Abstract
We describe here a comprehensive and reliable assay to test the functional significance of variants of unknown clinical significance (VUS) identified in the human breast cancer susceptibility gene, BRCA2. The assay is based on the ability of human BRCA2 to complement the loss of endogenous Brca2 in mouse embryonic stem cells. The procedure involves generation of a desired mutation in BRCA2 present in a bacterial artificial chromosome (BAC) and introduction of the BAC into ES cells engineered for the assay. These ES cells have one null and one conditional allele of Brca2. First, the effect of the BRCA2 variants on the viability of ES cells is tested by Cre-mediated deletion of the conditional allele. Subsequently, variants that result in viable ES cells are examined for their effect on known functions of BRCA2 using a variety of functional assays such as sensitivity to genotoxic agents, in vivo and in vitro proliferation, effect on homologous recombination and genomic stability. The method described herein allows for analysis of three to five sequence variants within 2–3 months. This approach can also be used for functional analysis of variants identified in other human disease genes that result in a phenotype detectable in ES cells.
Keywords: Breast cancer, BRCA2, variants of unknown clinical significance (VUS), missense mutation, mouse embryonic stem (ES) cells, functional assay, DNA repair, Bacterial artificial chromosome (BAC), HPRT1 minigene
1. Introduction
Identification of a number of human disease genes and advances in sequencing technologies have revolutionized the field of molecular diagnostics in the last decade (1). Among the genes that are routinely being tested are the breast cancer susceptibility genes BRCA1 and BRCA2. For BRCA1/2 mutation carriers the lifetime risk of developing breast or ovarian cancer is 80% and 37%, respectively. Therefore, it is not surprising that individuals with a family history of these cancers are now opting to know if they have a mutation in one of these genes. One of the inherent drawbacks of sequencing-based approaches is the determination of the actual risk associated with any variant identified in the gene. This is highlighted by the fact that more than 800 variants of BRCA1 and 1100 variants of BRCA2 are listed as variants of unknown clinical significance in the breast cancer information core (BIC) database (http://research.nhgri.nih.gov/projects/bic/). Segregation analysis in disease-afflicted families provides the most reliable information to distinguish between deleterious and neutral alterations identified in hereditary disease genes (2). However, there is an enormous need to have a functional assay to classify variants for which such information is not available because most mutations are rare and familial data are often insufficient (3).
The functional significance of nonsense and frame shifting mutations can be predicted with a high degree of confidence. In contrast, the functional significance of splicing, intronic, regulatory, and point mutations is more difficult to predict.
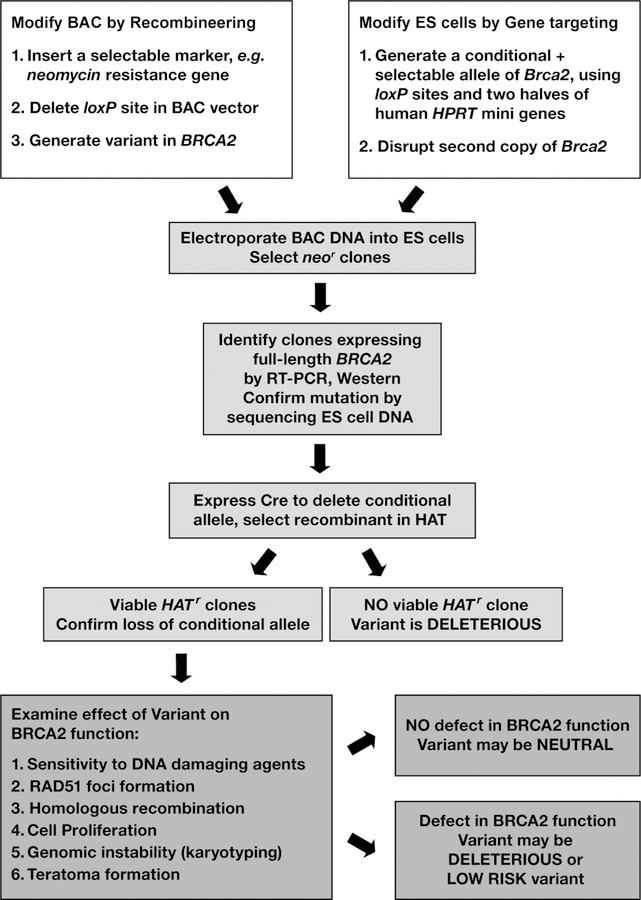
The functionality of some sequence variants can be tested in a complementation assay when a corresponding mutant gene is transfected into an established cell lines in which its endogenous counterpart is inactive (4). However, such complementation assays have two caveats. First, they usually rely upon delivery of a certain gene using cDNAs under the control of a heterologous promoter. This often results in a significantly higher expression level than the endogenous protein, which for many genes leads to apoptosis. In addition, cDNA expression vectors do not allow for testing mutations that affect splicing or map to regulatory regions. Second, established tumor cell lines are inherently prone to genomic instability and cell cycle deregulation, which sometimes complicates interpretation of the results (5). ES cells are more suitable for such studies because they retain the normal karyotype, even after extensive genetic manipulations, which makes them suitable for the functional analysis of DNA repair genes like BRCA2 (6). Instead of using cDNA expression vectors, BACs allow for gene expression at physiological levels under the control of its own promoter. More importantly, this allows examination of regulatory mutations as well as splicing and intronic alterations in addition to those that alter the protein-coding sequence. Desired mutations in BRCA2 can be rapidly generated in the BAC using the recombineering technology (7–10). We have demonstrated the suitability of mouse ES cells and BACs for the functional analysis of human mutations by analyzing sequence variants of the human BRCA2 (11). Using this approach, as outlined in Fig. 1, 5 variants can be tested in 2–3 months.
Figure 1. Overall scheme to evaluate functional significance of human BRCA2 variants.
Overview of the experimental design to examine the functional significance of BRCA2 variants in mouse ES-cells. Different steps are described in detail in the text.
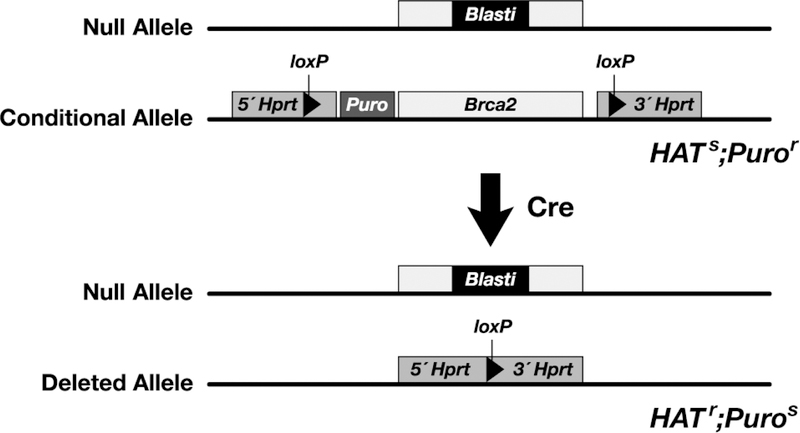
The functional assay is based on the observation that BRCA2 is essential for ES cell viability and DNA double strand break repair (12). The ability of BRCA2 variants to rescue the lethality of Brca2-deficient ES-cells and complement DNA repair defect is used to evaluate their functional significance. The assay utilizes a mouse ES cell line, PL2F7, in which one allele is functionally null and the other allele is rendered conditional (Fig. 2). The allele that is functionally null has an insertion of the Blasticidin resistance gene in exon 11. The conditional allele has two loxP sites upstream and downstream of Brca2. In addition, a puromycin resistance gene is targeted next to the loxP site upstream of Brca2 such that it would be deleted along with Brca2 after Cre-mediated recombination of the loxP sites. To allow for the selection of the recombinant clones, two halves of the human HPRT1 minigene are targeted along with the loxP sites. The HPRT1 minigene can be split into two fragments, a 5´fragment containing sequences from exons 1–2 and intron 2, and the 3´fragment containing intron 2 and exons 3–9 (13). A loxP site is present in the intron 2 in both the fragments. The two halves of HPRT1 can recombine in the presence of the Cre protein to generate a functional HPRT1 minigene and the recombinants can be selected in the presence of HAT. The PL2F7 ES cell line is derived from an Hprt-deficient ES cell line (AB2.2), which allows the use of HPRT1-mediated selection.
Figure 2. Generation of ES cells with a conditional allele of Brca2.
To generate a conditional allele of Brca2 and to select the recombinants, two loxP sites were targeted upstream and downstream of the gene along with the 3ór 5´ halves of the human HPRT1 minigene and a Puromycin resistance gene by homologous recombination. To disrupt the other allele of Brca2, a Blasticidin resistance gene was targeted to exon 11. The conditional allele of Brca2 can be deleted by transiently expressing Cre. ES cells become HATr and Puros. However, such cells do not survive because of loss of functional BRCA2.
When Cre is transiently expressed in PL2F7 cells, HATr clones do not survive due to the loss of BRCA2 (11). However, when a BAC clone expressing the wild type human BRCA2 is introduced into PL2F7 cells, viable HATr clones are obtained. BRCA2 variants can be generated in the BAC DNA and electroporated into PL2F7 cells before the conditional allele of Brca2 can be deleted. When no HATr colonies are obtained after Cre expression, the BRCA2 sequence variant is likely to be deleterious. Variants that result in viable ES cells are tested for their effect on the known functions of BRCA2 such as DNA repair, in vitro proliferation, RAD51 foci formation, homologous recombination, genomic stability, in vivo proliferation and differentiation by teratoma formation in mice (Fig.1). We describe here in detail various steps involved in generation of a BRCA2 variant in BAC, introducing the BAC into ES cells and performing various functional assays.
2. Materials
2.1. Bacterial strain
The E. coli SW102 strain harboring the defective lambda prophage and used for recombineering can be obtained from NCI recombineering resource (http://recombineering.ncifcrf.gov/). This strain is temperature sensitive and must be grown at 32°C.
2.2. Cell lines
2.3. Equipment
Standard laboratory equipments are used, some of which are described below:
An incubator set at 32 °C, a shaking incubator set at 32 °C and shaking water bath (200 rpm) set at 42 °C.
CO2 incubators.
Electroporator (Genepulser II with Pulse Controller II, BioRad) and cuvettes with 0.1 and 0.4 cm gap (BioRad).
High speed centrifuge and a refrigerated microcentrifuge.
PCR machine.
Spectrophotometer.
Microplate reader.
Coulter particle counter Z1 (Beckman Coulter).
Inverse microscope with differential interference device (Leica).
Hybridization oven (VWR).
UV crosslinker 1800 (Stratagene).
137Cs γ-irradiator.
2.4. Antibodies
Rabbit anti-Rad51 (Calbiochem).
Mouse anti- γ-H2AX antibody (Upstate).
FDAR (Fluorescein (FITC) AffiniPure Donkey Anti Rabbit IgG, Jackson Labs).
RDAM (Rhodamine (TRITC) AffiniPure Donkey Anti-Mouse IgG, Jackson Labs).
Rabbit Anti-BRCA2 (Ab-2, Calbiochem).
Goat anti-rabbit IgG-HRP (Santa Cruz).
2.5. Other reagents
BAC Clone RPCI-11 777 19I (for simplicity referred to as BAC777, 250 kb genomic fragment containing human BRCA2 in pBACe3.6 vector, in which loxP511 site was deleted and the second loxP site replaced for PGK-Neo, available upon request).
Plasmid DNA purification reagents (Qiagen).
Gel extraction kit or reagents to purify DNA from Agarose gel (Qiagen).
Expand High Fidelity (HiFi) PCR System (Roche).
D-PBS (Gibco):
DMEM without phenol red (Gibco)
Trypsin-EDTA (Invitrogen)
HAT supplement (Gibco).
HT supplement (Gibco).
G418 (Geneticin®, Gibco).
Puromycin (Sigma).
Prime-It® II Random Primer Labeling Kit (Stratagene).
LB Medium:
LB Agar:
M15 medium: To make 600 mL of M15 media, add 90 mL of Fetal Bovine Serum (HyClone ES cell quality, final conc. 15 %), 6 mL of 10 mM β-mercaptoethanol (Sigma, final conc. 0.1 mM) and 6 mL of 100 X GPS (glutamate, penicillin/streptomycin, Gibco) to 500 mL of Knockout-DMEM (Invitrogen).
ES-cell Lysis Buffer: 10mM Tris-HCl, pH 7.5, 10mM EDTA, pH 8.0, 10 mM NaCl, 0.5 % Sarcosyl,1 mg/mL Proteinase K.
1x TAE buffer:
1x TBE buffer:
TBS buffer:
RIPA buffer:
NuPAGE MOPS SDS Running buffer:
1x NuPAGE Transfer Buffer:
Antibody blocking solution:
2.6. Mice
Immunocompromised athymic nude mice (C3H/HeNCr-nu).
3. Methods
3.1. Generation of a desired mutation in BRCA2 in the BAC DNA by recombineering technology using a two-step “hit and fix” method.
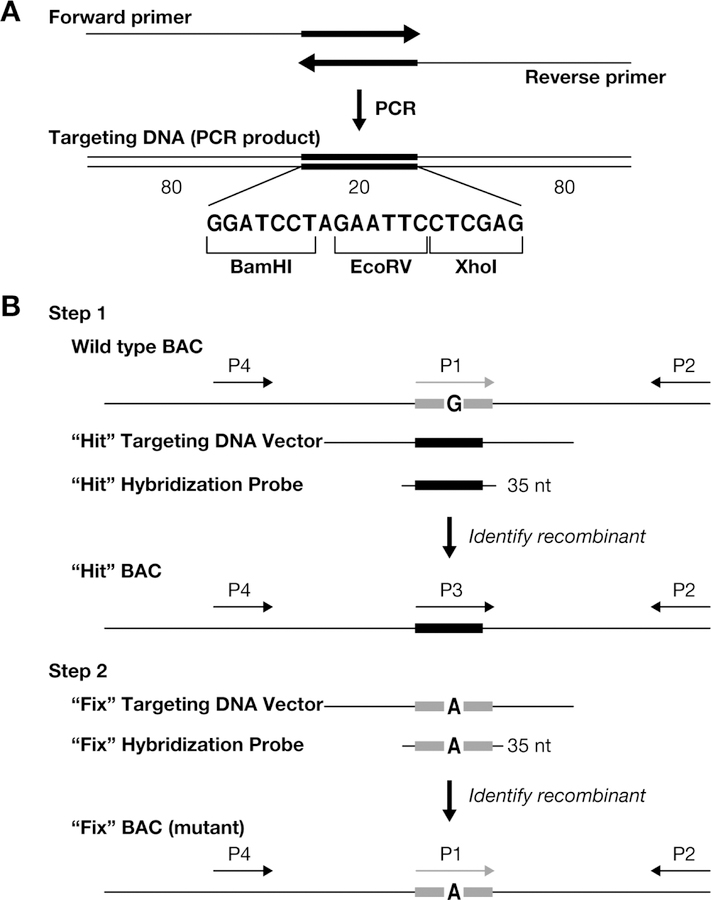
Use of oligonucleotides to generate subtle alterations in a BAC without the use of any selectable marker by recombineering in E.coli strains harboring a defective lambda prophage, such as DY380 or SW102, has been described in detail previously (9, 15, 16). Recombinants can be identified by a mismatch amplification mutation assay (17). While this approach is helpful, it can occasionally result in false positive clones. A two-step “hit and fix” method allows for a rapid generation of any mutation in BACs and recombinants can be easily identified using standard PCR-based methods (9). The two-step “hit and fix” procedure involves introduction of 20 unique nucleotides (unique heterologous sequence) to the site where a mutation has to be generated in the first “hit” step (Fig. 3).
Figure 3. Two-step “hit & fix” method to generate subtle mutations using single stranded short PCR product or oligonucleotides as targeting vector.
(a) The single-stranded oligonucleotides containing 160 bases of homology and 20 unique bases are generated by using two 100-mer oligonucleotides in a PCR reaction. The two 100-mer oligonucleotides have 20 complementary bases (in this case the 20 bp contains restriction sites BamHI, EcoRV, XhoI) at the 3´ end. The 180 bp PCR product can be denatured to obtain single-stranded oligonucleotides that can be used as the targeting construct. (b) Schematic representation of the two steps involved in the “hit and fix” method to generate subtle alterations (e.g. G to A) without the use of a selectable marker. In step 1, a 180-mer single-strand oligonucleotide is used to replace 20 nucleotides (gray box) around the target site with 20 heterologous nucleotides (black box). Recombinants can be identified by colony hybridization using an end-labeled 35-mer oligonuclotide that can specifically anneal only to the recombinant DNA. A primer set specific for the heterologous “hit” sequence (P3 and P2) can be used to confirm the presence of recombinant clones by PCR. A second primer set (P1 and P2) can be used as a control to amplify only the non-recombinant DNA. Generation of a correct recombinant clone can be confirmed by digesting the PCR product (~300–500 bp) produced using primers P2 and P4 with BamHI, EcoRV or XhoI. In step 2, the 20 nucleotides are restored to the original sequence, except for the desired mutation. Such clones can be identified by colony hybridization using a 35-mer oligonucleotide as a probe and further confirmed by PCR amplification using primers P1 and P2, by testing for loss of the restriction sites inserted in step 1, by digesting the PCR product of primers P2 and P4, and by sequencing. (Reproduced from Nature Protocols, 2009, 4:206–223)
The original sequence is then restored with the exception of the point mutation in the second “fix” step. In both steps, 180-bases-long single-stranded DNA fragments generated by PCR are used as targeting vectors. The 20 nucleotide sequence can be used for initial screening by PCR, however, the hybridization approach allows screening of more than 5,000 individual colonies from a single 15 cm agar plate (10). It also allows simultaneous screening of multiple mutations (“hit” step) by using the same hybridization probe. In addition, the “hit” cassette is designed to contain several restriction sites that facilitate rapid confirmation of the correct targeting.
3.1.1. Introduction of the BAC DNA into SW102 strain to provide the lambda recombination function
To prepare electro-competent SW102 cells, pick an isolated colony of SW102 strain of E.coli from a LB plate and grow overnight in 3 mL of LB at 32 °C.
Next morning add 1mL of the culture to 50 mL of LB in a 250 mL flask and grow at 32 °C to an OD600 of 0.50 – 0.60. Transfer 10 mL of the culture to a 50 mL Oak Ridge tube and centrifuge at 6000 x g in a pre-chilled rotor for 10 minutes at 1 °C.
Wash the cell pellet once with 10 mL of ice-cold water, resuspend in 1 mL of water and transfer to a chilled 1.5 mL tube. Centrifuge at 18,000 x g for 20–30 seconds at 1 °C.
Wash the cells two more times with 1 mL of ice cold water. Resuspend the cell pellet in water to a final volume of 50µL and keep on ice.
Mix 1 µL of BAC DNA (100 ng) with 50 µl of electro-competent E.coli cells and chill on ice for 5 minutes and then transfer into a 0.1 cm gap cuvette. Set the Gene Pulser at 1.8 kV, 25 µF capacitance and 200 ohm resistance.
Electroporate the BAC DNA into the cells and immediately add 1 mL of LB medium.
Grow cells at 32 °C for 1 hour. Spin down the cells and resuspend in 200µLof LB medium.
Plate the cells on an LB agar plate containing chloramphenicol (12 µg/ml). Incubate overnight at 32 °C.
Pick isolated colonies for recombineering. Confirm the integrity of the BAC DNA by digesting the BAC with a few restriction enzymes (e.g. BamHI, EcoRI, HindIII, EcoRV) and comparing the restriction pattern with the original BAC clone by running the two samples in parallel on a 0.8 % agarose gel. Make glycerol stocks and freeze at −80 °C.
3.1.2. Generation of a targeting cassette to introduce a desired mutation in a BAC
A targeting cassette to generate a point mutation in a BAC is synthesized by PCR using two 100-mer oligonucleotide primers overlapping by 20 nucleotides at their 3´-end. The resulting targeting vector contains two homology arms that are 80 bases in length flanking a 20-mer heterologous sequence in the middle (Fig. 3a). This heterologous sequence (5´-GGATCCTAGAATTCCTCGAG-3´) is the same for all “hit” targeting vectors allowing for a highly efficient and simultaneous screening of targeted clones of any number of mutated genes or regions. “Fix” targeting cassettes are composed of the same homology arms as the corresponding “hit” vectors, while the heterologous middle region of the “hit” vector is replaced with the final sequence including a desired mutation. At this point a 20–35-mer oligonucleotide encompassing this region can serve as a probe to differentiate “fix”- recombinants from the “hit” (Fig. 3b).
Set up the following reaction using the Expand High Fidelity (HiFi) PCR System (Roche, cat.# 11732641001). In general, use 6 µL of each 100-mer oligonucleotide (10 µM) and 10 µL of 2 mM dNTPs, 2 µL of HiFi Taq Polymerase (3.5 U/µL) in a 100µL PCR reaction. Conditions for the PCR cycle include an initial denaturation at 94 °C for 1 minute followed by 40 cycles of 94 °C for 30 sec, 55–60 °C for 30 sec and 72 °C for 30 sec and a final extension at 72 °C for 2 minutes.
Examine 1 µL of the reaction on a 1.0 −1.5 % agarose gel in 1xTAE buffer. A 180 bp product should be observed (see Note 1).
Purify the targeting vector using a Qiagen PCR Purification Kit and elute in 30 µL of Qiagen Elution Buffer or ethanol-precipitate and dissolve in 20–30 µL dH2O.
3.1.3. Induction of the lambda recombination genes and preparation of electrocompetent cells
Inoculate SW102 cells containing the BAC from a frozen glycerol stock or a single colony into 3 to 5 mL LB medium. Shake at 32 °C overnight.
Add 0.5 mL of the overnight culture to 35 mL of LB medium in a 250-mL (baffled) Erlenmeyer flask.
Grow cells at the 32 °C for about 2 hr. The cells are ready when the OD600 is between 0.5 and 0.6. It is important not to over-grow the cells.
Transfer 10 mL of the culture to a 50 mL Erlenmeyer flask and place that flask in the 42 °C water bath. Shake for 15 min at 200 rpm to induce the lambda recombination genes. Leave the remainder of the culture on ice (see Note 2).
Immediately after inducing for 15 min at 42 °C, rapidly cool the flask on ice with gentle swirling for 10–15 min. As a control, use 10 mL of uninduced cells and process them exactly as the induced cells. While the cells are on ice, pre-cool the centrifuge to 4 °C and chill two labeled 50 mL Oak Ridge tubes.
Transfer 10 mL of both the induced and uninduced cultures to the Oak Ridge tubes. Centrifuge for 10 min at 4600 x g at 4 °C. Carefully discard the supernatant.
Add 1 mL ice-cold distilled water to the cell pellet in the bottom of each tube and gently resuspend the cells with a large pipet tip (do not vortex). Add 30 mL ice-cold distilled water to each tube and mix gently. Centrifuge tubes again as in the previous step.
Decant the supernatant very carefully from the soft pellet in each tube and resuspend each cell pellet in 1 mL ice-cold distilled water. (Remove tubes from the centrifuge promptly. Because the pellet is very soft, care should be taken not to dislodge it, especially when processing multiple tubes).
Transfer the resuspended cells to prechilled microcentrifuge tubes. Centrifuge 30 to 60 sec at a maximum speed at 4 °C. Carefully aspirate the supernatant.
Wash the cells two more times with 1 mL of ice cold water.
Resuspend the cell pellet in the cold distilled water to a total volume of 50 µL.
3.1.4. Electroporating the targeting vector into SW102 cells containing the BAC
Chill the desired number of 0.1-cm electroporation cuvettes on ice.
In a 0.5 mL tube, mix appropriate volume of DNA (200 to 300 ng of salt-free PCR fragment) with 50 µL of electro-competent induced or uninduced cells. Leave the tubes on ice for 5 minutes.
Electroporate the DNA into the cells using 1.8 kV, 25 µF capacitance and 200 ohm resistance. Immediately add 1 mL LB medium to the cuvette and transfer the electroporation mix to sterile culture tubes and incubate the tubes with shaking at 32 °C for 1.5 hr.
Serially dilute the cell suspension in LB medium and plate 200 µL of 10−2 and 10−3 dilutions onto a 15 cm LB agar plate containing an appropriate antibiotic (Chloramphenicol at 12 µg/mL or Kanamycin at 25 µg/mL). Use sterile glass beads instead of a bacteriological spreader to achieve uniform distribution of colonies throughout the plate. Incubate agar plates for 18–22 h at 32 °C.
3.1.5. Identifying the recombinant clones
Pick the plate with approximately 3000–6000 colonies. Colony density should be such that individual colonies can be identified. Transfer the colonies to a charged nylon membrane (Hybond) by standard methods and hybridize with γ−32P-labelled oligonucleotide probe for 2–3 hours at 50 °C. Hybridizing for more than 4 hours results in high background. This oligonucleotide probe corresponds to theheterologous sequence that is being inserted in the “hit” step.
After positive clones are identified, they should be subcloned by a second round of hybridization to obtain a pure recombinant clone that does not contain the original non-recombinant BAC.
Confirm by testing for restriction sites present in the heterologous sequence or by sequencing.
Once the correctly targeted pure “hit” colonies are identified, repeat the targeting step (fix targeting) to replace the 20-nucleotide heterologous sequence with the desired mutation exactly as described above for the “hit” step.
Confirm the mutation by sequencing.
3.2. Electroporating the BAC DNA into ES cells
Prepare BAC DNA using QIAGEN Maxi Prep kit from bacteria cultured overnight at 32˚C in 250 mL LB medium according to a manufacturer’s protocol. Dissolve the DNA pellet in 100 µL dH20. The usual yield is between 50 – 150 µg of DNA.
Expand PL2F7 ES cells by growing them on SNL feeder cells in M15 medium using standard ES cell culturing techniques as described previously (18).
Split PL2F7 cells at 1:2 ratio one day prior to BAC electroporation. 1.0 X 107 cells are needed for each electroporation. The usual yield from one 80–90 % confluent 10 cm dish is 2.5–3 × 107 cells.
Change the M15 medium 3–4 hours prior to the electroporation.
Wash the plates 2 times with PBS, add 2 ml trypsin, incubate 15 min at 37°C, add 2 mL M15, disaggregate the cells by vigorous pipetting 25–30 times using a plastic disposable transfer pipette, pellet cells by centrifugation at 250 x g for 5 min, wash once with 10 mL PBS, pellet cells as before, resuspend in 1 ml PBS and count 20 µL aliquot using a Coulter counter. Adjust cell concentration to 1.1 × 107 cells/ml with PBS.
Transfer 0.9 mL cell suspension into an electroporation cuvette.
Add 25 µg BAC DNA. Mix by inverting several times. Leave the cuvette at room temperature for 5 minutes.
Set the electroporator (BioRad GenePulser II) at 230 V, 500 µF and electroporate the BAC into ES cells. The time constant should be between 5.6 and 7.3 msec.
Leave the cuvette at room temperature for 5 minutes. Transfer the electroporated cells to a 10 cm feeder plate containing 15 mL of M15 medium. After 36 hours, add G418 (180 µg/mL) to M15 medium.
Change media daily with fresh M15 medium containing G418 for 5 days. After 5 days add M15 without G418.
Once colonies become visible (usually takes 2–3 days), pick 24–36 colonies in a 96-well plate (one colony/well) for each BAC construct as described previously (18) (see Note 3).
Once cells are about 80 % confluent, split them into three 96-well plates (one plate with feeders and two gelatinized plates without feeders).
When cells are about 80 % confluent, freeze the plates with feeders using freezing media (60 % DMEM, 20 % FBS, 20 % DMSO; freshly prepared) as described previously (18).
Wash the plates without feeders twice with PBS, remove PBS and transfer at −80 ˚C.
Plates can be stored at −80 ˚C for up to 6 months.
3.3. Select BRCA2 BAC transgenic ES cell clones by Southern hybridization
Identify ES cells clones that have full-length BRCA2 gene by Southern blot analysis using probes from the 5´ and 3´ ends of the gene. Subsequently, test the “Southern positive” clones for BRCA2 expression by Western blot analysis. When a variant is predicted to result in no detectable protein product, RT-PCR should be performed to detect mRNA expression.
Remove one of the 96-well plates from step 14 of section 3.2, add 50 µL of ES-cell Lysis Buffer per well, incubate overnight at 60 ˚C.
2. Extract DNA, digest with EcoRI restriction enzyme as described previously (18)
Load the DNA samples on a 0.8 % agarose gel (1XTBE buffer). 96 DNA samples can be analyzed on a 20 × 25 cm2 gel with three rows of 36-wells. Run the gel long enough to separate 1.8 kb and 4.3 kb EcoRI fragments corresponding to 5´- and 3´-ends of BRCA2, respectively.
Transfer the DNA to a nylon membrane using Southern blotting procedure and hybridize the membrane with 32P-labelled both 5´- and 3´-BRCA2 probes (see Note 4) using standard procedures (19).
Identify ES cell clones positive for both probes. Usually 50 % of such clones express the BRCA2 protein.
3.4. Western Blot analysis to identify BRCA2 expressing clones
Thaw the 96-well plate containing frozen ES cells from step 14 of section 3.2 at 37 ˚C. Transfer Southern positive ES cells identified above into a 24-well plate with a feeder layer containing 2 mL M15 medium in each well. The entire well content including the oil should be transferred into 24-well plates. Incubate the plate until the wells become 80 % confluent. Change the M15 medium daily.
To obtain cells for the Western blot, trypsinize ES cells growing in 24-well plates with 250 µL trypsin-EDTA.
Add 250 µL M15 media, dissociate cells by pipetting and transfer 100 µL aliquot in a new 6-well plate pretreated with gelatin and containing 4 mL M15 medium. Freeze the rest of the cells in individual freezing vials after adding 400 µL of 2x freezing media.
Once the 6-well plate is almost 100 % confluent, lyse the cells for Western analysis.
Wash plates twice with PBS, add 200 µL RIPA buffer, scrape cells using a plastic scraper and transfer into 1.5 mL Eppendorf tubes.
Rotate the cell lysates at 4˚C for 40 minutes. Clear the lysates by centrifuging at 12,000 x g for 10 minutes.
Transfer the supernatant into a new tube and estimate the protein concentration in the lysates using the BCA method according to manufacturer’s procedure.
Prepare samples for Western blot. Take 35 µg protein, bring up the lysate volume to 15 µL with RIPA buffer, add 5.5 µL of 4x loading dye solution (Invitrogen), and 2 µL Sample Reducing Agent (Invitrogen).
Heat protein samples at 70 ˚C for 10 minutes and chill on ice.
Assemble a precast NuPAGE 4–12 % Bis-Tris gel cassette (Invitrogen) in a corresponding gel box with NuPAGE MOPS SDS Running buffer. Add 500 µL antioxidant in 200 mL running buffer in the upper chamber.
Load the samples and SeeBlue Plus 2 size marker (Invitrogen) in gel slots and run a gel at 200 V for I hour.
Set up protein transfer onto a PVDF membrane in 1x NuPAGE Transfer Buffer containing 10 % methanol according to manufacturer’s instructions. Transfer at 22 V overnight at 4 ˚C. Results are more consistent when one blot is transferred per transfer module.
Next day disassemble the transfer module, mark the wells and label the blot with a pencil. Soak the membrane in two changes of TBS.
Transfer the membrane into blocking buffer (1xTBS, 0.05 % Tween20, 5 % Milk) and incubate for 1 hour at room temperature with agitation.
Wash in 1xTBS, 0.05% Tween20 three times for 5 minutes.
Incubate the membrane with the primary antibody (Rabbit-anti-human BRCA2) in the blocking buffer overnight on a rocking platform at 4 ˚C.
Wash the blot 3 times for 30 minutes in 1xTBS, 0.05 % Tween20 at room temperature.
Incubate with the secondary antibody for 30 minutes at room temperature (HRP-labeled goat-anti-rabbit IgG antibody).
Wash 4 times 30 minutes in 1xTBS, 0.05 % Tween20 at room temperature.
Detect the chemiluminescent signal using ECL Plus system according to the manufacturer’s procedure. Identify “Western-positive” ES cell clones (see Note 5).
3.5. Excision of the conditional Brca2 allele
Thaw Western-positive ES cell clones in 6 cm feeder plates. Test at least two clones for each sequence variant. Select clones that express BRCA2 at a level comparable to the endogenous levels (e.g. in human ES cells).
Split 80 % confluent plates 1:2 one day prior to electroporation.
Electroporate 25 µg Pgk-Cre plasmid for each clone as described in steps 7–9 of section 3.2.
Plate 10 µL (105 cells) of the electroporated cells into 10 cm feeder plate.
Freeze the remaining unelectroporated cells into two freezing vials at approximately 5×106 cells per vial.
Select for recombinant clones in HAT containing M15 medium, 36 hours after electroporation. Maintain HAT selection for 5 days. Change HAT containing M15 medium everyday.
Replace HAT supplement with HT for two additional days.
Culture the cells in M15 medium for additional 3–4 days until colonies are ready to be picked. When a BAC containing wild-type BRCA2 is used, 1,000 −3,000 HATr colonies are obtained on one 10 cm dish. PL2F7 cells yield less than 10 colonies, which represent background colonies. Therefore, hypomorphic BRCA2 variants may yield less than 1,000 colonies depending on the mutation.
See Note 6 for an alternative approach.
3.6. Confirming the Cre-mediated loss of the conditional allele
Pick 12–24 colonies for each electroporated clone and transfer them into a 96-well feeder plate and expand them to one 96-well feeder plate and two 96-well plates without feeders as described above. Sensitivity of ES cell clones to puromycin can be used to determine the loss of the conditional allele. However, clones must be genotyped to confirm the loss of the conditional allele. Use one of the plates without feeders for genotyping by Southern analysis as described above in section 3.3.
Use EcoRV enzyme to digest the DNA and probe “144/145” for hybridization (see Note 7).
Identify clones that have lost the wild type (conditional) Brca2 allele (see Note 8)
Expand 3–5 correctly targeted HATr ES cell clones for each variant from a frozen 96-well feeder plate and freeze stocks in liquid nitrogen.
Proceed with functional analysis using two clones for each variant.
3.7. Functional analysis of ES cell clones expressing BRCA2 variants
3.7.1. Testing sensitivity to DNA damaging compounds
Test the DNA repair function of BRCA2 sequence variants by challenging them with various DNA-damaging compounds.
Seed 8,000 ES cells per well in a gelatinized 96-well plate. Clones with poor plating efficiency or proliferation defect may need to be seeded at a higher density to compensate for lower growth and seeding efficiency (see Note 9). Designate 7 rows for increasing drug concentrations with 3 columns for each clone being tested (triplicates). Leave the last row empty as a blank control.
18 hours after seeding the cells, replace the M15 for medium containing increasing doses of each of the DNA damaging compounds (see Note 10)
Maintain treatment for 3 days without changing the medium.
Estimate relative cell survival using XTT metabolic assay (20). Wash the plates twice with PBS. Leave the plates with the last change of PBS at 37 ˚C while mixing warm XTT solution with PMS. Replace PBS with 100 µL of XTT reagent per well.
Incubate for 2.5 – 3 hours at 37˚C, and then measure the absorbance at wavelength 450 nm for 0.1 seconds per well.
3.7.2. Testing sensitivity to ionizing radiation
To test the cell sensitivity to ionizing radiation, seed each tested clone in five 96-well plates at 8,000 cells per well, one column per clone, one plate per one radiation dose.
Next day expose each plate without changing media to a 137Cs source to achieve a designated γ-radiation dose (0, 0.5, 1, 2 and 4 Gy).
Measure cell survival 72 hours later using the XTT assay as described above (step 4 of section 3.7.1).
3.7.3. Testing sensitivity to UV light
Seed 32,000 ES cells (64,000 cells for mutant cells with severe proliferation defect) in gelatinized 24-well plates in triplicates.
18 hours later remove the medium and the plate lid, and irradiate with 0, 5, 10, or 15 J/m2 using a UV crosslinker (Stratagene) or any other suitable UV source. Replace the M15 medium.
72 hours later wash the plates twice with PBS, add 400 µL XTT reagent, incubate for 2.5–3 hours. Transfer 100 µL of developed color solution from each well into a 96-well plate for measurement as described above (step 4 of section 3.7.1).
3.7.4. Measuring in vitro proliferation
To determine the effect of a BRCA2 variant on in vitro proliferation, seed 3 sets of each clone at 50,000 cells per well in triplicates in 24-well feeder plates. Leave a few wells empty with only feeder cells as a background control.
After 24 hours, wash the plates twice with PBS, trypsinize with 200 µl trypsin-EDTA for 15 minutes at 37 ˚C, add 200 µL M15 media, pipette vigorously up and down 20 times to achieve a single cell suspension.
Count a 200 µL aliquot using a Coulter counter.
Repeat the procedure with the second and third set of cells on the second and third day after seeding, respectively.
To estimate the cell proliferation, subtract the average feeder cell count from each value and express the resulting numbers as multiples of the average cell number (or a concentration) recorded at the first day after seeding.
3.7.5. Measuring in vivo proliferation
To measure the in vivo proliferation, harvest ES cells from three confluent 10 cm plates. Resuspend ES cells in PBS at 108 cells/ml.
Inject 0.1 mLof this cell suspension (5 × 106 cells) subcutaneously in flanks of one-month-old immuno-compromised nude mice. 3–5 males and 3–5 females should be injected for each ES cell clone.
Monitor teratoma growth by measuring the length and width of the tumor for three weeks (or until the tumor reaches 1.5 cm in any dimention), starting on day 7 after injection. Calculate the tumor size as a product of 2 x length x width in cm3.
3.7.6. Evaluating genomic instability
To examine the effect of a BRCA2 variant on genomic stability, measure chromosomal aberrations by karytypic analysis.
Plate 3–5×106 ES cells in 6 cm gelatinized dishes without feeders.
Next day treat cells with 10 µg/mL colcemid for 1.5 hours to arrest cells at metaphase and prepare metaphase spreads using standard procedures (21).
Stain slides in a Giemsa solution as described elsewhere (22). Blindly score 200 well-spread metaphases containing at least 40 chromosomes for structural aberrations classified according to the ISCN scheme into chromosome and chromatid gaps, breaks and other anomalies (23).
3.7.7. Homologous recombination assay
To test ES cells directly for homologous recombination, electroporate them with 25 µg of linear Rosa26-Puro targeting vector (11) as described above (step 7–9 of section 3.2).
Select on puromycin for 5 days starting 36 hours after electroporation.
Pick 96 colonies for each cell line and expand them for Southern blot analysis as described above (step 10–14 of section 3.2), except that there is no need for a duplicate 96-well plate with feeders because these cells are not intended to be cultured any further.
Perform Southern blot analysis as described above (section 3.3), except that the DNA should be digested with SpeI and hybridized with the Rosa26-specifi probe. Clones that underwent a homologous recombination will reveal a 4.5 kb band and their proportion relative to the total number of clones indicates the efficiency of the homologous recombination.
3.7.8. RAD51 foci formation assay
BRCA2 is required for the recruitment of RAD51 to the sites of DNA double strand breaks (24). RAD51 foci at the sites of DNA damage can be visualized by immunofluorescence staining.
Seed 40,000 cells per well in gelatinized SonicSeal plastic slides (see Note 11).
48 hours later γ- irradiate the cells with 10 Gy.
Incubate cells at 37˚C for 6 hours.
Fix cells with 4 % paraformaldehyde for 5 minutes.
Wash twice with PBS and permeabilize in PBS-buffered 0.1% Triton X-100 solution for 10 minutes.
Wash twice with PBS and block overnight in antibody blocking solution at 4 ˚C.
Wash once with PBS for 5 minutes at room temperature, add rabbit anti-Rad51 antibody (PC130 diluted 1:200) and mouse anti γ-H2AX antibody (Upstate) diluted 1:1,000 in antibody blocking solution and incubate overnight at 4˚C.
Wash four times for 30 minutes with 1xPBS, 0.05 % Trion X-100.
Incubate with secondary antibodies FDAR and RDAM diluted in antibody blocking solution 1:100 and 1:150, respectively, for 2 hours at room temperature in the dark.
Wash as in step 6 of this section, stain with DAPI, and mount in Vectashield mounting medium.
Evaluate preparations under a fluorescent microscope equipped with a proper filter set and count the number of RAD51 foci per nucleus (see Note 12).
ACKNOWLEDGEMENTS
We thank Jiro Wada for help with illustrations. Research was sponsored by the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
4. Notes
The 180 bp band is quite frequently not a single clean band instead appears to be fuzzy. Such products have worked well in our hands and therefore should be used for targeting.
It helps to induce 20 mL of bacterial culture that yields 100 µL of electrocompetent cells. Although only 50µL are needed for each electroporation, it is good to have cells for an additional electroporation should something go wrong with the first set.
If very few G418r colonies are obtained after electroporating BAC DNA into ES cells, try linearizing the BAC DNA using a unique AscI restriction site. Digest DNA with AscI enzyme for 1 hour at 37 °C. Purify DNA by phenol : chloroform extraction. Precipitate DNA by adding 0.1 vol. of 3 M sodium acetate and 2 vol. of absolute ethanol. Spool DNA precipitated with ethanol and dissolve in 100 µL TE buffer.
Southern hybridization probes from the 5´ and 3´ ends of human BRCA2 can be generated by PCR using BAC 777 as template. To generate 5´ specific probe (581 bp) use primers: HB2UTR5F1, 5´-GAACTGCACCTCTGGAGCG-3´; HB2in1R1, 5´-AAGCACTCGAAACGTGGCTA-3 and for 3´ probe (287 bp) use primers: HB2ex25F1, 5´-GTGAGTAACCTTGTTCATAGGTG-3´ and HB2ex25R1, 5´-AATGACCTGTTGCTTACAGTG-3´.
If no full-length protein is detected by Western analysis of multiple independent ES cells containing a BRCA2 variant in a BAC, it may suggest that the mutant BRCA2 protein is unstable. In such cases, expression of the BRCA2 transcript can be examined by RT-PCR or Northern blot analysis.
- After electroporating the BAC into ES cells as described in section 3.2, pick G418r clones and expand them into three 96-well plates (one plate for RT-PCR analysis, one will be frozen as a master plate and third will be used to expand RT-PCR positive clones).
- Use one 96-well plate for expression analysis by RT-PCR to select clones with the full-length BAC. Use commercially available kits (such as, Rnaeasy-96 from Qiagen) to extract RNA from cells in 96-well plates. Lyse the cells in 100μLfor easy handling in 96- well plates and elute the RNA once with 40μL of water to ensure high concentration of RNA. Use Titan one step RT PCR kit (Roche) to perform RT-PCR using 1 µL of total RNA from the step above. For PCR, use primers specific to human BRCA2, hBRCA2_Ex11F, 5´-ACATGTCCCGAAAATGAGGA-3´ and hBRCA2_Ex18R, 5´-GCCGATCTTCTGCTTCTATCA-3´. The 1250 bp PCR product spans through exons 11–18 of BRCA2.
- Passage RT-PCR-positive ES cell clones from one of the 96-well plates (from a above) to a 24-well plate.
- Transduce 105 cells with adenovirus expressing Cre to delete the conditional allele and freeze the rest of the cells. In general, 106 ES cells are obtained from a 80–90 % confluent well of a 24-well plate. Resuspend one tenth of the cells in 100 μL of M15 media. For Adenoviral-Cre transduction, use MOI of 100. If the viral titer is 1010pfu/mL, use 1μL of the virus for 105 cells. After infection, bring up the volume of transduced cells to 1mL with M15 media and plate 10 μL and 100 μL in a 6 cm plate such that the numbers of cells plated will be 103 and 104, respectively.
- Select Adeno-Cre-transduced ES cells in the HAT media to select for recombinant clones.
- Pick clones, expand and genotype to confirm the of a conditional allele. If viable clones are obtained, perform drug sensitivity assay as described in section 3.7.
This approach is faster and saves about 4 weeks in evaluation time for each set of mutants. However, this approach has a disadvantage; if HATr clones are not obtained after Cre expression, it is difficult to conclude if the variant is deleterious or the clones failed to survive because of lack of expression of the full-length protein. In such cases, it is recommended to test clones individually to confirm protein expression. This disadvantage is off-set by the fact that 6–12 independent G418r & RT-PCR positive clones for each variant at a time can be tested easily. It is highly unlikely that none will express the full-length protein, unless reduction or lack of protein expression is linked to the mutation. We believe that the number of such cases will be relatively small and should not hinder the overall progress.
The Southern hybridization probe 144/145 (1250 bp) to genotype ES cells for loss of conditional allele can be generated by PCR using BAC 777 as template and PCR primers: PCR primers: SKS-144, 5´-TGTCATTGTGATGACATGCA-3´ and SKS-145, 5´-CAGTCACTCCTCCTCTTTTC-3´
Occasionally, genotyping of HATr ES cell clones show the presence of the conditional allele. This is likely to be due to a spontaneous trisomy of chromosome 5 containing the conditional Brca2 allele. Partial Cre-mediated recombination of only one of the conditional alleles will generate HATr clones, while the “unrecombined” allele will show the presence of the conditional allele.
For some hypomorphic mutants, we have observed cell numbers and OD values in the XTT assay to be significantly lower compared with controls. This is due to the severe growth retardation associated with a defect in BRCA2 function. In such cases, plating more cells than control samples compensates for lower seeding efficiency and growth rate.
- MMC (Mitomicin C): 0, 5, 10, 20, 30, 40, 60 ng/mL
- MMS (Methyl-methanesulfonate): 0, 5, 10, 15, 20, 30, 40µg/mL
- Cisplatin (cis-Diammineplatinum(II)dichloride): 0, 0.1, 0.2, 0.4, 0.5, 0.6, 0.8 µM
- MNNG (N-Methyl-N’-Nitro-N-Nitrosoguanidine): 0, 1, 2, 8, 15, 20, 30 µM
Mouse ES cells poorly adhere to glass slides or cover slips. Therefore, only plastic slides suitable for tissue culture can be used to grow ES cells for immunofluorescence.
Images of 50–100 nuclei should be taken and evaluated for the numbers of gamma H2AX and Rad51-positive foci. Rad51 foci not overlapping with gamma H2AX are likely to be unspecific and should be disregarded. Depending on the cell cycle stage, not all nuclei will show the foci.
References
- 1.Beaudet AL and Belmont JW (2008) Array-based DNA diagnostics: let the revolution begin. Annu Rev Med 59, 113–29. [DOI] [PubMed] [Google Scholar]
- 2.Thompson D, Easton DF and Goldgar DE (2003) A full-likelihood method for the evaluation of causality of sequence variants from family data. Am J Hum Genet 73, 652–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, et al. (2007) A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet 81, 873–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho MA, Couch FJ and Monteiro AN (2007) Functional assays for BRCA1 and BRCA2. Int J Biochem Cell Biol 39, 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S,et al. (2003) Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res 63, 8634–8647. [PubMed] [Google Scholar]
- 6.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, et al. (2000) Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 227, 271–278. [DOI] [PubMed] [Google Scholar]
- 7.Copeland NG, Jenkins NA and Court DL (2001) Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2, 769–779 [DOI] [PubMed] [Google Scholar]
- 8.Swaminathan S, Ellis HM, Waters LS, Yu D, Lee EC, Court DL et al. (2001) Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis 29, 14–21. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y and Sharan SK (2003) A simple two-step, ‘hit and fix’ method to generate subtle mutations in BACs using short denatured PCR fragments. Nucleic Acids Res 31, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharan SK, Thomason LC, Kuznetsov SG and Court DL (2009) Recombineering: a Homologous Recombination-Based Method of Genetic Engineering. Nat Protoc 4, 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuznetsov SG, Liu P and Sharan SK (2008) Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat Med 14, 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharan SK, Morimatsu M, Albrecht U, Lim D, Regel E, Dinh C, et al. (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–10. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez-Solis R, Liu P and Bradley A (1995) Chromosome engineering in mice. Nature 378, 720–724. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich G & Soriano P (1993) Insertional mutagenesis by retroviruses and promoter traps in embryonic stem cells. Methods Enzymol 225, 681–701. [DOI] [PubMed] [Google Scholar]
- 15.Warming S, Costantino N, Court DL, Jenkins NA and Copeland NG (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D, Ellis HM, Lee EC, Jenkins NA and Copeland NG, Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97, 5978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha RS, Zarbl H, Keohavong P and Thilly WG (1992) Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl 2, 14–20. [DOI] [PubMed] [Google Scholar]
- 18.Ramirez-Solis R, Davis AC and Bradley A (1993) Gene targeting in embryonic stem cells. Methods Enzymol 225, 855–78. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF and Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press; NY. [Google Scholar]
- 20.Scudiero DA , Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, et al. (1988) Evaluation of a Soluble Tetrazolium/Formazan Assay for Cell Growth and Drug Sensitivity in Culture Using Human and Other Tumor Cell Lines. Cancer Res 48, 4827–4833. [PubMed] [Google Scholar]
- 21.Barch MJ, Knutsen T and Spurbeck JL (ed.) (1997) The AGT Cytogenetics Laboratory Manual, Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 22.Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, et al. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitelman F (ed.) (1995) ISCN: An International System for Human Cytogenetic Nomenclature, Karger, Basel. [Google Scholar]
- 24.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, et al. (1998) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2, 317–328. [DOI] [PubMed] [Google Scholar]