Abstract
Liver cancers, hepatocellular carcinomas (HCCs), cholangiocarcinomas (CCs) and fibrolamellar HCCs (FL-HCCs), are among the most common cancers worldwide and are associated with a poor prognosis. Investigations of genes important in liver cancers have focused on Sal-like protein 4, SALL4, a member of a family of zinc finger transcription factors. It is a regulator of embryogenesis, organogenesis, pluripotency, can elicit reprogramming of somatic cells, and is a marker of stem cells. We found it expressed in normal murine hepatoblasts, normal human hepatic stem cells, hepatoblasts and biliary tree stem cells, but not in mature parenchymal cells of liver or biliary tree. It was strongly expressed in surgical specimens of human HCCs, CCs, a combined hepatocellular and cholangiocarcinoma, a FL-HCC and in derivative, transplantable tumor lines in immune-compromised hosts. Bioinformatics analyses indicated that elevated expression of SALL4 in tumors is associated with poor survival of HCC patients.
Experimental manipulation of SALL4’s expression results in changes in proliferation versus differentiation in human HCC cell lines in vitro and in vivo in immune-compromised hosts. Virus-mediated gene transfer of SALL4 was used to do gain and loss of function analyses in the cell lines. Significant growth inhibition in vitro and in vivo, accompanied by an increase in differentiation occurred with down-regulation of SALL4. Over-expression of SALL4 resulted in increased cell proliferation in vitro, correlating with an increase in expression of cytokeratin19 (CK19), EpCAM, and ATP-binding cassette-G2 (ABCG2).
SALL4’s expression is an indicator of stem cells, a prognostic marker in liver cancers, correlates with cell and tumor growth, with resistance to 5-FU, and its suppression results in differentiation and slowed tumor growth. SALL4 is a novel therapeutic target for liver cancers.
Keywords: hepatocellular carcinomas, cholangiocarcinomas, cancer stem cells, tumor-initiating cells, transcription factors
Introduction
Liver cancers, comprised primarily of hepatocellular carcinomas (HCCs), cholangiocarcinomas (CCs), and fibrolamellar HCCs (FL-HCCs), are the fifth most common cancer and the third leading cause of cancer mortality in the world(1). Cancers have a subpopulation of cancer stem cells (CSCs) or tumor-initiating cells (TICs), which have properties shared with normal stem cells(2,3). CSCs and TICs have highly aggressive phenotypes in oncogenesis and are resistant to chemotherapies and radiation therapies. Expression of membrane pumps, ATP-binding cassette-G2 (ABCG2), account for the resistance to chemotherapies and are responsible for elimination of DNA-binding dyes causing the cells to be displayed as a side fraction, a “side population (SP)”(4,5). Epithelial cell adhesion molecule (EpCAM), a key factor in the Wnt signaling pathway, was reported as a specific cell surface markers of human hepatic stem cells (hHpSCs), of some, but not all, subpopulations of human biliary tree stem cells (hBTSCs)(6–8) and liver TICs(9). CD133 (prominin), CD90 (Thy-1), CD44 (hyaluronan receptor), and CD13 (alanine aminopeptidase) have also been found in liver TICs(10–12). In parallel, CD133 and CD90 have been found on angioblasts or other mesenchymal cells tightly associated with hHpSCs(13), and so, some data discussing CD90 or CD133 may actually be interpreted as relevant to the mesenchymal cell components of the tumors. Several lines of evidence implicate genetic alternations during hepatocarcinogenesis, particularly the Wnt signaling pathway, p53 and alterations in matrix-degrading enzyme secretion(14–20).
SALL4, a homologue of the Drosophila homeotic gene spalt, is a zinc finger transcription factor required for proliferation and maintenance of pluripotency through interactions with OCT3/4, SOX2 and NANOG. It is found at high levels in embryonic stem cells (ESCs)(21–26), and is one of the genes capable of eliciting reprogramming of somatic cells to become induced pluripotent stem cells (iPSCs)(27,28). Mutations in SALL4 cause Okihiro syndrome, known as an autosomal dominant disorder and characterized by multiple organ defects(29). Recent studies have demonstrated that SALL4 is constitutively expressed in hematopoietic stem cells and a potent regulator of their expansion(30,31). SALL4 transgenic mice exhibit symptoms like myelodysplastic syndrome (MDS) and subsequently develop acute myeloid leukemia (AML). Primary AML and MDS patients have higher SALL4 expression levels than that in controls indicating that SALL4 plays a major role in leukemogenesis. Furthermore, SALL4 contributed to the maintenance of SP cells and chemosensitivity in leukemia by regulating the ABC drug transporter genes(31–33). Solid tumors, such as germ cell tumors, breast and alpha-fetoprotein (AFP)-producing gastric cancers also express SALL4(34–37). Taken together, these data suggest that SALL4 is a novel stem cell marker, a gene involved in embryogenesis and organogenesis and a putative stem cell gene associated with CSCs. We now report that SALL4 expression occurs in diverse liver cancers including HCCs, CCs and FL-HCCs, and that SALL4 increases growth and blocks differentiation in liver cancer cell lines.
Materials and Methods
Cell Proliferation and Chemo-resistance Assays
Liver cancer cell lines were infected with retroviruses or lentivirus at multiplicity of infection of 40 in the presence of 10 μg/mL protamine sulfate. After infection, cells were cultured for 3 days. Cells then were collected and isolated using a MoFlo™ fluorescence-activated cell sorter (FACS) (DAKO, Glostrup, Denmark). Then, 2×103 cells were seeded into 96 well plates and cultured in the presence or absence of 2 μg/ml 5-fluorouracil (5-FU) for 3 to 7 days. Cell proliferation was evaluated in triplicate using the Cell Counting Kit-8 (Dojindo Laboratory, Kumamoto, Japan). After incubation at 37ºC for 2 h, the absorbance at 450 nm was measured.
Immunohistochemistry
The tissues were embedded in paraffin and cut into 5 μm sections. After deparaffinization, antigen retrieval was performed with sodium citrate buffer for EpCAM, CK19, or ethylenediaminetetraacetic acid (EDTA) buffer (pH 8.0) for SALL4 in a steamer for 20 min. Endogenous peroxidases were blocked by incubation for 30 min in 0.3% H2O2. After blocking, primary antibodies (Supplementary Table 3) were applied at 4ºC overnight. M.O.M immunodetection kit (Vector Laboratories, Burlingame, CA) was used for detecting primary mouse anti-human SALL4 antibody on mouse xenotransplant FL-HCC tumor to avoid the inability of the anti-mouse secondary antibody to endogenous mouse immunoglobulins in the tissue. Sections were incubated for 30 min at room temperature with ImmPRESS peroxidase-micropolymer staining kits and 3,3’-diaminobenzidine substrate (Vector Laboratories). For double immunostaining, MACH2 peroxidase- and alkaline phosphatase-polymer detection kit, 3,3’-diaminobenzidine and Warp Red chromogen kit (Biocare Medical, Concord, CA) were used. Sections were lightly counterstained with hematoxylin.
Xenograft Transplantation
Each transplant consisted of 1×106 cells of each of the cell lines stably expressing-shRNA against SALL4 or luciferase suspended in 200 μl Dulbecco’s modified Eagle medium (DMEM) and Matrigel (1:1). The cells were transplanted into Non-obese diabetic, severe combined immunodeficient (NOD/SCID) mice (6-week-old, male) under anesthesia. Control and SALL4-knockdown cells were implanted into the subcutaneous space on the right and left sides of the backs of recipient mice, respectively. For 8 weeks, the mice were examined for tumor formation.
SALL4 Profiling Analyses in HCCs
SALL4 expression data were derived from cDNA microarray analysis of 139 HCC specimens described previously(38). The microarray data, with NCI’s Human Array-Ready Oligo Set microarray platform (GPL1528), are publically available at the Gene Expression Omnibus (GEO;http;://www.ncbi.nlm.nih.gov/geo) with accession numbers GSE1898 and GSE4024. High and low SALL4 groups were dichotomized according to the median SALL4 expression in tumors. Kaplan-Meier survival analysis was used to compare patient survival based on dichotomized SALL4 expression, using GraphPad Prism software 5.0 (GraphPad Software, San Diego, CA) with statistical P values generated by the Cox-Mantel log-rank test. Survival data linking to this cohort were kindly provided by Dr. Snorri Thorgeirsson at NCI.
Other material and methods can be found in the online supplement.
Results
SALL4 Expression in Human Normal Liver and Biliary Tree Tissues in situ and in vitro
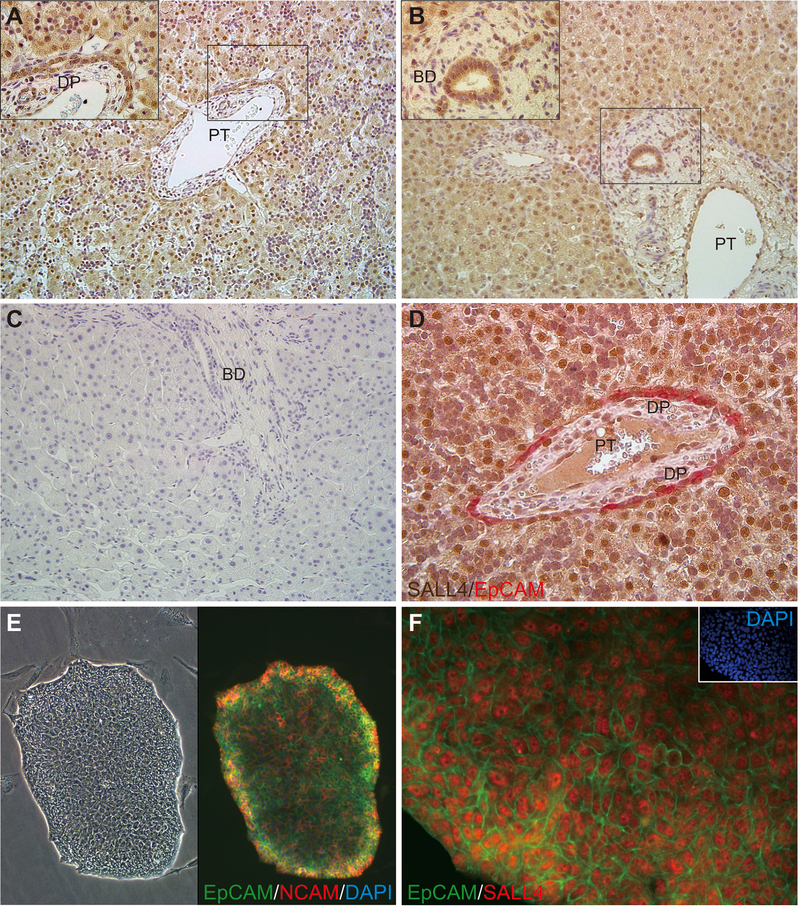
We have previously reported that SALL4 is expressed in murine hepatoblasts (mHBs) but not adult murine hepatocytes and plays a critical role in their differentiation(39). In these studies, we analyzed SALL4 expression in normal human liver tissues. Immunohistochemical analyses showed that SALL4 is diffusely expressed in the nuclei of liver cells from both fetuses and neonates. Neonatal hepatocytes were more weakly positive for SALL4 than parenchymal cells in fetal livers and some had lost SALL4 expression altogether. In contrast, SALL4 expression was not detected in mature hepatocytes and cholangiocytes in adult livers (Fig.1A–C). Double immunostaining of EpCAM and cytokeratin19 (CK19) show clearly that EpCAM and CK19 strongly co-stain the cytoplasm of ductal plate cells, now recognized to comprise hHpSCs, and human hepatoblasts (hHBs) in fetal and neonatal livers. It is found also in hBTSCs within peribiliary glands (PBGs), the stem cell niches of the biliary tree, in neonatal livers (Fig.S1A) and in adult livers(40). We found that SALL4 co-expressed with EpCAM+/CK19+ ductal plate cells, known to comprise hHpSCs (arrows), and the adjacent hHBs (arrowheads). It also was found in multiple subpopulations of hBTSCs within PBGs located within livers or biliary tree tissue from all donor ages and included cellular subpopulations that are EpCAM−/CK19+, EpCAM−/CK19−, EpCAM+/CK19− and EpCAM+/CK19+ cells. Shown are ones from fetal or neonatal livers (Figs.1D,S1A). We also found that SALL4, NCAM and EpCAM co-expressed in colonies of hHpSCs and in colonies of hBTSCs (Fig.1E–F,S1B). These results suggest that SALL4 is found only in early lineage stage parenchymal cells, such as hHpSCs, hBTSCs, hHBs, and to less extent in committed progenitors, but not in later lineage stages of parenchymal cells of either liver or biliary tree.
Figure 1.
Representative immunostaining of SALL4 and EpCAM expression in human normal livers and in a colony of hHpSCs. (A-D) Immunostaining of SALL4 expression during liver development. Fetal weeks gestation (A; 19 weeks, D; 16 weeks), neonatal (B; 4 months) and adult liver (C; 68 years) tissues. Sections were stained with an anti-SALL4 antibody (A-C) or antibodies against SALL4 and EpCAM (D). (E-F) A colony of hHpSCs. The colony was stained with antibodies against EpCAM and NCAM (E) or antibodies against EpCAM and SALL4 (F). Magnification ×200 (A-C), ×400 (D, F), ×100 (E). BD, bile duct; DP, ductal plate; PT, portal tract.
SALL4 Expression in Human Liver Cancers
We analyzed SALL4 expression in surgical specimens of non-cancerous liver tissue and in liver cancers. SALL4 was not detected in chronic hepatitis but faintly detected in bile ductules and in hepatocytes at the interface of parenchymal and stromal cells in liver cirrhosis (Fig.S2A–B). Seventeen of 20 HCC specimens were positive for SALL4 in the nuclei of the tumor cells, whereas 3 specimens showed no SALL4 expression. In some cases, biliary epithelial cells, presumptive hBTSCs, around the tumors expressed SALL4 (Figs.2A–C,S2C–D). Four of 5 CC specimens expressed SALL4. We found that SALL4 is expressed in combined hepatocellular and cholangiocarcinoma (HC-CC) and in a transplantable human tumor line derived from a FL-HCC (Fig.2D–F). Double immunostaining showed that SALL4+/EpCAM+/CK19+ cancer cells were observed in CC, which strongly expressed EpCAM and CK19 in serial sections (Fig.S2E–F). These results suggest that SALL4 expression indicates selection for stem cells as a minor cell population in normal tissue and cirrhotic tissues and as a dominant cell population in liver cancers.
Figure 2.
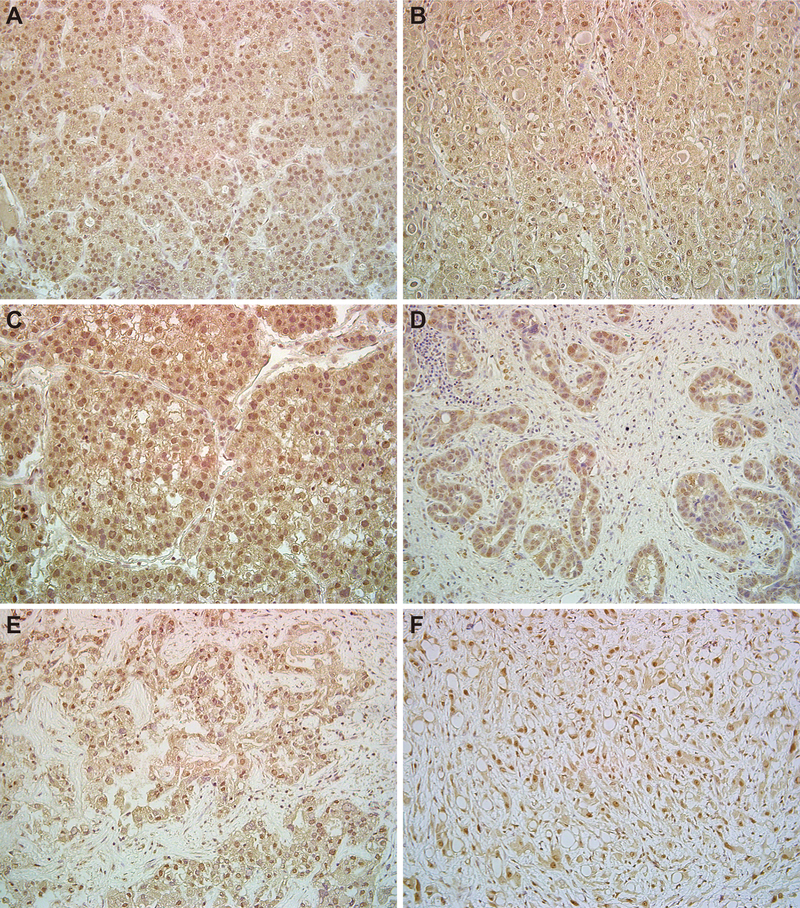
Representative immunostaining of SALL4 in surgical specimens of liver cancers and transplantable tumor lines of FL-HCC. HCC (A; T41, well-differentiated, B; T37, moderately-differentiated, C; T49, poorly-differentiated). CC (D; T5, poorly-differentiated). HC-CC (E; T45, moderately-differentiated). FL-HCC (F; poorly-differentiated). Magnification ×200.
SALL4 Expression in Human Liver Cancer Cell Lines
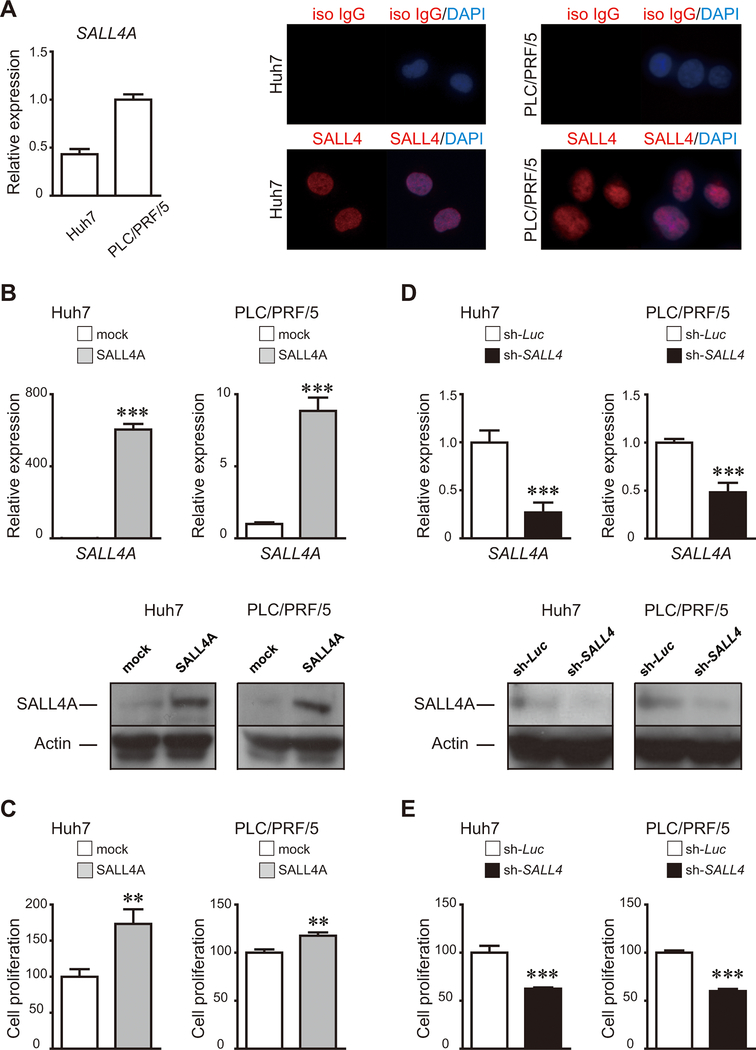
To investigate the functions of SALL4 in liver cancers, we used liver cancer cell lines, Huh7 and PLC/PRF/5 cells. The quantitative real time-polymerase chain reaction (qRT-PCR) analyses showed that both cell lines expressed SALL4A mRNA. SALL4 protein was also detected using immunocytochemistry (Fig.3A).
Figure 3.
SALL4 expression and the effects of SALL4 over-expression or knockdown on cell proliferation of liver cancer cells. (A) SALL4A mRNA and protein expression in liver cancer cells. (B, D) Expression of SALL4 mRNA and proteins in cultures derived from SALL4-over-expressing or SALL4-knockdown liver cancer cells. Cells infected with mock- or SALL4-expressing retroviruses, with shRNA against luciferase or SALL4-expressing lentiviruses were cultured for 3 days. (C, E) Cell proliferation assays of cells transduced by a SALL4-over-expressing retroviral vector or a SALL4-knockdown lentiviral vector were cultured for 7 days. Data are expressed as mean ± SD (triplicate samples,***p<0.001,**p<0.01).
Regulation of Cell Proliferation by SALL4
To examine whether SALL4 regulates tumor growth of liver cancer cell lines, we used a SALL4A-over-expressing retroviral vector(28). Over-expression of SALL4A was verified using qRT-PCR. Transduction of SALL4A into the cells significantly increased SALL4A mRNA and also protein levels by Western blots and immunocytochemistry (Figs.3B,S3). SALL4A-over-expressing liver cancer cells had enhanced cell proliferation (Fig.3C).
Next, we conducted SALL4 expression knockdown studies using a lentiviral vector expressing-short hairpin RNA (shRNA)(32, 39). Transduction efficiency was estimated using FACS revealing that the percentage of cells infected with lentiviruses expressing-shRNA against luciferase or SALL4 was more than 90% (Fig.S4). Transduction of shRNA into the cells significantly decreased both mRNA and protein production of SALL4 (Fig.3D). We observed growth inhibition in SALL4-knockdown liver cancer cells in culture (Figs.3E,S5). Therefore, SALL4 regulates the proliferative potential of liver cancer cell lines in vitro.
SALL4 Regulates Cell Proliferation through Cyclin D1 and D2 Expressions
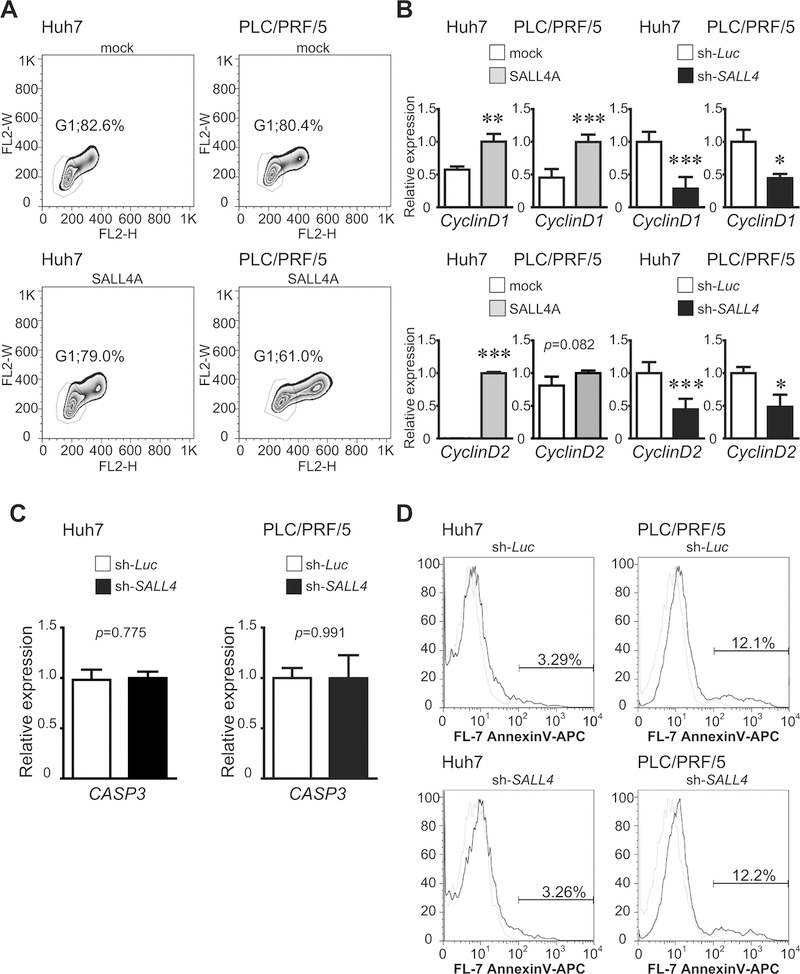
To analyze molecular mechanisms regulating SALL4-induced proliferation of liver cancer cell lines, cell-cycle analyses were examined. Cell-cycle analyses using flow cytometry showed that over-expression of SALL4 induced the decrease of the G1 phase in liver cancer cells (Fig.4A). Next, Cyclin D1 and D2 mRNA expressions were examined using qRT-PCR. Consistent with the flow cytometry analysis, Cyclin D1 and D2 levels were induced by SALL4A over-expression. In contrast, their levels were decreased by SALL4 knockdown (Fig.4B), implicating a correlation of Cyclin levels to those of cell proliferation. Though we also analyzed expression of cyclin inhibitors, significant changes were not observed (data not shown).
Figure 4.
Correlates of SALL4 over-expression or knockdown with respect to cell proliferation of liver cancer cells. (A) Cell-cycle analysis in SALL4-over-expressing liver cancer cells was estimated by flow cytometry. (B-C) Expression of Cyclin D1, Cyclin D2, and CASP3 in SALL4-over-expressing or SALL4-knockdown liver cancer cells. Cells transduced by a retroviral or lentiviral were cultured for 3 days. Cyclin D1, Cyclin D2, and CASP3 mRNA expression was detected using qRT-PCR. Data are expressed as mean ± SD (triplicate samples,***p<0.001,**p<0.01,*p<0.05). (D) Apoptosis in SALL4-knockdown liver cancer cells was estimated by FACS. Cells were cultured for 3 days and stained with allophycocyanin (APC)-conjugated anti-Annexin-V antibody.
To exclude the possibility that shRNA-knockdown of SALL4 expression inhibited cell proliferation by means of an induction of apoptosis, we analyzed the effect of viral infection on apoptosis of the liver cancer cell lines. The qRT-PCR analyses showed that caspase-3 (CASP3) expression, an early stage marker of apoptosis, did not change in SALL4-knockdown liver cancer cells (Fig.4C). Apoptosis was also evaluated using flow cytometric analyses. The number of Annexin-V+ cells did not change by SALL4 knockdown, suggesting that inhibition of cell proliferation was not due to apoptosis (Fig.4D).
SALL4 Expression is Inversely Correlated with Differentiation Markers
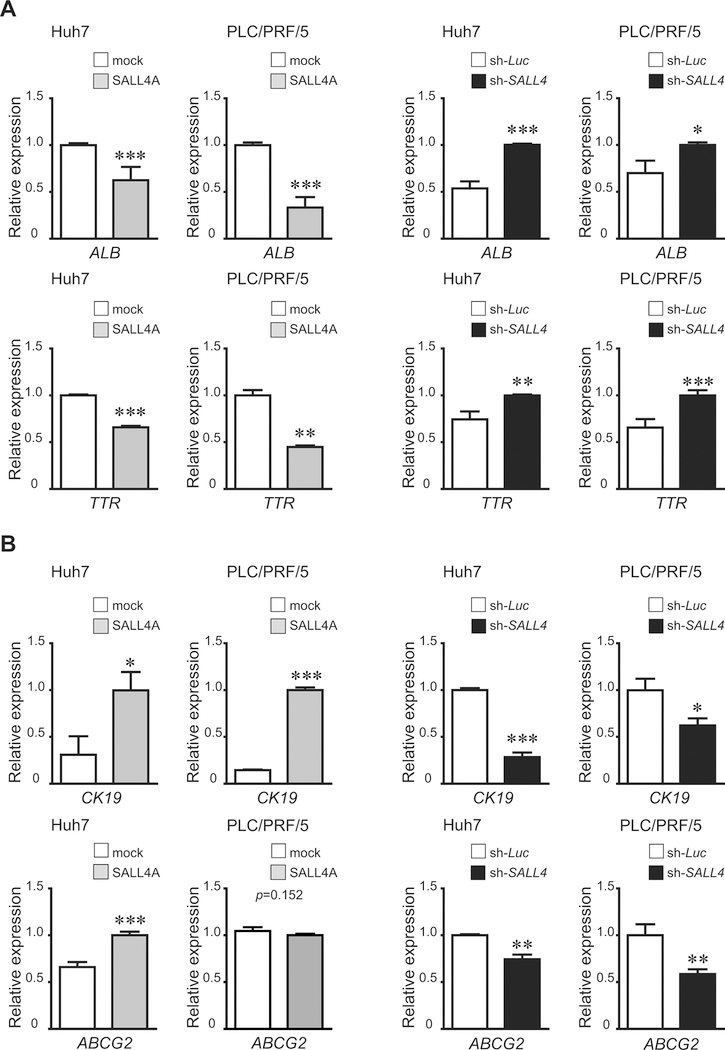
Given that hepatocytic maturation was suppressed by SALL4 over-expression in mHBs(39), we hypothesized that SALL4 could affect the differentiation of liver cancer cell lines. To explore this, we analyzed mRNA expression for hepatocytic differentiation marker genes using qRT-PCR. Expression of albumin (ALB), transthyretin (TTR), and UDP-glucuronosyltransferase-2B7 (UGT2B7) were suppressed by SALL4 over-expression. In contrast, their levels were significantly enhanced in SALL4-knockdown liver cancer cells (Figs.5A,S6A,S7). These results suggested that SALL4 inhibits hepatocytic differentiation in mHBs and also human liver cancer cell lines. Hepatocyte nuclear factor 4-alpha (HNF4α), a key transcriptional factor regulating differentiation of HBs into hepatocytes with acquisition of mature liver functions, did not decrease in SALL4-over-expressing liver cancer cells, indicating that SALL4 inhibits hepatocytic differentiation through a pathway independent of HNF4α (Fig.S6A). As shown above, CK19 and EpCAM are expressed in normal hHpSCs, hHBs, and cholangiocytes in livers of all donor ages but not adult hepatocytes, and EpCAM is also a TIC marker for liver cancer. Over-expression of SALL4 in liver cancer cells induced expression of CK19 and EpCAM (encoded by TACSTD1 gene), indicating a correlation between SALL4 and CK19. Down-regulation of SALL4 suppressed the expression of CK19 but not EpCAM in liver cancer cells. SALL4-over-expressing PLC/PRF/5 cells had up-regulated POU5F1 (OCT3/4) and CD90 (Figs.5B,S6B,S7). Similarly, ABCG2, a multidrug resistance gene found in normal hHpSCs as well as in CSCs and responsible for chemo-resistance, was significantly increased in SALL4-over-expressing Huh7 cells. In contrast, SALL4 knockdown of liver cancer cells resulted in lowered ABCG2 levels (Fig.5B). These results suggest that SALL4 either plays a role controlling maintenance of stemness and TIC marker genes or is a biomarker for stem cell phenotypic traits.
Figure 5.
Expression of hepatocytic differentiation (A) and stemness (B) genes in SALL4-over-expressing or SALL4-knockdown liver cancer cells. Cells transduced by a retroviral or lentiviral vector were cultured for 3 days. ALB, TTR, CK19 and ABCG2 mRNA expression was detected using qRT-PCR. Data are expressed as mean ± SD (triplicate samples,***p<0.001,**p<0.01,*p<0.05).
SALL4 Increases Expression of EMT Genes but does not Influence Cell Invasion
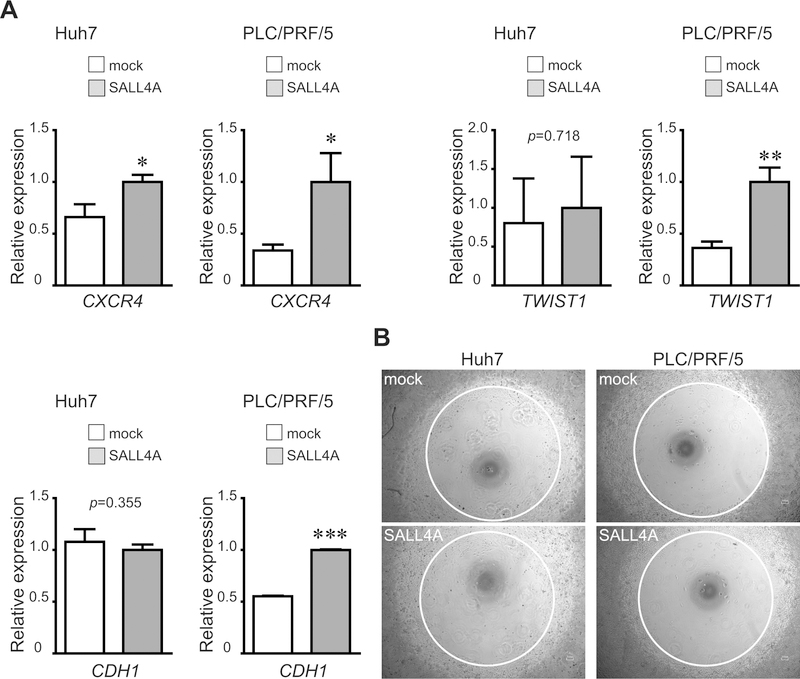
Epithelial–mesenchymal transition (EMT) phenomena occurs in invasion and metastasis of cancer cells and is also associated with the acquisition of stem cell-like characteristics. To investigate whether SALL4 regulates EMT, we analyzed its effects on EMT-related genes in liver cancer cell lines. The mRNA expression of CXCR4 and TWIST1, a direct transcriptional target of EMT inducers, was up-regulated by SALL4 over-expression. In contrast, another important EMT phenomenon, down-regulation of E-cadherin (encoded by the CDH1 gene) was not observed in SALL4-over-expressing liver cancer cells (Fig.6A) and nor were there significant changes in cell migration assays with the liver cancer cells (Fig.6B). These data suggest that cell migration and invasion of liver cancer cells are not directly affected by SALL4 even though some EMT-related genes are up-regulated.
Figure 6.
Expression of EMT-related genes and migration assays in SALL4-over-expressing liver cancer cells. (A) Cells transduced by an over-expressing retroviral vector were cultured for 3 days. CXCR4, TWIST1, and CDH1 mRNA expression was detected using qRT-PCR. Data are expressed as mean ± SD (triplicate samples,***p<0.001,**p<0.01,*p<0.05). (B) Migration assay in SALL4-over-expressing liver cancer cells.
SALL4 Expression is Correlated with Chemosensitivity
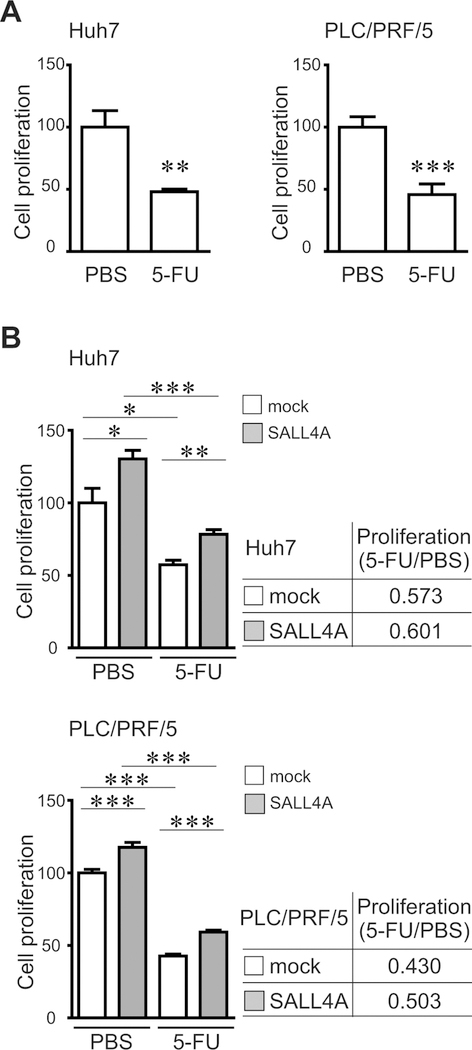
We previously reported that the oncostatin M (OSM) induced maturation of fetal hepatic cells(41). OSM induced hepatocytic differentiation of EpCAM+ liver CSCs into EpCAM-negative cells and increased chemosensitivity to 5-FU(42). As shown above, we have shown that over-expression of SALL4 suppressed hepatocytic differentiation and induced stem cell-like phenotype in liver cancer cells. We thus analyzed whether over-expression of SALL4 affects chemosensitivity of liver cancer cell lines. 5-FU treatment decreased cell proliferation in both lines. Cell survival and proliferation of liver cancer cells were induced by SALL4-over-expression with or without 5-FU. Interestingly, over-expression of SALL4 increased cell proliferation (5-FU/PBS) in liver cancer cells (Fig.7A–B). These results suggest that SALL4 expression results in selection of cells that are chemo-resistant.
Figure 7.
Chemo-resistance assays for SALL4-over-expressing liver cancer cells. Cells were transduced by a retroviral vector. Non-transduced (A) or transduced cells (B) were cultured in the presence or absence of 5-FU (2 μg/ml) for 7 days. The relative cell proliferation between PBS- and 5-FU-treated liver cancer cells is shown. Data are expressed as mean ± SD (triplicate samples,***p<0.001,**p<0.01,*p<0.05).
Down-regulation of SALL4 Inhibits Tumor Growth in Xenograft Transplantation
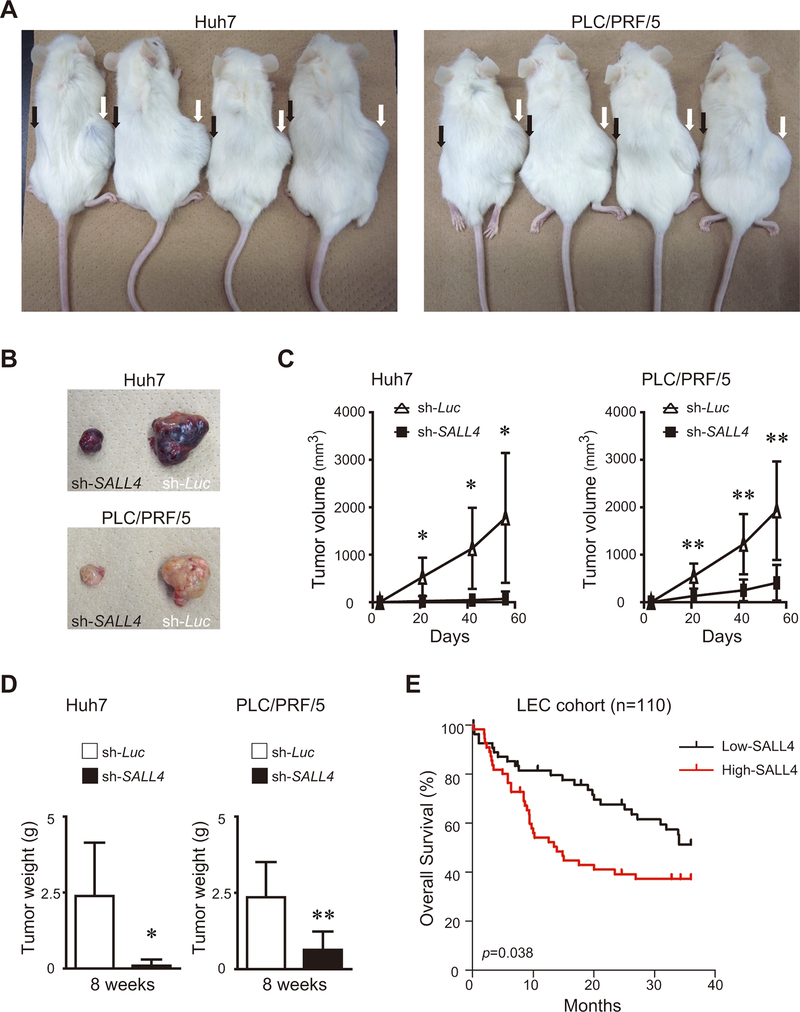
To determine whether SALL4 affects tumorigenicity of liver cancer cell lines, we generated stable liver cancer cells expressing-shRNA against luciferase or SALL4, and cells were transplanted into the subcutaneous space on the right versus left sides of immunodeficient mice, respectively. After 8 weeks, both control Huh7 and PLC/PRF/5 cells gave rise to subcutaneous tumors. In contrast, tumors derived from SALL4-knockdown liver cancer cells were significantly smaller than those of control cells (Fig.8A–C). The tumor weights were also smaller than those from control cells (Fig.8D). These results suggest that down-regulating SALL4 expression also inhibited growth of the tumors from liver cancer cell lines in vivo.
Figure 8.
Effect of SALL4 knockdown on xenograft tumor growth in vivo. (A) Control cells and SALL4-knockdown cells were implanted into recipient mice, respectively. White arrows show tumors derived from control cells and black arrows show tumors derived from SALL4-knockdown cells (Huh7 n=5, PLC/PRF/5 n=8). (B) Representative tumors derived from control versus SALL4-knockdown liver cancer cells at 8 weeks are shown. (C) The tumor growth curve over 8 weeks is shown. (D) The weight of the tumor at 8 weeks is shown. Data are expressed as mean ± SD (**p<0.01,*p<0.05). (E) Kaplan-Meier survival plot according to the relative level of SALL4 expression in HCC tumor samples, as determined by microarray analyses and with the use of the log-rank test. The median expression level was used to dichotomize low and high SALL4-expressing HCC tumors.
SALL4 Expression in HCC Clinical Specimens is Prognostic of Patient Survival (Bioinformatics Analyses)
We examined SALL4 expression in 139 HCC cases in a microarray data set published in Lee et al(38). A total of 110 cases with available expression and overall survival data were selected for survival analysis. We found that HCC patients with high SALL4 expression is significantly associated with shorter survival during the first 3 years of follow-up (p=0.038) (Fig.8E).
Discussion
Gene expression profiles and signaling pathways associated with self-renewal and differentiation are shared in normal stem cells and in CSCs(3). Accordingly, fully understanding these common molecular mechanisms that regulate self-renewal and differentiation is a necessary step towards novel therapeutic modalities for cancer. The only curative treatments for liver cancers are surgical resection and liver transplantation for early stage patients. However, most patients are diagnosed at advanced stages by which extant therapies are ineffective. For the treatment of advanced HCC patients with unresectable tumors, transcatheter arterial chemoembolization and systemic chemotherapy, including Sorafenib, are one of the options, but the effects are limited(14,17). Therefore, the identification of novel molecules which can become targets for future therapies is urgently needed.
SALL4 is required for cell proliferation and maintenance of pluripotency in several types of stem cells (e.g. ESCs) and in malignantly transformed stem cells (e.g. leukemia and breast cancer)(21–26). In addition, our prior investigations with mHBs revealed that inhibition of SALL4 contributes to cell differentiation(39). Hence, it seemed likely that SALL4 expression could be a factor in liver cancers in which the CSCs might have a shared gene profile to normal hHpSCs and/or to normal hBTSCs. This hypothesis became plausible when we found SALL4 expression in normal hHpSCs, hHBs, and with weaker expression in committed progenitors in human fetal and neonatal liver tissues, in stem cells in PBGs, the stem cell niches of human biliary tree tissue, and in various liver cancers (Figs.1–2). In recent publications it was reported that SALL4 is expressed in hepatoid gastric carcinoma but not in other liver cancer(36,37). We hereby report that SALL4 expression in liver cancers (and cancers of the biliary tree) can be detected by using EDTA buffers, rather than citrate buffers, for antigen retrieval. The mechanisms of antigen retrieval are poorly understood. It has been reported that antigen retrieval is needed for disruption of methylene-bridges during fixation, which cross-link proteins and therefore mask antigenic sites. Indeed we were not able to obtain clearly positive SALL4 staining in liver cancer tissues when we used citrate buffer (pH 6.0), the most popular buffer for antigen retrieval. Therefore we decided to use EDTA buffer (pH 8.0), because it has been reported that the pH of antigen retrieval solution remarkably affects the intensity of immunostaining(43). SALL4-positive cells were observed by using EDTA/pH8.0 rather than citrate buffer (Fig.S8). This indicates that the pH of the retrieval buffer and the presence of EDTA, the chelating agent, are important factors for masking the epitopes available for binding either by eliminating masking molecules and/or proper refolding of SALL4-specific epitopes to bind with antibody.
One of the main regulators of G1-S phase transition in the cell cycle, Cyclin D1 has been shown to have capabilities of carcinogenesis and progression in cancer through controlling cell proliferation(44). Moreover the strong relationship of tumorigenesis and self-renewal by Ras-Cyclin D2 activation has been elucidated in spermatogonial stem cells(45). With respect to SALL4’s effects on growth, recent studies revealed that Cyclin D1 has been shown to bind to SALL4 and works synergistically in transcriptional repression; Cyclin D1 is a downstream target of SALL4 in malignant cells and in ESCs(25,31,46). We found over-expressing SALL4 induced a shorter G1 phase, and there was a positive correlation between expression of SALL4 and Cyclin D1 and D2 in liver cancer cell lines. This suggest that SALL4 regulates cell proliferation either by selection of early lineage stage cells or by controlling G1-S transition through regulating expression of Cyclin D1 and D2 directly. Although SALL4 has been proposed to play a role in survival and apoptosis in leukemic cells(32), we did not observed any difference in apoptosis between control and SALL4-knockdown liver cancer cell lines (Fig.4), indicating that downstream targets for SALL4 may be different in liver cancer cells and leukemic cells.
Analyses of functions using models of liver cancer cell lines indicated that SALL4 over-expression leads to cells with enhanced phenotypic traits such as ABCG2 and CK19 expression, ones highly expressed in stem cells. SALL4 is associated also with CD90 (Thy-1), known to be highly expressed in mesenchymal cells tightly associated with the stem cell. In contrast, SALL4 knockdown provided evidence of slowed growth and more parenchymal cell differentiation. In summary, SALL4 expression is a marker of stem cells and early lineage descendants from those stem cells, implicating it as a marker of TICs. Its expression correlates with cell proliferation, survival and a minimally differentiated status in normal and in malignantly transformed cells.
Findings reported recently corroborate our own in that OSM induction or HNF4α gene transfer into liver cancer cells resulted in more differentiated cells with reduced tumor-initiating ability and enhancement of sensitivity to 5-FU(42,47). High levels of SALL4 correlate with growth and stemness features, and SALL4 suppression results in inhibition of growth, increased hepatocytic differentiation of cells, and reduced tumorigenicity (Figs.3–8).
SALL4 has been found in normal hHpSCs and hHBs, stem/progenitor cell populations found intrahepatically and associated with canals of Hering(6,48); both of these are positive for EpCAM and CK19, and the hHBs are positive also for AFP and for ALB. Interestingly, it is found strongly expressed in all of the subpopulations of hBTSCs, ones located with PBGs throughout the biliary tree and that comprise the most primitive stem cells identified (LGR5+/NCAM+/SOX17+/PDX1+/CK19+/EpCAM−/AFP−/ALB−); others with phenotypic traits identical to or similar to that of hHpSCs (LGR5+/NCAM+/EpCAM+/SOX17+/PDX1−/CK19+/AFP−/ALB−); and yet others with traits overlapping with those of hHBs (LGR5−/EpCAM+/SOX17−/PDX1−/ICAM-1+/CK19+/AFP++/ALB+−)(7,8,40). It is also found in stem/progenitor cells of human fetal but not adult pancreas (Oikawa, Wauthier and Reid, unpublished data).
SALL4 has also been identified as a novel molecule in reprogramming of somatic cells to become iPSCs(27,28). This background makes interpretable published bioinformatics analyses(49) in which there is no significant correlations between the expression of SALL4, EpCAM, AFP, or ALB in liver cancers. Rather, we found that it correlates with HCC patient’s prognosis since an increased SALL4 expression is associated with shorter survival in HCC patients (Fig.8). It should be noted that we have not yet done bioinformatics analyses relating SALL4 expression in survival of patients with CC; however, we hypothesize that it will be relevant to survival for patients with CC given that SALL4 expression is strong in all the subpopulations of normal hBTSCs. We interpret this to mean that high SALL4 expression indicates tumors enriched for CSCs, whether or not they express EpCAM, AFP or ALB. Thus, SALL4 is a reliable indicator of stem cell populations, whether normal or malignantly transformed, and its levels quantitatively indicate the proportion of the tissue comprised of those stem cells. Therefore, our findings corroborate those of others suggesting that SALL4 is indicative of aggressiveness and poor prognosis in liver cancers(9,38,50).
Taken together, SALL4 is an excellent target for identifying treatments for liver cancers. Suppression of SALL4 expression may contribute to inhibition of tumor growth by 1) attenuation of cell cycle progression via Cyclin D1 and D2; 2) reduction in stem cell traits and, thereby, allowing a more differentiated state; and 3) reduction in multidrug resistance genes with increased sensitivity to chemotherapies. Further analyses on SALL4 mediated mechanisms may provide a novel future therapeutic strategy against liver cancers.
Supplementary Material
Acknowledgments
We thank Prof. Yamanaka and Prof. Takahashi (Kyoto University, Kyoto, Japan) for the hSALL4 expression vector. We thank Dr. Derek Y. Chiang (UNC School of Medicine, Chapel Hill, NC) for preliminary bioinformatics analyses on SALL4, EpCAM, albumin, and alpha-fetoprotein, findings that indicated no significant relationships. Discussions with Dr. Oswaldo A. Lozoya (Duke University Medical Center, Durham, NC) made us aware of the fact that some published bioinformatics databases consist of data from tumors normalized to that from the so-called normal margins of the tumors; other studies of our own had revealed that the so-called “normal margins”, in fact, are not normal both with respect to the epithelial and mesenchymal cellular components. Dr. Lozoya’s counseling resulted in our searching for alternative ways to assess the relevance of SALL4 to liver tumors using bioinformatics analyses. With respect to the investigations within the United States, we thank Ms. Lucendia English for glassware washing and lab management. Various core services provided essential support including several of the histology cores, confocal microscopy core, and tissue culture core. No author has equity or a position in Vertex Pharmaceuticals or in Vesta Therapeutics, and none are paid consultants.
Financial Support: The labs in Japan were supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology in Japan (22790674 and 23689040). Lola M. Reid and associates were funded by sponsored research grants from Vertex Pharmaceuticals (Cambridge, MA) and Vesta Therapeutics (Bethesda, MD) and by an NCI grant (CA016086). Xin Wei Wang was supported by the Intramural Research Program of the Center for Cancer Research, the National Cancer Institute (Z01 BC 010876). Lance D. Miller was funded by a grant from the American Cancer Society (RSG-12198-01-TBG).
Abbreviations
- ABCG2
ATP-binding Cassette-G2
- AFP
alpha-fetoprotein
- ALB
albumin
- BD
bile duct
- CASP3
caspase-3
- CC
cholangiocarcinoma
- CK19
cytokeratin19
- CSCs
cancer stem cells
- DAPI
4’,6-diamidino-2-phenylindole
- DP
ductal plate
- EMT
epithelial–mesenchymal transition
- EpCAM
epithelial cell adhesion molecules
- FACS
fluorescent-activated cell sorter
- FL-HCC
fibrolamellar hepatocellular carcinoma
- 5-FU
5-fluorouracil
- hBTSCs
human biliary tree stem cells
- hHBs
human hepatoblasts
- HCC
hepatocellular carcinoma
- HC-CC
combined hepatocellular and cholangiocarcinoma
- HNF4α
hepatocyte nuclear factor 4-alpha
- hHpSCs
human hepatic stem cells
- PBGs
peribiliary glands
- PT
portal tract
- qRT-PCR
quantitative real-time polymerase chain reaction
- shRNA
short hairpin RNA
- TICs
tumor-initiating cells
- TTR
transthyretin
- UGT2B7
UDP-glucuronosyltransferase-2B7
Footnotes
Disclosures: There are no conflicts of interest to disclose.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics,2002.CA Cancer J Clin 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells.Nature 2001;414:105–111. [DOI] [PubMed] [Google Scholar]
- 3.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells.Annu Rev Cell Dev Biol 2007;23:675–699. [DOI] [PubMed] [Google Scholar]
- 4.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H,et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties.Hepatology 2006;44:240–251. [DOI] [PubMed] [Google Scholar]
- 5.Zen Y, Fujii T, Yoshikawa S, Takamura H, Tani T, Ohta T, Nakanuma Y. Histological and culture studies with respect to ABCG2 expression support the existence of a cancer cell hierarchy in human hepatocellular carcinoma.Am J Pathol 2007;170:1750–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, Moss N,et al. Human hepatic stem cells from fetal and postnatal donors.J Exp Med 2007;204:1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, Berloco PB,et al. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets.Hepatology 2011;54:2159–2172. [DOI] [PubMed] [Google Scholar]
- 8.Cardinale V, Wang Y, Carpino G, Mendel G, Alpini G, Gaudio E, Reid LM,et al. The biliary tree:a reservoir of multipotent stem cells.Nat Rev Gastroenterol Hepatol 2012;9:231–240. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H,et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features.Gastroenterology 2009;136:1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ,et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007;132:2542–2556. [DOI] [PubMed] [Google Scholar]
- 11.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW,et al. Significance of CD90+ cancer stem cells in human liver cancer.Cancer Cell 2008;13:153–166. [DOI] [PubMed] [Google Scholar]
- 12.Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, Akita H,et al. CD13 is a therapeutic target in human liver cancer stem cells.J Clin Invest 2010;120:3326–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Yao HL, Cui CB, Wauthier E, Barbier C, Costello MJ, Moss N,et al. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology 2010;52:1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma.Nat Genet 2002;31:339–346. [DOI] [PubMed] [Google Scholar]
- 15.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment.Nat Rev Cancer 2006;6:674–687. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res 2007;67:10831–10839. [DOI] [PubMed] [Google Scholar]
- 17.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology 2008;48:1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra L, Banker T, Murray J, Byers S, Thenappan A, He AR, Shetty K,et al. Liver stem cells and hepatocellular carcinoma.Hepatology 2009;49:318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marquardt JU, Factor VM, Thorgeirsson SS. Epigenetic regulation of cancer stem cells in liver cancer: current concepts and clinical implications. J Hepatol 2010;53:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis.Semin Liver Dis 2011;31:173–187. [DOI] [PubMed] [Google Scholar]
- 21.Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T,et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development.Development 2006;133:3005–3013. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y,et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1.Nat Cell Biol 2006;8:1114–1123. [DOI] [PubMed] [Google Scholar]
- 23.Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function.Proc Natl Acad Sci USA 2006;103:16319–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH,et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages.Cell Stem Cell 2008;3:543–554. [DOI] [PubMed] [Google Scholar]
- 25.Yuri S, Fujimura S, Nimura K, Takeda N, Toyooka Y, Fujimura Y, Aburatani H,et al. Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression.Stem Cells 2009;27:796–805. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells.PLoS One 2010;5:e10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong CC, Gaspar-Maia A, Ramalho-Santos M, Reijo Pera RA. High-efficiency stem cell fusion-mediated assay reveals Sall4 as an enhancer of reprogramming.PLoS One 2008;3:e1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts.Genes Cells 2009;14:683–694. [DOI] [PubMed] [Google Scholar]
- 29.Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, et al. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 2002;11:2979–2987. [DOI] [PubMed] [Google Scholar]
- 30.Aguila JR, Liao W, Yang J, Avila C, Hagag N, Senzel L, Ma Y. SALL4 is a robust stimulator for the expansion of hematopoietic stem cells.Blood 2011;118:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM, Lai R, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice.Blood 2006;108:2726–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D,et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells.Blood 2008;112:805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong HW, Cui W, Yang Y, Lu J, He J, Li A, Song D,et al. SALL4, a stem cell factor, affects the side population by regulation of the ATP-binding cassette drug transport genes.PLoS One 2011;6:e18372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao D, Humphrey PA, Allan RW. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors.Cancer 2009;115:2640–2651. [DOI] [PubMed] [Google Scholar]
- 35.Bard JD, Gelebart P, Amin HM, Young LC, Ma Y, Lai R. Signal transducer and activator of transcription3 is a transcriptional factor regulating the gene expression of SALL4.Faseb J 2009;23:1405–1414. [DOI] [PubMed] [Google Scholar]
- 36.Ushiku T, Shinozaki A, Shibahara J, Iwasaki Y, Tateishi Y, Funata N, Fukayama M. SALL4 represents fetal gut differentiation of gastric cancer, and is diagnostically useful in distinguishing hepatoid gastric carcinoma from hepatocellular carcinoma.Am J Surg Pathol 2010;34:533–540. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda H, Sato Y, Yoneda N, Harada K, Sasaki M, Kitamura S, Sudo Y,et al. alpha-Fetoprotein-producing gastric carcinoma and combined hepatocellular and cholangiocarcinoma show similar morphology but different histogenesis with respect to SALL4 expression.Hum Pathol 2012;in press. [DOI] [PubMed] [Google Scholar]
- 38.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A,et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med 2006;12:410–416. [DOI] [PubMed] [Google Scholar]
- 39.Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009;136:1000–1011. [DOI] [PubMed] [Google Scholar]
- 40.Wang YLG, Carpino G, Cui C, Dominguez-Bendala J, Wauthier E, Cardinale V,et al. Biliary Tree Stem Cells, Precursors to Pancreatic Committed Progenitors:Evidence for Possible Life-long Pancreatic Organogenesis.Stem Cell 2012;in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K,et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer.Embo J 1999;18:2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita T, Honda M, Nio K, Nakamoto Y, Yamashita T, Takamura H, Tani T,et al. Oncostatin m renders epithelial cell adhesion molecule-positive liver cancer stem cells sensitive to 5-Fluorouracil by inducing hepatocytic differentiation.Cancer Res 2010;70:4687–4697. [DOI] [PubMed] [Google Scholar]
- 43.Shi SR, Imam SA, Young L, Cote RJ, Taylor CR. Antigen retrieval immunohistochemistry under the influence of pH using monoclonal antibodies.J Histochem Cytochem 1995;43:193–201. [DOI] [PubMed] [Google Scholar]
- 44.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer.Nat Rev Cancer 2011;11:558–572. [DOI] [PubMed] [Google Scholar]
- 45.Lee J, Kanatsu-Shinohara M, Morimoto H, Kazuki Y, Takashima S, Oshimura M, Toyokuni S,et al. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation.Cell Stem Cell 2009;5:76–86. [DOI] [PubMed] [Google Scholar]
- 46.Bohm J, Kaiser FJ, Borozdin W, Depping R, Kohlhase J. Synergistic cooperation of Sall4 and Cyclin D1 in transcriptional repression.Biochem Biophys Res Commun 2007;356:773–779. [DOI] [PubMed] [Google Scholar]
- 47.Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL,et al. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene.Hepatology 2008;48:1528–1539. [DOI] [PubMed] [Google Scholar]
- 48.Zhang L, Theise N, Chua M, Reid LM. The stem cell niche of human livers: symmetry between development and regeneration.Hepatology 2008;48:1598–1607. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A,et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma.Cancer Res 2008;68:1451–1461. [DOI] [PubMed] [Google Scholar]
- 50.Yang XR, Xu Y, Yu B, Zhou J, Qiu SJ, Shi GM, Zhang BH,et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma.Gut 2010;59:953–962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.