this effect is due to changes in plasma/local levels of thyroid hormone (8, 20).
Status epilepticus (SE) is an insult to the CNS characterized by a prolonged seizure with loss of consciousness or by sequential seizures of short duration without full recovery of consciousness between them (21, 22). SE may lead to the development of temporal lobe epilepsy that is the most common type of epilepsy among adult subjects and represents ∼40% of all epilepsy cases (23, 24). SE causes a myriad of secondary acute seizure-related injuries, including peripheral and brain ischemia and hypoxia (25, 26) that may contribute to neuronal damage and epileptogenesis.
Taking in account that ischemia and hypoxia affect D2 and D3 expression and activities in the CNS, the present investigation tested whether SE modifies thyroid hormone metabolism/action in the brain and how it affects the hippocampal acute transcriptional response.
Material and Methods
Animals
Studies were performed at two sites: Mackenzie Presbyterian University (São Paulo, Brazil) and Rush University Medical Center (Chicago, IL). C57Bl/6J mice were purchased from Center for the Development of Experimental Models for Medicine and Biology, Federal University of São Paulo (São Paulo, Brazil) and from The Jackson Laboratory (Bar Harbor, ME). Astro-D2KO mice were created as described previously (27). Throughout the studies with Astro-D2KO mice, GFAP-Cre animals were used as controls. Only male mice were used, and at the time of the studies all mice were ~8 to 10 weeks old. Animals were maintained at 21 ± 2°C with a continuous 12-hour light: 12-hour dark cycle (lights on at 7:00 am) and ad libitum access to regular chow and water. All drugs and reagents, unless otherwise specified, were purchased from Sigma Chemical Co. (St. Louis, MO). All experiments were carried out in accordance with the Institutional Ethical Committee on Animal Use at Mackenzie Presbyterian University (no. 099/05/2013) and the Institutional Animal Care and Use Committee at Rush University Medical Center (no.15-033) (28).
Induction of SE
SE was induced by pilocarpine or lithium pilocarpine administration.
Pilocarpine-induced SE
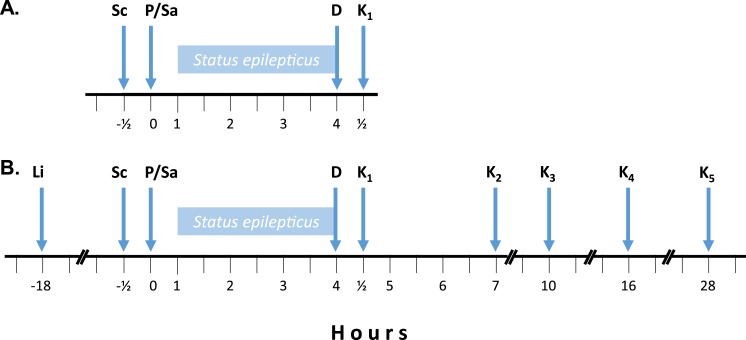
Pilocarpine is an alkaloid extracted from Jaborandi (Pilocarpus jaborandi) leaves that acts nonselectively on muscarinic acetylcholine receptors inducing SE (29). The anticholinergic scopolamine methyl bromide (Sigma; 1 mg/kg subcutaneously; indicated by Sc in Fig. 1A) was administered to all animals 30 minutes before, to minimize the peripheral cholinergic effects of pilocarpine. Mice were randomly divided in two groups and given pilocarpine hydrochloride (n = 13; 325 mg/kg IP; indicated by P in Fig. 1A) or 0.9% saline (n = 13; indicated by Sa in Fig. 1A). Animals that did not develop SE during the first hour received additional dose of 162.5 mg/kg pilocarpine; if 40 minutes after the second dose the animal still did not develop SE, another injection of 162.5 mg/kg pilocarpine was given, to a maximum of 3 injections. Three hours after the onset of SE, both groups received diazepam (10 mg/kg subcutaneously; indicated by D in Fig. 1A) and, 30 minutes later, animals were anesthetized with urethane (Sigma; 560 mg/kg IP) and euthanized by decapitation or transcardiac perfusion (indicated by K1 in Fig. 1A). For immunohistochemistry experiments, mice (n = 4) were perfused through the heart and the whole brain removed and fixed. For quantitative RT-PCR (RT-qPCR) experiments, mice (n = 9) brain structures were dissected, immediately frozen in liquid nitrogen, and stored at −80°C until RNA isolation.
Figure 1.
Protocol timeline. (A) Sc was given 30 minutes before P or Sa administration; D was given 3 hours after the onset of SE or an equivalent time in saline injected mice. (B) Same as in panel (A) except that Li was given 18 hours before P or Sa administration; here, animals were also euthanized at longer time points after D administration. D, diazepam; K1, animals euthanized 30 minutes after D administration; K2, animals euthanized 3 hours after D administration; K3, animals euthanized 6 hours after D administration; K4, animals euthanized 12 hours after D administration; K5, animals euthanized 24 hours after D administration; Li, lithium chloride; P, pilocarpine; Sa, saline; Sc, methyl scopolamine bromide.
Lithium pilocarpine–induced SE
Pilocarpine-induced SE is known for being associated with high mortality (30, 31), which in our hands reached ~65%. Therefore, in some experiments as indicated, mice were given lithium chloride (Sigma; 10 mEq/kg IP; indicated by Li in Fig. 1B) 18 hours before administration of pilocarpine or saline. This lowers the animal’s threshold for seizure and a much lower dose of pilocarpine (65 mg/kg IP) can be used, hence reducing mortality to ∼25%. Three hours after the onset of SE, both groups received diazepam (10 mg/kg subcutaneously; indicated by D in Fig. 1B) and, 30 minutes later, mice were euthanized by asphyxiation in a carbon dioxide chamber or by transcardiac perfusion after anesthesia with ketamine (100 mg/kg IP) and xylazine (5 mg/kg IP) (indicated by K1 in Fig. 1B). In some experiments, as indicated, mice were euthanized at different time points after diazepam administration: 3 hours (indicated by K2 in Fig. 1B), 6 hours (indicated by K3 in Fig. 1B), 12 hours (indicated by K4 in Fig. 1B), or 24 hours (indicated by K5 in Fig. 1B). We also evaluated mice undergoing reduced times of SE, in which case they were euthanized 15 or 30 minutes after the onset of SE.
In all cases, blood samples were collected from the right ventricle after the animals were anesthetized, serum separated, and stored at −80°C.
Perfusion and light microscopy immunohistochemistry for HIF-1α and D3
Mice were anesthetized with urethane (Sigma; 560 mg/kg IP) or with ketamine (100 mg/kg IP) and xylazine (5 mg/kg IP) and underwent transcardiac perfusion at a flow rate of 8 to 10 mL/min first with 0.1M phosphate buffer (PB; pH 7.4) followed by 4% paraformaldehyde (PF; pH 7.4) in 0.1M PB. Brains were removed and immersed in fixative solution of 4% PF for at least 24 hours at room temperature. Later, brains were immersed in 30% sucrose in 0.1M PB for 24 more hours at 4°C. Brains were then kept in 1% PF for at least 24 hours at 4°C. Coronal 50-µM-thick sections through the dorsal hippocampus (Bregma, −1.64) were obtained on a Vibratome (Leica Microsystems, Buffalo Grove, IL) and processed for immunohistochemistry to evaluate the HIF-1α and D3 immunoreactivity. The sections were washed in 0.1M PB three times for 10 minutes each, incubated in BLOXALL™ Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution (Vector Laboratories, Burlingame, CA) for 10 minutes to reduce endogenous peroxidase activity, and rinsed again in 0.1M PB three times for10 minutes each. Then, the sections were incubated in a mixture of 0.5% H2O2 and 0.5% Triton X-100 in 0.1M PB for 15 minutes to increase antibody penetration, and rinsed again in 0.1M PB three times for 10 minutes each. The nonspecific antibody binding was blocked by treatment with blocking serum [VECTASTAIN® Elite ABC Kit (rabbit IgG); Vector Laboratories] for 20 minutes followed by incubation in primary antibody. To detect HIF-1α immunoreactivity, the sections were incubated overnight at 4°C in an affinity-purified, rabbit, polyclonal antibody (1:50; catalog no. NB100-479B; RRID: AB_1521173; Novus Biologicals, Littleton, CO); to detect D3 immunoreactivity, the sections were incubated for 36 hours at 4°C in an affinity-purified, rabbit, polyclonal antibody (1:1000; catalog no. NBP1-05767; RRID: AB_1556282; Novus Biologicals). After the incubation with primary antibody, the sections were washed in 0.1M PB three times for 10 minutes each, incubated with biotinylated secondary antibody solution (rabbit IgG; Vector Laboratories) for 2 hours, and then rinsed again in 0.1M PB three times for 10 minutes each. This was followed by incubation with rabbit IgG (Vector Laboratories) for 60 minutes. After this incubation, the sections were washed in 0.1M PB three times for 10 minutes each, and the complex antigen-antibody was visualized using diaminobenzidine (DAB Peroxidase Substrate; Vector Laboratories). Finally, the sections were rinsed again in 0.1M PB and mounted onto glass slides, dried, dehydrated in ethanol from 80% to 100%, diaphanized in xylol, and coverslipped with Entellan (Merck, Kenilworth, NJ). Quantification of HIF-1α signal was performed in 20 slides of each group using Image Pro Plus 6.0 software (Media Cybernetics, Inc., Bethesda, MD) calibrated with digital color filters so that only immunostained areas were identified automatically and background staining excluded from the measurement. Results were expressed per area analyzed relative to the total area, with 0 being absence of staining and 1 staining of the entire area.
mRNA analysis by RT-qPCR
Total tissue RNA was extracted using either the RNeasy® Plus Universal Mini Kit (Qiagen, Germantown, MD) or RNeasy® Lipid Tissue Mini Kit (Qiagen) and digested with DNase using the RNase-Free DNase Set (Qiagen). RNA was quantified using the NanoDrop 2000 spectrophotometer (Thermo Fisher, Waltham, MA), and 1.0 µg of total RNA was used to synthesize cDNA using either the SuperScript® III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, Carlsbad, CA) in a Mastercycler® Gradient thermocycler (Eppendorf, Hauppauge, NY) or the Transcriptor First Strand cDNA Synthesis Kit (Roche) in a Mastercycler® pro thermocycler (Eppendorf). Genes of interest were analyzed (Eco™ Real-Time PCR System; Illumina/Step-one real-time PCR system; Step-one-plus real-time PCR system; Applied Biosystems, Foster City, CA) using either QuantiTect™ SYBR® Green (Qiagen) or PerfeCTa SYBR Green Fastmix Rox (Quantas, Beverly, MA). Standard curves were built with 3 to 4 points of serially diluted (factor of 5) mixed experimental and control group cDNA; r > 0.95 for all curves of hippocampus, >0.98 for all curves of amygdala, and >0.97 for all curves of prefrontal cortex; amplification efficiency of 50% to 100% for the hippocampus, 42% to 93% for the amygdala, and 69% to 102% for the prefrontal cortex. Relative expression of genes of interest was expressed as a function of the housekeeping internal control gene Cyclophilin A (Cyclo A). The PCR program consisted in 2 minutes at 50°C, a hot start of 95°C for 15 minutes, 40 to 50 cycles of 15 seconds at 94°C, 30 seconds at 60°C, 30 seconds at 72°C, followed by a melt curve of 15 seconds at 95°C, 15 seconds at 55°C and 15 seconds at 95°C. All primer sequences used in this study are listed in Supplemental Table 1. Mir128-1 mRNA analysis was performed using Taqman reagents (assay IDMm04238113_s1; catalog no. 4426961; Applied Biosystems) with the following conditions: 95°C for 20 seconds, followed by 40 cycles of 1 second at 95°C and 20 seconds at 60°C. Cyclo A was also used as an internal control gene, and there was no difference in its expression between the groups.
Microarray analysis
Total tissue RNA was extracted from hippocampus of Astro-D2KO mice and their control using the RNeasy® Lipid Tissue Mini Kit (Qiagen). The DNase treatment was performed using the RNase-Free DNase Set (Qiagen). RNA was quantified using a NanoDrop spectrophotometer, and RNA quality and integrity were analyzed by electrophoresis in a 1% denaturing agarose gel stained with ethidium bromide. Microarray analysis was performed by the Joslin Diabetes Center Genomics Core Laboratory (Boston, MA). Gene expression was evaluated using Affymetrix GeneChip® Mouse Gene 2.0 ST arrays (Affymetrix, Santa Clara, CA); gene expression data was preprocessed using Affymetrix Expression Console software. Differential expression analysis was performed with Affymetrix Transcriptome Analysis Console. One-way ANOVA was used to calculate P values for each fold change (linear); multitesting correction was then performed using the Benjamini-Hochberg Step-Up false discovery rate (FDR)–controlling procedure for all the expressed genes. The differentially expressed genes were considered statistically significant with an ANOVA P < 0.05; gene set enrichment analysis (GSEA) used the Broad Institute platform. Expression values for all genes from all hippocampus samples were used in the analysis; no filter was applied to eliminate genes with low expression. GSEA included calculation of enrichment scores, estimation of significance level of enrichment scores (nominal P value), and adjustment for multiple hypothesis testing including the normalized enrichment score and FDR. A nominal P value <1% was chosen to indicate significance.
Deiodinase activity and serum T3 and T4 levels
Deiodinase activities were assayed in tissue sonicates as previously described (32).
Serum T3 and T4 were measured using an enzyme-linked immunosorbent assay kit from CUSABIO (CSB-E05086m and CSB-E05083m; Atlanta, GA) using an in-house standard curve prepared with mouse charcoal-stripped serum (28). In sum, 35 µL of standard or samples were added in a 96-well plate. Then, 50 µL of conjugate reagent was subsequently added and the plate was incubated for 60 minutes at 37°C. Wells were aspirated and washed three times using 200 µL of wash buffer. Next, 50 µL of horseradish peroxidase–avidin reagent was added, and the plate incubated for 30 minutes at 37°C. Wells were aspirated and washed three times using 200 µL of wash buffer. Then, 50 µL of substrate A and B were added and the plate was incubated for 15 minutes at 37°C in the dark. Stop solution was added and the optical density measured within 10 minutes using a microplate reader set to 450 nm. Data were plotted and calculated using a four-parameter logistic curve fit.
Statistical analyses
All data were analyzed using PRISM software (GraphPad Software) and are expressed as mean ± SEM. Comparisons between two groups were analyzed with a two-tailed Student t test; comparisons between more than two groups were analyzed by one-way ANOVA followed by the Newman-Keuls multiple comparison test to detect differences between groups. P < 0.05 was considered statistically significant.
Results
SE in mice
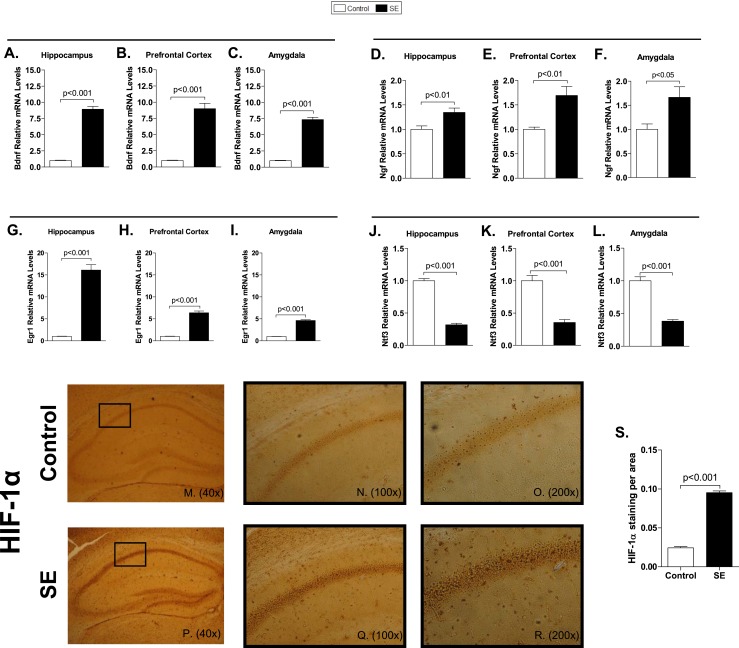
SE in mice initiated 30 to 40 minutes after pilocarpine administration (indicated by P in Fig. 1) and was interrupted 3 hours later with diazepam (indicated by D in Fig. 1). About 13% of the animals receiving pilocarpine did not exhibit SE and were considered later as indicated. Many genes are known to be affected by SE. For example, mRNA levels of Bdnf, Ngf, and Egr1 were elevated in the hippocampus, prefrontal cortex, and amygdala (Fig. 2A–2I), whereas Ntf3 was universally downregulated (Fig. 2J–2L) after 3 hours of SE in animals euthanized 30 minutes later. Seizure was stopped by diazepam (indicated by K1 in Fig. 1A). To evaluate whether the 3 hours of SE resulted in hypoxia, HIF-1α immunostaining was assessed in the hippocampal formation and showed an increase in all hippocampus, especially in the CA1 region (Fig. 2M–2R).
Figure 2.
mRNA levels of typical genes affected by seizures and HIF-1α staining in C57Bl/6J mice following 3 hours of SE. In all cases, SE was induced by pilocarpine administration, as in Fig. 1A. Bdnf mRNA levels in (A) hippocampus, (B) prefrontal cortex, and (C) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). Ngf mRNA levels in (D) hippocampus, (E) prefrontal cortex, and (F) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). Egr1 mRNA levels in (G) hippocampus, (H) prefrontal cortex, and (I) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). Ntf3 mRNA levels in (J) hippocampus, (K) prefrontal cortex, and (L) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). Values represent the mean ± SEM. (M–R) Low-power magnification images showing immunohistochemical analysis of HIF-1α in hippocampus (n = 4); (N, Q) ×100 and (O, R) ×200 magnifications of the CA1 field. (S) HIF-1α signal expressed per area in hippocampal CA1 field; values are mean ± SEM.
Deiodinase activity and serum T3 and T4 levels
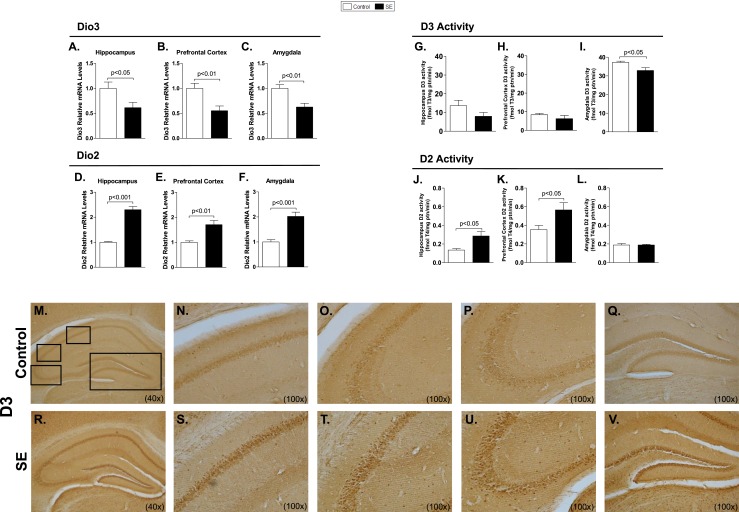
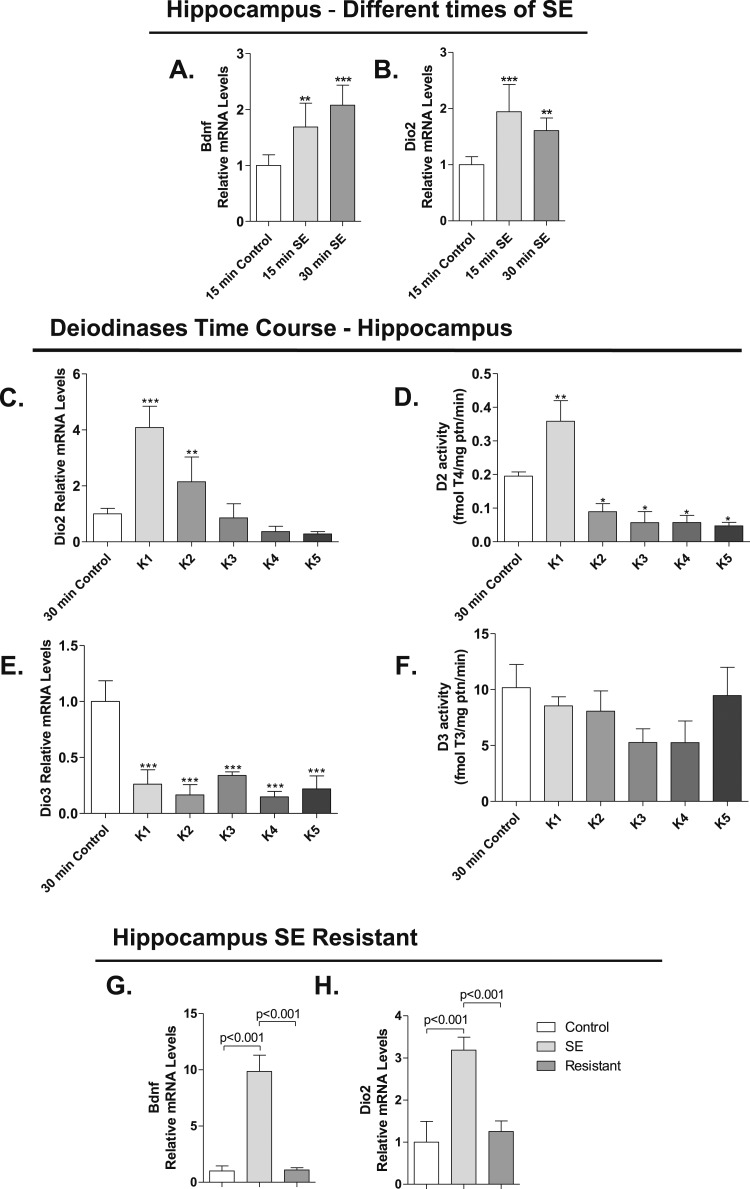
Dio3 mRNA levels were reduced in the hippocampus, prefrontal cortex, and amygdala (Fig. 3A–3C), whereas D3 activity remained stable, mildly reduced in the amygdala (Fig. 3G–3I). However, D3 immunostaining was enhanced, given its transit to the neuronal nuclei (Fig. 3M–3V). Dio2 expression increased in the three regions (Fig. 3D–3F) and there was activity only in the hippocampus and cerebral cortex (Fig. 3J–3L). Changes in Dio2 and Bdnf expression in the hippocampus occurred rapidly, after only 15 or 30 minutes of seizure initiation (Fig. 4A and 4B). Dio2 mRNA levels and activity returned to baseline shortly after diazepam was injected (Fig. 4C–4D). Notably, at later time points, D2 activity decreased markedly, to <50% of baseline levels (Fig. 4D). In contrast, Dio3 expression remained low throughout the observation period, but D3 activity never decreased (Fig. 4E–4F). Notably, neither Dio2 nor Bdnf mRNA levels was modified in a subset of mice that are refractory to pilocarpine-induced seizure (Fig. 4G–4H). This indicates that changes in Dio2 expression following SE are due to increased neuronal activity caused by seizures and not to pilocarpine or diazepam per se. Circulating T3 [control (n = 4): 0.35 ± 0.04 vs SE (n = 5): 0.36 ± 0.01 ng/mL] and T4 levels [control (n = 4): 64.53 ± 14.3 vs SE (n = 5): 73.55 ± 8.48 ng/mL] remained unaffected by 3 hours of SE.
Figure 3.
Deiodinase expression and activity and D3 staining in C57Bl/6J mice following 3 hours of SE. Dio3 mRNA levels in (A) hippocampus, (B) prefrontal cortex, and (C) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). Dio2 mRNA levels in (D) hippocampus, (E) prefrontal cortex, and (F) amygdala measured by RT-qPCR and using Cyclo A as internal control (n = 9). (G–L) SE was induced by lithium chloride and pilocarpine administration, as in Fig. 1B; animals were euthanized at K1. D3 activity in (G) hippocampus, (H) prefrontal cortex, and (I) amygdala (n = 5). D2 activity in (J) hippocampus, (K) prefrontal cortex, and (L) amygdala (n = 5). Values represent mean ± SEM. (M–V) SE was induced by pilocarpine administration, as in Fig. 1A. Low-power magnification images showing immunohistochemical analysis of D3 in hippocampus (n = 4): (M, R) ×40 magnification of hippocampus, (N–Q, S–V) high magnifications of the black boxes in (M) and (R), respectively: (N, S) ×100 magnification of CA1 field, (O, T) ×100 magnification of CA2 field, (P, U) ×100 magnification of CA3 field, and (Q, V) ×100 magnification of the dentate gyrus.
Figure 4.
Gene expression and deiodinases activity in mice after 3 hours of SE. In all cases, SE was induced by lithium chloride and pilocarpine administration, as in Fig. 1B. (A) Bdnf mRNA levels in hippocampus of C57Bl/6J mice undergoing different times of SE (15 and 30 minutes); measurements by RT-qPCR using Cyclo A as internal control; given the short duration of the SE, animals were not treated with diazepam (saline, n = 6; 15 minutes, n = 7; 30 minutes, n = 4). (B) Dio2 mRNA levels in hippocampus of C57Bl/6J mice undergoing different times of SE (15 and 30 minutes) measured by RT-qPCR and using Cyclo A as internal control; as before, SE animals were not treated with diazepam (saline, n = 7; 15 and 30 minutes, n = 5). (C) Dio2 mRNA levels in hippocampus of C57Bl/6J mice euthanized in different times (K1, K2, K3, K4, and K5) after diazepam injection measured by RT-qPCR and using Cyclo A as internal control (control, n = 5; K1, n = 5; K2, n = 4; K3, n = 3; K4, n = 4; K5, n = 4). (D) D2 activity in hippocampus of C57Bl/6J mice euthanized in different times (K1, K2, K3, K4, and K5) after diazepam injection (control, n = 4; K1, n = 4; K2, n = 4; K3, n = 3; K4, n = 4; K5, n = 4). (E) Dio3 mRNA levels in hippocampus of C57Bl/6J mice euthanized at different times (K1, K2, K3, K4, and K5) after diazepam injection measured by RT-qPCR and using Cyclo A as internal control (control, n = 4; K1, n = 5; K2, n = 4; K3, n = 2; K4, n = 4; K5, n = 4). (F) D3 activity in hippocampus of C57Bl/6J mice euthanized in different times (K1, K2, K3, K4, and K5) after diazepam injection (control, n = 4; K1, n = 2; K2, n = 4; K3, n = 3; K4, n = 4; K5, n = 4). (G) Bdnf mRNA levels in hippocampus of GFAP-Cre animals resistant to pilocarpine measured by RT-qPCR and using Cyclo A as internal control (control, n = 4; SE, n = 4; resistant, n = 4). (H) Dio2 mRNA levels in hippocampus of GFAP-Cre animals resistant to pilocarpine measured by RT-qPCR and using Cyclo A as internal control (control, n = 4; SE, n = 3; resistant, n = 3). Values represent the mean ± SEM *P < 0.05 vs control; **P < 0.01 vs control; ***P < 0.001 vs control.
Unbiased hippocampal gene expression in mice experiencing 3 hours of SE
To understand the role played by D2 in the modifications of CNS gene expression associated with SE, we focused on the hippocampus and performed an unbiased gene expression analysis using microarray of hippocampal RNA obtained 3 hours after seizure was stopped (indicated by K2 in Fig. 1B); a time chosen based on the time-course data (Fig. 5D–5F). For these experiments, we also used Astro-D2KO mice (and their CRE-GFAP controls), in which Dio2 is selectively inactivated in glial cells (27). Here also, circulating T3 [Astro-D2KO (n = 5): 0.34 ± 0.02 vs Astro-D2KO-SE (n = 4): 0.38 ± 0.01 ng/mL] and T4 levels [Astro-D2KO (n = 5): 60.39 ±1 2.47 vs Astro-D2KO-SE (n = 4): 76.98 ± 11.86 ng/mL] remained unaffected by 3 hours of SE.
Figure 5.
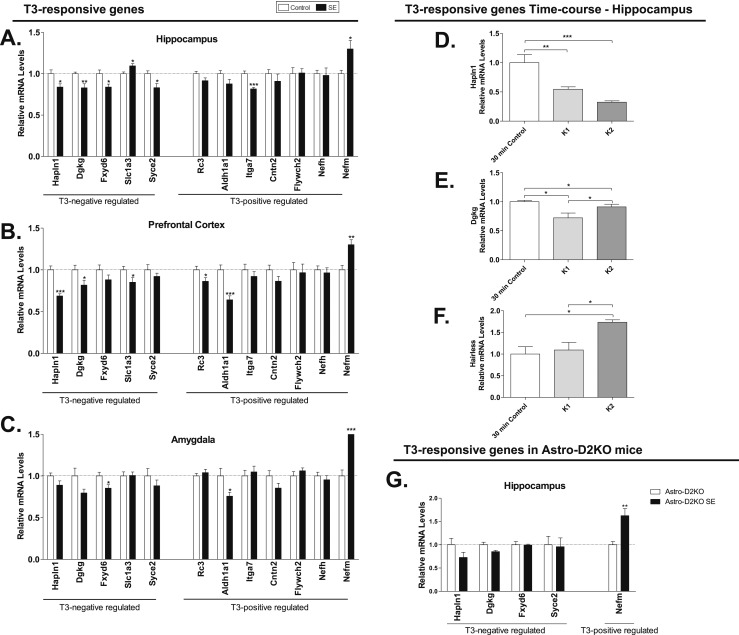
mRNA levels of T3-responsive genes in mice after SE. (A–C) SE was induced by pilocarpine administration, as in Fig. 1A. (D–G) SE was induced by lithium chloride and pilocarpine administration, as in Fig. 1B. T3-responsive gene mRNA levels in (A) hippocampus, (B) prefrontal cortex, and (C) amygdala of C57Bl/6J mice following pilocarpine-induced SE that were euthanized 30 minutes after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 9). (D–F) T3-responsive gene mRNA levels in hippocampus of C57Bl/6J mice following lithium pilocarpine–induced SE that were euthanized 30 minutes and 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (control, n = 5; K1, n = 5; K2, n = 4). (D) Hapln1, (E) Dgkg, (F) hairless, and (G) T3-responsive gene mRNA levels in hippocampus of Astro-D2KO mice following lithium pilocarpine–induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control. Values represent mean ± SEM *P < 0.05 vs control; **P < 0.01 vs control; ***P < 0.001 vs control.
The differential expression analysis of control vs control-SE hippocampus RNA indicated that in control-SE mice, 3934 transcripts were modified at a P value < 0.01, of which 1602 were up- and 2332 downregulated. If a threefold cutoff was used, these numbers were reduced to 128 and 3 (Supplemental Table 2), respectively, with the top 20 upregulated genes being involved in neuronal plasticity, response to stress, and inflammation (Table 1). The same analysis was done for Astro-D2KO vs Astro-D2KO-SE hippocampus RNA. In these animals, SE modified the expression of 4402 transcripts, of which 1696 were up- and 2706 downregulated. With the threefold cutoff, these numbers were reduced to 104 and 3 (Supplemental Table 3), respectively. The list of top five genes in both animals is very similar, but with markedly reduced fold induction in Astro-D2KO-SE animals (Table 1). It is also noticeable that genes that ranked high in the control-SE are in general ranked low in the Astro-D2KO-SE and vice versa (Table 1).
Table 1.
Hippocampal Genes Enriched in Both Control-SE and Astro-D2KO-SE
| Gene Symbol | Description | Control-SE | Astro-D2KO-SE | ||
|---|---|---|---|---|---|
| Rank | Fold | Rank | Fold | ||
| Npas4 | Neuronal PAS domain protein 4 | 1 | 46 | 3 | 23 |
| Hspa1b | Heat shock protein 1B | 2 | 37 | 1 | 28 |
| Hspa1a | Heat shock protein 1A | 3 | 36 | 2 | 26 |
| Fosb | FBJ osteosarcoma oncogene B | 4 | 27 | 5 | 18 |
| Fos | FBJ osteosarcoma oncogene | 5 | 18 | 6 | 14 |
| Inhba | Inhibin β-A | 6 | 17 | 13 | 9 |
| Ccl3 | Chemokine (C-C motif) ligand 3 | 7 | 16 | 4 | 22 |
| Egr1 | Early growth response 1 | 8 | 11 | 15 | 9 |
| Atf3 | Activating transcription factor 3 | 9 | 11 | 18 | 8 |
| Tnfrsf23 | Tumor necrosis factor receptor superfamily, member 23 | 10 | 11 | 21 | 7 |
| Egr4 | Early growth response 4 | 11 | 10 | 19 | 7 |
| Pcsk1 | Proprotein convertase subtilisin/kexin type 1 | 12 | 10 | 10 | 11 |
| Arc | Activity regulated cytoskeletal-associated protein | 13 | 9.4 | 24 | 6 |
| Nr4a1 | Nuclear receptor subfamily 4, group A, member 1 | 14 | 9.2 | 14 | 9.2 |
| Lilrb4 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 15 | 8.8 | 9 | 13 |
| Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | 19 | 7.6 | 7 | 13 |
| Serpinb2 | Serine (or cysteine) peptidase inhibitor, clade B, member 2 | — | — | 8 | 13 |
| Nts | Neurotensin | 27 | 6.1 | 11 | 10 |
| Gadd45g | Growth arrest and DNA-damage-inducible 45 γ | 20 | 7.0 | 12 | 10 |
Differential expression was at P < 0.01 and a fold change ≥3 or ≤−3 in control-SE (n = 4) and Astro-D2KO-SE (n = 4) hippocampus.
Although several common genes were affected by SE in both control and Astro-D2KO animals (2416 transcripts; Supplemental Table 4 for > 3-fold change), we noticed that a substantial number of genes (1518 transcripts) were only affected by SE in control animals, 6 of which previously identified as being responsive to T3 in cerebral cortex and/or striatum (i.e., Luzp1, Paqr6, Shh, Cirbp, Hapln1, and Nrgn) (33–35). Considering fold change > 2, this number dropped to 36 genes (Table 2). Using the same criteria, the 1986 transcripts that were affected by SE only in Astro-D2KO dropped to 28 genes (Table 3). A major difference between these lists is the presence of several mitochondrial-related genes in control-SE and several inflammation-related genes in the Astro-D2KO–SE samples (Tables 2 and 3).
Table 2.
Individual Transcripts Affected by SE in Hippocampus of Control-SE Mice
| Gene Symbol | Description | Fold Change | P |
|---|---|---|---|
| Genes Modified in Control-SE Hippocampus Only | |||
| Angiogenesis | |||
| n-R5s87 | Nuclear encoded rRNA 5S 87 | 3.46 | 0.003093 |
| Mir128-2 | microRNA 128-2 | 2.19 | 0.001731 |
| Cell cycle and proliferation | |||
| Cks2 | CDC28 protein kinase regulatory subunit 2; cyclin-dependent kinases regulatory subunit 2-like; predicted gene 12891; predicted gene 15452 | 2.73 | 0.002548 |
| Mnd1 | Meiotic nuclear divisions 1 homolog (Saccharomyces cerevisiae) | 2.62 | 0.002064 |
| Mir136 | microRNA 136; mmu-mir-136; mmu-mir-3071 | 2.2 | 0.006001 |
| Lcmt2 | Leucine carboxyl methyltransferase 2 | 2.19 | 0.004942 |
| Cdk6 | Cyclin-dependent kinase 6 | 2.15 | 0.008803 |
| Mir708 | microRNA 708 | 2.13 | 0.000156 |
| Cell death mechanisms | |||
| Mir376a | microRNA 376a | 2.32 | 0.009128 |
| Enzymes | |||
| Acat3 | Acetyl-coenzyme A acetyltransferase 3 | 2.59 | 0.002661 |
| Nanp | N-acetylneuraminic acid phosphatase | −2.17 | 0.001961 |
| Mitochondrial function | |||
| mt-Tk | Mitochondrially encoded tRNA lysine | 6.39 | 0.002554 |
| mt-Tt | Mitochondrially encoded tRNA threonine | 4.81 | 0.001726 |
| mt-Tt | Mitochondrially encoded tRNA threonine | 4.64 | 0.002216 |
| mt-Ts2 | Mitochondrially encoded tRNA serine 2; mitochondrially encoded tRNA leucine 2 | 3.85 | 0.003586 |
| mt-Ts2 | Mitochondrially encoded tRNA serine 2 | 3.29 | 0.007613 |
| mt-Ty | Mitochondrially encoded tRNA tyrosine | 2.1 | 0.00825 |
| Histones | |||
| Hist2h3b | Histone cluster 2, H3b; histone cluster 1, H3e; histone cluster 1, H3b; histone cluster 1, H3d; histone cluster 1, H3c; histone cluster 1, H3f; histone cluster 2, H3c2; histone cluster 2, H3c1 | 3.32 | 0.004662 |
| Hist1h3d | Histone cluster 1, H3d | 2.03 | 0.000865 |
| Inflammation | |||
| Stk40 | Serine/threonine kinase 40 | 2.77 | 0.00856 |
| Extracellular matrix | |||
| Gm13034 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 pseudogene | 2.53 | 0.000736 |
| Tinag | Tubulointerstitial nephritis antigen | 2.34 | 0.006487 |
| Dsp | Desmoplakin | −3.49 | 0.002388 |
| Glt8d2 | Glycosyltransferase 8 domain containing 2 | −2.09 | 0.000497 |
| Neural plasticity | |||
| Emp1 | Epithelial membrane protein 1 | 3.25 | 0.00215 |
| Neuroprotective mechanisms | |||
| Dnajb5 | DnaJ (Hsp40) homolog, subfamily B, member 5 | 2.12 | 0.00161 |
| Snord14e | Small nucleolar RNA, C/D box 14E; heat shock protein 8 | 2.01 | 0.001037 |
| Neurotransmitter signaling | |||
| Sstr2 | Somatostatin receptor 2 | 2.31 | 0.007373 |
| Gucy2c | Guanylate cyclase 2c | 2.16 | 0.002788 |
| Dok5 | Docking protein 5 | 2.08 | 0.004218 |
| Per2 | Period circadian clock 2 | 2.04 | 0.002296 |
| Ttc26 | Tetratricopeptide repeat domain 26 | −2.12 | 0.006375 |
| Ttc26 | Tetratricopeptide repeat domain 26 | −2.12 | 0.006375 |
| Slc22a8 | Solute carrier family 22 (organic anion transporter), member 8 | −2.45 | 0.007346 |
| Transcription factors | |||
| Hes5 | Hairy and enhancer of split 5 (Drosophila) | −2 | 0.001097 |
Differential expression was at a P < 0.01 and a fold change ≥2 or ≤−2 in control-SE (n = 4) hippocampus.
Table 3.
Individual Transcripts Affected by SE in Hippocampus of Astro-D2KO-SE Mice
| Gene Symbol | Description | Fold Change | P |
|---|---|---|---|
| Genes Modified in Astro-D2KO-SE Hippocampus Only | |||
| Apoptosis | |||
| Mir128-1 | microRNA 128-1 | 2.17 | 0.00109 |
| Enzymes | |||
| Arxes1 | Adipocyte-related X-chromosome expressed sequence 1; adipocyte-related X-chromosome expressed sequence 2 | 2.05 | 0.000814 |
| B3gnt5 | UDP-GlcNAc: β Gal β-1,3-N-acetylglucosaminyltransferase 5 | 2.03 | 0.000779 |
| Pi15 | Peptidase inhibitor 15 | 2 | 0.001314 |
| Nt5c1a | 5-nucleotidase, cytosolic IA | 2 | 0.001601 |
| Aspa | Aspartoacylase | −2.03 | 0.004198 |
| Extracellular matrix | |||
| Grem2 | gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) | 2.67 | 0.000002 |
| Hs3st5 | heparan sulfate (glucosamine) 3-O-sulfotransferase 5 | 2.08 | 0.00285 |
| Inflammation | |||
| Serpinb2 | Serine (or cysteine) peptidase inhibitor, clade B, member 2 | 13.01 | 0.000372 |
| Ctla2a | Cytotoxic T lymphocyte-associated protein 2 α | 2.6 | 0.00251 |
| Msr1 | Macrophage scavenger receptor 1 | 2.04 | 0.003938 |
| Il11 | Interleukin 11 | 2.02 | 0.007672 |
| Sbno2 | Strawberry notch homolog 2 (Drosophila) | 2.4 | 0.007635 |
| Membrane transporters | |||
| Slc10a6 | Solute carrier family 10 (sodium/bile acid cotransporter family), member 6 | 2.18 | 0.007324 |
| Neural plasticity | |||
| Zfp804a | Zinc finger protein 804A | 2.46 | 0.002032 |
| Gdnf | Glial cell line derived neurotrophic factor | 2.03 | 0.001031 |
| Mir344e | microRNA 344e | −2.36 | 0.000517 |
| Ptch1 | Patched homolog 1 | −2.13 | 0.000437 |
| Neurotransmitter signaling | |||
| Penk | Preproenkephalin; short chain dehydrogenase/reductase family 39U, member 1 | 2.47 | 0.004063 |
| Gfra1 | Glial cell line derived neurotrophic factor family receptor α 1 | 2.43 | 0.002592 |
| Smim3 | Small integral membrane protein 3 | 2.22 | 0.00379 |
| Other | |||
| Ccdc138 | Coiled-coil domain containing 138 | 2.22 | 0.008305 |
| Snord107 | Small nucleolar RNA, C/D box 107; SNRPN upstream reading frame | −2.38 | 0.001387 |
| Mir1839 | microRNA 1839; RIKEN cDNA 2900076A07 gene | −2.18 | 0.005299 |
| Oxidative stress | |||
| Gpx8 | Glutathione peroxidase 8 (putative) | 2.19 | 0.001493 |
| Transcription factor | |||
| Osr1 | Odd-skipped related 1 (Drosophila) | −2.01 | 0.001485 |
Differential expression was at P < 0.01 and a fold change ≥2 or ≤−2 in Astro-D2KO-SE (n = 4) hippocampus.
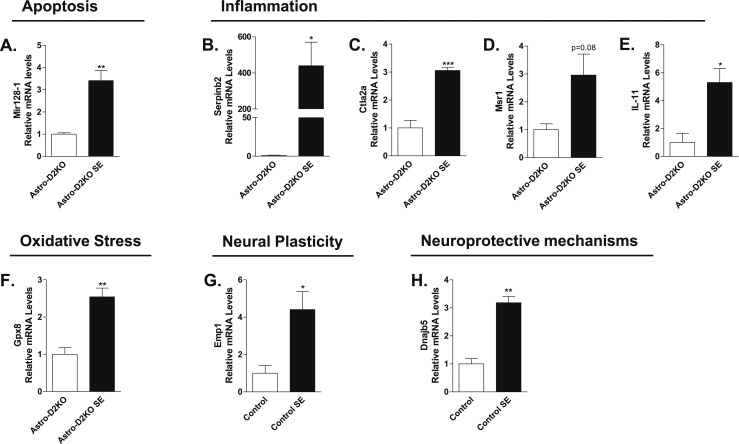
We also compared the complete set of genes modified by SE in control vs Astro-D2KO mice using GSEA at a nominal P value < 1% (Supplemental Table 5). GSEA revealed that 88 gene sets were enriched and 8 impoverished in control-SE, whereas in the Astro-D2KO-SE 96 gene sets were enriched and 5 impoverished. There are 28 enriched gene sets common to control and Astro-D2KO animals, representing those genes that are modified by SE independently of D2 expression (Supplemental Table 4). In contrast, of the 141 gene sets that were affected by D2 expression or lack thereof, 68 were enriched in the control animals only and 73 in the Astro-D2KO animals only (Supplemental Table 5). The top 10 gene sets enriched in control-SE include genes involved in response to oxidative stress, cell signaling, and homeostasis (Table 4). In contrast, the top gene sets enriched in Astro-D2KO-SE include transfer of phosphorus-containing groups and kinase activity (Table 4). These findings were confirmed by RT-qPCR of representative genes from each of these gene sets (Fig. 6A–6H).
Table 4.
GSEA of Genes Affected by SE in Control vs Astro-D2KO Mice
| Gene Ontology | Description | Total No. Genes in Set | ES | NES | Nominal P | FDR q Value |
|---|---|---|---|---|---|---|
| Enriched in control-SE animals | ||||||
| GO:0006979 | Response to oxidative stress | 40 | −0.39 | −1.35 | 0 | 0.276 |
| GO:0019932 | Second-messenger–mediated signaling | 140 | −0.47 | −1.34 | 0 | 0.276 |
| GO:0007267 | Cell–cell signaling | 369 | −0.42 | −1.35 | 0 | 0.277 |
| GO:0045597 | Positive regulation of cell differentiation | 24 | −0.68 | −1.36 | 0 | 0.279 |
| GO:0009991 | Response to extracellular stimulus | 33 | −0.65 | −1.37 | 0 | 0.281 |
| GO:0019725 | Cellular homeostasis | 133 | −0.48 | −1.37 | 0 | 0.281 |
| GO:0009887 | Organ morphogenesis | 139 | −0.43 | −1.37 | 0 | 0.282 |
| GO:0048878 | Chemical homeostasis | 139 | −0.48 | −1.37 | 0 | 0.284 |
| GO:0008284 | Positive regulation of cell proliferation | 128 | −0.49 | −1.38 | 0 | 0.291 |
| GO:0030234 | Enzyme regulator activity | 294 | −0.36 | −1.38 | 0 | 0.292 |
| Enriched in AstroD2KO-SE animals | ||||||
| GO:0016772 | Transferase activity transferring phosphorus-containing groups | 392 | −0.26 | −1.26 | 0 | 0.366 |
| GO:0004672 | Protein kinase activity | 261 | −0.31 | −1.3 | 0 | 0.371 |
| GO:0051239 | Regulation of multicellular organismal process | 139 | −0.45 | −1.29 | 0 | 0.373 |
| GO:0016301 | Kinase activity | 340 | −0.27 | −1.29 | 0 | 0.376 |
| GO:0016773 | Phosphotransferase activity alcohol group as acceptor | 309 | −0.29 | −1.31 | 0 | 0.38 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 72 | −0.37 | −1.34 | 0 | 0.381 |
| GO:0009607 | Response to biotic stimulus | 95 | −0.55 | −1.34 | 0 | 0.384 |
| GO:0031667 | Response to nutrient levels | 29 | −0.64 | −1.32 | 0 | 0.385 |
| GO:0006937 | Regulation of muscle contraction | 17 | −0.58 | −1.3 | 0 | 0.385 |
| GO:0008283 | Cell proliferation | 470 | −0.44 | −1.31 | 0 | 0.387 |
Comparison of [control (n = 4) vs control-SE (n = 4)] vs [Astro-D2KO (n = 4) vs Astro-D2KO-SE (n = 4)] animals at a nominal P < 0.01.
Abbreviations: ES, enrichment score; NES, normalized enrichment score.
Figure 6.
mRNA levels of representative genes identified through analysis of microarray data. All samples are from hippocampi of control-SE or Astro-D2KO-SE mice undergoing 3 hours of SE. In all cases, SE was induced by lithium chloride and pilocarpine administration, as in Fig. 1B. (A) Mir128-1 mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (B) Serpinb2 mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (C) Ctla2a mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (D) Msr1 mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE euthanized 3 hours after diazepam injection measured by RT-qPCR and using Cyclo A as internal control (n = 4). (E) IL-11 mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (F) Gpx8 mRNA levels in hippocampus of Astro-D2KO mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (G) Emp1 mRNA levels in hippocampus of control mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). (H) Dnajb5 mRNA levels in hippocampus of control mice following pilocarpine-induced SE that were euthanized 3 hours after diazepam injection, measured by RT-qPCR and using Cyclo A as internal control (n = 4). Values represent the mean ± SEM *P < 0.05 vs control with; **P < 0.01 vs control with; ***P < 0.001 vs control.
Expression of typical T3-responsive genes in the CNS of mice following SE
We used RT-qPCR to test whether induction of D2 activity by SE was associated with changes in the expression of typical T3-responsive genes. We looked at two sets of genes: five negatively and seven positively regulated by T3 in the CNS (35). In the hippocampus, SE reduced the expression of four genes negatively regulated by T3 (i.e., Hapln1, Dgkg, Fxyd6, and Syce2), whereas it increased the expression of one gene positively regulated by T3 (i.e., Nefm) (Fig. 5A). Two genes responded with the opposite behavior, Slc1a3 and Itga7, whereas other T3-responsive genes were not affected (Fig. 5A). A similar general pattern was observed in the prefrontal cortex (Fig. 5B) and amygdala (Fig. 5C), albeit the latter was only minimally responsive to T3. A time-course analysis of three representative genes in the hippocampus indicated a progressive decrease in Hapln1 mRNA and a time-dependent increase in hairless mRNA, whereas Dgkg mRNA levels went down by 30 minutes and recovered almost fully by 3 hours (Fig. 5D–5F). That the response to T3 was predominantly on the negatively regulated genes is reminiscent of the fact that negatively regulated genes by T3 tend to be more dependent of D2-generated T3 than on plasma T3 (34). Dependency on Dio2 activation was confirmed by studying those 5 T3-regulated genes in Astro-D2KO mice after 3 hours of SE (Fig. 5G). The four negatively regulated genes (i.e., Hapln1, Dgkg, Fxyd6, and Syce2) were no longer affected by 3 hours of SE, whereas the positively regulated gene (i.e., Nefm) remained responsive to 3 hours of SE (Fig. 5G).
Discussion
The current study revealed that SE in mice rapidly increases CNS Dio2 expression, which greatly modifies hippocampal gene expression profile and its response to SE (Tables 1–4). Within 30 minutes of SE interruption (indicated by K1 in Fig. 1A), there is doubling of Dio2 expression in the hippocampus, prefrontal cortex, and amygdala (Fig. 3D–3F) that is accompanied by a decrease in Dio3 mRNA levels (Fig. 3A–3C), albeit D3 activity was not modified within the observation time frame (Fig. 3G–3I). Whereas several T3-responsive genes in the CNS were affected as a result of the SE-induced Dio2 expression (Fig. 5), an unbiased analysis of the hippocampal transcriptome using mice with targeted disruption of Dio2 in astrocytes revealed a much broader effect of Dio2 presence (Tables 1–4). In fact, several genes and gene sets were modified by SE in mice with a functional D2 pathway only, which were mostly related to mitochondrial function and oxidative stress (Tables 1 and 2). In contrast, the profile of SE-induced genes in the animals with inactive Dio2 included peptidase inhibitors, cytokines, and genes involved in inflammation and apoptosis (Tables 1 and 3). This is reminiscent of recent findings that bleomycin-induced lung injury in mice causes Dio2 activation and local T3 production that promotes mitochondrial biogenesis, improved mitochondrial bioenergetics, and weakened mitochondria-regulated apoptosis in alveolar epithelial cells (36). It is notable that the relatively low overlap of the genes potentially regulated by Dio2 in this model and those reportedly regulated by T3 in other datasets. This may be due to differences in brain region, developmental stages, or the possibility that genes primarily affected by Dio2 are few compared with those affected by SE via other mechanisms.
Although pilocarpine binds to all muscarinic acetylcholine receptor subtypes (29), only the M1 receptor mediates seizures via its typical downstream signaling pathway, DAG and IP3. M1Rs are expressed both in neurons and astrocytes, mostly in the cerebral cortex and hippocampus. However, once seizure activity settles in, there is increased expression of cAMP-responsive genes that occurs most likely as a result of increased glutamate release (37–40). Notably, Dio2 is highly responsive to cAMP (6), hence the likely mechanism through which Dio2 expression is increased in the current study. CNS accumulation of cAMP requires and is secondary to seizure activity (41). Indeed, in the current study, seizure but not pilocarpine administration triggered Dio2 expression (Fig. 4H). The expression pattern of T3-responsive genes revealed increased T3-TR signaling in the brain. In addition, Dio2 mRNA increases 15 minutes into the SE (Fig. 4B). It is also noticeable that once SE is stopped with diazepam, the drop in D2 activity was faster than the drop in Dio2 mRNA (Fig. 4C and 4D), probably because the D2 ubiquitin ligase WSB-1 (42) is an HIF-inducible gene (43) and its induction is likely to accelerate D2 ubiquitination and degradation.
In coordination with the Dio2 induction, Dio3 mRNA levels decreased markedly (Fig. 4E), even though sample variability prevented a statistically significant decrease in D3 activity (Fig. 4F). Although expression of Dio2 and Dio3 are often coordinated, a reduction in Dio3 mRNA during hypoxia is unexpected given that Dio3 was shown to be upregulated in recently deceased individuals suffering from diseases associated with poor tissue perfusion (44), in postinfarction rat myocardium (45), in right ventricular hypertrophy (46), and in hypoxic neuroblastoma cells in culture (16, 17). Unfortunately, an analysis of the gene pathways activated by SE as seen in Tables 1–4 does not help identify the mechanism that is overriding the HIF-1α induction of Dio3. Nevertheless, D3 is seen concentrating in the nucleus of the neurons in the SE CNS (Fig. 3M–3V). This is reminiscent of our previous observations that insult to the CNS, such as ischemia, redirects D3 trafficking to the cell nucleus via induction of Hsp40, enhancing its ability to inactivate T3 and reduce T3-TR signaling (16, 17). In the case of SE animals, Hsp40 is induced by about fourfold in both control and Astro-D2 mice (Supplemental Tables 2 and 3).
The hippocampus of animals undergoing SE has been previously studied by microarray; the most affected gene pathways fall into the categories included in Tables 2 and 3. The consensus among these studies is that a robust response in gene expression is mounted to adapt and minimize damage resulting from SE (47–49). Therefore, it is important that, in the absence of a functional D2 pathway, as in the Astro-D2KO animals, the hippocampi of SE animals exhibited a unique gene expression profile that was quite different from control-SE animals. The control-SE animals activate gene pathways related to response to oxidative stress, followed by other gene sets involved in cell homeostasis (Table 4). In contrast, the Astro-D2KO-SE animals activate gene sets that reflect accelerated phosphorylation and kinase activity (Table 4). In fact, the list of top 20 genes upregulated in Astro-D2KO-SE include those connected with inflammation, apoptosis, and cell death, suggesting that a functional Dio2 pathway is critical for the CNS adaptive response that follows SE.
We conclude that thyroid hormone metabolism and action are affected in the CNS following SE in mice. Not only are traditional T3-responsive genes involved, but so also are a large number of genes that participate in the adaptive response mounted by the CNS following SE. The present studies were performed acutely, only a few hours after SE was terminated. Given that these animals could eventually develop permanent brain damage and spontaneous seizures (50), future studies should clarify whether the Astro-D2KO animals are more or less susceptible to developing a chronic epileptic state.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK65055 (to A.C.B.) and Fundação de Amparo à Pesquisa do Estado de São Paulo, Brazil, Grant 2015/13235-1 (to M.O.R.) and Grant 2013/09133-3 (to B.P.P.N.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- CNS
central nervous system
- Cyclo A
Cyclophilin A
- D2
type 2 iodothyronine deiodinase
- D3
type 3 iodothyronine deiodinase
- Dio2
type 2 iodothyronine deiodinase gene
- Dio3
type 3 iodothyronine deiodinase gene
- FDR
false discovery rate
- GSEA
gene set enrichment analysis
- PB
phosphate buffer
- PF
paraformaldehyde
- RT-qPCR
quantitative RT-PCR
- SE
status epilepticus
- TR
thyroid hormone receptor
References
- 1. Brent GA. Mechanisms of thyroid hormone action. J Clin Invest. 2012;122(9):3035–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gereben B, Zavacki AM, Ribich S, Kim BW, Huang SA, Simonides WS, Zeöld A, Bianco AC. Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. 2008;29(7):898–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohacsik P, Zeold A, Bianco AC, Gereben B. Thyroid hormone and the neuroglia: both source and target. J Thyroid Res. 2011;2011:215718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tu HM, Kim SW, Salvatore D, Bartha T, Legradi G, Larsen PR, Lechan RM. Regional distribution of type 2 thyroxine deiodinase messenger ribonucleic acid in rat hypothalamus and pituitary and its regulation by thyroid hormone. Endocrinology. 1997;138(8):3359–3368. [DOI] [PubMed] [Google Scholar]
- 5. Guadaño-Ferraz A, Obregón MJ, St Germain DL, Bernal J. The type 2 iodothyronine deiodinase is expressed primarily in glial cells in the neonatal rat brain. Proc Natl Acad Sci USA. 1997;94(19):10391–10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartha T, Kim SW, Salvatore D, Gereben B, Tu HM, Harney JW, Rudas P, Larsen PR. Characterization of the 5′-flanking and 5′-untranslated regions of the cyclic adenosine 3′,5′-monophosphate-responsive human type 2 iodothyronine deiodinase gene. Endocrinology. 2000;141(1):229–237. [DOI] [PubMed] [Google Scholar]
- 7. Canettieri G, Celi FS, Baccheschi G, Salvatori L, Andreoli M, Centanni M. Isolation of human type 2 deiodinase gene promoter and characterization of a functional cyclic adenosine monophosphate response element. Endocrinology. 2000;141(5):1804–1813. [DOI] [PubMed] [Google Scholar]
- 8. Burmeister LA, Pachucki J, St Germain DL. Thyroid hormones inhibit type 2 iodothyronine deiodinase in the rat cerebral cortex by both pre- and posttranslational mechanisms. Endocrinology. 1997;138(12):5231–5237. [DOI] [PubMed] [Google Scholar]
- 9. Kim SW, Harney JW, Larsen PR. Studies of the hormonal regulation of type 2 5′-iodothyronine deiodinase messenger ribonucleic acid in pituitary tumor cells using semiquantitative reverse transcription-polymerase chain reaction. Endocrinology. 1998;139(12):4895–4905. [DOI] [PubMed] [Google Scholar]
- 10. Steinsapir J, Harney J, Larsen PR. Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes. J Clin Invest. 1998;102(11):1895–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol Endocrinol. 2000;14(11):1697–1708. [DOI] [PubMed] [Google Scholar]
- 12. Bernal J, Guadaño-Ferraz A, Morte B. Perspectives in the study of thyroid hormone action on brain development and function. Thyroid. 2003;13(11):1005–1012. [DOI] [PubMed] [Google Scholar]
- 13. St Germain DL, Galton VA, Hernandez A. Minireview: defining the roles of the iodothyronine deiodinases: current concepts and challenges. Endocrinology. 2009;150(3):1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tu HM, Legradi G, Bartha T, Salvatore D, Lechan RM, Larsen PR. Regional expression of the type 3 iodothyronine deiodinase messenger ribonucleic acid in the rat central nervous system and its regulation by thyroid hormone. Endocrinology. 1999;140(2):784–790. [DOI] [PubMed] [Google Scholar]
- 15. Peeters R, Fekete C, Goncalves C, Legradi G, Tu HM, Harney JW, Bianco AC, Lechan RM, Larsen PR. Regional physiological adaptation of the central nervous system deiodinases to iodine deficiency. Am J Physiol Endocrinol Metab. 2001;281(1):E54–E61. [DOI] [PubMed] [Google Scholar]
- 16. Freitas BC, Gereben B, Castillo M, Kalló I, Zeöld A, Egri P, Liposits Z, Zavacki AM, Maciel RM, Jo S, Singru P, Sanchez E, Lechan RM, Bianco AC. Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest. 2010;120(6):2206–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jo S, Kalló I, Bardóczi Z, Arrojo e Drigo R, Zeöld A, Liposits Z, Oliva A, Lemmon VP, Bixby JL, Gereben B, Bianco AC. Neuronal hypoxia induces Hsp40-mediated nuclear import of type 3 deiodinase as an adaptive mechanism to reduce cellular metabolism. J Neurosci. 2012;32(25):8491–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Margaill I, Royer J, Lerouet D, Ramaugé M, Le Goascogne C, Li WW, Plotkine M, Pierre M, Courtin F. Induction of type 2 iodothyronine deiodinase in astrocytes after transient focal cerebral ischemia in the rat. J Cereb Blood Flow Metab. 2005;25(4):468–476. [DOI] [PubMed] [Google Scholar]
- 19. Lamirand A, Mercier G, Ramaugé M, Pierre M, Courtin F. Hypoxia stabilizes type 2 deiodinase activity in rat astrocytes. Endocrinology. 2007;148(10):4745–4753. [DOI] [PubMed] [Google Scholar]
- 20. Zou L, Burmeister LA, Styren SD, Kochanek PM, DeKosky ST. Up-regulation of type 2 iodothyronine deiodinase mRNA in reactive astrocytes following traumatic brain injury in the rat. J Neurochem. 1998;71(2):887–890. [DOI] [PubMed] [Google Scholar]
- 21. Wasterlain CG, Fujikawa DG, Penix L, Sankar R. Pathophysiological mechanisms of brain damage from status epilepticus. Epilepsia. 1993; 34(Suppl 1)S37–S53. [DOI] [PubMed] [Google Scholar]
- 22. McNamara JO. Cellular and molecular basis of epilepsy. J Neurosci. 1994;14(6):3413–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gastaut H, Gastaut JL, Gonçalves e Silva GE, Fernandez Sanchez GR. Relative frequency of different types of epilepsy: a study employing the classification of the International League Against Epilepsy. Epilepsia. 1975;16(3):457–461. [DOI] [PubMed] [Google Scholar]
- 24. Walczak TS. Neocortical temporal lobe epilepsy: characterizing the syndrome. Epilepsia. 1995;36(7):633–635. [DOI] [PubMed] [Google Scholar]
- 25. Sutter R, Dittrich T, Semmlack S, Rüegg S, Marsch S, Kaplan PW. Acute systemic complications of convulsive status epilepticus-a systematic review. Crit Care Med. 2018;46(1):138–145. [DOI] [PubMed] [Google Scholar]
- 26. Fabene PF, Merigo F, Galiè M, Benati D, Bernardi P, Farace P, Nicolato E, Marzola P, Sbarbati A. Pilocarpine-induced status epilepticus in rats involves ischemic and excitotoxic mechanisms. PLoS One. 2007;2(10):e1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fonseca TL, Correa-Medina M, Campos MP, Wittmann G, Werneck-de-Castro JP, Arrojo e Drigo R, Mora-Garzon M, Ueta CB, Caicedo A, Fekete C, Gereben B, Lechan RM, Bianco AC. Coordination of hypothalamic and pituitary T3 production regulates TSH expression. J Clin Invest. 2013;123(4):1492–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bianco AC, Anderson G, Forrest D, Galton VA, Gereben B, Kim BW, Kopp PA, Liao XH, Obregon MJ, Peeters RP, Refetoff S, Sharlin DS, Simonides WS, Weiss RE, Williams GR; American Thyroid Association Task Force on Approaches and Strategies to Investigate Thyroid Hormone Economy and Action American Thyroid Association Guide to investigating thyroid hormone economy and action in rodent and cell models. Thyroid. 2014;24(1):88–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turski WA, Cavalheiro EA, Schwarz M, Czuczwar SJ, Kleinrok Z, Turski L. Limbic seizures produced by pilocarpine in rats: behavioural, electroencephalographic and neuropathological study. Behav Brain Res. 1983;9(3):315–335. [DOI] [PubMed] [Google Scholar]
- 30. Curia G, Longo D, Biagini G, Jones RS, Avoli M. The pilocarpine model of temporal lobe epilepsy. J Neurosci Methods. 2008;172(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lévesque M, Avoli M, Bernard C. Animal models of temporal lobe epilepsy following systemic chemoconvulsant administration. J Neurosci Methods. 2016;260:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christoffolete MA, Ribeiro R, Singru P, Fekete C, da Silva WS, Gordon DF, Huang SA, Crescenzi A, Harney JW, Ridgway EC, Larsen PR, Lechan RM, Bianco AC. Atypical expression of type 2 iodothyronine deiodinase in thyrotrophs explains the thyroxine-mediated pituitary thyrotropin feedback mechanism. Endocrinology. 2006;147(4):1735–1743. [DOI] [PubMed] [Google Scholar]
- 33. Diez D, Grijota-Martinez C, Agretti P, De Marco G, Tonacchera M, Pinchera A, de Escobar GM, Bernal J, Morte B. Thyroid hormone action in the adult brain: gene expression profiling of the effects of single and multiple doses of triiodo-L-thyronine in the rat striatum. Endocrinology. 2008;149(8):3989–4000. [DOI] [PubMed] [Google Scholar]
- 34. Morte B, Ceballos A, Diez D, Grijota-Martínez C, Dumitrescu AM, Di Cosmo C, Galton VA, Refetoff S, Bernal J. Thyroid hormone-regulated mouse cerebral cortex genes are differentially dependent on the source of the hormone: a study in monocarboxylate transporter-8- and deiodinase-2-deficient mice. Endocrinology. 2010;151(5):2381–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hernandez A, Morte B, Belinchón MM, Ceballos A, Bernal J. Critical role of types 2 and 3 deiodinases in the negative regulation of gene expression by T3in the mouse cerebral cortex. Endocrinology. 2012;153(6):2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu G, Tzouvelekis A, Wang R, Herazo-Maya JD, Ibarra GH, Srivastava A, de Castro JPW, DeIuliis G, Ahangari F, Woolard T, Aurelien N, Arrojo E Drigo R, Gan Y, Graham M, Liu X, Homer RJ, Scanlan TS, Mannam P, Lee PJ, Herzog EL, Bianco AC, Kaminski N. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2018;24(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moore AN, Waxham MN, Dash PK. Neuronal activity increases the phosphorylation of the transcription factor cAMP response element-binding protein (CREB) in rat hippocampus and cortex. J Biol Chem. 1996;271(24):14214–14220. [DOI] [PubMed] [Google Scholar]
- 38. Lee B, Dziema H, Lee KH, Choi YS, Obrietan K. CRE-mediated transcription and COX-2 expression in the pilocarpine model of status epilepticus. Neurobiol Dis. 2007;25(1):80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beaumont TL, Yao B, Shah A, Kapatos G, Loeb JA. Layer-specific CREB target gene induction in human neocortical epilepsy. J Neurosci. 2012;32(41):14389–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hansen KF, Sakamoto K, Pelz C, Impey S, Obrietan K. Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing. Sci Rep. 2014;4(1):6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrendelli JA, Blank AC, Gross RA. Relationships between seizure activity and cyclic nucleotide levels in brain. Brain Res. 1980;200(1):93–103. [DOI] [PubMed] [Google Scholar]
- 42. Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeöld A, Capelo LP, Curcio-Morelli C, Ribeiro R, Harney JW, Tabin CJ, Bianco AC. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7(7):698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haque M, Kendal JK, MacIsaac RM, Demetrick DJ. WSB1: from homeostasis to hypoxia. J Biomed Sci. 2016;23(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peeters RP, Wouters PJ, Kaptein E, van Toor H, Visser TJ, Van den Berghe G. Reduced activation and increased inactivation of thyroid hormone in tissues of critically ill patients. J Clin Endocrinol Metab. 2003;88(7):3202–3211. [DOI] [PubMed] [Google Scholar]
- 45. Olivares EL, Marassi MP, Fortunato RS, da Silva AC, Costa-e-Sousa RH, Araújo IG, Mattos EC, Masuda MO, Mulcahey MA, Huang SA, Bianco AC, Carvalho DP. Thyroid function disturbance and type 3 iodothyronine deiodinase induction after myocardial infarction in rats a time course study. Endocrinology. 2007;148(10):4786–4792. [DOI] [PubMed] [Google Scholar]
- 46. Simonides WS, Mulcahey MA, Redout EM, Muller A, Zuidwijk MJ, Visser TJ, Wassen FW, Crescenzi A, da-Silva WS, Harney J, Engel FB, Obregon MJ, Larsen PR, Bianco AC, Huang SA. Hypoxia-inducible factor induces local thyroid hormone inactivation during hypoxic-ischemic disease in rats. J Clin Invest. 2008;118(3):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hatazaki S, Bellver-Estelles C, Jimenez-Mateos EM, Meller R, Bonner C, Murphy N, Matsushima S, Taki W, Prehn JH, Simon RP, Henshall DC. Microarray profile of seizure damage-refractory hippocampal CA3 in a mouse model of epileptic preconditioning. Neuroscience. 2007;150(2):467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hunsberger JG, Bennett AH, Selvanayagam E, Duman RS, Newton SS. Gene profiling the response to kainic acid induced seizures. Brain Res Mol Brain Res. 2005;141(1):95–112. [DOI] [PubMed] [Google Scholar]
- 49. Lukasiuk K, Pitkänen A. Large-scale analysis of gene expression in epilepsy research: is synthesis already possible? Neurochem Res. 2004;29(6):1169–1178. [DOI] [PubMed] [Google Scholar]
- 50. Cavalheiro EA, Leite JP, Bortolotto ZA, Turski WA, Ikonomidou C, Turski L. Long-term effects of pilocarpine in rats: structural damage of the brain triggers kindling and spontaneous recurrent seizures. Epilepsia. 1991;32(6):778–782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.