Abstract
Airway smooth muscle cells require coordinated protrusion and focal adhesion dynamics to migrate properly. However, the signaling cascades that connect these two processes remain incompletely understood. Glia maturation factor (GMF)-γ has been implicated in inducing actin debranching and inhibiting nucleation. In this study, we discovered that GMFγ phosphorylation at Y104 regulates human airway smooth muscle cell migration. Using high-resolution microscopy coupled with three-dimensional object–based quantitative image analysis software, Imaris 9.2.0, phosphomimetic mutant, Y104D-GMFγ, was enriched at nascent adhesions along the leading edge where it recruited activated neural Wiskott-Aldrich syndrome protein (N-WASP; pY256) to promote actin-branch formation, which enhanced lamellipodial dynamics and limited the growth of focal adhesions. Unexpectedly, we found that nonphosphorylated mutant, Y104F-GMFγ, was enriched in growing adhesions where it promoted a linear branch organization and focal adhesion clustering, and recruited zyxin to increase maturation, thus inhibiting lamellipodial dynamics and cell migration. The localization of GMFγ between the leading edge and focal adhesions was dependent upon myosin activity. Furthermore, c-Abl tyrosine kinase regulated the GMFγ phosphorylation–dependent processes. Together, these results unveil the importance of GMFγ phosphorylation in coordinating lamellipodial and focal adhesion dynamics to regulate cell migration.
Keywords: airway smooth muscle, cell migration, actin cytoskeleton, protein phosphorylation
Smooth muscle cell migration is critical for the development of airways, vasculature, gut, and bladder; however, it also contributes to the progression of diseases, such as asthma (1–4). Cell migration involves the extension of membrane protrusions (lamellipodia and filopodia) and attachment of adhesive structures (focal adhesions) to the extracellular matrix, which, together, generate mechanical tension to propel the cell through its environment (5, 6). The formation of the lamellipodia is established through dynamic actin network assembly that is regulated by the actin-related protein 2/3 (Arp2/3) complex (1, 7–9). Nucleation promoting factors (NPFs), such as neural Wiskott-Aldrich syndrome protein (N-WASP), Wiskott-Aldrich syndrome protein, and SCAR homolog complex (WASH), WAS/WASL interacting protein family member 1 (WIP) and WASP family member 1 (WAVE) (10–14) activate the Arp2/3 complex in motile cells (15–17).
Focal adhesions are large macromolecular complexes that attach cells to the extracellular matrix through the transmembrane integrins (6, 18, 19). Recent studies suggest that myosin activation is involved in focal adhesion assembly. Myosin activation may generate contractile force to facilitate recruitment of various proteins to focal adhesions (6, 20–22). Furthermore, focal adhesion dimension and maturation are important for determining migratory speed (23, 24). ABL proto-oncogene 1, a nonreceptor tyrosine kinase (c-Abl), regulates actin cytoskeletal reorganization essential for multiple cellular processes, such as cell migration (1, 25–27), proliferation (1, 28, 29), cytokinesis (30), smooth muscle contraction (9, 31, 32), and cancer metastasis (26). c-Abl expression is upregulated in asthmatic human airway smooth muscle cells (HASMCs) (31, 33). Inhibition of c-Abl by the inhibitor, imatinib, reduces airway hyperresponsiveness and remodeling in animal models of asthma (34), and relieves breathing difficulty of patients with severe asthma (4, 35).
In smooth muscle cells, c-Abl orchestrates actin reorganization by controlling glia maturation factor (GMF)-γ (36), which is a member of the ADF/cofilin depolymerizing factor superfamily (37–41). GMFγ is expressed in a variety of cell types, including airway smooth muscle cells (36, 37, 39, 40). Unlike its relative, cofilin, GMFγ does not interact directly with actin filaments, but rather binds specifically to two binding sites of the Arp2/3 complex to initiate actin branch disassembly and inhibit further nucleation (37, 41). Previous loss-of-function studies have revealed the importance of GMFγ as a regulator of lamellipodia and overall migration (37, 38, 40). Furthermore, our group has previously shown that GMFγ undergoes phosphorylation at Tyr-104 in smooth muscle upon contractile activation. Tyr-104 is positioned within the Arp2 binding motif of GMFγ (36). GMFγ phosphorylation leads to its dissociation from the Arp2/3 complex and regulates actin dynamics (36).
In this study, we investigated the role of GMFγ phosphorylation at Tyr-104 in the coordination of both lamellipodial and focal adhesion dynamics through remodeling of actin during cell migration. We used high-resolution microscopy coupled with the three-dimensional (3D) reconstruction software, Imaris 9.2.0, to identify quantitative changes in 3D morphology of individual focal adhesions and actin architecture based on GMFγ functionality, which would otherwise be insufficient with a method of two-dimensional (2D) immunosfluorescent intensity analyses or co-IP (18). We unexpectedly identified a shift in GMFγ localization to either the leading edge or to growing focal adhesions. Phosphorylated GMFγ was localized to nascent adhesions near the leading edge and enhanced lamellipodial dynamics. Nonphosphorylated GMFγ was recruited to focal adhesions, induced maturation, and inhibited cell migration. Furthermore, c-Abl regulates GMFγ spatial distribution, lamellipodial dynamics, and focal adhesion assembly.
Methods
Cell Culture
HASMCs were prepared from bronchi and adjacent tracheas of control subjects (who died from nonasthmatic causes) and patients (who died from severe asthma) obtained from the International Institute for Advanced Medicine (42). Nonasthmatic and asthmatic HASMCs were also obtained from Dr. Reynold A. Panettieri of Rutgers University (33). Human tissues were nontransplantable and consented by patients for research. This study was approved by the Albany Medical College Committee on Research Involving Human Subjects. Details are provided in the data supplement.
Reagents and Transfection
Detailed plasmid sequences, lentiviral shRNA, and primary antibody information are provided in the data supplement.
Transfections for migration assay were performed using FuGene HD Transfection Reagent (Promega Corporation). Transfections of cells for fixed- and live-cell confocal microscopy were performed with Neon electroporation and Neon reagents (MPK10025; ThermoFisher).
Western Blot and Co-IP
Cells lysed with 2× SDS sample buffer boiled for 5 minutes and separated onto SDS-PAGE, then electrotransferred to nitrocellulose paper. Membranes were blocked using 2% BSA in PBS for 1 hour and then probed with specific primary antibodies followed by horseradish peroxidase–conjugated secondary antibody (Fisher Scientific). Proteins were visualized using the Amersham Imager 600 (GE Healthcare). Additional Western blot and co-IP information are detailed in the data supplement.
Immunofluorescence Microscopy
Imaging for fixed cells and live cells was conducted on a Zeiss LSM 880 NLO confocal microscope with Fast Airyscan module (Carl Zeiss Microscopy) equipped with 63× oil 1.4 numerical aperture objective lens and collected through a 32-channel GaAsP detector as 0.2 Airy units per channel. Z-stack collecting was under Nyquist sampling and with the Fast Airyscan SR settings. Live-cell imaging of GMFγ-GFP–tagged mutants, Life-Act-RFP, and paxillin-mcherry constructs used the Fast Airyscan module on the Zeiss LSM 880 confocal microscope. Microscope software used is the Zen Black 2 edition to process images for the Airyscan. Time-lapse microscopy was achieved by using a Leica A600 microscope with a six-well incubator chamber hooked up to 5% CO2. Additional detailed methods for immunofluorescent microscopy are found in data supplement.
Image Analysis
Detailed descriptions for image analysis are found in the data supplement.
Statistical Analysis
All data were analyzed using GraphPad Prism version 6.00 software (Windows; GraphPad Software). A two-tailed, one-way ANOVA, followed by Tukey’s multiple comparisons test, was used for comparing time-lapse microscopy parameters, lamellipodial protrusion/retraction velocity, and events, comparing vinculin and zyxin area and number, all filament tracer experiments, and comparing the percent of GMFγ inside and outside adhesions (significance was determined by a P < 0.05). A two-tailed Student’s t test was used to determine a significance of P less than 0.05 for knockdown cell focal adhesion parameters, protein level expression, and “wound” closure rates. A two-tailed Student’s t test was used to determine a significance of P less than 0.05 for blebbistatin-treated cells. Box-and-whisker plots and bar graphs were used to represent data shown. “n” denotes the number of individual cells, experiments, or objects rendered, as stated in the text.
Results
Knockdown of GMFγ Inhibits Smooth Muscle Cell Migration
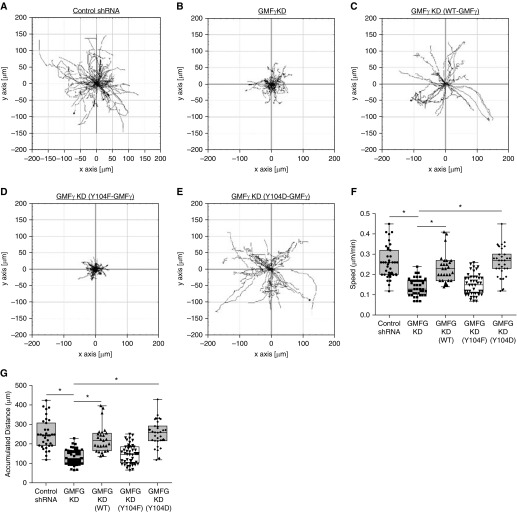
To interrogate the function of GMFγ, we generated stable GMFγ knockdown in HASMCs by using lentiviral particles encoding control or GMFγ shRNA. Cells were serum starved overnight before replating onto collagen-coated six-well plates in 10% FBS/F12 medium. Migration of control or GMFγ knockdown cells was monitored live by time-lapse microscopy and analyzed using the NIH ImageJ software. Cells undergoing cell proliferation over the course of the experiment were not chosen for analysis. GMFγ knockdown diminished the speed and accumulated distance of motile HASMCs (Figures 1A, 1B, 1F, and 1G and Movie E1 in the data supplement). Immunoblot analysis verified effective knockdown in cells expressing GMFγ shRNA by 80% (Figure E1A). We also performed a wound-healing assay to assess the effects of GMFγ knockdown on directed cell migration (Figure E1B). Loss of GMFγ led to a decrease in the ability of cells to close the scratch area in 12 hours as compared with control shRNA-expressing cells (Figure E1C). Furthermore, we re-expressed GMFγ in the knockdown cells using the experimental procedures as we previously described (36) (Figures E1D–E1F). Rescue of GMFγ restored the speed and distance of migratory cells (Figures 1A–1C, 1F, and 1G and Movie E1).
Figure 1.
(A–E) GMFγ phosphorylation at Y104 regulates smooth muscle cell migration. Time-lapse microscopy was used to track human airway smooth muscle cells (HASMCs) expressing control shRNA and GMFγ shRNA, as well as cells transfected with wild-type (WT)-GMFγ, Y104F-GMFγ, and Y104D-GMFγ plasmids. Images were taken every 10 minutes for 16 hours. Migration plots generated by Image J plugin display migration patterns for each cell type. (F and G) Graphical comparisons represent the calculated speed and accumulated distance for each cell type. Two-tailed, one-way ANOVA with Tukey’s post hoc test was used (*P < 0.05; control shRNA n = 51, GMFγ knockdown (KD) n = 41, WT-GMFγ n = 27, Y104F-GMFγ n = 45, Y104D-GMFγ n = 31 n = pooled cell numbers from four human donors without asthma). GMFG = glia maturation factor γ.
GMFγ Phosphorylation at Y104 Regulates Smooth Muscle Cell Migration
Because GMFγ has a role in regulating smooth muscle contraction, we evaluated the role of GMFγ phosphorylation at this residue in cell migration. We engineered EGFP-tagged non-phosphorylated mutant (Y104F) GMFγ (substitution of phenylalanine at Y104) and phosphorylation mimic mutant (Y104D) GMFγ (aspartic acid substitution at Y104) (36). These DNA constructs were transiently transfected into GMFγ knockdown cells (Figures E1D–E1F), and migration of live cells was monitored by time-lapse microscopy (Figures 1A–1E and Movie E1). The expression of Y104F-GMFγ in the knockdown cells did not recover cell migration (Figures 1D, 1F, and 1G). However, the expression of Y104D-GMFγ in the knockdown cells restored the speed and distance of motile cells (Figures 1E–1G). These results suggest that GMFγ phosphorylation contributes to the regulation of cell migration.
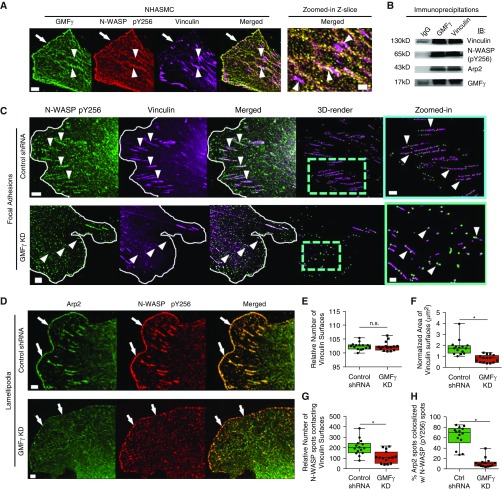
Knockdown of GMFγ Disrupts N-WASP (pY256) Spatial Distribution and Reduces Focal Adhesion Area
Because speed and distance were attenuated by knockdown of GMFγ, we evaluated the spatial distribution of GMFγ in smooth muscle cells with a focus on lamellipodia and lamella (dynamic cell front during migration). GMFγ was localized on the cell leading edge of the lamellipodia (Figure 2A). Moreover, merged z-slice confocal images showed that a pool of GMFγ was colocalized with vinculin, a marker of focal adhesions. To our knowledge, this is the first evidence of GMFγ localization to the focal adhesion compartment. In addition, active N-WASP has been implicated in cell migration (10, 13); we also examined the cellular location of N-WASP (pY256) in motile cells. N-WASP was distributed both in focal adhesions and the leading edge (Figure 2A). Furthermore, co-IP analysis was used to assess interaction of GMFγ with vinculin and N-WASP (pY256). Vinculin and N-WASP (pY256) were found in GMFγ immunoprecipitates (Figure 2B). Conversely, GMFγ and N-WASP (pY256) were found in vinculin precipitates (Figure 2B), suggesting an association of GMFγ with vinculin and N-WASP (pY256).
Figure 2.
Knockdown of GMFγ disrupts N-WASP (pY256) spatial distribution and reduces focal adhesion area. (A) HASMCs were plated on collagen-I–coated coverslips and immunostained for total GMFγ, N-WASP (pY256), and vinculin. Z-slice images were taken on a Zeiss LSM880 confocal microscope with the Fast Airyscan module using 488-, 561-, and 633-nm lasers. Scale bars: 5 μm and zoomed-in = 2 μm. Arrowheads point to focal adhesions and arrows point to the leading edge. Representative image from n = 10 individual cells. (B) Co-IP assay was performed on HASMCs using antibodies targeting endogenous GMFγ or vinculin. Co-IP samples were loaded and separated on an SDS-PAGE gel, where they were electrotransferred onto nitrocellulose paper and immunoblotted with primary antibodies for vinculin, N-WASP (pY256), Arp2, and GMFγ. Representative immunoblot from n = 4 independent experiments. (C) Control shRNA or GMFγ knockdown–expressing cells were immunostained for N-WASP (pY256) and vinculin. Z-slice images were taken on a Zeiss LSM880 confocal microscope with the Fast Airyscan module. Scale bar: 5 μm, zoomed-in = 2 μm. White line denotes the leading edge; arrowheads point to focal adhesions. (D) Control shRNA or GMFγ knockdown–expressing cells were immunostained for Arp2 and N-WASP (pY256). Arrows point to the leading edge. (E–G) Imaris 9.1.2 software was used to 3D render spots (control shRNA = 18,433, GMFγ KD = 21,163) and surfaces (control shRNA = 1,993, GMFγ KD = 2,121) for quantitative analysis of vinculin number, area, and percent N-WASP spots contacting vinculin surfaces from 10 individual cells (Figure E3 and Methods). (H) Imaris 9.1.2 software was used to quantify the percent Arp2 spots (control shRNA = 12,660, GMFγ KD = 16,962) colocalized with N-WASP (pY256) spots (control shRNA = 10,879, GMFγ KD = 14,640) from 10 individual cells. Student’s t test was used (*P < 0.05). Arp2 = actin-related protein 2; Co-IP = co-immunoprecipitation; Ctrl = control; NHASMC = nonasthmatic HASMC; n.s. = not significant; N-WASP = neural Wiskott-Aldrich syndrome protein.
To determine whether GMFγ affects focal adhesions and the actin-regulatory proteins, we assessed the effects of GMFγ knockdown on focal adhesion size and distribution of N-WASP (pY256). Focal adhesions within HASMCs span multiple Airyscan z-slices; therefore, to quantify morphological changes, we used the Imaris 3D object-based rendering software (Figures E2 and E3A–E3F and Methods) to reconstruct vinculin into 3D objects using the surface module because of its ability to accurately quantify focal adhesion size and distribution changes by surface area and distance transformation—two MatLab algorithms (18, 43) (Figure E3). We also reconstructed the punctate immunostaining of N-WASP (pY256) into spots using the spot module of the Imaris software. 3D surfaces and spots were reconstructed based on immunofluorescent intensity, surface area, and quality of rendering to identify morphological parameters and interaction between objects in 3D (18, 43). Knockdown of GMFγ caused a significant decrease in vinculin surface area without impacting individual vinculin numbers as compared with control cells (Figures 2C, 2E, and 2F). We also observed a significant decrease in the enrichment of N-WASP (pY256) with vinculin in GMFγ knockdown cells (Figures 2C and 2G). Altogether, these results suggest that GMFγ is localized within focal adhesions and that it may regulate focal adhesion growth through the recruitment of N-WASP (pY256).
We further examined whether N-WASP localization was also disrupted by GMFγ knockdown at the leading edge of the lamellipodia (Figures 2D and E3G–E3I). Knockdown of GMFγ led to a decrease in Arp2 colocalized with N-WASP (pY256) at the leading edge as compared with cells expressing control shRNA (Figures 2D and 2H).
GMFγ Phosphorylation at Y104 Impacts Focal Adhesion Dynamics in Live Cells
During migration, focal adhesions undergo dynamic changes to accommodate alterations in their surroundings (1). We next sought to determine if the phosphorylation state of GMFγ affects focal adhesion dynamics in live cells by observing mCherry-labeled paxillin, a focal adhesion–associated protein that also interacts with vinculin in both nascent and mature adhesions (21). Cells were cotransfected with wild-type (WT), Y104F, or Y104D-GMFγ and mcherry-paxillin. Labeled paxillin images of cells were monitored live by Fast Airyscan microscopy (Figures E4A–E4C and Movie E2). Images were taken every 30 seconds over the course of a 20-minute period with a sampling of four z-slices. Imaris software with surface module package was used to 3D reconstruct paxillin surface area based on fluorescent intensity, quality, area, and track duration (minimum of 300 s in length; see Methods). The surface area of each paxillin object (WT-GMFγ n = 1,489, Y104F-GMFγ n = 2,012, Y104D-GMFγ n = 2,387) was tracked over time for each sample (n = 10 individual cells). Representative regions of interest were graphed to demonstrate focal adhesion dynamics (Figures E4A–E4C). WT-GMFγ–expressing cells exhibited an initial increase in paxillin surface area, which peaked at an average of 6.73 μm2 (SD ±2.44 μm2), followed by a decrease in surface area (Figures E4A, E4D, and E4E). Several focal adhesions did display stability in our WT-GMFγ–expressing cells, but averaged a consistent area of 8 μm2 (SD ±1.67 μm2). Conversely, Y104F-GMFγ–expressing cells displayed large, stable paxillin focal adhesions, which remained at an average of 14.1 μm2 (SD ±6.22) throughout the duration of imaging (Figures E4B, E4D, and E4E). Unexpectedly, during the period of imaging, we did not observe turnover of any of these focal adhesions (Figure E4B). In addition, expression of Y104D-GMFγ revealed a similar trend to WT-GMFγ–expressing cells, but was limited in the growth of paxillin focal adhesions averaging a peak surface area of 3.36 μm2 (SD ±0.784; Figures E4C–E4E). However, there were several occurrences of focal adhesions that did not disassemble in the time frame, but again were limited to an average area of 3.41 μm2 (SD ±1.23; Figure E4D). Quantification analysis showed that average peak of paxillin surface area was higher in cells expressing Y104F-GMFγ than in cells expressing WT or Y104D-GMFγ (Figure E4E). These results suggest that GMFγ phosphorylation regulates the dynamics of focal adhesions.
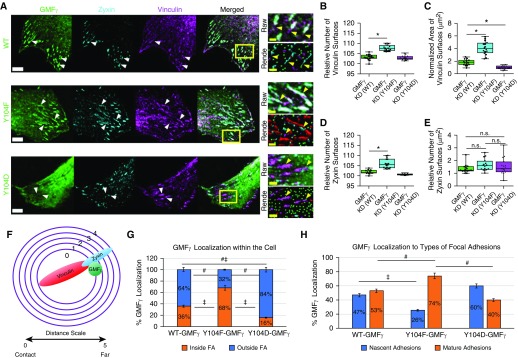
Phosphorylation at Y104 Regulates Focal Adhesion Clustering and GMFγ Distribution
Because we observed that knockdown of GMFγ decreased vinculin clustering, we asked whether the phosphorylation state of GMFγ affects focal adhesion assembly by expressing phosphorylation-modified GMFγ mutants in GMFγ knockdown (KD) cells. Cells expressing EGFP-tagged WT-, Y104F- or Y104D-GMFγ were plated on coverslips followed by immunostaining for vinculin and zyxin (Figure 3A). Zyxin is a marker for mature focal adhesions (44). Imaris software was used to analyze number and surface area of focal adhesions (Figure E5 and Methods). Expression of Y104F-GMFγ increased the number and area of vinculin surfaces as compared with cells expressing WT-GMFγ (Figures 3A–3C). In contrast, introduction of Y104D-GMFγ did not increase the number or area of vinculin surfaces when compared with cells expressing WT-GMFγ (Figures 3A–3C). In fact, expression of Y104D-GMFγ decreased the area of vinculin surfaces by 1 μm2, but this decrease was significant (Figures 3A–3C). Furthermore, expression of Y104F-GMFγ increased the number of zyxin surfaces without affecting the surface area of zyxin (Figures 3D and 3E). Y104D-GMFγ did not affect the number or area of zyxin surfaces (Figures 3D and 3E). These results suggest that lower phosphorylation of GMFγ at this residue promotes focal adhesion clustering and recruitment of zyxin to adhesions.
Figure 3.
Phosphorylation at Y104 regulates focal adhesion clustering and GMFγ distribution. (A) GMFγ knockdown cells were transfected with WT, Y104F, or Y104D-GMFγ GFP-tagged plasmids, then fixed and immunostained for zyxin and vinculin. Arrowheads point to focal adhesions. Scale bars: white bars 50 μm and yellow bars 2 μm. (B–E) Imaris 9.1.2 software was used to render GMFγ (WT = 49,304, Y104F = 45,524, Y104D = 27,178) spots, zyxin (WT = 2,228, Y104F = 3,390, Y104D = 755), and vinculin (WT = 4,482, Y104F = 7,751, Y104D = 3,800) surfaces from 10 individual cells to quantitate number and area of surfaces (Figure E3 and Methods). (F and G) Imaris 9.1.2 software was used to mask zyxin and vinculin surfaces followed by distance transformation algorithm to determine the percent of GMFγ inside or outside focal adhesions (FA). Distance transformation creates concentric circles around an object to measure distance based on the fluorescent intensity from other objects denoted by the distance scale (Methods). (H) Masked zyxin and vinculin channels were used to separate GMFγ localization within focal adhesions, and nascent adhesions contain only vinculin, whereas mature adhesions contain both vinculin and zyxin. A one-way ANOVA was used for statistical analysis with a Tukey’s post hoc test for between-group comparisons, *P < 0.05, #P < 0.05, and ‡P < 0.05.
We also examined the spatial distribution of WT and mutant GMFγ in HASMCs (Figures 3F and E5A–E5E and Methods). Imaris software was used to separate populations of GMFγ based on the proximity to vinculin and zyxin markers within lamellipodia (Figure 3F). We unexpectedly found that phosphorylation state of GMFγ affected its distribution in focal adhesions. Approximately 68% of Y104F-GMFγ were found inside focal adhesions as compared with 36% of WT-GMFγ. In contrast, 16% of Y104D-GMFγ were localized inside focal adhesions (Figure 3G). Moreover, 32% of Y104F-GMFγ, 64% of WT-GMFγ, and 84% of Y104D-GMFγ were positioned outside of focal adhesions (Figure 3G).
Next, we used Imaris software to further analyze detailed distribution of GMFγ and its mutants within individual focal adhesions (Figures 3F and E5A–E5E and Methods). This method allowed for separation of GMFγ populations associated with vinculin or zyxin reconstructed surfaces, nascent and mature adhesions, respectively. A total of 47% of WT-GMFγ localized with nascent adhesions, whereas 53% of WT-GMFγ contacted mature adhesions (Figure 3H). However, Y104F-GMFγ largely (74%) associated with mature adhesions with only 26% Y104F-GMFγ within nascent adhesions (Figure 3H). Moreover, 60% of Y104D-GMFγ localized to nascent adhesions with 40% of Y104D-GMFγ within mature adhesions (Figure 3H). These results suggest that nonphosphorylated GMFγ largely contacts zyxin-associated mature adhesions, whereas phosphorylated GMFγ localizes with nascent vinculin-only adhesions.
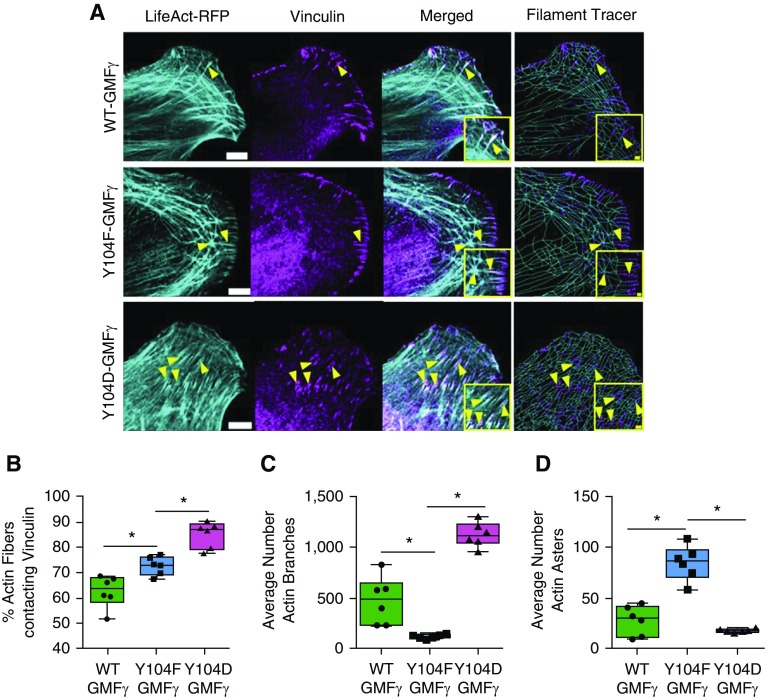
Actin Architecture and Connection of Actin Fibers to Vinculin and Are Regulated by GMFγ Phosphorylation at Y104
Because GMFγ phosphorylation was found to affect focal adhesion clustering, we asked whether GMFγ Y104 phosphorylation affects actin architecture by assessing the effects of nonphosphorylated or phosphorylated mutant on F-actin structure in lamellipodia. Cells were cotransfected with constructs for WT-, Y104F-, or Y104D-GMFγ and LifeAct-RFP plasmid, which generates a 17-amino acid peptide to visualize F-actin, followed by immunostaining for vinculin. Fast Airyscan microscopy and Imaris software with filament tracer package were used to evaluate actin architecture (Methods). In cells treated with WT-GMFγ, actin fibers displayed both a linear structure and an actin meshwork within lamellipodia. In contrast, actin fibers of cells expressing Y104F-GMFγ showed asterisk-like topology characterized by nucleation centers and arm-like radially orientated F-actin strands. Moreover, cells expressing Y104D-GMFγ displayed a majority of actin meshwork architecture (Figure 4A). Furthermore, the expression of Y104F-GMFγ increased actin fibers contacting vinculin (Figure 4B), reduced actin fiber branching (Figure 4C), and enhanced the occurrence of actin asters (Figure 4D). Moreover, the expression of Y104D-GMFγ increased vinculin-associated actin fibers (Figure 4B) and actin fiber branches (Figure 4C), and reduced actin asters (Figure 4C). These findings suggest that GMFγ phosphorylation at this residue regulates the reorganization of the actin cytoskeleton and the interaction of actin fibers with focal adhesions.
Figure 4.
Actin architecture is regulated by GMFγ phosphorylation at Y104. (A) GMFγ knockdown cells were transfected with WT, Y104F, or Y104D-GMFγ and LifeAct–red fluorescent protein (RFP) (pseudocolored cyan) plasmids, fixed, and immunostained for vinculin. Images were taken using a Zeiss LSM880 confocal with the Fast Airyscan module. Scale bars: 5 μm, merged inset = 2 μm from n = 10 individual cells per expression plasmid. Arrowheads point to actin fibers and focal adhesions. (B–D) Imaris software with the Filament Tracer package was used to trace actin fibers and analyze actin fibers contacting vinculin, as well as branch and aster morphology (Methods). Rendered vinculin (WT = 1,518, Y104F = 1,137, Y104D = 780) surfaces are in magenta, actin fibers (WT = 40,070, Y104F = 29,600, Y104D = 31,064) are in cyan, and “actin asters” or beginning points are labeled yellow. A one-way ANOVA with a Tukey’s post hoc test for between-group comparisons was used for statistical analysis. *P < 0.05.
GMFγ Phosphorylation at Y104 Modulates Lamellipodial Dynamics
During migration, cells undergo cyclic extension and retraction of lamellipodia to facilitate cell movement (1, 5, 8). We also evaluated the role of GMFγ phosphorylation in lamellipodial dynamics. Cells expressing WT-, Y104F-, or Y104D-GMFγ were transfected with LifeAct-RFP to monitor changes in lamellipodial dynamics using a Fast Airyscan microscope. To analyze lamellipodial dynamics, we used an ImageJ plugin known as “automated detection and analysis of protrusions” (45) (Methods). Expression of Y104F-GMFγ severely impaired protrusion velocity, as well as retraction velocity, as compared with cells expressing WT-GMFγ (Figures E6A–E6C and Movie E3). Moreover, actin asters were also observed, but were dynamic in live cells expressing Y104F-GMFγ (Figure E6A and Movie E3). However, expression of Y104D-GMFγ recovered protrusion velocity and retraction velocity (Figures E6A–E6C and Movie E3). Furthermore, expression of Y104F-GMFγ reduced protrusion events and enhanced retraction events as compared with cells expressing WT-GMFγ. However, expression of Y104D-GMFγ restored these events (Figures E6A, E6D, and E6E). Together, these data suggest that the phosphorylation of GMFγ can modulate the actin cytoskeleton to promote efficient lamellipodial dynamics.
Myosin Activation Regulates the Recruitment of GMFγ to Focal Adhesions
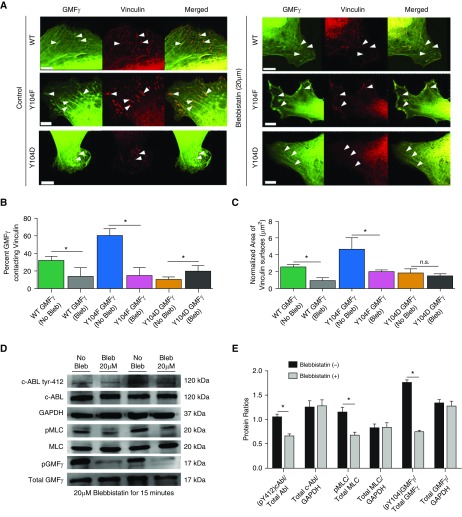
As described previously here, GMFγ phosphorylation affects its distribution in focal adhesions (Figure 3). Because myosin activity has been implicated in the recruitment of focal adhesion–associated proteins and focal adhesion morphology (6, 21, 22), we examined whether myosin activation influences GMFγ distribution. Cells expressing WT-GMFγ, Y104F-GMFγ, or Y104D-GMFγ were seeded on coverslips, followed by treatment with blebbistatin, a small-molecule inhibitor of myosin II ATPase activity (46). Cells were then immunofluorescently stained for vinculin (Figure 5A). Approximately 30% of WT-GMFγ contacted vinculin, whereas 60% of Y104F-GMFγ and around 10% of Y104D-GMFγ associated with vinculin (Figure 5B). Treatment with blebbistatin reduced the populations of WT-GMFγ and Y104F-GMFγ contacting vinculin. Moreover, blebbistatin treatment increased the population of Y104D-GMFγ contacting vinculin (Figure 5B). Furthermore, treatment with blebbistatin diminished the area of vinculin surfaces in cells expressing WT-GMFγ and Y104F-GMFγ, but not Y104D-GMFγ (Figures 5B and 5C). These results suggest that myosin activity affects phosphorylation-dependent GMFγ recruitment to focal adhesion and focal adhesion morphology.
Figure 5.
Myosin activation regulates the recruitment of GMFγ to focal adhesions. (A) GMFγ knockdown cells were transfected with WT, Y104F, or Y104D-GMFγ plasmids overnight. Cells were then trypsinized and replated onto collagen-I–coated coverslips for 2 hours. After 2 hours, cells were treated with 20 μM blebbistatin for 15 minutes, then fixed and immunostained for vinculin. Scale bar = 10 μm; arrowheads point to focal adhesions. (B and C) Imaris 9.1.2 software was used to render vinculin (WT = 1,119, Y104F = 1,801, Y104D = 1,827) surfaces and GMFγ spots (WT = 14,916, Y104F = 14,475, Y104D = 10,723) for quantification of vinculin area and percent GMFγ spots contacting vinculin surfaces. Student’s t test was used for statistical analysis comparing no treatment to blebbistatin for each individual mutant GMFγ (n = 10 cells). *P < 0.05. (D) HASMCs were grown to confluence in 60-mm cell culture–treated dishes, then subjected to 20 μM (−/−) blebbistatin treatment for 15 minutes. Cells were harvested using 1× SDS sample buffer containing 1× protease and phosphatase inhibitor, scraped, and boiled for 5 minutes. Samples were run on SDS-PAGE, then electrotransferred onto nitrocellulose paper and immunoblotted for c-Abl tyr-412, total c-Abl, GAPDH, pMLC, MLC, GMFγ tyr-104, and total GMFγ. Western blots from n = 4 individual experiments were imaged using the GE Amersham 600 and analyzed using IQTL software. (E) Quantification of phospho:total protein ratio normalized to GAPDH was graphed. Student’s t test was used to compare the effect of blebbistatin treatment. *P < 0.05. Bleb = blebbistatin; c-ABL = a nonreceptor tyrosine kinase.
Because c-Abl is known to catalyze GMFγ Y104 phosphorylation (36), we assessed the role of myosin activity in c-Abl phosphorylation at Y412, an indication of c-Abl activity as well as GMFγ phosphorylation at Y104 (25). Treatment with blebbistatin inhibited both c-Abl phosphorylation and GMFγ phosphorylation (Figures 5D and 5E). Moreover, we verified that blebbistatin treatment reduced myosin light chain-20 phosphorylation (Figures 5D and 5E).
c-Abl Regulates the Accumulation of GMFγ to Focal Adhesions of Migrating Cells
Next, we assessed whether c-Abl affects the spatial distribution of GMFγ in motile cells. Cells expressing control shRNA and c-Abl knockdown cells were generated by the methods previously described (30), and immunostained for GMFγ, N-WASP (pY256), zyxin, and vinculin. c-Abl knockdown resulted in increases in the number and surface area of vinculin (Figures E7A, E7C, and E7D) and zyxin (Figures E7B, E7E, and E7F). Furthermore, we used Imaris software with the surface module package to analyze the localization of GMFγ and N-WASP (pY256) in vinculin or zyxin adhesions (Methods). Knockdown of c-Abl enhanced localization of GMFγ spots with vinculin surfaces from 30% to 48% (Figure E7G). Localization of N-WASP (pY256) spots with vinculin surfaces was also higher in c-Abl knockdown cells (Figure E7H). Moreover, contact of GMFγ and N-WASP (pY256) with zyxin was higher in c-Abl knockdown cells compared with control cells (Figures E7I and E7J).
GMFγ Phosphorylation Is Involved in Faster Migration of HASMCs
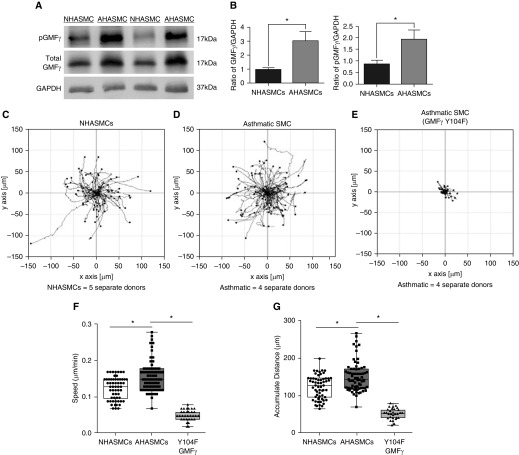
Because airway smooth muscle cell motility has been implicated in airway remodeling of asthma (1, 3, 4), and our present results demonstrate a role of GMFγ phosphorylation in cell migration, we assessed whether GMFγ and its phosphorylated form are altered in asthma. Immunoblot analysis showed that total and phosphorylated GMFγ was upregulated in asthmatic HASMCs (Figures 6A and 6B). Moreover, speed and accumulated distance were increased in asthmatic HASMCs (Figures 6C, 6D, 6F, and 6G). More importantly, the expression of Y104F-GMFγ inhibited migration of asthmatic HASMCs (Figures 6E–6G).
Figure 6.
Phosphorylated GMFγ is upregulated in asthmatic HASMCs (AHASMCs) and contributes to its enhanced migratory phenotype. (A) Normal HASMC and airway smooth muscle cells isolated from five different donors with asthma were grown to confluence in 60-mm cell culture–treated dishes, then harvested with 1× SDS sample buffer containing 1× protease and phosphatase inhibitor, scraped, and boiled for 5 minutes. Samples were run on SDS-PAGE, then electrotransferred onto nitrocellulose paper and immunoblotted for phospho-GMFγ (custom antibody; Methods), total GMFγ, and GAPDH. Western blots were imaged using the GE Amersham 600 and analyzed using IQTL software. (B) Quantification of phospho:total protein ratio normalized to GAPDH was graphed. Student’s t test was used to compare normal versus asthma GMFγ expression. *P < 0.05. (C–E) Cells were serum starved overnight then replated onto collagen-I–coated six-well dishes in 10% FBS medium. Time-lapse microscopy was used to track NHASMC and AHASMC migration with a start time 2 hours after seeding. Images were taken every 10 minutes for 16 hours. Migration plots were generated using an ImageJ plugin to trace individual cells migratory pattern. (F and G) Graphical comparisons represent the calculated speed and accumulated distance for each cell type. Two-tailed, one-way ANOVA with Tukey’s post hoc test was used (*P < 0.05; NHASMC n = 45, AHASMC n = 57, Y104F-GMFγ–expressing AHASMC n = 49; n represents pooled cell numbers from four donors without and five human donors with asthma).
Discussion
GMFγ is highly expressed in HASMCs (36). Our present study suggests that GMFγ is an important molecule that regulates cell migration. GMFγ deficiency reduced active N-WASP recruitment to focal adhesions and inhibited focal adhesion formation (Figure 2). This may be because GMFγ can bind the C terminus of N-WASP (37) to facilitate their interaction. N-WASP and its associated proteins are known to modulate focal adhesion assembly (12). Furthermore, GMFγ deficiency reduced the activation of the Arp2/3 complex in the leading edge. Again, this could be due to the ability of GMFγ to interact with Arp2 and N-WASP (37).
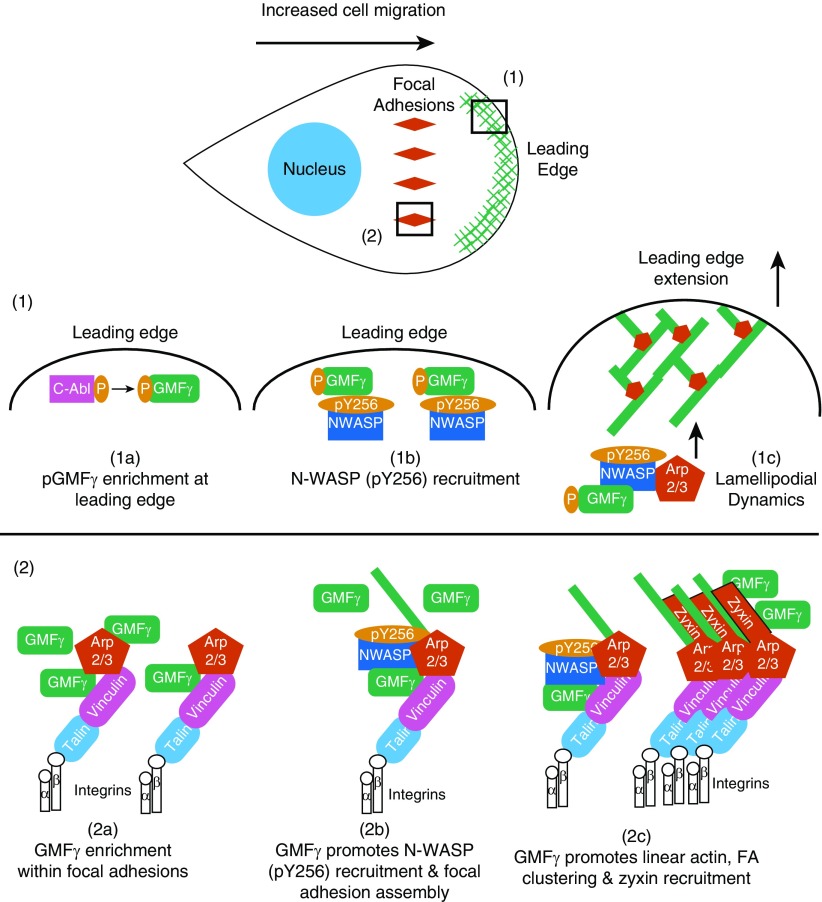
Our experimental and quantitative results suggest a model for how GMFγ phosphorylation can regulate lamellipodial and focal adhesion dynamics during airway smooth muscle cell migration (Figure 7). Localization of phospho-GMFγ at the leading edge increased the recruitment of N-WASP (pY256) to promote Arp2/3-mediated actin branching at the cell front, as indicated by presence of actin branched organization within protrusions (Figures 4 and 7), which may increase lamellipodial extension (Figure E6). Phosphorylation at Y-104 promotes dissociation of GMFγ from Arp2 (36). Thus, it is possible that the released GMFγ may facilitate N-WASP recruitment to the leading edge. Several studies have identified an important connection between branched actin and the formation of nascent adhesions in an Arp2/3-dependent manner (16, 17, 19). We hypothesized that GMFγ phosphorylation may promote a switch in actin organization to regulate focal adhesion morphology (Figure 7). We observed a change in actin organization between branched and linear actin as nonphosphorylated GMFγ was enriched in mature focal adhesions (Figures 3 and 4). Our results suggest that nonphosphorylated GMFγ promotes aster actin formation that may enhance focal adhesion maturation and stability, leading to a dramatic reduction in cell migratory speed (Figures 1, 3, and 4). This is because focal adhesion maturation and size affect cell migration (23). In addition, aster actin may change membrane structure (43) and inhibit protrusion extension and cell migration (Figures 1 and 4 and Figures E4 and E6). The formation of geodesic-actin organization (actin asters) has previously been reported in cells undergoing topological stress (47, 48), and could arise through actin nodes containing the formin, disheveled-associated activator of morphogenesis 1 (DAAM1), the cross-linker filamin A (FlnA) and myosin II filaments (49).
Figure 7.
Model: phosphorylation state of GMFγ dictates its localization and functionality to regulate cell migration. (1a) At the leading edge, cellular cues trigger the enrichment of phosphorylated GMFγ. (1b) There, phospho-GMFγ recruits N-WASP (pY256) to the leading edge to enhance actin reorganization through Arp2/3 activation. (1c) Increased actin remodeling leads to increased protrusion extension and enhances lamellipodial dynamics. (2a) Upon myosin activation, nonphosphorylated GMFγ becomes enriched within focal adhesions, which includes talin and integrins, as well as many other proteins. (2b) Nonphosphorylated GMFγ recruits N-WASP (pY256) and increases linear actin formation and focal adhesion assembly. (2c) Nonphosphorylated GMFγ promotes actin reorganization, focal adhesion clustering, and recruitment of zyxin to enhance focal adhesion maturation. Sustained mechanical tension will increase c-Abl activation within focal adhesions, leading to phosphorylation of GMFγ, thus liberating it from Arp2/3 and returning GMFγ to the leading edge.
Our studies on phosphomimetic Y104D-GMFγ suggest that increased actin fiber branching separates focal adhesions into smaller units within protrusions of migrating cells. Recently, a study found that focal adhesions undergo splitting events into multiple focal adhesion units of uniform width that are laterally associated through actin filaments generated by actin-assembly proteins, adenomatosis polyposis coli (APC) protein and vasodilator-stimulated phosphoprotein (VASP) (18). These events were found to occur under increased actin tension driven by myosin II activity (18).
During cell migration, c-Abl phosphorylation and activation at the leading edge can be initiated by β1 integrin and growth factor (1). In this article, phosphorylated GMFγ localized to the leading edge. c-Abl knockdown inhibits GMFγ phosphorylation. Thus, β1 integrin and growth factors are important initiators for GMFγ-mediated processes during migration.
The molecular mechanism for the directed translocation of phosphorylated GMFγ between the leading edge and focal adhesions remains unknown. Here, we demonstrate that the enrichment of nonphosphorylated GMFγ within focal adhesions is driven by myosin II activity, as blebbistatin treatment attenuated GMFγ enrichment with vinculin (Figure 5). Myosin activation is also involved in the recruitment of other focal adhesion–associated proteins (6). Myosin activation may induce filament contraction and promote protein accumulation in the adhesive structure. However, other possibilities may also exist. One study in macrophages suggests that GMFγ may play a role in β1 integrin recycling back to the leading edge through interaction with syntaxin-4 (STX4) and syntaxin-binding protein 4 (STXBP4), two proteins involved with vesicle trafficking and fusion to the plasma membrane (40). In addition, previous studies have revealed that cofilin-1, a homologous actin depolymerizing factor to GMFγ, interacts with high affinity to membrane phospholipids PI(4,5)P2, PI(3,4)P2, and PI(3,4,5)P3 through electrostatic interactions (50). Phospholipid composition and clustering on vesicles may recruit and bind phosphorylated GMFγ, thus transporting GMFγ to the leading edge. Furthermore, a homologous actin depolymerizing factor, actin-binding protein 1 (Abp1), was found to regulate vesicle trafficking by interacting with distinct golgi membrane regions (39). Abp1 was shown in yeast to enhance actin polymerization by interacting with both N-WASP and Arp2/3 (39). Abp1 can also bind endocytic proteins, such as myosin Vb and synaptojanin, to connect vesicle recycling with dynamic cortical actin (39). Moreover, Arp2/3-mediated actin polymerization on the endosomal membrane surface was critical for integrin recycling back to the plasma membrane (7). Future studies are needed to test these possibilities in the context of cell migration.
We additionally observed that c-Abl activation was myosin dependent, as treatment with blebbistatin decreased c-Abl phosphorylation at Y-412, a marker of c-Abl catalytic activity, as well as GMFγ phosphorylation at Y-104 (Figure 5). c-Abl and its isoform, Arg, have been reported to interact with integrins and focal adhesion–associated proteins (14, 26), as well as to promote actin formation (27). Our image analysis showed a dramatic shift in localization of Y104D-GMFγ outside of adhesions, suggesting that c-Abl phosphorylation might liberate GMFγ from the adhesions upon increased mechanical tension. c-Abl may act as a switch to promote changes in actin organization needed to control focal adhesion growth and subsequent actin branch formation within protrusions by regulating GMFγ localization during cell migration (Figure 7).
Our results suggest that GMFγ phosphorylation at Y-104 is involved in asthma pathogenesis, as evidenced by higher expression of phosphorylated and total GMFγ, and inhibition of asthmatic cell migration by nonphosphorylated GMFγ mutant. Furthermore, our previous studies show that c-Abl expression is higher in airway smooth muscle cells from patients with asthma and from animal models of asthma (31, 33). Thus, the c-Abl–GMFγ pathway may contribute to increased airway smooth muscle cell migration during asthma progression.
In summary, our study provides novel insight into the critical role of GMFγ phosphorylation in regulating lamellipodial and focal adhesion dynamics, important for directed HASMC migration (Figure 7). Phosphorylation of GMFγ by c-Abl nonreceptor tyrosine kinase and myosin contractility are determinants for GMFγ spatial localization to the leading edge and growing focal adhesions, respectively (Figure 7). Localization of phospho-GMFγ at the leading edge leads to increased N-WASP (pY256) enrichment, Arp2 activation, and increased protrusion extension, whereas localization of nonphosphorylated GMFγ within focal adhesion promotes N-WASP (pY256) recruitment, actin reorganization, focal adhesion clustering, and recruitment of zyxin (Figure 7). Thus, we propose that GMFγ localization and phosphorylation state is an important modulatory switch that controls actin organization and focal adhesion dynamics to promote cell migration.
Supplementary Material
Acknowledgments
Acknowledgment
The authors acknowledge Dr. Gabrielle Fredman (Albany Medical College) for assisting in editing the manuscript, the Albany Medical College Imaging Core and Dr. J. Mazurkiewicz (Albany Medical College) for helping with the Zeiss LSM 880 and Imaris software.
Footnotes
Supported by National Heart, Lung, and Blood Institute/National Institutes of Health grants HL-110951, HL-113208, and HL-130304 (D.D.T.), and by American Heart Association predoctoral fellowship 16PRE31430001 (B.D.G.).
Author Contributions: B.D.G. performed and analyzed the majority of experiments; B.D.G., G.L., A.C.R., and R.W. generated DNA constructs and shRNA knockdown cells; B.D.G., K.T., and M.B. performed imaging and Imaris analysis; B.D.G. wrote manuscript; B.D.G., K.T., M.B., and D.D.T. reviewed and edited manuscript; D.D.T. supervised the study.
This article has a data supplement, which is accessible from this issue's table of contentsat www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0352OC on February 27, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Tang DD, Gerlach BD. The roles and regulation of the actin cytoskeleton, intermediate filaments and microtubules in smooth muscle cell migration. Respir Res. 2017;18:54. doi: 10.1186/s12931-017-0544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salter B, Pray C, Radford K, Martin JG, Nair P. Regulation of human airway smooth muscle cell migration and relevance to asthma. Respir Res. 2017;18:156. doi: 10.1186/s12931-017-0640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gizycki MJ, Adelroth E, Rogers AV, O’Byrne PM, Jeffery PK. Myofibroblast involvement in the allergen-induced late response in mild atopic asthma. Am J Respir Cell Mol Biol. 1997;16:664–673. doi: 10.1165/ajrcmb.16.6.9191468. [DOI] [PubMed] [Google Scholar]
- 4.Kaminska M, Foley S, Maghni K, Storness-Bliss C, Coxson H, Ghezzo H, et al. Airway remodeling in subjects with severe asthma with or without chronic persistent airflow obstruction. J Allergy Clin Immunol. 2009;124:45–51.e1–4. doi: 10.1016/j.jaci.2009.03.049. [DOI] [PubMed] [Google Scholar]
- 5.Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- 6.Burridge K, Guilluy C. Focal adhesions, stress fibers and mechanical tension. Exp Cell Res. 2016;343:14–20. doi: 10.1016/j.yexcr.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizarro-Cerdá J, Chorev DS, Geiger B, Cossart P. The diverse family of arp2/3 complexes. Trends Cell Biol. 2017;27:93–100. doi: 10.1016/j.tcb.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 9.Tang DD. The dynamic actin cytoskeleton in smooth muscle. Adv Pharmacol. 2018;81:1–38. doi: 10.1016/bs.apha.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol. 2013;24:258–266. doi: 10.1016/j.semcdb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2014;156:195–207. doi: 10.1016/j.cell.2013.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramesh N, Massaad MJ, Kumar L, Koduru S, Sasahara Y, Anton I, et al. Binding of the WASP/N-WASP-interacting protein WIP to actin regulates focal adhesion assembly and adhesion. Mol Cell Biol. 2014;34:2600–2610. doi: 10.1128/MCB.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Y, Leong NT, Jiang T, Tangara A, Darzacq X, Drubin DG. Switch-like Arp2/3 activation upon WASP and WIP recruitment to an apparent threshold level by multivalent linker proteins in vivo. eLife. 2017;6:pii:e29140. doi: 10.7554/eLife.29140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MM, Lapetina S, MacGrath SM, Sfakianos MK, Pollard TD, Koleske AJ. Regulation of actin polymerization and adhesion-dependent cell edge protrusion by the Abl-related gene (Arg) tyrosine kinase and N-WASp. Biochemistry. 2010;49:2227–2234. doi: 10.1021/bi901721u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaney KF, Li R. Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr Opin Cell Biol. 2016;42:63–72. doi: 10.1016/j.ceb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duleh SN, Welch MD. Regulation of integrin trafficking, cell adhesion, and cell migration by WASH and the Arp2/3 complex. Cytoskeleton (Hoboken) 2012;69:1047–1058. doi: 10.1002/cm.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrels B, Serrels A, Brunton VG, Holt M, McLean GW, Gray CH, et al. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat Cell Biol. 2007;9:1046–1056. doi: 10.1038/ncb1626. [DOI] [PubMed] [Google Scholar]
- 18.Young LE, Higgs HN. Focal adhesions undergo longitudinal splitting into fixed-width units. Curr Biol. 2018;28:2033–2045.e5. doi: 10.1016/j.cub.2018.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chorev DS, Moscovitz O, Geiger B, Sharon M. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat Commun. 2014;5:3758. doi: 10.1038/ncomms4758. [DOI] [PubMed] [Google Scholar]
- 20.Maiuri P, Rupprecht J-F, Wieser S, Ruprecht V, Bénichou O, Carpi N, et al. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 21.Zhou DW, Lee TT, Weng S, Fu J, García AJ. Effects of substrate stiffness and actomyosin contractility on coupling between force transmission and vinculin-paxillin recruitment at single focal adhesions. Mol Biol Cell. 2017;28:1901–1911. doi: 10.1091/mbc.E17-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DH, Wirtz D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013;27:1351–1361. doi: 10.1096/fj.12-220160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D-H, Wirtz D. Predicting how cells spread and migrate: focal adhesion size does matter. Cell Adhes Migr. 2013;7:293–296. doi: 10.4161/cam.24804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woodring PJ, Hunter T, Wang JY. Regulation of F-actin–dependent processes by the Abl family of tyrosine kinases. J Cell Sci. 2003;116:2613–2626. doi: 10.1242/jcs.00622. [DOI] [PubMed] [Google Scholar]
- 26.Smith-Pearson PS, Greuber EK, Yogalingam G, Pendergast AM. Abl kinases are required for invadopodia formation and chemokine-induced invasion. J Biol Chem. 2010;285:40201–40211. doi: 10.1074/jbc.M110.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleary RA, Wang R, Waqar O, Singer HA, Tang DD. Role of c-Abl tyrosine kinase in smooth muscle cell migration. Am J Physiol Cell Physiol. 2014;306:C753–C761. doi: 10.1152/ajpcell.00327.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia L, Wang R, Tang DD. Abl regulates smooth muscle cell proliferation by modulating actin dynamics and ERK1/2 activation. Am J Physiol Cell Physiol. 2012;302:C1026–C1034. doi: 10.1152/ajpcell.00373.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Mercaitis OP, Jia L, Panettieri RA, Tang DD. Raf-1, actin dynamics, and abelson tyrosine kinase in human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2013;48:172–178. doi: 10.1165/rcmb.2012-0315OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Tang DD. c-Abl tyrosine kinase regulates cytokinesis of human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2014;50:1076–1083. doi: 10.1165/rcmb.2013-0438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleary RA, Wang R, Wang T, Tang DD. Role of Abl in airway hyperresponsiveness and airway remodeling. Respir Res. 2013;14:105. doi: 10.1186/1465-9921-14-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Cleary RA, Wang T, Li J, Tang DD. The association of cortactin with profilin-1 is critical for smooth muscle contraction. J Biol Chem. 2014;289:14157–14169. doi: 10.1074/jbc.M114.548099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao G, Panettieri RA, Tang DD. MicroRNA-203 negatively regulates c-Abl, ERK1/2 phosphorylation, and proliferation in smooth muscle cells. Physiol Rep. 2015;3:e12541. doi: 10.14814/phy2.12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rhee CK, Kim JW, Park CK, Kim JS, Kang JY, Kim SJ, et al. Effect of imatinib on airway smooth muscle thickening in a murine model of chronic asthma. Int Arch Allergy Immunol. 2011;155:243–251. doi: 10.1159/000321261. [DOI] [PubMed] [Google Scholar]
- 35.Cahill KN, Katz HR, Cui J, Lai J, Kazani S, Crosby-Thompson A, et al. Kit inhibition by imatinib in patients with severe refractory asthma. N Engl J Med. 2017;376:1911–1920. doi: 10.1056/NEJMoa1613125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Cleary RA, Wang R, Tang DD. Glia maturation factor-γ phosphorylation at Tyr-104 regulates actin dynamics and contraction in human airway smooth muscle. Am J Respir Cell Mol Biol. 2014;51:652–659. doi: 10.1165/rcmb.2014-0125OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goode BL, Sweeney MO, Eskin JA. Gmf as an actin network remodeling factor. Trends Cell Biol. 2018;28:749–760. doi: 10.1016/j.tcb.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poukkula M, Hakala M, Pentinmikko N, Sweeney MO, Jansen S, Mattila J, et al. GMF promotes leading-edge dynamics and collective cell migration in vivo. Curr Biol. 2014;24:2533–2540. doi: 10.1016/j.cub.2014.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poukkula M, Kremneva E, Serlachius M, Lappalainen P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 2011;68:471–490. doi: 10.1002/cm.20530. [DOI] [PubMed] [Google Scholar]
- 40.Aerbajinai W, Liu L, Zhu J, Kumkhaek C, Chin K, Rodgers GP. Glia maturation factor-gamma regulates monocyte migration through modulation of beta1-integrin. J Biol Chem. 2016;291:8549–8564. doi: 10.1074/jbc.M115.674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luan Q, Nolen BJ. Structural basis for regulation of Arp2/3 complex by GMF. Nat Struct Mol Biol. 2013;20:1062–1068. doi: 10.1038/nsmb.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Wang R, Gannon OJ, Rezey AC, Jiang S, Gerlach BD, et al. Polo-like kinase 1 regulates vimentin phosphorylation at ser-56 and contraction in smooth muscle. J Biol Chem. 2016;291:23693–23703. doi: 10.1074/jbc.M116.749341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritzsche M, Li D, Colin-York H, Chang VT, Moeendarbary E, Felce JH, et al. Self-organizing actin patterns shape membrane architecture but not cell mechanics. Nat Commun. 2017;8:14347. doi: 10.1038/ncomms14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uemura A, Nguyen TN, Steele AN, Yamada S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys J. 2011;101:1069–1075. doi: 10.1016/j.bpj.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barry DJ, Durkin CH, Abella JV, Way M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J Cell Biol. 2015;209:163–180. doi: 10.1083/jcb.201501081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolega J. Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun. 2004;320:1020–1025. doi: 10.1016/j.bbrc.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 47.Bermudez JY, Montecchi-Palmer M, Mao W, Clark AF. Cross-linked actin networks (CLANs) in glaucoma. Exp Eye Res. 2017;159:16–22. doi: 10.1016/j.exer.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Entcheva E, Bien H. Mechanical and spatial determinants of cytoskeletal geodesic dome formation in cardiac fibroblasts. Integr Biol. 2009;1:212–219. doi: 10.1039/b818874b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo W, Lieu ZZ, Manser E, Bershadsky AD, Sheetz MP. Formin daam1 organizes actin filaments in the cytoplasmic nodal actin network. PLoS One. 2016;11:e0163915. doi: 10.1371/journal.pone.0163915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J. 2010;98:2327–2336. doi: 10.1016/j.bpj.2010.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.